Abstract
Ebola virus is a deadly pathogen responsible for a frequent series of outbreaks since 1976. Despite various efforts from researchers worldwide, its mortality and fatality are quite high. For antiviral drug discovery, the computational efforts are considered highly useful. Therefore, we have developed an 'anti-Ebola' web server, through quantitative structure–activity relationship information of available molecules with experimental anti-Ebola activities. Three hundred and five unique anti-Ebola compounds with their respective IC50 values were extracted from the ‘DrugRepV’ database. Later, the compounds were used to extract the molecular descriptors, which were subjected to regression-based model development. The robust machine learning techniques, namely support vector machine, random forest and artificial neural network, were employed using tenfold cross-validation. After a randomization approach, the best predictive model showed Pearson's correlation coefficient ranges from 0.83 to 0.98 on training/testing (T274) dataset. The robustness of the developed models was cross-evaluated using William’s plot. The highly robust computational models are integrated into the web server. The ‘anti-Ebola’ web server is freely available at https://bioinfo.imtech.res.in/manojk/antiebola. We anticipate this will serve the scientific community for developing effective inhibitors against the Ebola virus.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10291-7.
Keywords: Ebola virus, Machine learning, Prediction algorithm, Random forest, QSAR, Web server
Introduction
Ebola virus (EBOV) is a member of Filoviridae family also known as Zaire ebolavirus, on the basis of the origin country, i.e., Democratic Republic of Congo (formerly Zaire). EBOV is responsible for thousands of deaths due to its periodic outbreaks since 1976. According to the World Health Organization (WHO), the fatality rate of the EBOV outbreak varies from 25 to 90% (https://www.who.int/news-room/fact-sheets/detail/ebola-virus-disease). EBOV cases are mainly found in the region of sub-Saharan Africa and pass-through animals like a bat, other nonhuman primates or any patient infected with EBOV. As per WHO, the EBOV outbreak is classified under level 3 emergency due to its high mortality and fatality.
EBOV is a negative-stranded, enveloped, non-segmented and helical single-stranded RNA with 19-kb nucleotides. It constitutes eight structural and one nonstructural proteins. The structural proteins include the nucleoprotein (NP), glycoprotein (GP), soluble glycoprotein (sGP), RNA-dependent RNA polymerase (L) and four virion proteins (VP24, VP30, VP35, VP40) [1]. As EBOV is an RNA virus, thus the development of effective antivirals against EBOV is a very challenging task. Currently, Favipiravir, Remdesivir, ZMapp and INMAZEB are the four most commonly used anti-Ebola agents for the treatment of EBOV infection. Among them, Favipiravir and Remdesivir are the ‘experimental’ category drugs that inhibit the viral polymerases while the ZMapp is the mixture of the three monoclonal antibodies, which are directed against the surface glycoproteins [2, 3]. INMAZEB, also known as REGN-EB3, is a mixture of three monoclonal antibodies, namely atoltivimab, maftivimab and odesivimab. It is the first USFDA-approved therapeutics in 2020 against EBOV infection. The Favipiravir (6-fluoro-3-hydroxy-2-pyrazinecarboxamide) and Remdesivir (GS-5734) are in use as the broad-spectrum antiviral drugs. Initially, the Favipiravir was used to treat influenza virus, but now has been used against EBOV [4]. Likewise, anti-Ebola drug Remdesivir was also repurposed to inhibit murine hepatic virus (MHV), Middle East respiratory syndrome (MERS-CoV), severe acute respiratory syndrome (SARS-CoV) and Nipah virus (NiV) [5].
Numerous computational studies are reported in the literature to highlight the use of machine learning in drug development against various pathogens. Todeschini R et al. described the importance of molecular descriptors in the process of designing the efficient drugs [6, 7]. Hansch C et al. explained the importance of physicochemical parameters in the quantitative structure–activity relationship (QSAR) analysis [8]. Matta CF explored the role of biophysical and biological properties in the formulation of QSAR models [9]. Toussi CA et al. design the Ser/Thr-protein kinase inhibitors by using machine-trained elastic networks [10]. However, our group previously implemented the machine learning approaches to develop computational methods to predict the antiviral compounds against various viruses like flaviviruses, Nipah virus and coronaviruses as AVCpred [11], anti-Flavi [12] and anti-Nipah [13] and anti-corona [14], respectively. Recently, we have developed a comprehensive repository of experimentally validated repurposed drugs against 23 viruses (including Ebola virus) responsible for causing epidemics/pandemics [15].
