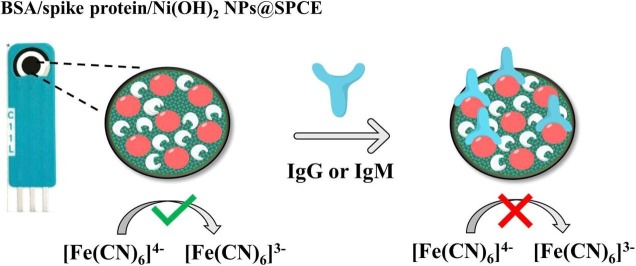
Graphical abstract
Keywords: SARS‐CoV‐2, Spike protein, SARS‐CoV‐2-specific viral antibody, Screen-printed carbon electrode, Nickel hydroxide nanoparticles, Biodevice
Abstract
As a promising approach for serological tests, the present study aimed at designing a robust electrochemical biosensor for selective and quantitative analysis of SARS‐CoV‐2-specific viral antibodies. In our proposed strategy, recombinant SARS-CoV-2 spike protein antigen (spike protein) was used as a specific receptor to detect SARS‐CoV‐2-specific viral antibodies. In this sense, with a layer of nickel hydroxide nanoparticles (Ni(OH)2 NPs), the screen-printed carbon electrode (SPCE) surface was directly electrodeposited to ensure better loading of spike protein on the surface of SPCE. The differential pulse voltammetry (DPV) showed signals which were inversely proportional to the concentrations of the antibody (from 1 fg mL−1 L to 1 µg mL−1) via a specific and stable binding reaction. The assay was performed in 20 min with a low detection limit of 0.3 fg mL−1. This biodevice had high sensitivity and specificity as compared to non-specific antibodies. Moreover, it can be regarded as a highly sensitive immunological diagnostic method for SARS‐CoV‐2 antibody in which no labeling is required. The fabricated hand-held biodevice showed an average satisfactory recovery rate of ~99-103% for the determination of antibodies in real blood serum samples with the possibility of being widely used in individual serological qualitative monitoring. Also, the biodevice was tested using real patients and healthy people samples, where the results are already confirmed using the enzyme-linked immunosorbent assay (ELISA) procedure, and showed satisfactory results.
1. Introduction
Renamed as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Coronavirus disease 2019 (COVID-19), has been regarded as an emerging human infection which causes fatal pneumonia in humans [1]. The disease is highly contagious and has a rate of much faster transmission than SARS and MERS [2]. This has led to an epidemic of the disease around the world, so that, until April 12th, 2021, more than 136.78 million COVID-19 cases have been reported globally, including more than 2.95 million deaths [3].
There are several test methods for coronavirus; most of which are based on molecular or serological tests. Molecular tests look for signs of active infection which can only help diagnose current cases of Coronavirus [4]. The tests try to find viral genes in nose or throat swab and, then, the samples will be tested by polymerase chain reaction (PCR) [5], [6]. This test has been widely used around the world after the release of the Coronavirus genetic code in early January [7]. SARS-CoV-2 serological tests detect antibodies produced by the body to fight the virus. While immunoglobulin M (IgM) antibodies can be produced during the early stages of the infectious disease (between 4 and 10 days), immunoglobulin G (IgG) response is produced later (around 14 days) [4]. Moreover, not only the mentioned test has the capability of detecting the active infections, but also it can determine the past infections. In this sense, it can be because of the fact that the body can retain antibodies against pathogens [8], [9]. Moreover, serological tests are especially useful in diagnosing cases with mild or no symptoms [10], [11]. Moreover, donors for convalescent plasma therapy can be identified by serology testing which can be applied in order to model the course of the pandemic and understand the antibody responses mounted upon SARS-CoV-2 infection and vaccination [12], [13], [14]. Therefore, the development of sensitive immunological diagnostic methods which are capable of detecting viral antibodies in clinical samples is highly significant for a fast and precise diagnosis of SARS-CoV-2-specific antibodies.
During recent decades, different kinds of serological assays have been developed in order to measure antibody responses to pathogens in bodily fluids, i.e. blood serum or plasma i.e. enzyme-linked immunosorbent assay (ELISA) [15], [16], [17]. As compared to conventional immunoassays, electrochemical biosensors are popular due to their ease of use, feasibility of miniaturization and the subsequent portability, low cost, rapid response times, high sensitivity and compatibility [18], [19]. Therefore, biosensors have held great potential for the next-generation detection strategy.
As structural proteins, spike, envelope, matrix, and nucleocapsid are encoded by SARS-CoV-2 [20], [21]. In this sense, the spike protein has been significantly considered as a diagnostic antigen due to the following features: being the basic transmembrane protein of the virus, being immunogenic and also, exhibiting amino acid sequence diversity among coronaviruses [21]. Consequently, it would have the potential to enable the specific detection of lgM/lgG antibody [22].
Regarded as an encouraging tool for point-of-care diagnostics, a SPCE has provided “lab-to-market” capability for many sensors [23]. In addition, the mentioned electrode is also capable of providing an inexpensive kit and also disposable devices in order to simply and rapidly detect and also quantitatively analyze the biomolecules in a sample matrix [24], [25], [26]. Different nanomaterial matrices and nanocomposites can be applied to enhance the biosensor’s performance features and also to immobilize biomolecules onto the electrode surfaces [27]. Electrodeposition of Ni(OH)2 NPs on the surface of carbon electrodes can be regarded as an attractive strategy because of the fact that its preparation is direct, fast and easier. Moreover, Ni(OH)2 NPs have some excellent advantages; such as electrocatalytic properties, cost-effectivity, high porosity, electro-inactivity in physiological pH solutions and the ability to interact with biomolecules [28], [29]. Besides, it is able to maintain the spike protein-specific antigen bioactivity.
