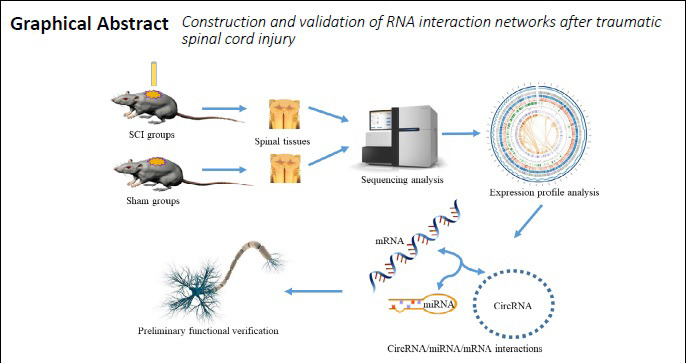
Keywords: bioinformatics, circRNA/miRNA/mRNA network, circular RNA, gene, KLF4, miR-135b-5p, spinal cord injury, trauma
Abstract
Differential expression of non-coding RNA after traumatic spinal cord injury (TSCI) is closely related to the pathophysiological process. The purposes of this study were to systematically profile and characterize expression of circular RNA (circRNA) in the lesion epicenter of spinal tissues after TSCI, and predict the structure and potential function of the regulatory circRNA/miRNA network. Forty-eight C57BL/6 mice were randomly and equally assigned to two groups: one subjected to TSCI at T8–10 with an Allen’s drop impactor, and a second subjected to laminectomy without TSCI. Spinal cord samples were stained with hematoxylin and eosin, sequenced, and validated. RNA-Seq, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes analysis, and network analyses (Targetscan and miRanda) were used to predict and annotate the circRNA/miRNA/mRNA network. Luciferase reporter, quantitative reverse transcription polymerase chain reaction, and western blot assays were used to profile expression and regulation patterns of the network in mouse models of TSCI. Hematoxylin-eosin staining revealed severe damage to the blood-spinal cord barrier after TSCI. Differentially expressed circRNA and miRNA profiles were obtained after TSCI; differentially expressed circRNAs, which were abundant in the cytoplasm, were involved in positive regulation of transcription and protein phosphorylation. miR-135b-5p was the most significantly downregulated miRNA after TSCI; circRNAAbca1 and KLF4 were predicted to be its target circRNA and mRNA, respectively. Subsequently, the circAbca1/miR-135b-5P/KLF4 regulatory axis was predicted and constructed, and its targeted binding was verified. After inhibiting circAbca1, GAP43 expression was upregulated. Differential expression of circRNAs might play an important role after TSCI. circAbca1 plays a neuroinhibitory role by targeted binding of the miR-135b-5P/KLF4 axis. The identified circRNA/miRNA/mRNA network could provide the basis for understanding pathophysiological mechanisms underlying TSCI, as well as guide the formulation of related therapeutic strategies. All animal protocols were approved by the Research Ethics Committee of West China Hospital of China (approval No. 2017128) on May 16, 2017.
Chinese Library Classification No. R452; R363; R364
Introduction
Traumatic spinal cord injury (TSCI) is a type of central nervous system injury with high mortality and morbidity (Chen et al., 2020; Jiang et al., 2020; Ray et al., 2020). Damage often leads to loss and death of nerve tissues, causing sensory and motor dysfunction (Fouad et al., 2011; Holmes, 2017). Motor vehicle collision is the most common cause of TSCI. However, with the global population aging, the proportion of TSCI due to falls has increased from 16% in 2000 to 30% in 2012 (Cristante et al., 2012). Peripheral nerve fibers project from the spinal cord to relay signals to and from muscles and organs. However, unlike peripheral nerves, neural connections in the spinal cord are rarely repaired after injury; accordingly, almost all patients with TSCI experience pain throughout their lifetime. Current understanding of the underlying mechanisms of TSCI remain limited and there is no effective treatment for this condition (Tran et al., 2018). Therefore, it is necessary to better understand the cellular and molecular pathophysiological mechanisms following TSCI to identify new therapeutic strategies that enhance axonal regeneration and functional recovery.
Modern biological techniques have confirmed that 98% of the human genome comprises RNAs that do not code for protein – most are called noncoding ribonucleic acids (ncRNAs). ncRNAs play key roles in the regulation of genetic programming, epigenetics, transcription, and translation in many pathophysiological processes (Geng et al., 2018). Circular RNAs (circRNAs), a recently discovered subtype of the ncRNA family, have a closed-loop structure with no 5′ to 3′ polarity or polyadenylated tail. Compared with linear RNAs, circRNAs have a more stable structure and are highly expressed (Zhang and Xin, 2018). Studies have found that circRNAs such as circRim2, circEl2, circPlxnd1, and circDym are abundant and function in the nervous system (Rybak-Wolf et al., 2015; Wang et al., 2018a). However, further investigation is required to predict and verify the functions of many newly identified circRNAs. MicroRNAs (miRNAs) are evolutionarily conserved, single-stranded ncRNAs composed of 18–23 nucleotides. miRNAs regulate gene expression at the post-transcriptional level, either by degradation or inhibiting translation, by binding to the 3′ UTR of target mRNAs (Ameres and Zamore, 2013). circRNA/miRNA/mRNA networks have been shown to play important regulatory roles in several studies (van Rossum et al., 2016; Han et al., 2018; Kleaveland et al., 2018). However, roles of the circRNA/miRNA/mRNA network in TSCI remain to be systematically investigated. Our present study employed high-throughput sequencing of miRNAs and circRNAs to screen for significantly expressed miRNAs following TSCI. After matching these miRNAs with differentially expressed circRNA profiles, signaling pathway and function analysis were performed, and the resulting predictions were used to construct an interaction network. After verifying the predicted targeted binding relationships, we preliminarily validated the interaction axes in vitro.
The aim of this study was to predict and construct a circRNA/miRNA/mRNA network that plays a potential regulatory role after TSCI.
Materials and Methods
Animals
All animal protocols were approved by the Research Ethics Committee of West China Hospital, China (No. 2017128) on May 16, 2017. Animal experiments met the requirements of the ARRIVE Guidelines (https://arriveguidelines.org/). Forty-eight adult male mice provided by the Experimental Animal Center of Sichuan University (license No. SCXK (Chuan) 2018-026), specific pathogen-free, 8 weeks of age, body weight 18–22 g, were used in animal experiments. Animals were housed in a clean facility under controlled conditions of 22–24°C, relative humidity of 30–50%, and a 12-hour light/dark cycle.