Furthermore, various computational approaches have been tried to identify repurposed or novel leads against EBOV. Anantpadma M et al. developed Bayesian machine learning models and identified three active molecules, namely tilorone, pyronaridine and quinacrine against EBOV [16]. Kwofie SK et al. used pharmacoinformatics and molecular docking approach to prioritize 19 compounds against EBOV after screening 7675 natural products [17]. Zhao Z et al. used a molecular dynamics approach to screen all FDA-approved drugs and finalized 15 potent drug candidates against EBOV [18]. Ekins et al. integrated Bayesian machine learning models to filter out potential lead compounds against EBOV [19]. However, most of the drug repurposing approach was done by various in vitro and in vivo assays, e.g., minigenome assay [20], GIP/HIV core pseudovirus with firefly luciferase reporter gene [21], HIV pseudovirions with high-throughput assay [22] and many more. However, any dedicated web server to identify the promising drug candidates is not available in the literature. In the current study, we have developed a machine-learning-based pipeline named 'anti-Ebola' for the identification of inhibitors against Ebola virus.
Methods
Data collection
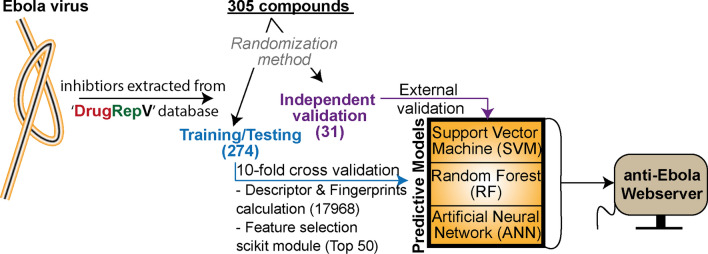
The anti-Ebola predictor was developed using the data of EBOV inhibitors available from our recently published ‘DrugRepV’ database [15]. There are 868 compounds reported in this database, which were experimentally validated for anti-Ebola activities. However, we have selected only those molecules whose antiviral activities are given in terms of IC50/EC50 so as to develop regression-based models. Further, we used strict quality control filters like IC50/EC50 uniqueness, SMILES, assays, etc., to finalize our dataset. Finally, we obtained 305 unique inhibitors with the respective half-maximal inhibitory concentration (IC50/EC50) values from our database [15]. The IC50/EC50 values were converted into the negative logarithm of half-maximal inhibitory concentration (pIC50) using formula:
| 1 |
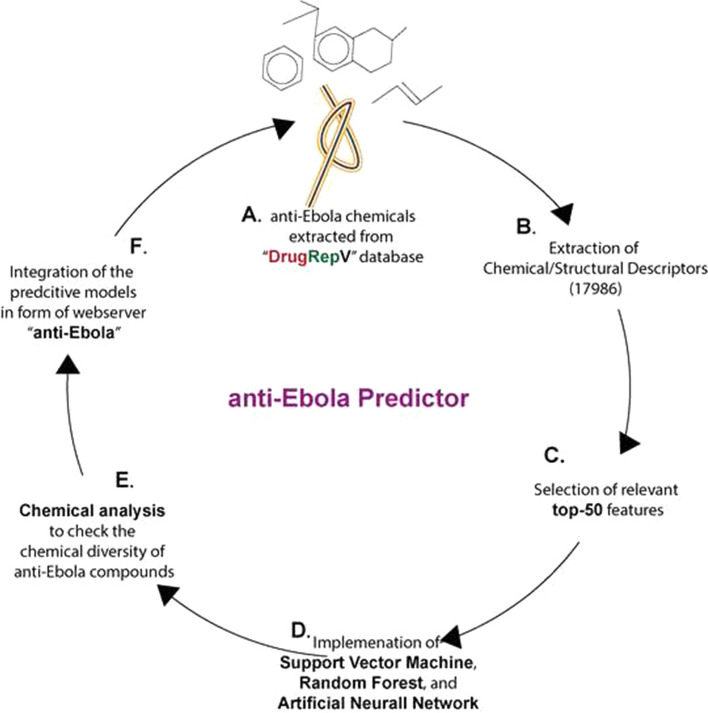
where IC50 is in the form of dimensionless activity that can be approximated numerically as molar concentrations. The higher pIC50 indicates exponentially greater potency. The pIC50 is used for the designing of various regression-based prediction algorithms [12, 13, 23]. Overall methodology of the anti-Ebola is available in Fig. 1.