Herein, we design a label-free electrochemical sensor made by using the spike protein, as a specific-receptor in order to detect lgM/lgG antibody. The spike protein was immobilized on the SPCE modified with Ni(OH)2 NPs as a nanotool for the ultra-sensitive selective quantitative analysis. Moreover, bovine serum albumin (BSA) was applied in order to block the non-specific sites. Cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS) and DPV were used to investigate the proposed biodevice and to analyze the lgM/lgG antibody using [Fe(CN)6]4−/3− as the redox probe. When lgM/lgG antibody is specifically attached to the spike protein, the [Fe(CN)6]4−/3− electron-transferring can be disturbed, as shown in the CV, DPV and EIS curves. The suggested biodevice can have the following advantages: i.e. speed response, good mechanical and chemical stability, simple fabrication process, low cost and possibility of in-situ testing and on-site screening of samples. This novel biodevice can simply and selectively detect lgM/lgG antibody followed by a successful detection of antibodies in serum samples. Heretofore, as far as we know, no electrochemical biosensor has been reported for the detection of lgM/lgG-specific viral antibody based on spike protein and Ni(OH)2 NPs.
2. Experimental section
2.1. Materials
Spike protein antigen and antibodies was purchased from cusabio Company (https://www.cusabio.com). We prepared 1 mg.ml−1 of antibodies, then diluted with phosphate-buffered (PB) solutions. Sulfuric acid (H2SO4), acetic acid (CH3COOH), sodium acetate (C2H3NaO2), disodium hydrogen phosphate (Na2HPO4), monosodium dihydrogen phosphate (NaH2PO4), nickel(II) nitrate hexahydrate Ni(NO3)2·6H2O, and all other reagents were purchased from Sigma-Aldrich and Merck Co. LLC (USA) and used without further purification. In order to prepare the PB solutions, 0.1 M Na2HPO4 and NaH2PO4 were applied. Besides, a solution containing 5 mM K3Fe(CN)6/K4Fe(CN)6 with a ratio of 1:1 as a redox probe in 0.1 M KCl was used for the experiments. In this regard, in order to prepare the solutions deionized water was used. It is worth mentioning that room temperature (25 ± 0.5 °C) was used for the experiments.
2.2. Apparatus and procedures
The EIS, DPV and CV techniques were used to record the electrochemical data. In this sense, μ-AUTOLAB electrochemical system type III and FRA2 board computer, controlled by Potentiostat/Galvanostat (Eco-Chemio, Switzerland) with NOVA software, was used. Bought from Dropsens (Spain), A SPCE was applied as a three electrode planar based on a graphite working electrode with 2 mm in diameter, a carbon counter electrode, and a silver pseudo-reference electrode. We executed the EIS analysis within the frequency range of 0.1–100 kHz with 5 mV amplitude and with a bias potential of 0.20 V. The DPV measurements were performed by scanning the potential of − 0.2 to 0.7 V with modulation time of 50 ms and modulation amplitude of 20 mV. In order to measure the pH, a Metrohm pH meter (model 780 pH/mV meters) was applied. Moreover, morphological, structural and chemical analyses of the samples were performed using a FESEM (TESCAN, Mira III LMU, Czech Republic) equipped with an EDS probe.
2.3. Construction of the biosensor
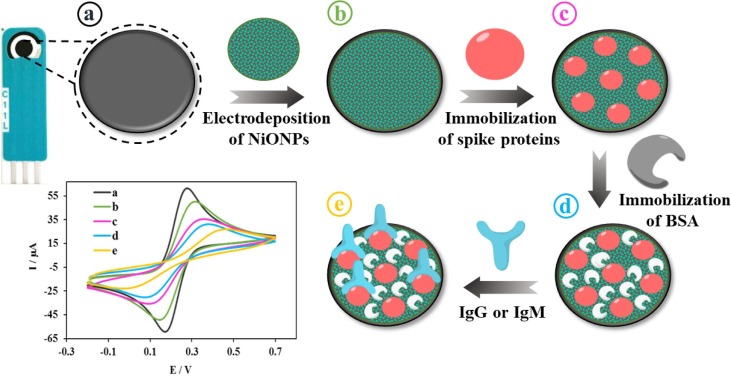
The fabrication procedure of the biosensor, as a biodevice for detecting SARS‐CoV‐2 antibodies, is shown in schematic 1. Before measurements, in order to electrochemically activated the SPCE surface, we carefully dropped 6 μL H2SO4 (2 M) on the surface of SPCE and, then, a voltage of 1.5 V during 150 s by chronoamperometry technique was applied. Afterwards, distilled water was used to wash the clean electrode which was then dried under N2 gas. Next, in order to achieve the NiO NPs film, we aimed at performing the controlled electrodeposition of the NiO NPs by a simple technique as an efficient platform. For this in-situ synthesis, 6 μL of a solution containing 0.1 M NiNO3 in acetate buffer (pH = 4.0) was dropped on the surface of SPCE and, then, the cycling process was applied in the potential scanning between 0 to − 1 V for 20 cycles at scan rate of 50 mV s−1. In the next step, in order form the nickel hydroxide layer, it was in a range of 0 to + 0.8 V at a scan rate of 100 mV s−1 during 20 scans that the CV was performed in a 0.1 M NaOH. The electrodeposited Ni(OH)2 NPs film was applied onto the SPCE surface as a substratum in order to load spike protein molecules. Next, 6 μL of spike protein (10 µg mL−1) in PB solutions (pH = 7.4), as a receptor of IgM/IgG, was casted onto the Ni(OH)2 NPs@SPCE surface during 2 h in order to immobilize it via the amine groups in the protein structure by the formation of Ni(OH)2 NPs-NH2 covalent bond. Subsequently, PB solutions was used to wash the electrode in order to remove the non-bonded molecules and, then, it was dried under N2 gas. In the last step, we used 6 μL of BSA (1%) solution to cover the electrode surface for 30 min. Accordingly, the available active sites will be blocked and also the non-specific adsorption will be avoided. Then, we washed the obtained spike protein/Ni(OH)2 NPs@SPCE, as a biodevice, with PB solutions which can be directly applied for the detection experiments or stored at 4 °C to later use (Scheme 1 )
Scheme 1.
Schematic of the step-by-step preparation of biodevice. a) bare SPCE; b) electrodeposition of Ni(OH)2 NPs on the SPCE surface; c) Immobilization of spike protein on the Ni(OH)2 NPs/SPCE surface; d) Immobilization of BSA on the spike protein/Ni(OH)2 NPs/SPCE surface and e) Immobilization of IgG or IgM on the BSA/spike protein/Ni(OH)2 NPs/SPCE surface.