Construction of a mouse SCI model and tissue acquisition
Mice were randomly divided into six groups (eight mice per group): three underwent SCI (SCI1, SCI2, and SCI3), and the remaining three underwent a sham operation with laminectomy only (Sham1, Sham2, and Sham3). Mouse models were established as previously described (Wang et al., 2018c). Briefly, after anesthesia with pentobarbital sodium (30 mg/kg), tissue layers were incised, exposed, and localized to the thoracic T8–10 segment. In sham groups, vertebral plates were removed before suture. In SCI groups, contusion injuries were created using an Allen’s drop impactor with a drop weight of 6 g and drop height of 60 mm. At the moment of injury, the mouse tail was lifted high or swung violently, and the lower limbs convulsed violently, which were regarded as criteria for the successful induction of injury induction. The wound was then closed by suturing (Xie et al., 2018). Tissues of the lesion epicenter were extracted from seven randomly selected mice per group and combined as one sample for sequencing and qRT-PCR. Tissues from the lesion epicenter of the remaining mice in each group were used for hematoxylin and eosin (HE) staining. Spinal cord samples were collected separately after 5 days. After incision, spinal cord tissue in the lesion epicenter was dissected and subjected to RNA extraction by adding TRIzol (Invitrogen, Carlsbad, CA, USA), followed by chloroform. The middle layer was extracted after centrifugation, and isopropanol was added. The resulting mixture was centrifuged and the supernatant was discarded. Ethanol was added and the mixture was further centrifuged, followed by drying of the precipitate. RNase-free water was added to dissolve the resulting pellet.
HE staining of tissues at the site of spinal cord injury
At 5 days post-SCI, mice were administered anesthesia and a previously described protocol was used to harvest and stain spinal cord tissues (Wang et al., 2018c). Briefly, 4% paraformaldehyde was perfused into mice for fixation, and the lesion epicenter of spinal tissues was extracted and placed in 4% paraformaldehyde overnight, followed by dehydration, paraffin embedding, and sectioning. After xylol dewaxing, samples were dehydrated in an ethanol gradient, stained with hematoxylin, differentiated with 1% hydrochloric acid, and stained with eosin. After dehydration using an ethanol gradient, samples were immersed in xylol and mounted in neutral gum. Tissue integrity was compared between SCI and sham groups under a light microscope (Olympus Corporation, Tokyo, Japan).
Sequencing library construction and sequencing analysis
Total RNA was extracted from spinal cord tissue and measured with a spectrophotometer (Invitrogen) according to the ratio of optical densities at 260 nm and 280 nm (OD260/OD280). To generate a sequencing library for circRNA, total ribosomal RNA was first removed. Purified and recovered residual RNA was randomly broken into short fragments using fragmentation reagents. Fragmented circRNA was used as a template to synthesize single-stranded cDNA with a six-base random primer, before further synthesis into double-stranded cDNA using dNTPs, RNaseH, and DNA polymerase I. Double-stranded products were purified using AMPure XP beads (Beckman Coulter, Brea, CA, USA), and the sticky ends of DNA were repaired to blunt ends using T4 and Klenow DNA polymerases. After adding adenosine at the 3′ end, a linker was added. Subsequently, AMPureXP beads were used for fragment selection. Finally, PCR amplification was performed to establish a sequencing library. The Hiseq2000/2500 platform (Illumina, San Diego, CA, USA) was used for sequencing with a read length parameter of 2 × 150 bp. The miRNA library was treated with a TruSeq Small RNA Sample Prep Kit (Illumina) and sequenced with an Illumina Hiseq 2000/2500. Library sequencing was performed as single-ended 1 × 50 bp reads.
Expression profile analysis and differential expression data collation
After obtaining the original sequencing data, differential expression was identified and analyzed. RNA circularization can be divided into exon circularization and intron circularization, both have reverse cleavage sites that are used to identify the sequence of the circRNA. CIRCExplorer was used to predict the circRNA sequence, and TopHat was used for alignment. circRNA positional relationship classification was used for annotation, and the distribution of gene expression values was calculated and compared between different samples to confirm their biological reproducibility. Finally, statistical analysis of differential gene expression in different tissue samples was performed. The coincidence of circRNA expression in SCI and Sham groups was analyzed using a Venn diagram, and circRNAs with low expression were excluded using the following conditions: P < 0.05 and |log2(fold change)| ≥ 1. A t-test was used for comparison, and differential expression factors were arranged in descending order. Selected miRNA sequences were compared with those of known miRNAs in the miRBase 21.0 database (http://www.mirbase.org/) and used for RNA secondary structure prediction. Finally, miRNAs of different tissue samples were statistically compared, and those that were significantly expressed after injury were selected.
Gene Ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed circRNA
The results of differential expression analysis were further analyzed for function and signaling pathway enrichment. In GO enrichment analysis (Gene Ontology, 2015), the basic unit is the term, with each term corresponding to an attribute. GO enrichment analysis first maps all significant differentially expressed genes to terms in the GO database (http://www.geneontology.org/), and then calculates the number of genes for each term and employs a hypergeometric test. Enriched genes were compared with the whole genome background with P ≤ 0.05 as the threshold. GO terms that were significantly enriched among the significantly differentially expressed genes were screened, and the main biological functions of differentially expressed genes were predicted. GO term enrichment analysis results represent biological processes, cellular components, and molecular functions. KEGG (http://www.genome.jp/kegg/) (Kanehisa et al., 2017) the main public database of pathways, is commonly used for pathway enrichment analyses. This hypergeometric test identifies significantly enriched pathways, representing the most important biochemical metabolic and signal transduction pathways associated with differentially expressed genes.
