Abstract
The COVID-19 pandemic challenges have been only partially addressed so far. The pathogenicity of SARS-CoV-2 is considered the combination of severe and high infectivity. Herd immunity is attained when a critical proportion of the population is immune, providing the virus with fewer chances to spread locally. To overcome the rising tide of the COVID-19 pandemic, efficacious and safe vaccines providing defensive and long-lasting immunity responses are urgently needed. Vaccines that induce virus-neutralizing antibodies with great affinity can optimally fight against infection. Worldwide, over 120 novel vaccine candidates, including live-attenuated, inactivated, viral-vectored nonreplicating and replicating, peptide- and protein-based, and nucleic acid-based approaches are in the process of preclinical and clinical trials (phase 1–4). In addition to comprehensive safety assessments and immune responses, precise clinical management is also important for trials of vaccines. The recent emergence of different variants of SARS-CoV-2 is becoming a new threat for the world and a challenge for scientists to introduce the most influential vaccine against COVID-19. The possibility of natural and vaccine-induced immunity in variants finds it necessary to establish next-generation vaccines, which generate general neutralization against existing and future variants. Here, we summarize the cellular and humoral responses of SARS-CoV-2, current progress in vaccination development, the antibody titer response of available phase 4 vaccinations in vaccinated populations of different countries worldwide, and the success and challenges ahead of vaccine development.
Keywords: SARS-CoV-2, COVID-19, Vaccination, Antibody titer, Cellular immunity, Adaptive immunity, SARS-CoV-2 variants
1. Introduction
The pandemic of COVID-19 presents significant problems that are managed only in part by the nations. SARS-CoV-2 pathogenicity is a combination of severe and high infectivity. This is further enhanced because, unlike SARS-CoV-1 and MERS-CoV, it is spread by symptomatic patients and can be more reliably confined, asymptomatic, and pre-symptomatic to transmit the virus [1], [2]. To minimize COVID-19 harm, the main efforts are focused on isolation, physical distance, and several more infection prevention steps, such as designing the best interventions to prevent viral transmission [3], [4]. Scientific observation and comprehension of and the capacity to propagate the biological processes of the virus are essential. Based on that understanding, realistic policies should have at least three urgencies: firstly, to maintain hygiene and physical detachment; secondly, to optimize the geography and time-bound viral control, to emphasize viral control locally and to minimize propagation anywhere it is possible; and thirdly, to improve the global population's immunity effectively. SARS-CoV-2 may be diagnosed by detecting viral RNA from a nose-swab or saliva, nucleic acid tests (NAT), or screening for viral protein antigens [5], [6]. In infected people, the findings are, on average, only positive for a short period, until 14 days after the start of symptoms [7], [8]. In addition, positive NAT results do not help to determine whether or not the infected individual is immune. Serologic checks are also essential since the different forms of antibodies in the blood that last for months or even years may be detected [1], [9].
Likely, the planet cannot afford to allow most people to get SARS-CoV-2 infections because the potential risk will be huge. Current data show that the pandemic outbreak of COVID-19 infects only low populations (usually within a single-digit) in countries that take successful viral propagation steps [10]. To prevent the pandemic spread of disease outbreaks, the amount of reproductive disease must stay below 1, implying that an individual infected is transmitting on average to 1. It is doubtful that the infection would collapse spontaneously. Further outbreaks are predicted if protection precautions are discontinued. It may take more than one year substantially before most people become immune from infection. As mentioned above, the extent to which natural infection triggers immunity and how long it will guard against reinfection would be determined.
Innate immune stimulation and antigen-related responses of B and T cells are involved in the immune response to SARS CoV-2 [11]. Virus-neutralizing immunity, which is a principle that refers to the overwhelming majority of viral infections under which people are given solid immune defense owing to infection or vaccines, is mostly used to shield them from viral infections. Therefore, vaccines for protective immune response induction need to be developed urgently, particularly via SARS-CoV-2-specific virus-neutralizing antibodies. Although successful vaccinations are globally required for at least 1–2 years, vaccination can still be the fastest and most cost-efficient strategy to achieve comprehensive immune defense. When a critical percentage of the population is resistant, so-called “herd immunity” is achieved, leaving the infection with fewer opportunities to replicate locally. This will happen if >90% of people are immune. However, as soon as “only” 60–70% of the population has been immune, broad immunization is beneficial, as relative simplicity is necessary to prevent viral transmission. In addition, future evolving microbes can increase vaccine production and usage to achieve herd immunity more quickly in the event of more disease-controlled outbreaks [12], [13], [14].
2. Cellular immunity to SARS-CoV-2
Case studies of a limited number of patients indicate that the ratio of CD38+, HLA-DR+T cells rises within the first 7–10 days of indications of COVID-19 and starts returning to baseline about day 20 [15]. SARS-CoV-2-specific perforin 1 and granzyme cells are expressed on in-vitro viral antigens restimulation. In some studies, but not others, the growth in the proportion of T-cells SARS-CoV-2 seems to be related to disease incidence, a major unresolved issue that could impact vaccine production [16], [17]. Severe illnesses were also connected to a more significant decrease in the peripheral CD4+, and CD8+ T cell counts relative to non-serious diseases, which suggested an association between disease gravity and cellular immune response [18].
T-cell responds to peptides extracted from SARS-CoV-2 spike glycoprotein is analyzed by Braun et al. [19] to classify epitope-specific CD4+ T cells utilizing recognition signal induction. In 15 (83%) of 18 COVID-19 patients, the stimulated T cells HLA-DR + and CD38+, which were glycoprotein-specific, could be detected. In addition, in 24/68 control volunteers (healthy), T cells responsive to spike glycoprotein had been detected in particular. While the function of these current viral-reactive cells is unclear, the existence and absence of these cells are hypothesized that could lead to the various medical indexes of COVID-19.
In ten patients with COVID-19, Grifoni et al. [16] employed expectation algorithms for identifying viral sensitive T cells. In seven (70%) COVID-19 patients, CD4+ T-cell responding virus was found. In contrast, T-cell response virus-specific CD8+ was observed in all 10 COVID-19 patients, thus showing that most individual patients will produce SARS-CoV-2T-cell responses. The CD4+ T cell response was primarily made up of Th1-cells associated with high levels of IFNα production and the propension for structural glycoprotein spike, membrane, and nucleocapsid proteins (in that order). The IFNα and the tumor necrosis factor (TNF) α developed in CD8+ T-cell response, reflecting the distorted response to Th1 cells. The immunodominance patterns varied from Th1 cell reaction, but structured proteins were also preferred to non-structural proteins. The non-exposed donors were also found with SARS-CoV-2 peptide reactive cells (6/10) and CD8+ T cells (4/11).
In 42 patients with unresolved controls utilizing an overlapping peptide pool approach, except for ORF1, Peng et al. [20] analyzed T-Cell responses in 42 patients recovering from COVID-19. Their findings showed that the responses of CD4+ T, including CD8+ T cells, with IFNγ, IL-2, and TNFα, were primarily tilted toward Th1 cells and the spiked glycoprotein were immunodominant. The intensity and the width of the immune reaction in patients with serious disorders were improved relative to patients with moderate diseases in both studies. However, a few peptides were addressed more often than others. A more thorough assessment of T-cell response in 203 COVID-19 patients showed that the active cytotoxic heterogeneity of T-cells was present in acute infections. At the same time, the memory phenotype of viral T-cells assessed mostly during the convalescent process was poly-functional, both CD4+ T cell and CD8+ T cells expressing IFNγ, IL-2, and TNFα. T cell responses were particularly noticeable in people who had recovered from mild COVID-19 with no detectable SARS-CoV-2 antibody response [21].