2.4. Pretreatment of real samples
Due to the use of the spike protein as an IgM/IgG-specific antigen, the proposed method allows us to detect IgM/IgG in biological fluids without interfering with other materials in real samples. The human blood serum sample was provided from a local hospital at Ilam city and, then, various concentrations of IgM/IgG were added. Next, it was ultrafiltrated by being loaded in a centrifugal filtration tube at 5000 rpm for 30 min and, then, was analyzed by standard addition method under optimized condition. Also, serum samples of real patients and healthy individual samples, the results of which have already been confirmed using the standard PCR method, were obtained from a clinical laboratory.
3. Results and discussion
3.1. Characterization of the biodevice
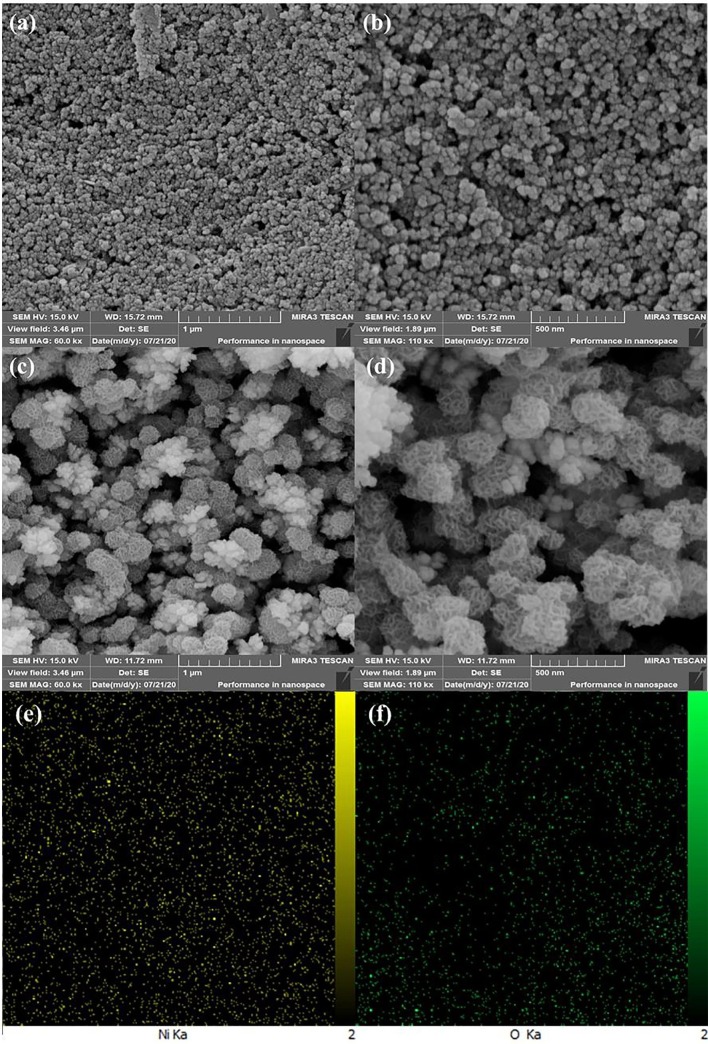
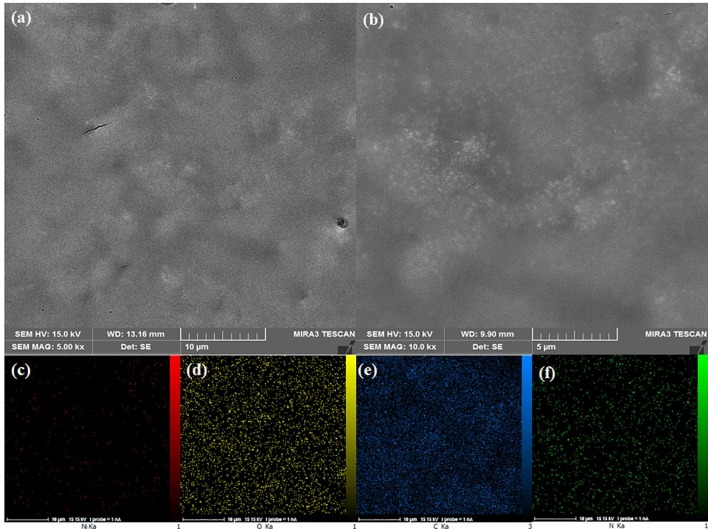
The FESEM technique was used to study the morphology of the electrodeposited Ni(OH)2 NPs film. The FESEM images of bare SPCE displayed that the surface of SPCE consists of a uniform layer of carbon particles (Fig. 1 a, b). When the Ni(OH)2 NPs were electrodeposited, a layer of Ni(OH)2 NPs were formed uniformly around each carbon particles (Fig. 1c, d). Moreover, the high-magnification FESEM image revealed that a hierarchical structure of Ni(OH)2 NPs were formed around each carbon particles (Fig. 1d), which can provide a substrate with high surface area and high hydroxyl groups for better immobilization of biological species like spike protein. Furthermore, the EDS mapping of the Ni(OH)2 NPs@SPCE shows the uniform distribution of Ni and O elements in the structure, confirming the successful formation of Ni(OH)2 nanostructures (Fig. 1e). The FESEM images of spike protein/Ni(OH)2 NPs@SPCE clearly confirm that the protein is well covered the electrode surface due to the interaction with Ni(OH)2 NPs and the flower-like structure of Ni(OH)2 NPs seems to have disappeared (Fig. 2 a, b). Moreover, the EDS mapping of the spike protein/Ni(OH)2 NPs@SPCE shows the uniform distribution of Ni, O, C, and N elements in the structure, confirming the successful immobilization of the protein on the surface of Ni(OH)2 nanostructures (Fig. 2 c-f).
Fig. 1.
The FESEM images of SPCE (a and b), Ni(OH)2 NPs@SPCE (c and d), EDS mapping of the Ni(OH)2 NPs@SPCE (e and f).
Fig. 2.
The FESEM images of spike protein/Ni(OH)2 NPs@SPCE (a and b), EDS mapping of the spike protein/Ni(OH)2 NPs@SPCE (c-f).