miRNA/circRNA interaction prediction analysis
The results of differential expression analysis were comprehensively analyzed to predict targeted interaction relationships. Targetscan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/) (Zhang et al., 2014) software were used to predict miRNA target genes and miRNA/circRNA interactions. Targetscan predicts complete sequence complementation information by seed matching in the seed region. miRanda constructs a scoring matrix to predict target information by base complementation (A=U, G=C). Default parameters were used in Targetscan, and a Max_Energy threshold < –10 was set during miRanda analysis. Overlapping results obtained by these two algorithms were considered the final result of this interaction analysis. Differential circRNA analysis was performed using the default difference threshold, |log2(fold change)| ≥ 1, and P ≤ 0.05. According to the number of reverse-cleaved back-splicing sites of the circRNA, accuracy was high and the proportion of false positives was low. The threshold for the TargetScan value was set to 50, such that the higher the value, the higher the combination rate. The threshold set was -10 for miRanda, as smaller values allow for higher combination rates. Finally, a miRNA/circRNA interaction map was drawn using Cytoscape to visualize the predicted interaction network information.
qRT-PCR quantitation
To verify sequencing results, total RNA extracted from tissue was subjected to PCR detection. qRT-PCR was used to quantitate expression of circRNAAbca1 (circAbca1), mmu-miR-21a-5p_R+1, KLF4, GAPDH, and U6 (Table 1). A reverse transcription reagent kit (TUREscript 1st Stand cDNA Synthesis Kit, Aidlab, Beijing, China) was used for reverse transcription, and cDNA was used as a template to prepare a 20-μL reaction system for PCR in an ABI PRISM 7500 RT-PCR System (Applied Biosystems, Foster City, CA, USA). SYBR green dye was used to detect amplification in real time. GAPDH was used as an internal reference for circRNA and mRNA, while U6 was used as an internal reference for miRNA expression analysis. Pre-denaturation at 95°C was performed for 3 minutes, followed by 40 cycles of 95°C for 10 seconds and annealing/extension at 58°C for 30 seconds. miRNA amplification conditions were pre-denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 2 seconds and annealing/extension at 60°C for 30 seconds. The experiment was repeated three times and data were analyzed using the 2–ΔΔCt method (Wang et al., 2018c).
Table 1.
Primers used in qRT-PCR
| Gene | Sequence (5′–3′) | |
|---|---|---|
| circRNA-Abca1 | Forward: | CTG ATC TCT GTA CGC CTG A |
| Reverse: | GGA GTT GGA TAA CGG AAG C | |
| mmu-miR-21a- | Forward: | GGC GGT ATG GCT TTT CAT TC |
| 5p_R+1 | Reverse: | TGT CGT ATC CAG TGC AGG GTC CGA GGT ATT |
| CGC ACT GGA TAC GAC TCA CAT | ||
| KLF4 | Forward: | TTC TCA TCT CAA GGC ACA C |
| Reverse: | CAC ACT TCT GGC ACT GAA | |
| GAPDH | Forward: | GGT GAA GGT CGG TGT GAA CG |
| Reverse: | CTC GCT CCT GGA AGA TGG TG | |
| U6 | Forward: | CTC GCT TCG GCA GCA CA |
| Reverse: | AAC GCT TCA CGA ATT TGC GT | |
GAPDH: Glyceraldehyde-phosphate dehydrogenase; KLF4: Kruppel-like factor 4; qRT-PCR: quantitative real-time polymerase chain reaction; SCI: spinal cord injury; U6: RNU6-1.
Luciferase reporter assay
To verify the predicted targeted binding relationship, 293T cells were seeded into 96-well plates at 70% confluence and transfected with the experimental plasmid pMIR-REPORT Luciferase-circAbca1 (WT) (LC Science, Houston, TX, USA) or pMIR-REPORT Luciferase-circAbca1 (MUT1 + MUT2) (LC Science) after 24 hours. Subsequently, a luciferase reporter plasmid (Firefly and Renilla) (Invitrogen) was transfected with 0.25 μL of microRNA transfection reagent (Invitrogen) for 48 hours in each well. A dual-luciferase reporter assay (E1910; Promega, Madison, WI, USA) was then performed with 50 μL of pre-mixed Stop & Glo Reagent (Invitrogen). Results were recorded at 2-second intervals.
Cell culture and transfection
The targeted binding relationship was preliminarily verified in vitro. Primary C57BL/6 mouse dorsal root ganglion cells (PriCells, Wuhan, China) were cultured at 37°C and 5% CO2 in Neurobasal medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 1% B-27™ serum-free supplement (50X, Thermo Fisher Scientific) and 100 kU/L of penicillin and streptomycin (Thermo Fisher Scientific). Cultured cells were transfected with circAbca1 inhibitor (5′-CCA ACT CAA TGT CAG CTG T-3′) or the corresponding controls (Ribobio, Guangzhou, China). Next, 120 μL of 1× riboFECT™ Buffer was diluted with 5 μL and 20 μM siRNA stock solution, and then 12 μL riboFECT™ Reagent (Ribobio, Guangzhou, China) was added and mixed at room temperature for 10 minutes. The mixture was added to medium without penicillin and streptomycin, and then added to cells for an additional 48 hours of culture.
Western blot analysis
After transfection, cell samples were collected and total protein was extracted. Proteins were incubated with cold lysis buffer (Solarbio, Beijing, China), centrifuged, concentrated, electrophoresed, and subsequently transferred onto polyvinylidene fluoride membranes. After blocking with phosphate-buffered saline containing 5% non fat milk overnight at 4°C (Wang et al., 2018c), membranes were incubated with antibodies against growth-associated protein 43 (GAP43; rabbit monoclonal, 1:1000; Abcam, Cambridge, UK) and GAPDH (rabbit monoclonal, 1:2500; Abcam) overnight at 4°C. The following day, membranes were treated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit IgG, 1:5000; Proteintech, Wuhan, China). An enhanced West Pico ECL Substrate kit (Solarbio) was utilized to perform chemiluminescent detection. GAP43 expression was quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA) and normalized to GAPDH.
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad Software, La Jolla, CA, USA) and SPSS 20.0 software (IBM, Armonk, NY, USA). Data are expressed as mean ± standard deviation (SD). Two-sample t-test, one-way analysis of variance, and Dunnett’s test were used to assess statistical significance. P-values < 0.05 were considered statistically significant.