Fig. 1.
Overall methodology used to develop anti-Ebola predictor
Data preparation
The chemical name was used to extract the chemical information like simplified molecular-input line-entry system (SMILES), which was then converted to 3D-SDF using obabel software [24]. Finally, the 3D-SDF is used to calculate the molecular descriptor and fingerprints.
For running the machine learning algorithm, the overall dataset (305) was divided into training/testing (T274) and independent validation (V31) datasets using randomization approaches in six sets [13, 25, 26].
PaDEL descriptor
The 3D-SDF structures were used for the calculation of 1D, 2D and 3D molecular descriptors as well as fingerprints. The PaDEL software is used for calculation of all the 17,968 descriptors available in the software [27]. Further, to take only relevant features and to rule out the possibility of overfitting of the model, we performed feature selection.
Feature selection
Feature selection is an important step to extract the most relevant features, remove irrelevant features and help to achieve high accuracy of the developed models [28, 29]. The feature selection was done using the support vector regression (SVR) implemented using libsvm using a parameter to control the number of support vectors. Finally, we extracted the most relevant 50 features out of 17,968 descriptors (Supplementary Table S2).
Ten fold cross-validation
The tenfold cross-validation was used to develop the predictive models. In the tenfold cross, training/testing (T274) was divided equally into ten sets. Initially, the nine datasets were combined for training and the remaining one set for testing to finally calculate the model performance. Likewise, all the sets get a chance to become the testing set; however, the average performance of all the testing sets represents the overall performance of the model. Further, the performance of the developed model was cross-evaluated using the independent dataset, which was not used during training and testing.
Machine learning techniques
In the current study, we implemented the three types of MLTs, i.e., support vector machine, random forest and artificial neural network techniques to develop predictive models.
Support vector machine is a supervised machine learning method which is used for both regression and classification-based problems. SVM constructs a set of hyperplanes which can be used to detect the regression/classification task. It is very effective for high-dimensional spaces [30]. Different kernel functions can be used as a decision function. The main objective of the SVM is to find the hyperplane in N-dimensional (N is the number of features) space which identifies the data points. Random forest is an ensemble machine learning technique and has been extensively used for both classification and regression problems. It functions by making decision trees from the training dataset, and the output would be in the form of mean prediction [31]. Artificial neural network is the organization of the connected units/nodes generally known as artificial neurons, which is analogous to the neurons in the human brain. The neural networks consist of input layer, output layer and hidden layers, which are used to transform the input to the reasonable output [32].
Performance measure
The performance of the developed model was analyzed through Pearson’s correlation coefficient (PCC), mean absolute error (MAE) and root mean absolute error (RMSE).
| 2 |
| 3 |
| 4 |
In eqns (2), (3) and (4), n, and are the size of the test set, predicted and actual efficiencies of Ebola inhibition, respectively.
Applicability domain
The robustness of the developed model was evaluated using William’s plot. William’s plot depicts the relationship between standardized residuals and leverage. The leverage (h) is set as a warning threshold (h*) of 3*p/n; in it the p is 1 + the number of finally used descriptors and n is the size of the training dataset. However, the standardized residuals threshold was ± 3σ [33]. The predictive model was robust if most data points lie within the warning threshold [13].
Chemical analysis
We performed the analysis of the anti-Ebola compounds to check their chemical diversity. The diversity was checked by the multidimensional scaling (MDS) with a similarity score of 0.4. The cluster map was constructed through ChemmineR software [34]. Further, the chemical dendrogram was formed using the Scaffoldhunter software through the chemical Fingerprints [35].
Web server
The best performing predictive models are implemented in the form of web server 'anti-Ebola.' The front end of the web server is designed using HTML, CSS and PHP while the backend of the web server is constructed using python, perl and javascript.
Results
Performance of QSAR models
Among the six randomized training/testing (T274) datasets, the best QSAR model displayed a PCC of 0.83, 0.98 and 0.95 for SVM, RF and ANN machine learning techniques, respectively, on the best performing dataset (Table 1). Cross-validation of the training/testing dataset was done using independent validation (V31) dataset and showed the PCC values of 0.65, 0.62 and 0.64 for SVM, RF and ANN correspondingly (Table 1). The performance of all the remaining five training/testing and independent validation datasets is provided in Supplementary Table S1.
Table 1.