The FTIR spectra of Ni(OH)2 NPs@SPCE shows a small peak appeared about 650 cm−1 that can be corresponded to the presence of Ni-O bending vibration [30] (Fig. 3 a). A broad peak at around 3447 cm−1 can be attributed to the vibrational mode of O–H stretching, which relates to the vibration of OH groups and intercalated H2O molecules located in inter-lamellar spaces of Ni(OH)2 [30]. The other vibration modes can be related to the SPCE layer or acetate groups trapped in the deposited layers. The FTIR spectra of spike protein/Ni(OH)2 NPs@SPCE shows some differences compared to the Ni(OH)2 NPs@SPCE (Fig. 3b). The broad and sharp peaks around 3426 cm−1 can be related to the N–H or O–H stretching vibrations in the protein. The sharp peaks at around the 2203 cm−1 and 1084 cm−1 can be corresponded to the C = N and C-N vibrational modes, respectively, [31]. The Ni-O bending vibration are appeared about 659 cm−1 which has shifted to higher wavenumber compared to the Ni(OH)2 NPs@SPCE, which can be due to its interaction with proteins. The FTIR results well show the presence and interaction of proteins to the surface.
Fig. 3.
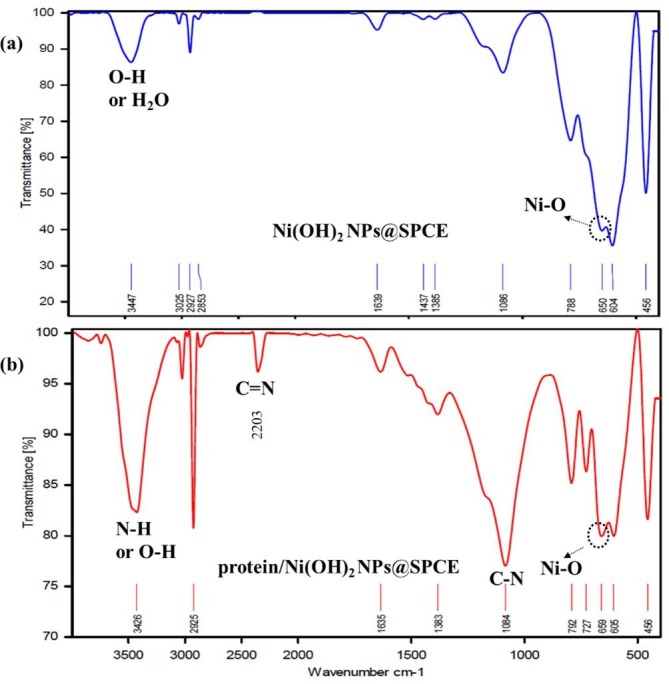
The FTIR spectra of Ni(OH)2 NPs@SPCE (a) and The FTIR spectra of spike protein/Ni(OH)2 NPs@SPCE (b).
The EIS technique was used to study the stepwise fabrication processes of biodevice. In this sense, the EIS can be regarded as an applicable method in order to obtain electrical information in a broad range of frequency and also to monitor the modified electrode traits29. In this technique, due to the importance of the charge transfer resistance (Rct), the variations in Rct amounts for modified electrodes at each stage of biodevice fabrication process were investigated. After electrodeposition of the Ni(OH)2 NPs nanofilm on the SPCE surface, Rct value increased from 0.350 kΩ for the bare SPCE (curve a) to 1.320 kΩ for the Ni(OH)2 NPs@SPCE (curve b). Since the Ni(OH)2 NPs were formed of metal oxide with the negative partial charge, the Rct increases because of the repulsion between the Ni(OH)2 NPs and the anion redox probe, indicating the successful metal oxide layer’s attachment. The Ni(OH)2 NPs with the very high surface to volume and metal sites which possess the capability to interact with the spike protein, due to the presence of the abundant amine groups in the spike protein structure, can make a suitable substrate, resulting in the spike protein’s immobilization onto the electrode surface via Ni(OH)2 NPs-NH2 covalent bond23. Afterwards, with the immobilization of spike protein, as a capture molecule, on the Ni(OH)2 NPs@SPCE, the Rct was increased and spike protein hindered the transmission of electrons due to the steric/conformational restrictions (curve c, Rct = 2.824 kΩ). In addition, when BSA solution was dropped onto spike protein/Ni(OH)2 NPs@SPCE, the Rct similarly increased further (curve d, Rct = 3.550 kΩ). Finally, by incubating IgM/IgG as a target onto the sensing platform surface, Rct was increased, representing a successful connection of IgM/IgG to spike protein via the specific interaction between spike protein molecules and IgM/IgG via Fab fragment [32] (e, Rct = 5.945 kΩ). These results indicated that the interaction of IgM/IgG with spike protein at the electrode surface is well done (Fig. 4 A).
Fig. 4.
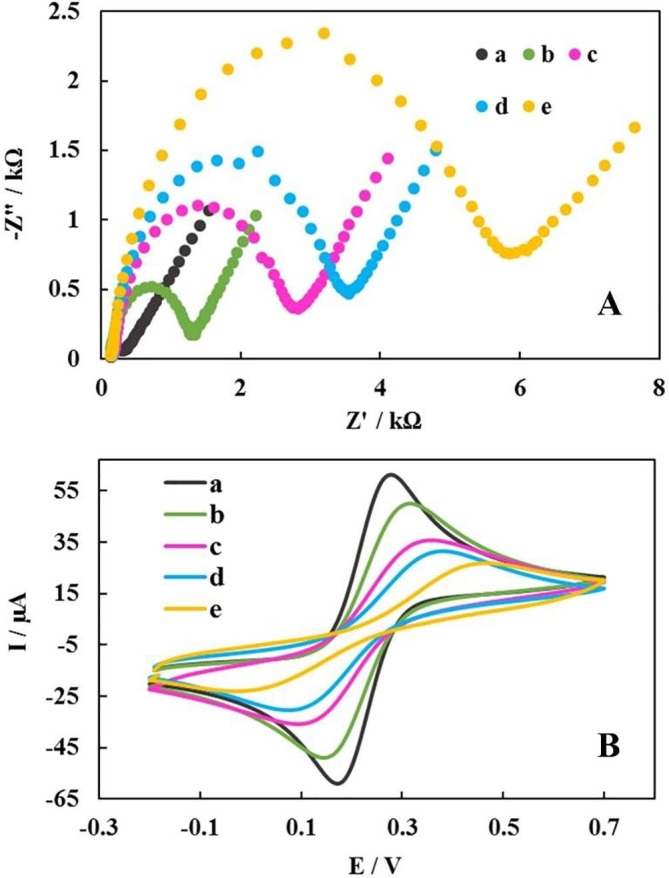
The resulted from EIS (A) and CV (B) from various stages of fabrication of the modified electrode in the probe solution containing 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1:1) and 0.1 M KCl with a scan rate of 100 mV S−1: SPCE (a), Ni(OH)2 NPs@SPCE (b), spike protein/Ni(OH)2 NPs@SPCE (c), BSA/spike protein/Ni(OH)2 NPs@SPCE (d) and IgM or IgG/BSA/spike protein/Ni(OH)2 NPs@SPCE (e).