Results
Establishment of SCI mouse models and expression profiles of circRNAs
To explore expression profiles of circRNAs in the lesion epicenter after SCI, a mouse model of SCI was established using an Allen’s drop method. HE-stained sections of SCI and sham groups were observed by microscopy. Nuclei were stained dark blue by hematoxylin, while cytoplasm appeared pink and collagen fibers were pale pink after eosin staining. The spinal cord structure in sham groups was clear with intact and unruptured meninges, and a centered central tube. Moreover, there was no bleeding or signs of damage (Figure 1A). In contrast, the spinal cord structure of SCI groups exhibited significant damage and rupture, meningeal defects, and central tube offset deformation. Extensive red blood cell diffusion and inflammatory cell infiltration caused by vascular rupture were also observed (Figure 1B). A cluster map and volcano plot show differentially expressed circRNAs after TSCI (Figure 1C and D).
Figure 1.
Establishment of SCI mouse models and expression profiles of circRNA.
Light microscopy of HE-stained spinal cord samples at 5 days post-surgery. (A) Sham group (scale bars: 100 μm, left; 50 μm, right). (B) SCI group (scale bars: 100 μm, left; 50 μm, right). (C) Hierarchical clustering analysis of circRNAs differentially expressed between sham and SCI groups. Red pixels correspond to increased gene abundances in the indicated sample, whereas blue pixels indicate decreased gene abundances; fold-change > 2.0 and P < 0.05. (D) Volcano plot showing significantly differentially expressed circRNAs; red pixels represent differentially expressed genes, while gray pixels indicate genes with no difference; fold-change > 2.0 and P < 0.05. HE: Hematoxylin and eosin; SCI: spinal cord injury.
Enriched GO terms and KEGG pathways of differentially expressed circRNAs
To explore major biological functions and significantly enriched pathways, GO and KEGG analyses were performed. Differential expression profiles of circRNAs were further screened according to their expression level and log2(fold change). Enriched terms for differentially expressed genes identified by GO term analysis were used to determine their major biological functions. The results of GO enrichment analysis showed that the highest degree of enrichment was associated with ‘negative regulation of plasminogen activation’, ‘peptide cross linking’, ‘positive regulation of the endothelial cell apoptotic process’, and ‘macropinosome’ (Figure 2A). Analysis of significantly enriched KEGG pathways revealed the most important biochemical metabolic pathways and signal transduction pathways encoded by differentially expressed genes included ‘complement and coagulation cascades’, ‘glycerolipid metabolism’, ‘one carbon pool by folate’, ‘pentose phosphate pathway’, ‘bladder cancer’, and ‘P53 signaling pathway’ (Figure 2B).
Figure 2.

Enriched GO terms and KEGG pathways of differentially expressed circRNAs in SCI mice.
GO enrichment and KEGG pathway analysis of mRNAs originating from the host genes of altered circRNAs. (A) Comparison of enriched GO terms between sham and SCI groups. (B) Comparison of enriched KEGG pathways between sham and SCI groups. Rich factor: Ratio of the number of differentially expressed genes identified by KEGG pathway analysis to the total number of genes in the KEGG pathway; the larger the Rich factor value, the greater the level of enrichment. Dot size represents the number of enriched genes (the larger the dot, the more enriched genes). A change in the color of the dot from blue to red represents a decrease in P value. circRNA: Circular RNA; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; SCI: spinal cord injury.
Prediction of interactions between differentially expressed miRNA/circRNAs
To investigate regulatory roles of the targeted interaction axis after SCI, we predicted the miRNA/circRNA axis using bioinformatic analyses. After TSCI, differential expression profiles of circRNAs and miRNAs were screened according to their expression and log2(fold change). miR-135b-5p was significantly downregulated (P < 0.05), as it satisfied the significance threshold of |log2(fold change)| ≥ 1. Indeed, miR-135b-5p showed the most significant reduction in expression after SCI (log2(fold change) = –1.01) (Figure 3A). High-throughput sequencing revealed significant upregulation (P < 0.05), of circAbca1 after TSCI (Figure 3B). Differentially expressed circRNAs were further screened based on their binding targets. In total, six differentially expressed circRNAs that potentially bind miR-135b-5p were screened, including circAbca1, circRNA5953, circRNA15386, circRNA3868, circRNA6769, and circRNA9429 (Figure 3C). Based on comprehensive analysis of backsplice junction loci, TargetScan, and miRanda scores, we identified circAbca1 as a key circRNA that interacts with miR-135b-5p to influence pathophysiological processes after TSCI. Reverse-screens identified miRNAs that bind to circAbca1, including miR-135b-5p, miR-23a-5p, miR-135a-5p, miR-155-5p, and miR-138-5p, which also target the bound mRNAs. Together with circAbca1, these miRNAs/mRNAs were used as the core for constructing the circRNA/miRNA interaction network after TSCI (Figure 3D).
Figure 3.
circRNA–miRNA–mRNA regulatory interaction network analysis.
(A, B) Sequencing results of miR-135b-5p and circAbca1. FPKM: Fragments per kilobase of exon model per million reads mapped. Data are expressed as the mean ± SD (n = 3). *P < 0.05, **P < 0.01 (two-sample t-test). (C) miR-135b-5p and targeted circRNAs. (D) circ-Abca1 and targeted miRNAs-mRNAs. Interaction networks were predicted using Targetscan software. circRNA: Circular RNA; miRNA: microRNA; SCI: spinal cord injury.
Validation of differentially expressed ncRNAs and confirmation of their relationship
To verify the accuracy of high-throughput sequencing and confirm the circAbca1/miR-135b-5p relationship, we performed qRT-PCR and fluorescent reporter assays. qRT-PCR was used to validate expression of miR-135b-5p and circAbca1, which showed significant differential expression following library sequencing. qRT-PCR results were consistent with our sequencing results (Figure 4A and B). We predicted a targeted binding site for circAbca1 and miR-135b-5p (Figure 4C), and constructed a luciferase reporter plasmid (Figure 4D) to validate the targeted binding relationship. Luciferase activity of the miR-135b-5p mimic group was significantly lower than that of the negative control group before mutation (P < 0.05), but this difference disappeared post-mutation. These results confirm that circAbca1 can bind to miR-135b-5p (Figure 4E), consistent with our predicted results.
Figure 4.
Validation of differential non-coding RNA expression and confirmation of relationships among non-coding RNAs.