T-cell responses of 108 vaccinated patients were tested by an IFNγ-enzyme-linking immunospot and cellular cytokine staining by Zhu et al. [22]. They observed minimum or inexistent T-cell pre-vaccination reactions to SARS-CoV-2 spike glycoprotein in all patients, meaning the community has no cross-reactive T-cell immunity. Instead, there appears to be a degree of interactivity between SARS-CoV-1 and SARS-CoV-2T-cell responses. T follicular helper (Tfh) cells react by building germ centers, and co-initiating and cytokines to B cells, for developing robust humoral immunity [23]. An autopsy examination of people who have died of COVID-19 showed that the depletion of germ centers and the lack of BCL6+ Tfh cells indicated that the lack of active Tfh response would potentially prevent a deficiency in robust antibody response to SARS-CoV-2. Fig. 1 shows antibody response against SARA-CoV-2 [24]. However, the CD4+ T-cell Response to SARS-CoV-2 analysis observed more significant Tfh cells for patients with serious illness than patients with moderate disease increased proportions. Furthermore, the research mentioned the canonical expressions of Tfh-gene (for example, CXCL13, IL21, and BTLA) in cells endowed by SARS-CoV-2-specific CD4+ T-cells suggesting that SARS-CoV-2 infectious diseases lead to the generation of Tfh cells. Furthermore, the increased T-helper 17 (Th17) cell count is frequently associated with risk factors for severe COVID-19 infection. There is evidence that Th17 cell accumulation in the lungs may lead to chronic COVID-19 infection [23], [24], [25].
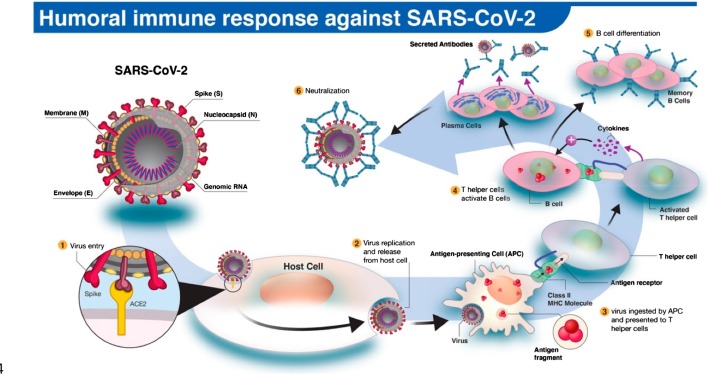
Fig. 1.
The human antibody response to SARS-CoV-2 infection. (1) The SARS-CoV-2 virus enters the host cell via interaction between viral spike (S) and host angiotensin-converting enzyme 2 (ACE2) proteins. (2,3) Following replication and release from the host cells, a subset of viruses will be engulfed and digested by antigen-presenting cells (APCs) like macrophages or dendritic cells. (4) Fragmented SARS-CoV-2 antigen(s) will be presented to T helper cells, which in turn will interact and activate B cells. (5) Activated B cells will proliferate and differentiate into plasma or memory B cells with high-affinity binding receptors for the original SARS-CoV-2 antigen. Plasma cells secrete their SARS-CoV-2-specific receptors in the form of IgM, IgG, or IgA antibodies. (6) Antibody-mediated neutralization occurs when SARS-CoV-2-specific antibodies bind to viral antigen(s) and prevent virus interaction and entry into host cells. Reprinted from Ref. [24] with permission under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/). Copyright © 2020 The authors. Licensee MDPI, Basel, Switzerland.
Numerous recent results showed that T cell numbers were statistically significantly reduced for COVID-19 patients, with more trials reporting the functional complications of the residual T cells [26]. However, the above studies examining SARS-CoV-2-specific cell-mediated immune responses did not report comparable results, whereas the responses to CD4+ T-cells are more robust than those of CD8+ T-cell reactions. Distinctions in the duration of studies may lead to contradictory outcomes, different meanings of mild or severe diseases, and other factors [27]. Laing et al. [28] reported a study aimed at defining the immune signature, which could be used to reference clinical care and evaluation in patients with COVID-19. Furthermore, the authors identified various additional functionality that could be distinguished from COVID 19 patients, both recovered and non-patient controls, in terms of the production of SARS-CoV-2-specific humoral and cell-mediated immune responses. Such clinical benefits, such as inhibiting inflammatory cytokine production, maybe gained if targeted therapies are reversed or minimized.
Overall, accumulating evidence suggests that CD4+ T-cell and CD8+ T-cell responses occur within 1–2 weeks of the initiation of the symptoms and contain primarily Th1 cytokines in certain patients infected with SARS-CoV-2. The occurrence of CD4+ T cells targeting spike glycoprotein associates with a neutral antibody, indicating that the reaction of T cells could also vary between persons with varying intensities of the disease [16]. Furthermore, two small findings show that some people exposed to SARS-CoV-2 developed specific T-cell memory responses in the absence of a particular antibody, implying that SARS-CoV-2 can trigger cellular immunity in the lack of a motor immune response. Cellular immunity's contribution to COVID-19 defense is not yet clear, but a controlled immune response consisting of high levels of neutralization antibodies and Th1-biased T cells is likely to be optimum [14], [21]. There is some indication that CD8+ T-cell repairs were better in mild-disease patients than those with severe disease. However, the function of the CD8+ T-cell response in protecting against COVID-19 is still not apparent. Further studies would be required to evaluate this theory on the cellular immune response to the vaccines SARS-CoV-2 and COVID-19. In certain but not all phase experiments, cell immunity was tested by COVID-19 vaccines; this hypothesis cannot, however, be thoroughly addressed.
3. Antibody responses
In most COVID-19 affected persons, IgM and IgG antibodies are detectable within 1–2 weeks of initiation of symptoms. There is an understandable connection between neutralizing antibodies, antigen-specific T cells, and the disorder's progression and clinical results. High levels of neutralizing antibodies are being identified in convalescent individuals correlating to T cell responses, specifically in CD4+ T cells, and appear to have some advantages in clinical practice with plasma convalescent [29]. Recent findings show that the extent of antibody neutralization is positively linked to the seriousness of the disease [30]. Although the reaction of antibodies to other 'common cold' coronaviruses [31] declines within weeks after infection in most persons infected by SRS-CoV-2, the extent of the neutral reaction of asymptomatic individuals is not only smaller. Still, it often decreases more quickly than that of symptomatic persons [32].
The primary objective of coronavirus neutralization is the S antigen, consisting of domains S1 and S2. The RBD that connects with the ACE2 cellular receiver is membrane distal (S1) S2, a proximal membrane that functions in the fusion of membranes. Furthermore, 88% of the S protein of SARS-CoV and SARS-CoV-2, both with strong affinity, is identical with the ACE2 protein [33]. Therefore, SARS-CoV-2 can be cross-neutralized Antibodies binding to RBD S1 inhibit their association with ACE2. In contrast, those binding to other S1 and S2 regions will inhibit an S-protein and block membrane fusion, respectively (Fig. 2 ) [34].
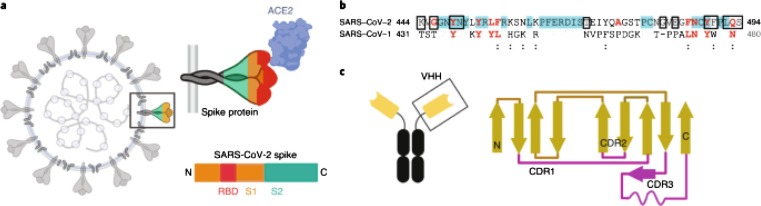
Fig. 2.
a) Spike protein (composed of S1 and S2 subunits) of SARS-CoV-2. S1 contains the RBD. Using the RBD, the trimeric spike molecule binds to ACE2 on human cells. b) RBD residues, and c) Camelids have antibodies that are dimers of a single chain. The constant region is in black and the variable region in yellow. Reprinted from Ref. [34] with permission from Springer Nature. License Number: 5102850860955.