The CV was used as another useful technique for step-by-step evaluation of the fabrication processes of the sensor in the [Fe(CN)6]4−/3− solution as the redox probe. First, the bare SPCE’s CV was recorded (curve a, ΔE = 14 mV). Next, by the electrodeposition of Ni(OH)2 NPs onto the surface of SPCE, we had a decrease in the peak current (curve b, ΔE = 19 mV). By attaching the spike protein onto the Ni(OH)2 NPs@SPCE, the current experienced a dramatic decrease along with the ΔE value increment (curve c, ΔE = 34 mV). Accordingly, it led to the inhabitation of the electron transfer (ET) between redox probe and the modified electrode’s surface. This electrochemical change would be related to the successful attachment of spike protein onto the Ni(OH)2 NPs@SPCE resulting in steric/conformational restrictions. Similarly, a decrease in current was observed when BSA was incubated on the surface spike protein/Ni(OH)2 NPs@SPCE (curve d, ΔE = 38 mV). As expected, the incubation of the target onto the surface of biodevice resulted in steric/conformational restrictions which then brought about the inhibition of the ET (curve e, ΔE = 47 mV). These results accorded with the EIS results and confirmed the successful preparation of the sensing interface and also the capability of the biodevice in measuring the IgM/IgG (Fig. 4B).
3.2. Optimization study of the experiment
Some parameters should be optimized in order to obtain a steady signal of the biodevice such as pH, concentration and incubation time must be optimized.
Optimization of pH: Due to the fact that spike protein and antibodies are biological molecules, a neutral environment is necessary so that these molecules are not destroyed. Therefore, all experiments were performed in a PB solutions with pH 7.4.
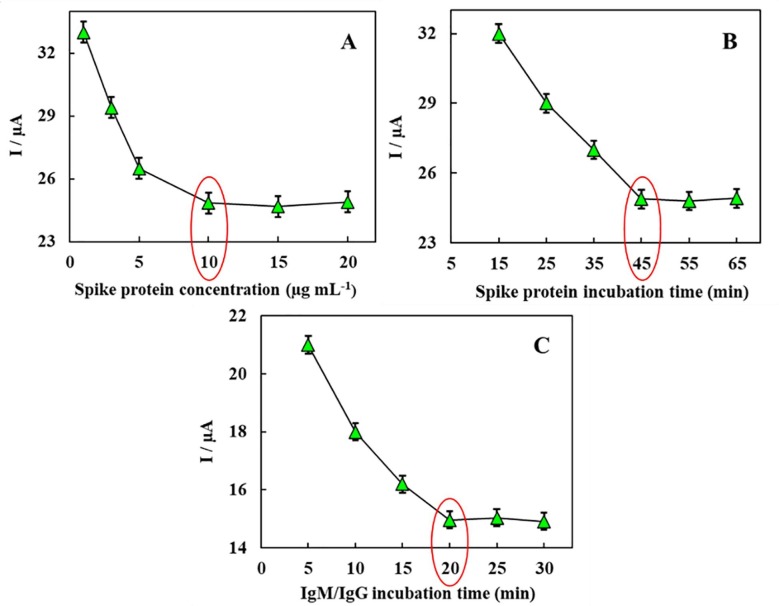
Optimization of the concentration of spike protein: In order to get the best response from the biodevice, analyzing the effect of spike protein concentration as the receptor molecule is very significant. Different concentrations of spike protein were incubated in the range of 1 µg mL− 1 to 20 µg mL− 1 on the surface of the modified SPCE for 45 min. The DPV response in the [Fe(CN)6]4−/3− solution as the redox probe was applied to monitor each electrode signal. As it is evident from Fig. 5 A, response signal growing was stopped specifically when the concentration of spike protein was over 10 µg mL−1; after which a plateau was obtained. In this sense, it indicated a maximum adsorptive quantity of spike protein on the electrode surface, therefore, 10 µg mL−1 was selected as the optimum concentration in subsequent experiments.
Fig. 5.
Optimization of analytical conditions. (A) Optimization of spike protein antigen concentration from 1 µg mL−1 to 20 µg mL−1, (B) Effect of binding time between spike protein antigen and Ni(OH)2 NPs from 15 min to 65 min, (C) Effect of binding time between antibody and immobilized antigen from 5 min to 30 min, (each measurement was performed 3 times and the RSD averaged 1.5%).
Optimization of the incubation time of spike protein: The connection time between spike protein and Ni(OH)2 NPs@SPCE was optimized. The tested incubation time varied from 15 min to 65 min. It was observed that, regarding the DPV response signals, there is no obvious change after 45 min, indicating that the adsorptive of spike protein on Ni(OH)2 NPs@SPCE reached the saturated condition at 45 min. Accordingly, the immobilization time of spike protein was set as 45 min. (Fig. 5B).
Optimization of the incubation time of IgM/IgG: The time required to form the spike protein-IgM/IgG complex was optimized to achieve the highest response of biodevice in the shortest time. DPV experiments were used to investigate the incubation time for the interaction of 10 pg mL−1 of IgM/IgG and spike protein for different time periods from 5 to 30 min. As can be seen in Fig. 5C, it is after 20 min, that we reached the maximum response; illustrating the surface saturation with the maximum number of antibody molecules. Accordingly, 20 min was selected as the optimum incubation time.