(A, B) Expression of circAbca1 and miR-135b-5p validated by qRT-PCR. Data are expressed as the mean ± SD, n = 3, ***P < 0.001 (two-sample t-test). (C) Annotation of the miR-135b-5p target site in circAbca1 using bioinformatics analysis. (D) Construction of WT circAbca1-bearing luciferase plasmids. (E) Relative luciferase activity of 293T cells transfected with WT or MT circAbca1 vectors and miR-135b-related vectors. MT: Mutant type; NC: negative control; qRT-PCR: quantitative real time polymerase chain reaction; WT: wild type.
Validation of functional target gene expression and confirmation of the network relationship
To validate expression of Kruppel-like factor 4 (KLF4) and verify the miR-135b-5p/KLF4 relationship, qRT-PCR and fluorescent reporter assays were performed. Subsequently, the potential neuroinhibitory function of circAbca1 was determined by western blot assay. qRT-PCR was used to validate KLF4 expression, while high-throughput sequencing revealed significant upregulation (P < 0.05) of miR-135b-5p after TSCI (Figure 5A), consistent with our qRT-PCR results (Figure 5B). The Luc-KLF4 3´UTR-WT plasmid (Figure 5C) was synthesized to validate the targeted binding relationship. The binding site for KLF4 and miR-135b-5p was predicted by Targetscan software (Figure 5D). Luciferase reporter assay results indicated significantly decreased (P < 0.05) luciferase activity in 293T cells co-transfected with MT vector or miR-135b mimics (Figure 5E).
Figure 5.
Validation of functional target gene expression and confirmation of the network relationship.
(A) Sequencing results for KLF4. (B) Expression of KLF4 validated by qRT-PCR. Data are expressed as the mean ± SD (n = 3), *P < 0.05, ***P < 0.001 (two-sample t-test). (C) Construction of KLF4-bearing luciferase plasmids. (D) Targetscan software predicted KLF4 as a target gene of miR-135b. (E) Relative luciferase expression of WT and MT KLF4 vectors co-transfected with miR-135b-5p vectors. (F) qRT-PCR was conducted to detect RNA expression of circAbca1, miR-135b, and KLF4 in mouse spinal dorsal root ganglion neurons following transfection with the circAbca1 inhibitor or scramble circ-NC. Data are expressed as the mean ± SD, n = 3, **P < 0.005, ***P < 0.001 (one-way analysis of variance and Dunnett’s test). (G) Western blot assay was performed to detect protein expression of GAP43 in mouse spinal dorsal root ganglion neurons following transfection with the circAbca1 inhibitor. circ-NC: Scramble circRNA negative control; KLF4: Kruppel-like factor 4; Mock: equal volume of phosphate-buffered saline was added in the blank group; MT: mutant type; UTR: untranslated region; WT: wild type.
To confirm the correlation between circAbca1, miR-135b-5p, and KLF4 in primary mouse dorsal root ganglion spinal neurons, they were transfected with circAbca1 inhibitors. Transfection efficiency and expression of miR-135b and KLF4 were detected by qRT-PCR. The results revealed that silencing of circAbca1 increased miR-135b-5p expression, but decreased KLF4 expression (Figure 5F). To further explore the potential neuroprotective functions of circAbca1 inhibition, a western blot assay was conducted. The results show that GAP43 expression was upregulated after circAbca1 silencing (Figure 5G).
Discussion
TSCI is mainly caused by traffic accidents and falling from high places, but can also be caused by vascular lesions, tumors, and iatrogenic injuries, which often lead to paralysis, unconscious movement, urinary and bowel incontinence, and depression (Cristante et al., 2012). Trauma causes neuronal death and damage to nerve connections, thereby disrupting the upstream and downstream excitation of nerves (Schwab et al., 2018; Song et al., 2018). Physical trauma usually leads to a prolonged secondary injury cascade (Tran et al., 2018) that starts with the expansion of tissue damage from the lesion epicenter; the process is divided into acute, subacute, and chronic phases (Sofroniew, 2018). The subacute phase (from 3 days to 2 weeks post-injury) is considered the best target for future cell-based therapeutic strategies (Rowland et al., 2008).
Increasing evidence suggests that non-coding RNAs play an important role in the progression of TSCI (Ning et al., 2014; Kleaveland et al., 2018). Although more than 70% of human genes are transcribed, less than 2% are translated into proteins; most of the remaining genes are transcribed into ncRNAs (Djebali et al., 2012). ncRNAs can be divided into two major categories according to their intrinsic functions: housekeeping ncRNAs, which include nuclear small RNA, ribosomal RNA, and handling RNA; and regulatory ncRNAs, which include miRNAs, circRNAs, and lncDNAs (Qu et al., 2015). Currently, the most comprehensively studied regulatory ncRNA is miRNA, which are widely distributed in various tissues and body fluids (Ameres and Zamore, 2013; Stoicea et al., 2016). This is particularly obvious in the nervous system, where miRNAs can act as regulatory or even hormone-like factors to affect communication and information transmission between target cells through autocrine or paracrine pathways, which has considerable effects on neurophysiology and neural regeneration (Kleaveland et al., 2018). circRNAs are a new class of ncRNA molecules that play important regulatory roles in cell biology; currently, study of circRNAs is still in an initial stage (Qi et al., 2015; Ren et al., 2019). Unlike other linear structures of ncRNAs, circRNAs exhibit a unique enclosed-loop structure and are more stable than linear RNA (Salzman et al., 2013). In the nervous system, circRNAs are expressed in large quantities and highly involved in nerve development, degeneration, and axon growth (Veno et al., 2015; You et al., 2015). Currently, it is believed that the most important role of circRNAs is their activity as miRNA-adsorbing sponges to exert post-transcriptional regulatory functions. For example, recent studies found that the lncCyrano/miR-7 interaction network regulates neural activity (Li et al., 2017; Kleaveland et al., 2018; Zhang et al., 2018).