High levels of antibodies to nucleoprotein (N) – the most abundant viral protein – are produced with normal SARS-CoV-2 immune reactions [35], [36]. Although N antibodies do not neutralize the virus, defenses against mouse hepatitis virus, a coronavirus of mice, have been documented. Notably, they were IgG2a, which indicated they could protect through Fc-controlled effector functions instead of neutralizing the virus directly [29]. In addition, several experiments have shown that IgA's S-Peak responses are older and more pronounced than IgM's, making IgA a possible appeal for antibody tests. However, the mechanical foundation for S-specific IgA induction is still unclear [37], [38].
The longevity of the SARS-CoV-2 antimicrobial reaction is still unclear. Even then, subsequent randomized trials of SARS-CoV patients have recorded significant deterioration from 1 year and 2 years following infection in the neutralizing titers of antibodies [39], [40]. The study shows a relatively rapid loss of antibodies against the 229E seasonal coronavirus [29]. There are no SARS-CoV-2 or other human coronaviruses immune correlates of defense. Therefore, it is not obvious enough to guard against infection by neutralizing antibodies. The production of successful COVID-19 vaccines requires establishing specific correlations [18].
4. Vaccines for COVID 19
Researchers are working harder to create and deliver vaccinations throughout the world to prevent the spread of COVID-19. To combat COVID-19, various vaccines are under production, including inactivated vaccines, vaccines for nucleic acids, vector vaccines for adenovirus, and vaccines for recombinants. Inactivated viruses are rendered uninfected physically or chemically and are desirable. They display several immune recognition viral proteins, have stable conformation-dependent antigenic epitopes expression, and can easily be generated in large amounts. Traditionally, purified inactivated viruses have been a vital antidote to combat virus outbreaks, including influenza. Here is an overview of these vaccinations and new advances in clinical trials of these vaccine candidates [41]. Phase 1 vaccines against COVID-19 are summarized in Table 1 . Phase 2 vaccines against COVID-19 are summarized in Table 2 . Table 3 summarizes phase 3 and phase 4 vaccines.
Table 1.
Phase 1 Vaccines against COVID-19 (Data was extracted from Gavi. Available online at: https://www.gavi.org/vaccineswork/covid-19-vaccine-race [Last accessed: July 8, 2021]).
| Vaccine type | Institute | Country |
|---|---|---|
| DNA vaccine | Providence health & services | USA |
| Live attenuated vaccine | Codagenix/serum institute of india | India |
| Viral vector vaccine | Vaxart | USA |
| Viral vector vaccine | Ludwig-maximilians university of munich | Germany |
| Viral vector vaccine | City of hope | USA |
| Protein subunit vaccine | Vaxine | Australia |
| Protein subunit vaccine | University hospital tübingen | Germany |
| RNA vaccine | Imperial college | UK |
| DNA vaccine | Symvivo | Australia |
| Viral vector vaccine (non-replicating) | Altimmune, inc | Maryland, USA |
| RNA vaccine | Chulalongkorn university | Thailand |
| DNA vaccine | Entos pharmaceuticals inc | Canada |
| Protein subunit vaccine | Adimmune corporation | Taiwan |
| Viral vector vaccine | Shenzhen geno-immune medical institute | China |
| Protein subunit vaccine | University of queensland | Australia |
| DNA vaccine | Universtiy of sydney | Australia |
| RNA vaccine | Providence therapeutics | Canada |
| Viral vector vaccine | Bharat biotech international limited | India |
| RNA vaccine | Glaxosmithkline | UK |
| Protein subunit vaccine | Sk bioscience co., ltd | South korea |
| Viral vector vaccine | Gritstone oncology | US |
| Protein subunit vaccine | Walter reed army institute of research | US |
| Inactivated vaccine | Organization of defensive innovation and research | Iran |
| Whole virus vaccine | Meissa vaccines, inc. | USA |
| Viral vector vaccine | The scientific and technological research council of turkey | Turkey |
| Protein subunit vaccine | Jiangsu rec-biotechnology | China |
| Inactivated vaccine | Kocak farma | Turkey |
| Viral vector (non-replicating) | Tetherex pharmaceuticals corporation | US |
| Virus like particle | Radboud university | The Netherlands |
| RNA vaccine | Senai cimatec | Brazil |
| Protein subunit vaccine | University of queensland & csl limited | Australia |
| Viral vector vaccine | Merck/dohme/iavi | USA |
| Viral vector vaccine | Merck/institute pasteur | USA |
Table 2.
Phase 2 Vaccines for COVID-19. (Data was extracted from Gavi. Available online at: https://www.gavi.org/vaccineswork/covid-19-vaccine-race [Last accessed: July 8, 2021]).
| Vaccine type | Institute | Country |
|---|---|---|
| Viral vector vaccine | Beijing wantai biological pharmacy | China |
| Protein subunit vaccine | West china hospital of sichuan university | China |
| RNA vaccine | Arcturus/duke-nus | USA/Singapore |
| Protein subunit vaccine | Medigen | Taiwan |
| Inactivated vaccine | Erciyes university | Turkey |
| Protein subunit vaccine | Razi vaccine and serum research institute | Iran |
| Protein subunit vaccine | Guangdong provincial center for disease control and prevention | China |
| RNA vaccine | Moderna/national institute of allergies and infectious diseases | US |
Table 3.
Phase 3 and 4 vaccines against COVID-19. (Data was extracted from Gavi. Available online at: https://www.gavi.org/vaccineswork/covid-19-vaccine-race [Last accessed: July 8, 2021]).
| Vaccine type | Institute | Country |
|---|---|---|
| Phase 3 vaccines | ||
| Inactivated vaccine | Beijing institute of biological products | China |
| Inactivated vaccine | Wuhan institute of biological products | China |
| Inactivated vaccine | Bharat biotech international ltd | India |
| Viral vector vaccine | Gamaleya research institute | Russia |
| Viral vector vaccine | Janssen/johnson&johnson | USA |
| Protein subunit vaccine | Novavax | USA |
| Protein subunit vaccine | Anhui zhifei longcom biopharmaceutical & institute of microbiology, chinese academy of sciences | China |
| DNA vaccine | Zydus cadila | India |
| Inactivated vaccine | Chinese academy of medical sciences | China |
| Inactivated vaccine | Research institute for biological safety problems | Kazakhstan |
| RNA vaccine | Curevac | Germany |
| Protein subunit vaccine | Sanofi pasteur/GSK | France |
| Protein subunit vaccine | Instituto finlay de vacunas | Cuba |
| Protein subunit vaccine | Vector institute | Russia |
| Protein subunit vaccine | The center for genetic engineering and biotechnology of Cuba | Cuba |
| RNA vaccine | People's liberation army (pla) academy of military sciences/ suzhou abogen biosciences/walvax biotechnology | China |
| Inactivated vaccine | Shenzhen kangtai biological products/ beijing minhai biotechnology co., ltd. | China |
| Phase 4 vaccines | ||
| RNA vaccine | Moderna | USA |
| Viral vector vaccine | Astrazeneca/university of oxford | UK |
| Rna vaccine | Pfizer/biontech | Germany |
| Inactivated vaccine | Sinovac | China |
5. Antibody titer after vaccination
5.1. Texas
BNT162b2 is a nucleoside-modified vaccine of the spike glycoprotein (S) perfusion against SARS-CoV-2. Vaccinations conferred 95% effectiveness towards coronavirus infection in a prospective, placebo-controlled clinical study comprising about 44,000 respondents of Covid-19. SARS-CoV-2 strains, which had been initially found in the UK, South Africa, and Brazil, have spread worldwide with variations in the S gene. To study the impact of BNT162b2 on neutralization, Liu et al. [42] bring about S mutations from each lineage into USA-WA1/2020. Infectious viral titers above 107 plaque formers per millilitre were produced from all mutant viruses. Plaques that were smaller than those made by the other viruses were made by the B.1.1.7 spike and B.1.351 spike viruses.