3.3. Performance study of the biodevice
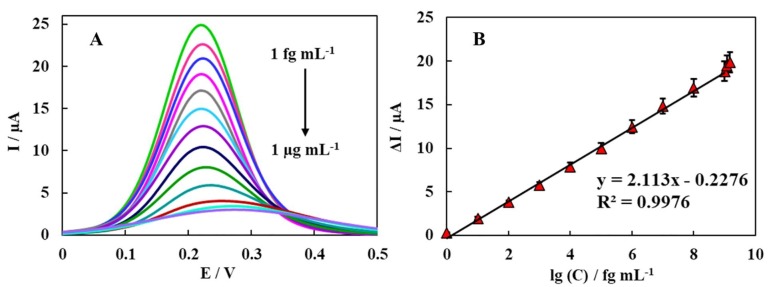
The feasibility of proposed biodevice based on BSA/Ni(OH)2 NPs@SPCE was investigated using DPV technique in [Fe(CN)6]4−/3− with scanning range from 0 to 0.5 V. Under optimal conditions, the dynamic response of the biodevice to different concentrations of IgM / IgG was evaluated in the range of 1 fg mL−1 to 1 µg mL−1. When the concentration of IgM/IgG was increased, the relative DPV current as a signal response was decreased, due to formation of IgM/IgG-spike protein complex on the surface of the proposed biodevice that significantly restrain the electrochemical reaction, consequently, the ET decreases. The regression equation was attained as ΔI (µA) = -2.113 log [IgM/IgG] (fg mL−1) + 22.76 (R2 = 0.9976) (Fig. 6 B). Thus, this biodevice can be measured quantitatively with IgM/IgG. Moreover, the LOD was calculated to be 0.3 fg mL−1 of IgM/IgG (S/N = 3) that was significantly lower than that of the ELISA method (Amanat et al. 2020). It is worth mentioning that the electrochemical sensor, as being user-friendly and having a low cost, can be regarded as more appropriate in areas with limited resources and expertise. Moreover, the mentioned method with its high sensitivity, fast response and its capability of miniaturization can be regarded as an excellent way of detecting IgM/IgG. A comparison of the proposed biodevice performance with some other method for SARS‐CoV‐2 detection are summered in Table 1 .
Fig. 6.
DPV responses of the designed biodevice after incubation with different of the IgM/IgG solutions with concentrations of 0, 1 fg mL−1, 10 fg mL−1, 100 fg mL−1, 1 pg mL−1, 10 pg mL−1, 100 pg mL−1, 1 ng mL−1, 10 ng mL−1, 100 ng mL−1, 1 µg mL−1, 1.2 µg mL−1 (out of the linear range) and 1.5 µg mL−1 (out of the linear range) (from top to bottom) n = 3. Calibration curve of DPV signal vs log C IgM/IgG (fg mL−1).
Table1.
Comparison of performance of the biodevice in this work with other method.
| Ref. | Receptor | Measurement | Assay time | Target | Method |
|---|---|---|---|---|---|
| [33] | Spike protein | Quantitative | 5 min | SARS-CoV-2 antibodies | Electrochemical impedance detection system |
| [34] | Nucleocapsid protein | Quantitative | – | IgG and IgM | DNA-assisted nanopore sensing |
| [35] | Nucleoprotein | Qualitative | – | IgG | ELISA |
| [36] | Nucleoprotein | Quantitative | 60 s | SARS-CoV-2 antibodies | Rapid ELISA detection |
| This work | Spike protein | Quantitative | 20 min | IgG and IgM | Electrochemical immunosensor |
3.4. Reproducibility, selectivity and stability of the method
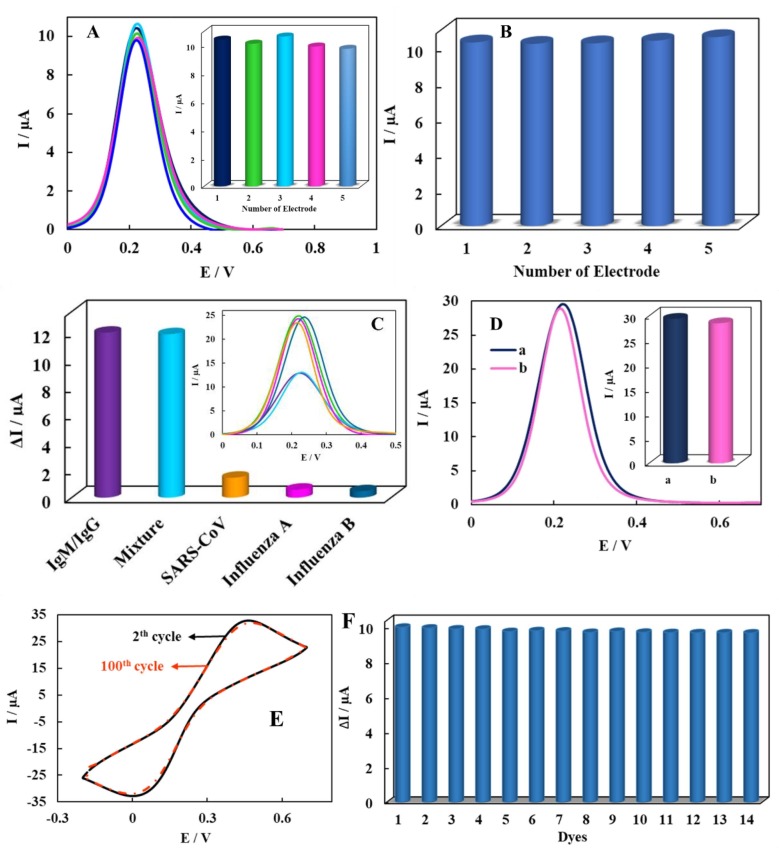
In designing the biodevice, reproducibility, selectivity and stability parameters can be regarded as key factors in improving the performance. To investigate the immunosensor’s reproducibility, five modified electrodes were used to detect 100 pg mL−1 of IgM/IgG. The RSD measurements were 3.1%; indicating the desirable reproducibility of the biodevice (Fig. 7 A) Moreover, for five repeated measurements of 100 pg mL−1 of IgM/IgG, the repeatability of the sensor was evaluated. The obtained RSD value was 1.4% (Fig. 7B); confirming that the designed biodevice has excellent repeatability. For this purpose the DPV response was used in the [Fe(CN)6]4−/3− solution as the redox probe.
Fig. 7.