Table depicting the performance of training/testing (T274) and independent validation data set (V31) for the support vector machine, random forest and artificial neural network
| Ebola | Training/Testing dataset | Independent Validation dataset | |||||
|---|---|---|---|---|---|---|---|
| Algorithm | Dataset | MAE | RMSE | PCC | MAE | RMSE | PCC |
| SVM | T274 + V31 | 0.33 | 0.47 | 0.83 | 0.48 | 0.66 | 0.65 |
| RF | T274 + V31 | 0.19 | 0.28 | 0.98 | 0.52 | 0.63 | 0.62 |
| ANN | T274 + V31 | 0.23 | 0.29 | 0.95 | 0.76 | 0.97 | 0.64 |
*MAE, mean absolute error; RMSE, root mean absolute error; PCC, Pearson’s correlation coefficient; SVM, support vector machine; RF, random forest; ANN, artificial neural network
Applicability domain
While plotting William’s plot, we found that most of the data points of both training/testing and validation data lie within the warning threshold, showing that the developed model is robust. We found the h* is 1.21, 1.25 and 1.18, while the 3σ is 2.0, 1.9, 1.0, respectively, for SVM (Fig. 2a), RF (Fig. 2b) and ANN (Fig. 2c). Both the h* and the 3σ were plotted as a warning threshold in William’s plot. William's plot shows the relationship between standardized residuals and leverage (Fig. 2).
Fig. 2.
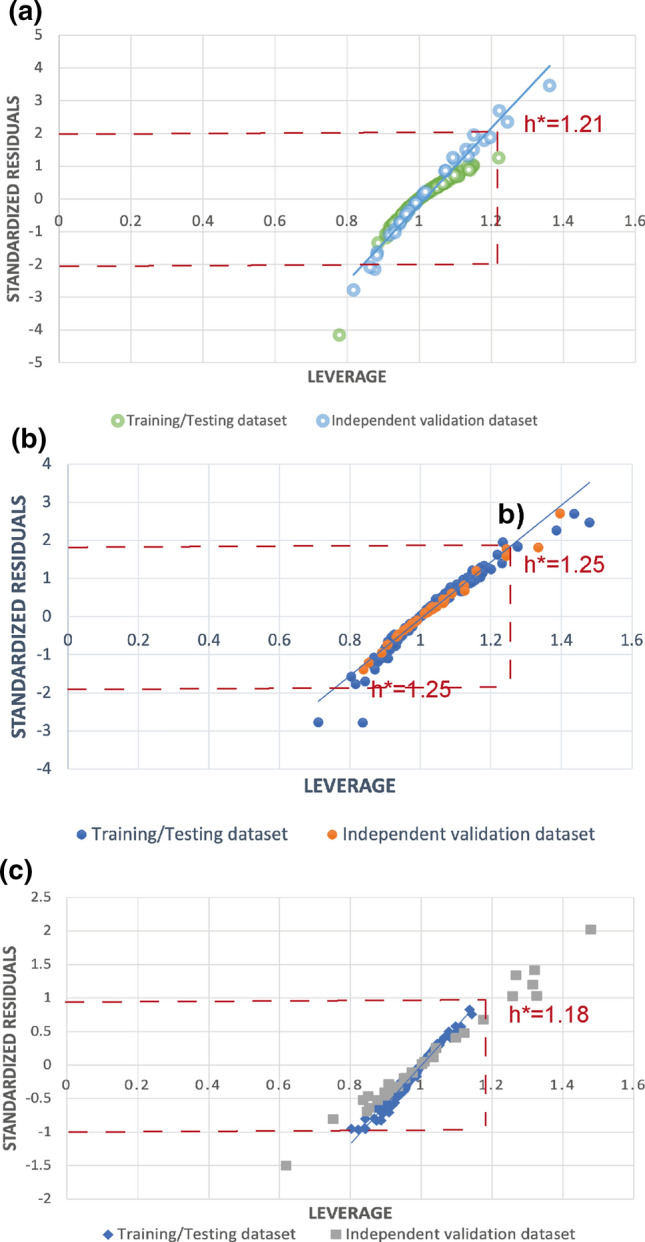
Applicability domain of the anti-Ebola compounds presented by William’s plot. a random forest, b support vector machine, c artificial neural Network
Chemical analysis
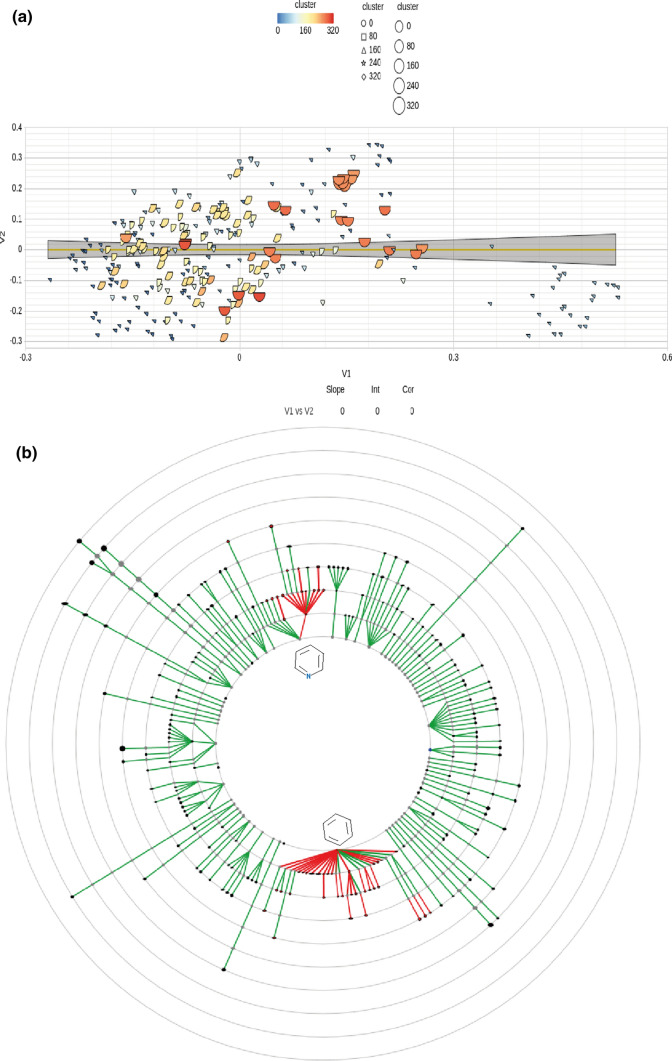
We performed an analysis of the anti-Ebola chemicals to explore the chemical variability. For the same, we used the multidimensional scaling (MDS) whose distance matrix was calculated by ‘all-against-all’ comparison of compounds through atom pair similarity measures (Fig. 3a). Further, the generated similarity scores were transferred into the distance values through the cmdscale method. The cluster map shows the diversity up to 320 clusters with the similarity cutoff of 0.4. Further, the chemical dendrogram was also constructed to check the details of the chemical scaffolds using the EstateNumericalFingerprint (largest fragment, deglycosilated) physicochemical properties. It showed that the highest number of the molecules, i.e., 55, comes under the parent chemical with benzene ring (Fig. 3b). Furthermore, 32 molecules consisted of pyridine parent molecules. Remaining information of all the anti-Ebola molecules is provided in Fig. 3b.