The 50% plaque reduction neutralization tests (PRNT50) were conducted using 20 serum samples collected from 15 respondents in the seminal study 2 or 4 weeks after the second 30 μg dose of BNT162b3 (the first three weeks). So, the neutralization of B.1.1.7-spike and P.1-spike viruses are theoretically equal in comparison with the neutralization of US-WA1/2020 and neutralization of the B.1.351-pike virus being stable but smaller. Their results often indicate lower titers of neutralization for the complete range of spike mutations B.1.351 compared to those of viruses with either of them. The discovery also indicates that the neutralization of mutations resulting in amino acid substituting location K417N, E484K and N501Y is more potent than removing the N-terminal domain of the spike protein from 242 to 244. The research limits require the possibility of mutations that trigger spike activity rather than antigenicity to change neutralization. Thus, each neutralization trial with a particular target virus is special, and care is needed to view correlations between neutralizing titers from various tests. The defense often entails the immunization of T-cells and CD8+ T-cells for the vaccination of BNT162b2, which identifies many variants.
5.2. Minnesota
In the first scientific report released last week in Nature Medicine, the contagious B117 was first detected in the British variants, and B1351, first found in South Africa, was isolated in the nasal swabs of symptomatic COVID-19 patient’s investigators at the Pasteur Institute in Paris. B117 and B1351 are, like several other new variants, more contagious than historically dominant varieties, causing concerns that the immune system could escape normal and vaccine-induced conditions. Researchers analyzed a SARS-CoV-2 vulnerability form from specimens of serum from 58 individuals with an existing D614G reference strain (Pseudovirus) infection and 19 persons who had obtained two doses in the preceding 6 weeks of Pfizer/BioNTech mRNA COVID-19 vaccine. Both B117 and D614G were neutralized in antibodies to patients who healed from the virus in the last 9 months. However, after 9 months of collection, specimens demonstrated a six-fold decrease of antibody levels with 40% unneutralized B1351 samples. Such antibodies may also be defended against B117 by people completely vaccinated against COVID-19 but less against B1351 relative to D614G. Yet after the second blast, antibodies against B1351 were 14% smaller than those against D614G, whereas the antibodies' cumulative response increased. Infrequently in vaccination nasal swabs, SARS-CoV-2 antibodies have been witnessed. Thus, rapidly transmitting strains of SARS-CoV-2 have obtained incomplete antibodies that are most common in persons with low levels of antibody resistance to natural infection or vaccine. The findings suggest that the probability of immunizations may improve with B1.351, but not B.1.1.7.
The researchers also indicated that previous experiments have shown that the modern mRNA vaccine COVID-19 still prevents virus strains but has a 5–10 times lower effectiveness than D614G compared with B1351. The researchers have indicated that it is necessary, rather than using laboratory-engineered pseudovirus used in most previous studies, to test different antibodies against virus strains with real, clinical viral isolates. They recommended enhanced testing of the function of immune reactions following vaccination among vaccinated individuals with and without prior COVID-19 pathogens and more extended follow-up periods. The second dose of the Pfizer Cominarty [sic] vaccine, which was linked to a significant rise in antibody neutralization and an expanded strain of cross-reactive antinutrients, also underlines the relevance of the research. Finally, the findings show that the lack of inter-reactivity toward new evolving viral strains involves suboptimal or deteriorating antibody reactions. The second report, led by researchers at the University of KwaZulu-Natal in Durban, South Africa, reported in Nature, compared antibody responses of live B1351 and SARS-CoV-2 from the first and second pandemic waves 1 month after the appearance of illness using plasma of patients hospitalized by COVID-19. In South Africa now, B1351 is the causative agent. None of the 14 first-wave plasma donors had B1351, which all six second-wave donors had been compromised. First-wave plasma donor antibodies neutralized the SARS-CoV-2 reference strain, but its efficiency was low towards B-1351, with a 15.1-fold decline in neutralization relative to second-wave donor antibodies. Although second-wave plasma donor antibodies successfully protected both B1351 and the reference strain, they were 2,3 times less efficient against the reference strain than first-wave donor antibodies. Neither of the second-wave donors within the first wave was affected by the comparison strain. The researchers said that it might assist in addressing vaccine production by learning how antibodies to one strain defend themselves against another. With successful neutralization by the plasma evoked in [B1351] first wave virus detected, vaccines based on sequences of [variants] might maintain their action against other SARS-CoV-2 lines in circulation [43].
5.3. Missouri
Currently, the Food and Drug Administration (FDA) licensed for emergency use two vaccines against SARS-CoV-2 involving messenger RNA (mRNA) technology. Phase 3 studies found that the indicative infection after two doses given in three to four weeks separately was over 90% efficient. The primary patients in these studies were those without prior SARS-CoV-2 infection. In the US, there are more than 26 million coronavirus disease cases (Covid-19), and in recent research, elevated seropositivity levels have been found. Therefore, it is essential to identify the immune response to vaccination in people with prior SARS-CoV-2 infections. Bradley et al. [44] identified antibody levels in 36 health employees, who had a clinical verification of SARS-CoV-2 infections 30–60 days before receiving the vaccine, and 152 medical staff, without a record of SARS-CoV-2 infections. In the course of a Children's Mercy Kansas City clinical trial, biospecimen from vaccine responders were collected, and their implementation was examined and accepted by the institutional control board for Children's Mercy. Published informed consent was revoked when participants automatically registered after a research newsletter was reviewed and asked questions. They found that after the first dose of the vaccine, antibody titers of both respondents were elevated against spike proteins by a multiplex bead-binding test measuring IgG. Six participants had antibody levels of unprecedented SARS-CoV-2 matching those of recently infected participants; these six participants might have experienced misdiagnosed infection. Following the first dose of the vaccine, participants newly contaminated had higher antibody titers of S1, S2, and the receptor-binding domain than those without an infection background.
Researchers used in vitro test to determine probable SARS-CoV-2-neutralization antibodies in the blood by antibodies blockage of the ACE2 receptor as a proxy to identify virus-neutralizing antibodies. As predicted, in the community with no history of COVID-19 infection, suppressing antibodies were imperceptible in the baseline and were observable at different levels in the previously infected and misdiagnosed group. After primary immune, they discovered that seropositive participants had higher levels of blocking antibodies than seronegative participants. Three weeks following a single immunization, individuals with a SARS-CoV-2 or seropositive infection were shown to have elevated production of antibodies than people without an infection background to four SARS-CoV-2 antigens and elevated numbers of neutralizing antibodies. However, additional investigations are essential for the duration of antibody reactions and other defensive immunity steps. Protective immunity following vaccination cannot accurately be calculated, and modification of successful immunization programs cannot be reliably prescribed without immune correlations for safety for SARS-CoV-2 vaccinations in humans.