(A) The histogram and DPV response of reproducibility investigation of the biodevice for 100 pg mL−1 of IgG/IgM on the five different modified electrodes, (B) the histogram of the repeatability investigation of the biodevice for five repeated measurements of 100 pg mL−1 of IgG/IgM, (C) the histogram and DPV response of then biodevice after incubation with IgM/IgG and some off-target species such SARS-CoV influenza A and B antibodies and a mixture of them, (D) the histogram and DPV response of the control electrode, (E) the recorded CVs of the 2th and 100th cycles related to the IgM or IgG/BSA/spike protein/Ni(OH)2 NPs@SPCE in electrolyte solution with scan rate of 100 mV s−1 and (F) the histogram of the long-term stability of the biodevice in present of 100 pg mL−1 of IgG/IgM.
In investigating each sensing method, the ability to identify the target species from other off-target ones is a key factor to evaluate the assay protocol. In order to confirm the biodevice selectivity, its response was studied against other unrelated antibodies, such as severe acute respiratory syndrome coronavirus (SARS-CoV), influenza A and B antibodies, with more 103-fold concentration than the 100 fg mL−1 IgM/IgG. The biodevice responses for the off-target were significantly lower than the IgM/IgG, which can be related to the sensor’s selectivity toward its respective antibody (Fig. 7C). Moreover, the control electrode on which BSA protein was immobilized on the surface instead of the specific antigen was tested. We witnessed no significant responses for the control electrodes clarifying the fact that there would be no non-specific adsorption on the biodevice (Fig. 7D). The results clarify that the proposed platform had appropriate selectivity. Thus, the immunosensor possesses good specificity for detecting IgM/IgG. For this purpose the DPV response was used in the [Fe(CN)6]4−/3− solution as the redox probe.
Another fascinating feature of this strategy was its stability to detect IgM/IgG. Thus, the biodevice stability was evaluated by performing CVs in electrolyte solution with scan rate of 100 mV s−1. The results showed that the peak separation was remained unchanged with only 3% reduction in peak current intensity after 100 periods; indicating the high stability of the modified electrode (Fig. 7E). Additionally, to evaluate the long-term stability of the provided sensor, the biodevice was incubated with 100 pg mL−1 IgM/IgG and stored at 4 °C. The biodevice showed only around 3% change in the current even after 14 days; indicating good stability of the assay (Fig. 7F). For this purpose the DPV response was used in the [Fe(CN)6]4−/3− solution as the redox probe.
3.5. Real sample analysis
Under optimal conditions, each sample was incubated separately on the biodevice surface and, accordingly, the measurement was performed by DPV technique. Besides, in order to investigate their repeatability, each sample was tested three times. Antibodies levels in serum sample were estimated using calibration curves and the relevant data are summarized in Table 2 . As shown in Table 2, the recovery shows that the fabricated biodevice can be correctly used in the serum sample.
Table 2.
Measurement of IgM/IgG in blood serum samples with proposed biodevice.
| Added | Founded | Recovery (%) | RSD (%) |
|---|---|---|---|
| 0 | 0 | – | – |
| 1 fg mL−1 | 1.01 | 101 | 2.8 |
| 1 pg mL−1 | 0.99 | 99 | 3.0 |
| 1 ng mL−1 | 1.02 | 102 | 2.9 |
| 1 µg mL−1 | 1.03 | 103 | 3.1 |
(Each sample is measured three times)
Also, for the feasibility of biodevice in the detection of SARS-CoV-2 antibodies in biological samples, the detection performance of biodevice was tested using clinical samples and the results obtained are summarized in Table 3 and compared with the ELISA test. The results showed possible application of the biodevice to detect SARS-CoV-2 antibodies in blood serum samples that exhibited great promise as a reliable nanotool for the detection of SARS-CoV-2 antibodies in biological sample.
Table 3.
Detection of SARS-CoV-2 antibodies in blood serum samples with proposed biodevice.
| Patients | Biodevice test | ELISA test |
|---|---|---|
| #1 | + | + |
| #2 | + | + |
| #3 | + | + |
| #4 | + | + |
| #5 | + | + |
| #6 | – | – |
| #7 | – | – |
| #8 | – | – |
| #9 | – | – |
| #10 | – | – |
4. Conclusion
A biodevice was constructed based on the SPCE modified with spike protein/Ni(OH)2 NPs for the quantitative detection of IgM/IgG for serological tests. This strategy displayed spike protein’s immobilization as a receptor element of IgM/IgG by chemisorption bond onto the modified electrode surface. In this sense, it aimed at improving the biodevice stability and making a good sensitivity and selectivity to the target detection. Moreover, under optimal conditions, the electrochemical biodevice was capable of accurately and readily detecting and determining the IgM/IgG in human blood serum at a remarkable detection limit in less than 20 min with broad dynamic range and without sacrificing its specificity. Moreover, disposable SPCEs reduce the cost of the assay, have the capacity of miniaturization and can be regarded as a tool for point-of-care diagnostics. The results demonstrate that the fabricated biodevice is capable of being used as a promising clinical tool, which can be used for the serological tests and human diseases diagnosis at the early stages with satisfactory results.
5. Compliance with ethical standards
All experimental protocols were approved by the Experimentation Ethics Committee of Ilam University of Medical Sciences (Code: IR. MEDILAM. REC.1399.210). The clinical samples were provided from a local clinical laboratory. Informed consent was obtained from all participants included in the study.
CRediT authorship contribution statement
Zeinab Rahmati: Resources, Investigation, Writing – original draft, Data curation, Visualization, Writing - review & editing. Mahmoud Roushani: Supervision, Conceptualization, Methodology, Validation, Funding acquisition, Writing - review & editing. Hadi Hosseini: Resources, Investigation, Writing – original draft, Data curation, Visualization, Writing - review & editing. Hamzeh Choobin: Validation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.C.P.E.R.E. Novel, The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China, Zhonghua Liu Xing Bing Xue Za Zhi= Zhonghua Liuxingbingxue Zazhi. 41 (2020) 145. [DOI] [PubMed]
- 2.Fani M., Teimoori A., Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol. 2020;15(5):317–323. [Google Scholar]
- 3.W.H.O. Coronavirus, https://covid19. who. int, (2020).
- 4.Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G. Vogel, New blood tests for antibodies could show true scale of coronavirus pandemic, Sci. Mag. AAAS, DC, USA. (2020).