In previous studies, moderate SCI was performed using a modified Allen’s weight drop apparatus (Xie et al., 2018). Excessive damage leads to death or paralysis of mice, but low-intensity damage leads to quick recovery, which reduces observation time. We found that a parameter setting of 6 g/60 mm caused ideal damage. Mice exposed to this condition lose lower extremity motor function within 2 days and gradually recover within 3–21 days after injury (Xie et al., 2018). In the present study, high-throughput sequencing of differentially expressed circRNAs and miRNAs was performed at 5 days post-TSCI. Day 5 post-injury represents the initial period of the subacute phase and scar formation; posttranscriptional regulation may play an important role in pathophysiological processes of the subacute phase. Subsequently, differential expression profiles of circRNAs and miRNAs following subacute injury were obtained.
GO and KEGG analysis of host genes related to differentially expressed circRNAs after TSCI was performed from a macroscopic viewpoint. The results revealed that differentially expressed genes were: 1) mainly involved in transcriptional regulation and protein phosphorylation; 2) abundant in cytoplasm, nucleus, exosomes, and plasma membranes; and 3) play a role in protein, adenosine triphosphate, and metal ion binding. Interestingly, some classical pathways were significantly differentially enriched; for example, pentose-phosphate, calcium signaling, p53 signaling, and transforming growth factor beta (TGF-β) signaling pathways. Activation of the pentose-phosphate pathway reduces neuronal oxidative stress (Mashima et al., 2018), while p53 pathway activation drives motor neuron degeneration (Simon et al., 2017) and the calcium signaling pathway regulates synaptic activity (Brini et al., 2014). The TGF-β signaling pathway is one of the most important signaling pathways regulating scar formation and axon regeneration after TSCI, as observed in our previous study (Liu et al., 2018; Wang et al., 2018c).
To determine the specific research core-ncRNA, miR-135b-5p was identified as the most significantly downregulated miRNA after TSCI. miRNA-135b-5p can regulate tumor proliferation, migration, and apoptosis in gastric carcinogenesis, cervical cancer, colorectal cancer, mammary carcinomas, and osteosarcoma (Bhinge et al., 2014; Valeri et al., 2014; Chu et al., 2018). miRNA-135b-5p also promotes hypoxia-induced vascular endothelial cell injury (Yang et al., 2018), chondrocyte proliferation (Wang et al., 2018b), pathological cardiac hypertrophy (Chu et al., 2018), and odontoblast-like differentiation (Song et al., 2017). Most importantly, in the nervous system, miR-135b-5p plays a neuroprotective role by targeting GSK3β3 (Duan et al., 2018; van Battum et al., 2018), and can regulate neuroectoderm formation through TGF-β/bone morphogenic protein signaling (Bhinge et al., 2014). miR-135b-5p can also participate in construction of the network and become its core. It was found that the ncRNA interaction network, made up of lncRNAGAS8-AS1-miR-135b-5p, lncRNAGAS5-miR-135b-5p, and CDR1as-miR-135b-5p, can interfere with cancer progression (Xue et al., 2017; Chen et al., 2019a, b). Our study predicted that miR-135b-5p is not just a part of the ncRNA/mRNA interaction network, but at its core. Among all differentially expressed circRNAs capable of binding to miR-135b-5p, circAbca1 exhibited the highest loop index and highest binding rate, and thus was selected for axial prediction.
KLF4 is an evolutionarily conserved zinc finger-containing transcription factor that plays key roles in diverse biological processes including proliferation, differentiation, cell growth, and apoptosis (Yuan et al., 2020). KLF4 may inhibit axon regeneration and suppress the migration and maturation of palingenetic neurons (Moon et al., 2018). Most importantly, KLF4 is the target gene of miR-135b-5p and regulated by the upstream component miR-135b-5p in gastric cancer cells (Chen et al., 2020a, b). Based software prediction, sequencing analysis, and literature retrieval, KLF4 was selected as a downstream component in the axial prediction and a complete targeting circAbca1/miR-135b-5p/KLF4 interaction axis was established. Gene expression was verified by PCR, while the results of luciferase reporter assays confirmed that both KLF4 and circAbca1 are target genes of miR-135b. Preliminary verification using circ-inhibitor transfection and detection of functional proteins revealed upregulated expression of miR-135b-5p and downregulated expression of KLF4 following circ inhibition, indicating that circAbca1 could dysregulate the expression of downstream RNAs. In addition to the axial annotation, GAP43 (Li et al., 1996), which plays a critical role in axon growth and regeneration, was also detected. Our results indicated upregulation of GAP43 following transfection with a circAbca1 inhibitor, consistent with KLF4 downregulation.
In conclusion, the present study preliminarily verified the structure and function of the circAbca1/miR-135b-5p/KLF4 interaction axis, and found that circAbca1 inhibitors play a neuroprotective role via this interaction network. However, the present study had some limitations. circRNAs commonly bind to several miRNAs; for example, circHIPK3 simultaneously binds to ten miRNAs (miR-654, miR-29b, miR-338, miR-193a, miR-152, miR-193a, miR-379, miR-584, miR-124, and miR-29a) (Zheng et al., 2016). Moreover, miRNAs bind to several mRNAs (Li et al., 2017; Zhang et al., 2018). Therefore, blocking only circRNA is insufficient to confirm network relationships. Indeed, the regulatory process likely involves a network rather than a single thread, and multiple targeted mRNAs bound by the same miRNA may play synergistic or antagonistic roles in the pathophysiological process after SCI through multiple pathways. Interestingly, miR-135b-5p can also bind to the TGF-β receptors TGF-βR1 and TGF-βR2 (Li et al., 2015; Wang et al., 2018b). Previously, we confirmed that the TGF-β signaling pathway is important for regulation of scar formation and axon regeneration after TSCI (Liu et al., 2018; Wang et al., 2018c). In subsequent studies, we will focus on the biological roles and mechanisms of the circAbca1/miR-135b-5p/KLF4 interaction network induced by TSCI in this study. Specifically, we will explore and validate the pathological and physiological functions associated with subacute inflammation. We hope that new ncRNA interaction networks and related factors, which can help elucidate the mechanisms underlying TSCI and reveal therapeutic targets and prognostic indicators, can be eventually discovered.
Additional file: Open peer review report 1 (95.6KB, pdf) .
Acknowledgments:
The authors express their thanks to Dr. Jia-Nan Chen and Dr. Hong-Fei Li, Shandong University, China and Dr. Yan-Lin Su, Huazhong University of Science and Technology, China for their help in establishing animal models.