5.4. Maryland
Some analysts have suggested unestablished schemes due to present constraints in the manufacture and delivery of COVID-19 vaccines. Those with COVID-19 are believed to have defensive immunity and memory response for at least 6 months. However, among those recently infected with SARS-CoV-2, either no retroactive or optimal vaccine dosing therapies were investigated. It was evaluated whether pre-COVID-19 health workers could mount a single dose of mRNA COVID-19 vaccine in their memory response [45]. Blood samples of vaccinated healthcare professionals were taken on days 0 (baseline), 7 and 14 after vaccination. The IgG spike trimer has been checked with ELISA and has been updated from a test such that half-maximum binding titers have been interpreted. The corresponding half-maximum binding titers reflect the plasma dilution, which completes the maximum binding of a specified control that reaches saturation by 50%. ID99 (the 99% inhibitory dosage, the maximum dilution, with 98% of cells protected) was also screened for day 0 and 14 samples from vaccines through live virus neutralization. Each sampling day was comparable to the Ab-negative group between each previous Ab-positive group (asymptomatic and symptomatic). 3151 of the 3816 medical staff registered in a serosurvey report were approached at random, and 59 were registered as volunteers: 17 in an Ab-negative group, 16 in an asymptomatic group, and 26 in the asymptomatic group. The proportion of females in the Ab-negative population was 71%, asymptomatic 75 and symptomatic 88%. In both asymptomatic and symptomatic (208, 29 364 and 34 033) and symptomatic (302, 32 301 and 35 460) classes, the average reciprocal half-maximum binding titers in all 0, 7 and 14 days were higher in comparison with the Ab-negative groups. At 0 and 14 days, the median virus neutralization titers ID99 were higher than the Ab-negative.
5.5. New York
In avoiding SARS-CoV-2 symptomatic inflammation in individuals despite preceding coronavirus disease 2019 (Covid-19), there was strong effectiveness of 2 vaccinations of SARS-CoV-2 spike messenger RNA (mRNA) vaccines [46]. Researchers have a limited snapshot of antibody reactions in 110 respondents with or without previously reported SARS-CoV-2 immunity and 67 seronegative participants. A 2-step immuno-sorbent assay was used to calculate SARS-CoV-2 spike IgG and expressed as a region under the curve. The replicate analysis during the first dose showed that most respondents in SARS-CoV-2 IgG responded in a dynamic and relatively low way within 9–12 days following vaccination. In contrast, the SARS-CoV-2 baseline antibody participants quickly established uniform, high-antibody titers in days following vaccination before their first injection.
The titer of antibody within vaccinated patients with previous vaccines was 10–45 times higher, with an antibody dose of 10–16 times higher than the immune dose of vaccine patients without pre-existing at the same time, after their first vaccine dose. While the antibody titers of the vaccines were raised three-fold following a second vaccine dose without previous protection, the COVID-19 patients, who had obtained a second vaccine dose, did not experience an improvement in antibody titers. In the dynamics of antibody responses caused by vaccines Pfizer and Moderna after the first dose, no significant gap was found. The current study is a descriptive survey in which not all researchers have provided antibody specimens in some subsequent periods. Continuing follow-up trials will show if these early immune response variations are sustained over a longer period of time. Furthermore, the rate of local injection-site-related and systemic reactions after the first dose was compared in 230 participants; 149 seronegative respondents. Overall, there were no adverse reactions to either vaccine. 159/230 (69%) participants completing the PARIS study recorded such after-vaccine effects. The most prominent were generalized signs (distress, swelling, and erythema), which appeared at the time of vaccination and recovered naturally with similar incidence regardless of serostatus within days of immunization. Vaccinated patients with pre-established immunity have a greater level of systemic adverse effects than people lacking acquired immunity. As there has been a preference survey and only respondents with accessible information have been investigated, vigilance is required before a complete data set can be analyzed, especially adverse outcomes after the first and second vaccine dose.
5.6. South California (BNT162b2 (Pfizer–BioNTech)
Messenger RNA (mRNA) vaccinations against SARS-CoV-2 are very promising for the thwarting of infection transmission. However, the supply chain challenges have led to questions about whether administering single doses instead of administering double doses could suffice for some people, even those recovering from previous infections. Emergent immune information has proposed potential alternative vaccine options for previously compromised patients, including anti-viral antibodies and the presence of virus-specific T-cells. Smaller trials have shown that people with a previous infection may normally have immunity that can be increased enough with one dose instead of the double dose of the vaccine given. For this reason, researchers tested SARS-CoV-2-specific antibody reactions in a large and varied dataset of health workers after the first and second doses of mRNA vaccine. They contrasted person's responses without previous proof of infection with documented previous infection of people [47].
They registered medical professionals in Southern California from a large academic hub. The vacuum receiver (n = 1090) with at least an antibody assay blood sample aged 41.9 ± 12.2 years: 981 vaccine recipients were presented with baseline (pre-vaccine) samples including 78 pre-infection SARS CoV-2 samples; 525 (35 pre-infective) samples were given after dose 1; and 239 (11 pre-infected) samples were presented after dose 2. A total of 217 people (ten of whom had previously been infected) provided blood tests three times. Measured amounts of antibodies at three points: before and up to 3 d following dose 1; 7–21 d following dosage 1; and 7–21 d following dose 2. Because the timing of an initial blood draw for antibody testing could sneak the combination of spike glycoprotein-specific IgG with early vaccine reaction, they have been using an IgG (IgG (N)) nuclear protein-specific denoting prior exposure SARS-CoV-2 while acknowledging minor cross-creation potentials with another coronavirus. Since BNT162b2 is a vaccine that only provides mRNA for spike protein, the intended elicited response is to produce antibodies IgG (S-RBD) and not antibody IgG (N). In addition, the long-term marker and predictor of post-infectious disease are also established for the use of IgG (N) antibodies [30]. Therefore, they have established the frequency and timing of the earlier SARS-CoV-2 outbreak in relation to the first vaccination date, based on evidence recorded in the health reports, the existence of an IgG (N) antibody at baseline pre-vaccination, the knowledge collection of the self-reported survey. Both instances of discrepancies in the previous infection with SARS-CoV-2 have been subjected to manual medical arbitration, including examining a medical chart to prove positive SARS-CoV-2 polymerase chain reaction or antibody tests.
As predicted, individuals with an SRS-CoV-2 infection had higher antibody levels at all times (P to 0.001) both for IgG (N) (representing pre-infection reaction) and IgG (S‐RBD) (representing a response, either to previous infections or vaccines). Compared with infections-naive persons who obtained a single vaccine dosage, IgG (S-RBD) amounts in baseline were marginally less for previously affected individuals. Moreover, after a single dose and infection-neutralized individuals with two doses, the S-RBD (IgG) amount was not statistically distinct between previously infected persons. Similar findings were identified in a sensitivity examination, which included those people who had antibody immunoassays at all three times. In particular, the pre-infected had IgG (S-RBD) at all times higher than those without pre-infection. No IgG (S-RBD) amounts were shown to vary between pre-infected people after one vaccine dose and pre-infected ones after a couple of doses.
Researchers tested IgG (S-RBD) values at or above 4160 AU ml−1 for substitute steps in antibody neutralization, as that equates to a 0.95 likelihood of receiving a plaque reduction ID50 dilution. Such percentages were notable for individuals that had historically been infected with a single dose below the proportions for individuals infected with two doses (P < 0.001); there were no two-dose intergroup variations. They also have a binding inhibition test of Angiotensin-converting 2 (ACE2) which is well related to the SARS-CoV-2 PRNT system and has strong relationships with the threshold of the IgG (S-RBD) assay. It was discovered that ACE2 binding was slightly higher in people who had previously been exposed than in those who were infectious during the single vaccine dose, with no difference between groups after the second vaccine dose. Time-shifted tests showed a little distinction between individual binding ACE2, after one dose of previous SARS-CoV-2, and individual infection-naive after two doses (94.3% compared to 97.8%, P = 0.52).