Fig. 3.
Chemical analysis of anti-Ebola compounds. a Scatter plot showing the diversity of the 305 anti-Ebola compounds, b chemical dendrogram of the anti-Ebola compounds showing the chemical side chain similarity among them
Web server
The web server 'anti-Ebola' is freely available at: https://bioinfo.imtech.res.in/manojk/antiebola. It contains the predictor, where the input query can be provided in the form of a SDF and the output displayed as a tabular form with information of SMILES, predicted IC50 in μM along with its structure. To make our web server more informative, we have also provided the important drug-like properties of the input query. We used filter-it software to calculate these drug-likeness properties. It includes the drug-likeness properties, namely Lipinski acceptor, Lipinski donor, H-bond acceptors, H-bond donor, molecular weight, logP, rotatable and rigid bonds, formal charges and molecular formula. The H-bond acceptor shows the number of hydrogen bond acceptors; it includes an aromatic N with no connected H atoms, no amide nitrogen and which doesn’t possess any positive charge; an aliphatic N with no connected H atoms as well as no positive charge on it; any O atom without any positive charge; and a thionyl sulfur atom. The H-bond donor shows the number of hydrogen bond donors and includes any H bonded to a N; any H bonded to an O; and any H bonded to a S. Lipinski acceptor refers to the Lipinski H-bond acceptor like any N or O atom which may or may not be connected to any H atom. Lipinski donor denotes the Lipinski H-bond donor e.g., each H-atom connected to N or O. Here, Lipinski’s rule of five is the rule of thumb to determine the drug likeness of a compound. It indicates whether the compound has certain biological, chemical, pharmacological activities appropriate for human consumption.