Footnotes
Conflicts of interest: None declared.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81874002 (to LL); the Science and Technology Support Project of Sichuan Province of China, Nos. 2018SZ0159 (to LL), 2018SZ0246 (to XLH); the Innovation and Entrepreneurship Project of Sichuan Technology Gallery of China, Nos. 2019JDRC0100 (to JL), 2020JDRC0054 (to WZW); the National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, China, Nos. Y2018B22 (to LL), Z20192013 (to JL) and West China Hospital Postdoctoral Research and Development Fund, No. 2019HXBH068 (to JL). The funding bodies played no role in the study design, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Institutional review board statement: All animal protocols were approved by the Research Ethics Committee of West China Hospital of China (approval No. 2017128) on May 16, 2017. The experimental procedure followed the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Ronak Reshamwala, Griffith University, Australia.
Funding: This study was supported by the National Natural Science Foundation of China, No. 81874002 (to LL); the Science and Technology Support Project of Sichuan Province of China, Nos. 2018SZ0159 (to LL), 2018SZ0246 (to XLH); the Innovation and Entrepreneurship Project of Sichuan Technology Gallery of China, Nos. 2019JDRC0100 (to JL), 2020JDRC0054 (to WZW); the National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, China, Nos. Y2018B22 (to LL), Z20192013 (to JL) and West China Hospital Postdoctoral Research and Development Fund, No. 2019HXBH068 (to JL).
P-Reviewer: Reshamwala R; C-Editor: Zhao M; S-Editors: Wang J, Li CH; L-Editor: Song LP; T-Editor: Jia Y
References
- 1.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 2.Bhinge A, Poschmann J, Namboori SC, Tian X, Jia Hui Loh S, Traczyk A, Prabhakar S, Stanton LW. MiR-135b is a direct PAX6 target and specifies human neuroectoderm by inhibiting TGF-beta/BMP signaling. EMBO J. 2014;33:1271–1283. doi: 10.1002/embj.201387215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brini M, Cali T, Ottolini D, Carafoli E. Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Mao M, Jiang J, Zhu D, Li P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019a;12:3869–3879. doi: 10.2147/OTT.S207938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Shi XY, Cheng J, Ye YC, Zhang ZW, Li XH, Sun HT. Protective effect of human umbilical cord blood mesenchymal stem cells on the rat’s blood-brain barrier after traumatic brain injury. Zhongguo Zuzhi Gongcheng Yanjiu. 2020a;24:3947–3952. [Google Scholar]
- 6.Chen N, Yin D, Lun B, Guo X. LncRNA GAS8-AS1 suppresses papillary thyroid carcinoma cell growth through the miR-135b-5p/CCND2 axis. Biosci Rep. 2019b;39:BSR20181440. doi: 10.1042/BSR20181440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Gao Y, Gao S, Song D, Feng Y. MiR-135b-5p promotes viability, proliferation, migration and invasion of gastric cancer cells by targeting Kruppel-like factor 4 (KLF4) Arch Med Sci. 2020b;16:167–176. doi: 10.5114/aoms.2019.87761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu Q, Li A, Chen X, Qin Y, Sun X, Li Y, Yue E, Wang C, Ding X, Yan Y, Zahra SM, Wang S, Jiang Y, Bai Y, Yang B. Overexpression of miR-135b attenuates pathological cardiac hypertrophy by targeting CACNA1C. Int J Cardiol Heart Vasc. 2018;269:235–241. doi: 10.1016/j.ijcard.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Cristante AF, Barros Filho TE, Marcon RM, Letaif OB, Rocha ID. Therapeutic approaches for spinal cord injury. Clinics. 2012;67:1219–1224. doi: 10.6061/clinics/2012(10)16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan Q, Sun W, Yuan H, Mu X. MicroRNA-135b-5p prevents oxygen-glucose deprivation and reoxygenation-induced neuronal injury through regulation of the GSK-3beta/Nrf2/ARE signaling pathway. Arch Med Sci. 2018;14:735–744. doi: 10.5114/aoms.2017.71076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouad K, Krajacic A, Tetzlaff W. Spinal cord injury and plasticity: opportunities and challenges. Brain Res Bull. 2011;84:337–342. doi: 10.1016/j.brainresbull.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 13.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Holmes D. Spinal-cord injury: spurring regrowth. Nature. 2017;552:S49. doi: 10.1038/d41586-017-07550-9. [DOI] [PubMed] [Google Scholar]
- 17.Jiang JP, Liu XY, Zhao F, Zhu X, Li XY, Niu XG, Yao ZT, Dai C, Xu HY, Ma K, Chen XY, Zhang S. Three-dimensional bioprinting collagen/silk fibroin scaffold combined with neural stem cells promotes nerve regeneration after spinal cord injury. Neural Regen Res. 2020;15:959–968. doi: 10.4103/1673-5374.268974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li GL, Farooque M, Holtz A, Olsson Y. Increased expression of growth-associated protein 43 immunoreactivity in axons following compression trauma to rat spinal cord. Acta Neuropathol. 1996;92:19–26. doi: 10.1007/s004010050484. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Liang H, Bai M, Ning T, Wang C, Fan Q, Wang Y, Fu Z, Wang N, Liu R, Zen K, Zhang CY, Chen X, Ba Y. miR-135b promotes cancer progression by targeting transforming growth factor beta receptor II (TGFBR2) in colorectal cancer. PLoS One. 2015;10:e0130194. doi: 10.1371/journal.pone.0130194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li TR, Jia YJ, Wang Q, Shao XQ, Lv RJ. Circular RNA: a new star in neurological diseases. Int J Neurosci. 2017;127:726–734. doi: 10.1080/00207454.2016.1236382. [DOI] [PubMed] [Google Scholar]