Researchers have also studied post-vaccine symptomology in conjunction with antibody reaction tests. In the previously infected organism, they found that after dose 1, reactogenicity is significantly more common than infection-naïve individuals. However, the substantial symptoms of dose 2 have not differentiated between groups. In time-shifted analyses, people infected became more reactogenic than previously infected after dose 2. Fever and chills were more common after the first dose among previously infected vaccine recipients, where after the second dose, headache, dizziness, or lightheadedness were more likely among infectious individuals. Reactogenicity often improved in analyses of shifts from dose 1 to dose 2 in those infected and lower in those previously infected. Ultimately, vaccine-induced antibody responses were produced by individuals previously infected with SARS-CoV-2 after a single dose of the mRNA BNT162b2 (Pfizer—BioNTech) vaccine, close to those shown during two-dose immunization given to individuals who had been infected with the virus. The outcomes of smaller trials that showed elevated amounts of baseline anti-S antibodies and after a single mRNA vaccine dose, correlated with those without previous exposure, were seen in a large and representative cohort of health care professionals [48], [49] and observed similar results after 1st and 2nd doses of vaccine. The neutralizing ability of prompted antibodies was further tested using a high-performance ACE2 inhibition substitute test. In a larger population, it was found that a second vaccinal dose did not give significantly greater benefits to previously infected persons over one single dose in antibody neutralization efficacy, similar to those reported in a smaller study that directly measured antibody neutralization in 59 volunteer workers. Therefore, data show that for persons with previous SARS-CoV-2 infection, a single dose of Pfizer-BioNTech vaccine is adequate when an anti-S antibody level reaction is considered and when the findings of the ACE2 inhibition analysis show a possible neutralizing capacity of elected antibodies are examined.
The sample restrictions include a 21-d timeline for measuring antibodies after any vaccine dose. Further detail about the supposed length of protection obtained from providing one dose versus one double dose of vaccination may also be provided by longer follow-up. T-cell response measures can help shed light on the potential for T-cell memory increases in formerly infected people with a single dose against a double dose of vaccine [48]. Further experiments are needed to decide if a particular vaccine time duration will optimize effectiveness and protection in previously infected persons. Larger cohorts are used to assess anomalies between population and therapeutic subgroups that show variations in antibodies following the vaccine, such that the statistical powers will be enough [50]. The single-dose reaction was numerically comparable but statistically substantially lower than that of an antibody response in two doses in infectious persons if potential neutralization was measured using the IgG (S-RBD) threshold of >4160 AU mla−1. By using this conservative >4160 AU ml−1 threshold, which corresponds to a 95% likelihood of highly neutralized antibody titer, statistical similarities of smaller subsets are vulnerable to extreme values. Notably, in time-shifting analyses after vaccine dose 1 and dose 2, no major difference was noted in the surrogate ACE-2 inhibition of binding infection among individuals with and without pre-infection. Despite experimental discrepancies between the IgG (S-RBD) examination stage and the ACE2 inhibition tests, these replacement steps indicate that the achievement of neutralizing ability levels is substantially identical. Some changes in the responses to antibodies can even be linked to the variability of historically infectious people, like the timing and seriousness of previous diseases. While circulating antimicrobial activities only are not conclusive immune status measurements, successive serological assessments for natural or vaccinated acquaintances are known to be well linked to efficient protecting immunity [51], and our outcomes demonstrate their potential utility in guiding the use of the vaccine in both previously infected and infected diseases. The findings also provide tentative proof for the intermediate relationship between vaccination interventions that are inspired by public health and immunological help. If validated, a single dose of vaccine will optimize the advantage of a restricted vaccine supply by supplying individuals with a reported background and a full timeline vaccine schedule for infect-neutral individuals.
5.7. Italy
It is not known whether people who recovered from SARS-CoV-2 should also be vaccinated. A few study trials have demonstrated a slightly greater response from vaccines previously compromised with SARS-CoV-2 than previously uninfected vaccines. Anichini et al. [52] included 100 medical staff with a reported background of SARS-CoV-2 infection in an observational cohort study, including 38 previously infected (9 men and 29 women). The mean age of those that have already been affected was 35.1 years. They have studied 62 participants (25 men and 37 women) who had not been affected previously. These people's average age was 44.7. The messenger BNT162b2 (Pfizer–BioNTech) RNA vaccination was received by both classes of participants. Serum samples were collected 10 days after the first dose from participants previously infected and 10 days after the second dose from the previously infected participants. Relevant IgG anti-SARS-CoV-2 Spike was then tested via a chemiluminescence microparticle immunoassay. Samples of previously infected participants and previously uninfected participants had demonstrated no substantial difference between the circulation of the IgG anti-spike antibiotic titers. In only one previously infected participant, circulating anti-spike IgG antibodies were not identified, and no normal SARS-CoV-2 antibody response was recorded.
Relevant anti-SARS-CoV-2 neutralizing antibodies were also analyzed for the same serum samples. A disparity was found between the samples of pre-infected participants and those previously infected. There were no significant variations between the processors of the newly infected participants and previously infected participants by age or sex. The previously infected participants were classified by period from diagnosis to vaccine into three groups: 1–2 months (8 participants); 2–3 months (17 participants); and 3 months; (12 participants). This categorization did not involve the previously affected patient with circulatory anti-spike IgG antibodies. IgG means circulation differs from the 1 to 2-month vaccination group to 21,450 arbitrary units per ml. There was no more substantial gap between the vaccinated category of participants for more than 2 months or 3 months and the vaccinating group for more than 3 months.
There are further differences between the three groups in neutralizing the antibodies, with geometric mean titers ranging from 437 to 559 vaccinated 1 to 2 months after infection with 694 vaccinated for more than 2 months or 3 months after infection. Although these results show that the booster reaction was more effective when the vaccine was given more than 3 months after diagnosis, there is insufficient evidence for a definite inference. After administering a second dose of the vaccine in pre-infected patients, the most interesting results were that a slightly smaller neutralizing titer than the titer after just one vaccine dosage in pre-infected patients. The effects on host transmission of the virus are not apparent from neutralizing antibodies titers. These results prove that after a single dose of vaccine, SARS-CoV-2 humoral response is higher than that of previously uninfected subjects given second doses in individuals with a history of SARS-CoV-2 infection.
5.8. France
Recent studies have shown that immunocompetent seropositive SARS-CoV-2 adults will need just 1 dose rather than 2 doses of RNA vaccine, but not older adults. Older adults residing in nursing homes are more likely than younger, healthy adults to have a severe COVID-19 immunity response. Blain et al. [53] contrasted IgG amounts in nursing homes, with or without COVID-19, following a single dose of BNT162b2 (Pfizer-BioNTech) vaccine. The analysis was authorized by the Hospital Review Board of the University of Montpellier, and the participants were given valid informed consent. All patients were subjected to blood tests for SARS-CoV-2 nucleocapsid (N) protein levels for six weeks following the conclusion of the breakdown. The IgG antibody levels against the SARS-CoV-2 spike (S) and N proteins were measured quantitatively by all inhabitants three weeklong later. Out of the 102 residents, there were 60 without previous SARS-CoV-2 infections; 36 of them with a positive RT-PCR outcome and Seropositive to SARS-CoV-2 N-protein IgG; and 6 of those who had a positive RT-PCCR outcome or seropositive SRSCoV-2 N-positive IgG (72.2%). All 36 previously COVID-19 residents were seropositive for S-protein IgG after 1 vaccine dose versus 29 of 60 without previous COVID-19 residents (49.2%). The average amount of S-protein IgG was 40 000 AU/mL or greater for those without previous COVID-19, of which the median is 22 801–1 000 AU/mL against 48,0 AU/mL. Another inhabitant with a good RT-PCR outcome measured seronegative N-protein IgG, having a robust S-protein IgG level. Five citizens have been found to be seropositive to N-protein IgG, despite the fact that previous RT-PCR results had been consistently negative. The S-protein IgG antibody was present in all 5 of these inhabitants. The values for S-protein IgG antibodies were substantially more significant than the 60 without previous COVID-19 of the seven residents with positive RT-PCR results or seropositive for N-protein IgG and were not statistically significantly different from the 36 persons with positive RT-PC results who were seropositive to N-protein IgG.