Case study
We have checked the utility of our web server by predicting the IC50/EC50 values of the already identified promising hits from other studies. We used an anti-Ebola SVM predictive model to predict anti-EBOV activity of these lead molecules. For example, Zheng et al. identified Indinavir, Maraviroc, Abacavir, etc. as good anti-EBOV compounds [18]. Interestingly, our predictive model also predicts high inhibition efficacy of Indinavir (IC50 0.03uM), Maraviroc (IC50 0.30uM), Abacavir (IC50 1.27uM). Likewise, Anantpadma A et al. identified three effective anti-EBOV drugs, namely Tilorone, Pyronaridine and Quinacrine with Kd values of 0.73 uM, 7.34uM and 7.55 uM [16]. These three lead molecules also show potential inhibition efficacy by our ‘anti-Ebola’ web server such as Tilorone (IC50 1.95uM), Pyronaridine (IC50 0.50uM) and Quinacrine (IC50 0.002uM). Thus, these findings further validate the utility of our prediction algorithm.
Discussion
Ebola is a dreadful pathogen, which is responsible for causing epidemics in the past, with a high mortality rate [36]. There is a need for developing effective anti-Ebola agents. In this endeavor, intervention of the computational approaches would accelerate the research in the field [16]. Therefore, in the current study, we provided machine learning-based prediction models to identify novel and effective anti-Ebola compounds. Apart from that, we also analyzed the chemical diversity of the available Ebola inhibitors.
We implement three MLTs like SVM, RF and ANN to develop effective predictive models. SVM, RF and ANN are the machine learning techniques that work on different principles. For example, the SVM is a nonlinear algorithm, RF works with a decision tree group of algorithms, and the ANN is a neural networks-based algorithm. Various researchers have used these techniques in numerous studies [37–40]. Likewise, we had also used these techniques to develop predictive algorithms like QSPpred [25], VIRsiRNApred [41], AVP-IC50Pred [42], anti-flavi [12] and many more. For the development of the high-quality predictive models, we extracted the highly relevant features out of the 17,968 (1D, 2D, 3D and fingerprints) features from the available anti-Ebola compounds. Among the three MLTs, the PCC of the SVM, RF and ANN ranges from 0.83 to 0.98. Further, we checked the robustness of the developed models by constructing William's plot (applicability domain). Further, we implemented the developed models in the form of a web server named ‘anti-Ebola’ (https://bioinfo.imtech.res.in/manojk/antiebola/). The implementation of the predictive models in the form of a web server makes them easily accessible for the users. Apart from that, we analyzed the chemical diversity of the available EBOV inhibitors. We noticed that the available anti-Ebola molecules showed high chemical diversity. However, the highest (55) amount of the molecules are derivatives of the benzene parent compound, followed by the 32 molecules which are the derivative of the pyridine heterocyclic ring. This is an important approach based on the implementation of the MLTs on the available experimentally validated anti-Ebola molecules. Thus, our study would be very important for identification of the new and promising anti-Ebola agents. Researchers can use our web server to identify the promising repurposed drug candidates also.
Few researchers performed computational studies for the identification of repurposed drugs against EBOV. These computational studies include the use of Bayesian machine learning models, molecular simulations, molecular docking, etc. [16, 17, 19]. These studies used different datasets as input like natural products, FDA-approved drugs and small active molecules from repositories. However, our study is different from these approaches, as we have incorporated three different MLTs for the prediction of anti-EBOV agents. For the development of the predictive models, we used the experimentally validated anti-EBOV compounds which are chemically diverse. Furthermore, our predictive models are incorporated as a web server which is not available with any of the previously published computational approaches for EBOV.
The frequent outbreaks of EBOV with high mortality and fatality rate are serious concerns worldwide. As EBOV is a dangerous infectious pathogen and comes under the Biosafety Level-4 (BSL-4) category, it requires a highly specialized laboratory to work. Therefore, designing an anti-Ebola agent is a challenging task. Thus, the intervention of computational approaches would be of great help in speeding up the identification of effective EBOV inhibitors. In this endeavor, we have developed the machine learning-based QSAR regression model 'anti-Ebola.' We will update the web server on a yearly basis or whenever a significant amount of data is available. Thus this 'anti-Ebola' web server would be helpful to researchers to predict Ebola inhibitors and the antiviral therapeutic development.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
This work was supported by the grants from the CSIR-Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR) (OLP0501, OLP0143 and STS0038).