- 6.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipsitch M., Kahn R., Mina M.J. Antibody testing will enhance the power and accuracy of COVID-19-prevention trials. Nat. Med. 2020;26(6):818–819. doi: 10.1038/s41591-020-0887-3. [DOI] [PubMed] [Google Scholar]
- 9.Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., Wang M. Presumed asymptomatic carrier transmission of COVID-19. Jama. 2020;323(14):1406. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W., McAdam A.J. Laboratory diagnosis of COVID-19: current issues and challenges. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krammer F., Simon V. Serology assays to manage COVID-19. Science (80-.) 2020;368(6495):1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 13.Calvet G., Aguiar R.S., Melo A.S.O., Sampaio S.A., De Filippis I., Fabri A., Araujo E.S.M., de Sequeira P.C., de Mendonça M.C.L., de Oliveira L. Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 2016;16(6):653–660. doi: 10.1016/S1473-3099(16)00095-5. [DOI] [PubMed] [Google Scholar]
- 14.Lipsitch M., Swerdlow D.L., Finelli L. Defining the epidemiology of Covid-19—studies needed. N. Engl. J. Med. 2020;382(13):1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- 15.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan W., Lu Y., Zhang J., Wang J., Dan Y., Tan Z., He X., Qian C., Sun Q., Hu Q. Viral kinetics and antibody responses in patients with COVID-19. MedRxiv. 2020 [Google Scholar]
- 17.Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19) MedRxiv. 2020 [Google Scholar]
- 18.Valipour A., Roushani M. Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: electrochemical oxidation of riboflavin was used as redox probe. Biosens. Bioelectron. 2017;89:946–951. doi: 10.1016/j.bios.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 19.Shahdost-fard F., Roushani M. Designing an ultra-sensitive aptasensor based on an AgNPs/thiol-GQD nanocomposite for TNT detection at femtomolar levels using the electrochemical oxidation of Rutin as a redox probe. Biosens. Bioelectron. 2017;87:724–731. doi: 10.1016/j.bios.2016.09.048. [DOI] [PubMed] [Google Scholar]
- 20.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 21.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2019;395(2020):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward S., Lindsley A., Courter J., Assa’ad A. Clinical testing for Covid-19. J. Allergy Clin. Immunol. 2020;146(1):23–34. doi: 10.1016/j.jaci.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahdost-fard F., Roushani M. Architecting of a biodevice based on a screen-printed carbon electrode modified with the NiONP nanolayer and aptamer in BCM-7 detection. Colloids Surfaces B Biointerfaces. 2020;190 doi: 10.1016/j.colsurfb.2020.110932. [DOI] [PubMed] [Google Scholar]
- 24.Roushani M., Shahdost-fard F. Impedimetric detection of cocaine by using an aptamer attached to a screen printed electrode modified with a dendrimer/silver nanoparticle nanocomposite. Microchim. Acta. 2018;185:214. doi: 10.1007/s00604-018-2709-6. [DOI] [PubMed] [Google Scholar]
- 25.Arduini F., Amine A., Majorani C., Di Giorgio F., De Felicis D., Cataldo F., Moscone D., Palleschi G. High performance electrochemical sensor based on modified screen-printed electrodes with cost-effective dispersion of nanostructured carbon black. Electrochem. Commun. 2010;12(3):346–350. [Google Scholar]
- 26.Caratelli V., Ciampaglia A., Guiducci J., Sancesario G., Moscone D., Arduini F. Precision medicine in Alzheimer’s disease: An origami paper-based electrochemical device for cholinesterase inhibitors. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112411. [DOI] [PubMed] [Google Scholar]
- 27.Antuña-Jiménez D., González-García M.B., Hernández-Santos D., Fanjul-Bolado P. Screen-Printed Electrodes Modified with Metal Nanoparticles for Small Molecule Sensing. Biosensors. 2020;10:9. doi: 10.3390/bios10020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahdost-fard F., Roushani M. The use of a signal amplification strategy for the fabrication of a TNT impedimetric nanoaptasensor based on electrodeposited NiONPs immobilized onto a GCE surface. Sensors Actuators B Chem. 2017;246:848–853. [Google Scholar]
- 29.Roushani M., Rahmati Z., Hoseini S.J., Fath R.H. Impedimetric ultrasensitive detection of chloramphenicol based on aptamer MIP using a glassy carbon electrode modified by 3-ampy-RGO and silver nanoparticle. Colloids Surfaces B Biointerfaces. 2019;183 doi: 10.1016/j.colsurfb.2019.110451. [DOI] [PubMed] [Google Scholar]
- 30.Roushani M., Hosseini H., Hajinia Z., Rahmati Z. Rationally designed of hollow nitrogen doped carbon nanotubes double shelled with hierarchical nickel hydroxide nanosheet as a high performance surface substrate for cortisol aptasensing. Electrochim. Acta. 2021;388 [Google Scholar]
- 31.Nie P., Shen L., Luo H., Li H., Xu G., Zhang X. Synthesis of nanostructured materials by using metal-cyanide coordination polymers and their lithium storage properties. Nanoscale. 2013;5(22):11087. doi: 10.1039/c3nr03289b. [DOI] [PubMed] [Google Scholar]
- 32.S. Pomplun, Targeting the SARS-CoV-2-spike protein: from antibodies to miniproteins and peptides, RSC Med. Chem. (2021). [DOI] [PMC free article] [PubMed]
- 33.Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., Wang X., Wei X., Zheng S.W., Lenhart B.J., Xu P., Li J., Pan J., Albrecht H., Liu C. Multiplex Quantitative Detection of SARS-CoV-2 Specific IgG and IgM Antibodies based on DNA-assisted Nanopore Sensing. Biosens. Bioelectron. 2021;181 doi: 10.1016/j.bios.2021.113134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tozetto-Mendoza T.R., Kanunfre K.A., Vilas-Boas L.S., Espinoza E.P.S., Paião H.G.O., Rocha M.C., de Paula A.V., de Oliveira M.S., Zampelli D.B., Vieira J.M., Jr Nucleoprotein-based ELISA for detection of SARS-COV-2 IgG antibodies: Could an old assay be suitable for serodiagnosis of the new coronavirus? J. Virol. Methods. 2021;290 doi: 10.1016/j.jviromet.2021.114064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Tan Y., Fu Q., Lin M., He J., He S., Yang M., Chen S., Zhou J. Reciprocating-flowing on-a-chip enables ultra-fast immunobinding for multiplexed rapid ELISA detection of SARS-CoV-2 antibody. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112920. [DOI] [PMC free article] [PubMed] [Google Scholar]