This preliminary research indicates that the single-dose vaccine BNT162b2 could be appropriate in the nursing homes previously diagnosed with COVID-19 based on RT-PCR findings to achieve a high degree of S-protein IgG antibodies. This is in accordance with the findings from IgG's previously published COVID-19 antibody spike trimer and neutralization titers. In addition, a second dose in individuals without a history of infection will help determine if S-protein IgG antibody levels are required even before the second vaccine dose. This could reduce potentially harmful effects associated with a reactogenicity and save valuable vaccine levels in previously affected patients. The research limitations include small sample size, potential loss of representability, and the lack of neutralization tests.
5.9. India
This research has been conducted to assess IgA and IgG serum titers in the earliest receiver of the SARS-CoV-2 spike antigen. These four people were community professionals and were also a vaccination target. The vaccine staff tested Antibodie amounts for no more than 80 days after the first dose. Baseline studies were negative for nucleocapsid (N) SARS-CoV-2 and spike (S) antigens. The serum amounts of the IgG spike-driven gradually increased exponentially after the first dose, until it ended at 18–21 days. After the second dose, a comparable increase occurred to plateau after seven days. In the remainder of the follow-up cycle, the IgG values plateaued at around 80% of the peak values for about 20–50 days. IgA levels showed a common pattern, peaking in both the first and second vaccine doses simultaneously as IgG. However, after the full titer, the reduction in titer with IgA was slightly faster than the reduction in titer with IgG.
Therefore, after the first injection, the IgA antibody levels fell to around half the titer at the peak reaction. It peaked and then plateaued at around 40% of the maximum dose in 50 days of the second injection after the booster dose. In addition to the SARS-CoV-2 vaccine, this trend of activation by IgG/IgA is in keeping with the serum half-life of the different immunoglobulin isotypes of 21–28 days compared with 5–6 days for IgA and IgG, respectively. The rapid decline in IgA serum levels is comparable to the findings of a study in Spanish health care professionals with a three-month follow-up on antibodies response in natural SARS-CoV-2 infection.
Another research revealed that, while the serum IgA is dropping rapidly against the spiky antigen after normal infection, the concentration of mucosal IgA persists for a longer time and can contain more neutralizing dimeric IgA molecules, which may have up to 15 times greater strength than IgA monomer. In this research, vaccination-induced and anti-specific antigens IgAs were not tested on mucosal surfaces individually. Serum IgA may cause mucosal shape, transducing the mucosal site or secreting IgA secreting plasma plasmas with a molecular surface profile that guides them to the mucosal surfaces. Another option is that B lymphocytes can undergo isotype changes in the mucosa to secrete IgA. The recent study indicates that more research is required to decide on IgA induction and its dissemination on the mucosal sites after mRNA vaccine administration. The fact that the spike antigen serum IgG continues to survive after vaccination will show that two doses of the mRNA vaccine are used for long-term protection. The usefulness of this measurement can also be seen as a biomarker for vaccine response. Secondly, these studies suggest that spiky IgAs are induced by these vaccinations, thus preventing the spread of the virus rather than only symptoms or infections. It is worth noting that the serum IgA dose decreases more quickly than the equivalent IgG amounts against the spike antigen. However, both IgG and IgA are quicker than the primary reaction following the booster dose [54].
5.10. Connecticut (New Haven)
Antibody levels were tracked by volunteers from a continuing serology analysis performed by healthcare workers after being immunized. Subjects supplied 3 cc of blood through vacutainer tube venipuncture; serum was removed at −80 °C and preserved before enzyme-related immuno-sorbents were checked (ELISA). The Yale University Human Research Board examined the studies, and the Yale University Institutional Advisory Committee approved the legal examination [55]. Coronavirus vaccination-sensitive serum levels of IgG rise exponentially and hit a peak around 18–21 days after the first vaccination dose. After the second vaccine injection, the serum IgG rose more and peaked about seven days later; it remained high (78% of peak values) for the next 20–50 days. COVID-19 mRNA also evoked antigen-specific spike IgA with identical inductive kinetics and time for maximum amounts following the first and second vaccine dose. The spike antigen-related IgA amounts, however, decreased substantially more quickly than the amount of IgG. The spike-specific IgA decreased between 1st and 2nd shots to an average of 50%, with a maximum level of 38% in 50 days after the second one.
IgG and IgA antibodies were induced and deteriorated in reaction to the new COVID-19 mRNA vaccine, compatible with the documented serum 1⁄2 lives of various immunoglobulin isotypes; gamma 21–28 days and alpha 5–6 days. The gradual depletion of the serum IgA levels is also compatible with the decline in a Spanish hospital after three months of follow-up in the course of natural diseases among health staff. Local concentrations on mucosal surfaces continue long, including dimeric isoforms with powerful neutralizer potential, 15x largely above the monomeric IgA amounts, whereas serum IgA values decrease rapidly after infection [56], [57].
This research concentrated on serum IgA clonally associated with IgA in mucosa but did not test antigen-specific IgA mucosal concentration caused by the vaccine. Serum IgA can be transduced or recirculated to the mucosal surface by IgA that secretes plasmablasts with a profile of the mucosal homing. However, remote B-cells can often experience isotype-class change with distinct kinetics in the mucosal micro-environment [58]. These results highlight the existing information deficit in the synthesis and delivery of IgA caused by the vaccine at COVID-19 mRNA sites. In sum, the COVID 19 mRNA vaccine recipients' longitudinal serology illustrates critical immune and vaccine reaction follow-up challenges. After vaccination, the persistence of IgG spike-specific serum is a good sign that vaccines can respond effectively long-term and clinically. In addition to IgG, the data show COVID-19 mRNA vaccines, which may be essential in reducing mortality and infection and producing antigen-specific IgA. However, the 'recall' response for IgG and IgA is considerably shorter than the primary response. Spikes specific IgA serum levels deteriorate substantially faster than spike-specific IgG levels.
6. Vaccines and SARS-CoV-2 variants
Globally, there are several variants of SRS-CoV-2. Variants of SARS-CoV-2 are listed as variants of interest, variants of concern and variants of great consequence by the Centers for Disease Control and Prevention (CDC). Three latest variants (B.1.1.7 (also recognized as VOC-202012/01), 501 Y.V2 (B.1.351) and P.1 (B.1.1.28.1)) have raised concerns within their countries quickly. The first cases of B.1.1.3, 501 Y.V2, and P.1 variants were reported in the UK, South Africa, and Brazil. B.1.1.3 and 501 Y.V2 strains are reported with 23 mutations while P.1 strain is reported with 35 mutations. In comparison, 17 amino acids are found changed in all three reported variants. All three variants show N501Y mutation, which switches asparagine (N) amino acid to tyrosine (Y) at location 501 in the spike protein receptor-binding domain. In addition, the 501 Y.V2 and P.1 variants have two additional domain mutations, K417N/T and E484K, which are receptor binding domain mutations. These mutations improve their binding affinity with the ACE2 receptor. The impacts on virus transmissibility, disease incidence, rate of reinfection (i.e., escape from natural immunity) and vaccine efficacy are four major issues arising from the advent of new variants. Two other strains (B.1.427 & B.1.429) have also reported in California [59].