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Beniac DR, Booth TF. Structure of the Ebola virus glycoprotein spike within the virion envelope at 11 Å resolution. Sci Rep. 2017;7:46374. doi: 10.1038/srep46374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JS, Adhikari NKJ, Kwon HY, et al. Anti-Ebola therapy for patients with Ebola virus disease: a systematic review. BMC Infect Dis. 2019;19:376. doi: 10.1186/s12879-019-3980-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller MA, Richard Stiehm E. Passive Immunity in Prevention and Treatment of Infectious Diseases. Clin Microbiol Rev. 2000;13:602–614. doi: 10.1128/cmr.13.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guedj J, Piorkowski G, Jacquot F, et al. Antiviral efficacy of favipiravir against Ebola virus: A translational study in cynomolgus macaques. PLoS Med. 2018;15:e1002535. doi: 10.1371/journal.pmed.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lo MK, Feldmann F, Gary JM, et al (2019) Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci Transl Med 11:eaau9242. 10.1126/scitranslmed.aau9242 [DOI] [PMC free article] [PubMed]
- 6.Todeschini R, Consonni V (2009) Molecular Descriptors for Chemoinformatics: Volume I: Alphabetical Listing / Volume II: Appendices, References. John Wiley & Sons
- 7.Todeschini R, Consonni V (2009) Molecular Descriptors for Chemoinformatics, 2 Volume Set: Volume I: Alphabetical Listing / Volume II: Appendices, References. Wiley-VCH
- 8.Hansch C, Leo A, Pomona College Albert Leo (1995) Exploring QSAR.: Fundamentals and applications in chemistry and biology. Amer Chemical Society
- 9.Matta CF. Modeling biophysical and biological properties from the characteristics of the molecular electron density, electron localization and delocalization matrices and the electrostatic potential. J Comput Chem. 2014;35:1165–1198. doi: 10.1002/jcc.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toussi CA, Haddadnia J, Matta CF. Drug design by machine-trained elastic networks: predicting Ser/Thr-protein kinase inhibitors’ activities. Mol Divers. 2021;25:899–909. doi: 10.1007/s11030-020-10074-6. [DOI] [PubMed] [Google Scholar]
- 11.Qureshi A, Kaur G, Kumar M. AVCpred: an integrated web server for prediction and design of antiviral compounds. Chem Biol Drug Des. 2017;89:74–83. doi: 10.1111/cbdd.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajput A, Kumar M. Anti-flavi: A Web Platform to Predict Inhibitors of Flaviviruses Using QSAR and Peptidomimetic Approaches. Front Microbiol. 2018;9:3121. doi: 10.3389/fmicb.2018.03121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajput A, Kumar A, Kumar M. Computational Identification of Inhibitors Using QSAR Approach Against Nipah Virus. Front Pharmacol. 2019;10:71. doi: 10.3389/fphar.2019.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajput A, Thakur A, Mukhopadhyay A, et al. Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning. Comput Struct Biotechnol J. 2021;19:3133–3148. doi: 10.1016/j.csbj.2021.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajput A, Kumar A, Megha K, et al. DrugRepV: a compendium of repurposed drugs and chemicals targeting epidemic and pandemic viruses. Brief Bioinform. 2021;22:1076. doi: 10.1093/bib/bbaa421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anantpadma M, Lane T, Zorn KM, et al. Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads. ACS Omega. 2019;4:2353–2361. doi: 10.1021/acsomega.8b02948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwofie SK, Broni E, Teye J, et al. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput Biol Med. 2019;113:103414. doi: 10.1016/j.compbiomed.2019.103414. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, Martin C, Fan R, et al. Drug repurposing to target Ebola virus replication and virulence using structural systems pharmacology. BMC Bioinformatics. 2016;17:90. doi: 10.1186/s12859-016-0941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekins S, Freundlich JS, Clark AM, et al (2015) Machine learning models identify molecules active against the Ebola virus. F1000Res 4:1091. 10.12688/f1000research.7217.3 [DOI] [PMC free article] [PubMed]