- 23.Liu R, Wang W, Wang S, Xie W, Li H, Ning B. microRNA-21 regulates astrocytic reaction post-acute phase of spinal cord injury through modulating TGF-beta signaling. Aging. 2018;10:1474–1488. doi: 10.18632/aging.101484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mashima K, Takahashi S, Minami K, Izawa Y, Abe T, Tsukada N, Hishiki T, Suematsu M, Kajimura M, Suzuki N. Neuroprotective role of astroglia in parkinson disease by reducing oxidative stress through dopamine-induced activation of pentose-phosphate pathway. ASN Neuro. 2018;10:1759091418775562. doi: 10.1177/1759091418775562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon BS, Bai J, Cai M, Liu C, Shi J, Lu W. Kruppel-like factor 4-dependent Staufen1-mediated mRNA decay regulates cortical neurogenesis. Nat Commun. 2018;9:401. doi: 10.1038/s41467-017-02720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning B, Gao L, Liu RH, Liu Y, Zhang NS, Chen ZY. microRNAs in spinal cord injury: potential roles and therapeutic implications. Int J Biol Sci. 2014;10:997–1006. doi: 10.7150/ijbs.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 28.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Ray SK. Modulation of autophagy for neuroprotection and functional recovery in traumatic spinal cord injury. Neural Regen Res. 2020;15:1601–1612. doi: 10.4103/1673-5374.276322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren C, Liu J, Zheng B, Yan P, Sun Y, Yue B. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif Cells Nanomed Biotechnol. 2019;47:3359–3367. doi: 10.1080/21691401.2019.1649273. [DOI] [PubMed] [Google Scholar]
- 31.Rowland JW, Hawryluk GW, Kwon B, Fehlings MG. Current status of acute spinal cord injury pathophysiology and emerging therapies: promise on the horizon. Neurosurg Focus. 2008;25:E2. doi: 10.3171/FOC.2008.25.11.E2. [DOI] [PubMed] [Google Scholar]
- 32.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 33.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab JM, Maas AIR, Hsieh JTC, Curt A. Raising awareness for spinal cord injury research. Lancet Neurol. 2018;17:581–582. doi: 10.1016/S1474-4422(18)30206-0. [DOI] [PubMed] [Google Scholar]
- 35.Simon CM, Dai Y, Van Alstyne M, Koutsioumpa C, Pagiazitis JG, Chalif JI, Wang X, Rabinowitz JE, Henderson CE, Pellizzoni L, Mentis GZ. Converging mechanisms of p53 activation drive motor neuron degeneration in spinal muscular atrophy. Cell Rep. 2017;21:3767–3780. doi: 10.1016/j.celrep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sofroniew MV. Dissecting spinal cord regeneration. Nature. 2018;557:343–350. doi: 10.1038/s41586-018-0068-4. [DOI] [PubMed] [Google Scholar]
- 37.Song HL, Zhang X, Wang WZ, Liu RH, Zhao K, Liu MY, Gong WM, Ning B. Neuroprotective mechanisms of rutin for spinal cord injury through anti-oxidation and anti-inflammation and inhibition of p38 mitogen activated protein kinase pathway. Neural Regen Res. 2018;13:128–134. doi: 10.4103/1673-5374.217349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Z, Chen LL, Wang RF, Qin W, Huang SH, Guo J, Lin ZM, Tian YG. MicroRNA-135b inhibits odontoblast-like differentiation of human dental pulp cells by regulating Smad5 and Smad4. Int Endod J. 2017;50:685–693. doi: 10.1111/iej.12678. [DOI] [PubMed] [Google Scholar]
- 39.Stoicea N, Du A, Lakis DC, Tipton C, Arias-Morales CE, Bergese SD. The MiRNA journey from theory to practice as a CNS biomarker. Front Genet. 2016;7:11. doi: 10.3389/fgene.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran AP, Warren PM, Silver J. The biology of regeneration failure and success after spinal cord injury. Physiol Rev. 2018;98:881–917. doi: 10.1152/physrev.00017.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, Paone A, Cascione L, Sumani KM, Veronese A, Fabbri M, Carasi S, Alder H, Lanza G, Gafa’ R, Moyer MP, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Battum EY, Verhagen MG, Vangoor VR, Fujita Y, Derijck A, O’Duibhir E, Giuliani G, de Gunst T, Adolfs Y, Lelieveld D, Egan D, Schaapveld RQJ, Yamashita T, Pasterkamp RJ. An image-based miRNA screen identifies miRNA-135s as regulators of cns axon growth and regeneration by targeting kruppel-like factor 4. J Neurosci. 2018;38:613–630. doi: 10.1523/JNEUROSCI.0662-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. doi: 10.1186/s13059-015-0801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang JJ, Liu C, Shan K, Liu BH, Li XM, Zhang SJ, Zhou RM, Dong R, Yan B, Sun XH. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018a;8:3408–3415. doi: 10.7150/thno.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R, Xu B, Xu H. TGF-beta1 promoted chondrocyte proliferation by regulating Sp1 through MSC-exosomes derived miR-135b. Cell Cycle. 2018b;17:2756–2765. doi: 10.1080/15384101.2018.1556063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Liu R, Su Y, Li H, Xie W, Ning B. MicroRNA-21-5p mediates TGF-beta-regulated fibrogenic activation of spinal fibroblasts and the formation of fibrotic scars after spinal cord injury. Int J Biol Sci. 2018c;14:178–188. doi: 10.7150/ijbs.24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie W, Yang SY, Zhang Q, Zhou Y, Wang Y, Liu R, Wang W, Shi J, Ning B, Jia T. Knockdown of MicroRNA-21 promotes neurological recovery after acute spinal cord injury. Neurochem Res. 2018;43:1641–1649. doi: 10.1007/s11064-018-2580-1. [DOI] [PubMed] [Google Scholar]
- 49.Xue Y, Ni T, Jiang Y, Li Y. Long noncoding RNA GAS5 inhibits tumorigenesis and enhances radiosensitivity by suppressing miR-135b expression in non-small cell lung cancer. Oncol Res. 2017;25:1305–1316. doi: 10.3727/096504017X14850182723737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang S, Yin J, Hou X. Inhibition of miR-135b by SP-1 promotes hypoxia-induced vascular endothelial cell injury via HIF-1alpha. Exp Cell Res. 2018;370:31–38. doi: 10.1016/j.yexcr.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 51.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan Y, Park J, Feng A, Awasthi P, Wang Z, Chen Q, Iglesias-Bartolome R. YAP1/TAZ-TEAD transcriptional networks maintain skin homeostasis by regulating cell proliferation and limiting KLF4 activity. Nat Commun. 2020;11:1472. doi: 10.1038/s41467-020-15301-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21. doi: 10.1186/s13045-018-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