- 20.Edwards MR, Pietzsch C, Vausselin T, et al. High-Throughput Minigenome System for Identifying Small-Molecule Inhibitors of Ebola Virus Replication. ACS Infect Dis. 2015;1:380–387. doi: 10.1021/acsinfecdis.5b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Cui R, Li G, et al. Teicoplanin inhibits Ebola pseudovirus infection in cell culture. Antiviral Res. 2016;125:1–7. doi: 10.1016/j.antiviral.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng H, Lear-Rooney CM, Johansen L, et al. Inhibition of Ebola and Marburg Virus Entry by G Protein-Coupled Receptor Antagonists. J Virol. 2015;89:9932–9938. doi: 10.1128/JVI.01337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalliokoski T, Kramer C, Vulpetti A, Gedeck P. Comparability of Mixed IC50 Data – A Statistical Analysis. PLoS ONE. 2013;8:e61007. doi: 10.1371/journal.pone.0061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Boyle NM, Banck M, James CA, et al. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajput A, Gupta AK, Kumar M. Prediction and analysis of quorum sensing peptides based on sequence features. PLoS ONE. 2015;10:e0120066. doi: 10.1371/journal.pone.0120066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur A, Rajput A, Kumar M. MSLVP: prediction of multiple subcellular localization of viral proteins using a support vector machine. Mol Biosyst. 2016;12:2572–2586. doi: 10.1039/c6mb00241b. [DOI] [PubMed] [Google Scholar]
- 27.Yap CW. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 28.Hira ZM, Gillies DF. A Review of Feature Selection and Feature Extraction Methods Applied on Microarray Data. Adv Bioinformatics. 2015;2015:198363. doi: 10.1155/2015/198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajput A, Thakur A, Sharma S, Kumar M. aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 2018;46:D894–D900. doi: 10.1093/nar/gkx1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes C, Vapnik V. Mach Learn. 1995;20:273–297. doi: 10.1023/a:1022627411411. [DOI] [Google Scholar]
- 31.Petkovic D, Altman R, Wong M, Vigil A. Improving the explainability of Random Forest classifier - user centered approach. Pac Symp Biocomput. 2018;23:204–215. doi: 10.1142/9789813235533_0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain AK, Mao J, Mohiuddin KM. Artificial neural networks: a tutorial. Computer. 1996;29:31–44. doi: 10.1109/2.485891. [DOI] [Google Scholar]
- 33.Fechner N, Jahn A, Hinselmann G, Zell A. Estimation of the applicability domain of kernel-based machine learning models for virtual screening. J Cheminform. 2010;2:2. doi: 10.1186/1758-2946-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Y, Charisi A, Cheng L-C, et al. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer T, Kriege N, Humbeck L, et al. Scaffold Hunter: a comprehensive visual analytics framework for drug discovery. J Cheminform. 2017;9:28. doi: 10.1186/s13321-017-0213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahai JI. The Ebola Pandemic in Sierra Leone. Cham: Palgrave Macmillan; 2017. [Google Scholar]
- 37.Jovic A, Bogunovic N. Electrocardiogram analysis using a combination of statistical, geometric and nonlinear heart rate variability features. Artif Intell Med. 2011;51:175–186. doi: 10.1016/j.artmed.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 38.You H, Ma Z, Tang Y, et al. Comparison of ANN (MLP), ANFIS, SVM and RF models for the online classification of heating value of burning municipal solid waste in circulating fluidized bed incinerators. Waste Manag. 2017;68:186–197. doi: 10.1016/j.wasman.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 39.Yu S, Tao J, Dong B, et al. Development and head-to-head comparison of machine-learning models to identify patients requiring prostate biopsy. BMC Urol. 2021;21:80. doi: 10.1186/s12894-021-00849-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirsadeghi L, Haji Hosseini R, Banaei-Moghaddam AM, Kavousi K. EARN: an ensemble machine learning algorithm to predict driver genes in metastatic breast cancer. BMC Med Genomics. 2021;14:122. doi: 10.1186/s12920-021-00974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qureshi A, Thakur N, Kumar M. VIRsiRNApred: a web server for predicting inhibition efficacy of siRNAs targeting human viruses. J Transl Med. 2013;11:305. doi: 10.1186/1479-5876-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qureshi A, Tandon H, Kumar M. AVP-IC50 Pred: Multiple machine learning techniques-based prediction of peptide antiviral activity in terms of half maximal inhibitory concentration (IC50) Biopolymers. 2015;104:753–763. doi: 10.1002/bip.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.