7. COVID-19 vaccines: Successes and challenges ahead
Adequate genomic surveillance, standard variation nomenclature, and a repository of variants and serum vaccine samples are needed to tackle the problems of the new SARS-CoV 2 variants. Still, a particular need is to provide a protective correlation to enhance the potency of vaccines generated in current variants. Moreover, recurring clinical trials with any variation may take so much time that even new variants may appear after these clinical trials [60]. As the immune response needed for the prevention of mild illness may vary from the serious disease, protective correlates may have to be stratified based on the seriousness of the disease. To accomplish this objective, there are four main criteria. First, openness and the free exchange of data should be the responsibility of all vaccine developers of SARS-CoV-2 for current or completed efficacy tests. Secondly, an advisory committee should be formed to study current and proposed tests to find protective correlations with each effective vaccine (preferably under the umbrella of WHO). Third, various vaccinations trials can be conducted to quickly identify an animal model, test or marker as a protective reference to correct discrepancies in the corresponding study plans. Finally, a central database should be developed to gather data on each effective vaccination and have broader samples to measure different values as protective correlations and verify whether a correlate found in a single study is accurate in other studies. It is critical and urgent to identify a correlation of security for individual investigators or vaccine creators to be left to an uncoordinated separate study. The correlation between the defense of mild and extreme SARS-CoV-2 infection would provide a substantial basis for certain decisions and remove barriers to the vision of the globally-focused SARS-CoV-2 management through widespread successful immunization [61].
8. Conclusion
The continuous production of simultaneous vaccines will compensate for the knowledge gap. In addition, certain fundamental, translation, and pre-clinical evidence in coronavirus science shapes the favorable ground for rapid development along with massive scientific efforts. It is essential to investigate the genetic drivers for SARS-CoV-2, defining in-depth targets of the humoral and cell-based immune response at the epitope levels, characterize the repertoire of B-cell receptors and T-cell receptors elicited by infection and vaccination, and establish long-term endurance and maintaining the SARS-CoV-2. COVID-19 vaccinations are promising because high levels of antibody neutralizing affinity are induced, and other immune responses with a chance of improving the epidemic are comparatively poor in numbers. However, the effectiveness and reliability of the vaccination can only partly be predicted. The introduction of various vaccine varieties, including other objective antigens, remains justified and could increase efficacy. Vaccines that induce neutralizing antibodies on the broadest possible scale are best used to improve the SARS-CoV-2 ratio of the population. Furthermore, vaccinations are realistic in the achievement of flock protection. In contrast, broad-based natural infections tend to be too dangerous for humans and the economy, even in countries with the less stringent virus spreading controls, which could trigger immunity in far greater fractions than are known to and predicted in the world. Therefore, the vaccine COVID-19 is highly urgent. If the vaccination has proved effective and secure, it should be registered to guarantee that the environment is equipped for present and potential outbreaks of SARS-CoV-2.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Consejo Nacional de Ciencia y Tecnología (CONACYT) is thankfully acknowledged for partially supporting this work under Sistema Nacional de Investigadores (SNI) program awarded to Hafiz M.N. Iqbal (CVU: 735340).
References
- 1.Speiser D.E., Bachmann M.F. COVID-19: Mechanisms of vaccination and immunity. Vaccines. 2020;8(3):404. doi: 10.3390/vaccines8030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat S.A., Bashir O., Bilal M., Ishaq A., Dar M.U.D., Kumar R., et al. Impact of COVID-related lockdowns on environmental and climate change scenarios. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilal M., Iqbal H.M.N. Recent advances in therapeutic modalities and vaccines to counter COVID-19/SARS-CoV-2. Human Vaccines Immunotherap. 2020;16(12):3034–3042. doi: 10.1080/21645515.2020.1794685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng V.C.C., Wong S.-C., Chen J.H.K., Yip C.C.Y., Chuang V.W.M., Tsang O.T.Y., Sridhar S., Chan J.F.W., Ho P.-L., Yuen K.-Y. Escalating infection control response to the rapidly evolving epidemiology of the coronavirus disease 2019 (COVID-19) due to SARS-CoV-2 in Hong Kong. Infect. Control Hosp. Epidemiol. 2020;41(5):493–498. doi: 10.1017/ice.2020.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. Weissleder, H. Lee, J. Ko et al., COVID-19 diagnostics in context, Sci. Trans. Med. 12 (546) (2020). [DOI] [PubMed]
- 7.Tan W., Lu Y., Zhang J., et al. Viral kinetics and antibody responses in patients with COVID-19. MedRxiv. 2020 [Google Scholar]
- 8.N.G. Davies, S. Abbott, R.C. Barnard, et al., Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England MedRxiv, (2021) 2020-12.
- 9.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., Zepeda S., di Iulio J., Bowen J.E., Montiel-Ruiz M., Zhou J., Rosen L.E., Bianchi S., Guarino B., Fregni C.S., Abdelnabi R., Foo S.-Y., Rothlauf P.W., Bloyet L.-M., Benigni F., Cameroni E., Neyts J., Riva A., Snell G., Telenti A., Whelan S.P.J., Virgin H.W., Corti D., Pizzuto M.S., Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.G. Vogel, First antibody surveys draw fire for quality, Bias (2020). [DOI] [PubMed]
- 11.I. Thevarajan, T.H. Nguyen, M. Koutsakos, et al., Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19, Nature Med. 26 (4) (2020) 453-455. [DOI] [PMC free article] [PubMed]
- 12.Gates B. Responding to Covid-19—a once-in-a-century pandemic? New England J. Med. 2020;382(18):1677–1679. doi: 10.1056/NEJMp2003762. [DOI] [PubMed] [Google Scholar]
- 13.Murchu O.E., Byrne P., Walsh K.A., et al. Immune response following infection with SARS-CoV-2 and other coronaviruses: A rapid review. Rev. Med. Virol. 2021;31(2) doi: 10.1002/rmv.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallais F., Velay A., Nazon C., Wendling M.-J., Partisani M., Sibilia J., Candon S., Fafi-Kremer S. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without seroconversion, France. Emerg. Infect. Dis. 2021;27(1):113–121. doi: 10.3201/eid2701.203611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuri-Cervantes L., Pampena M.B., Meng W., et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. BioRxiv. 2020 [Google Scholar]
- 16.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 18.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. The Lancet. 2020 doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Müller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 20.Y. Peng, A.J. Mentzer, G. Liu et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19, Nature Immunol. 21 (11) (2020) 1336-1345. [DOI] [PMC free article] [PubMed]
- 21.Sekine T., Perez-Potti A., Rivera-Ballesteros O., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F.C., Li Y.H., Guan X.H., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. The Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crotty S. Follicular helper CD4 T cells (Tfh) Annu. Rev. Immunol. 2011;29(1):621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 24.Ghaffari A., Meurant R., Ardakani A. (COVID-19 serological tests: how well do they actually perform? Diagnostics. 2020;10(7):453. doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y., Liu J., Zhang D., et al. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diao B., Wang C., Tan Y., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laing A.G., Lorenc A., Del Barrio I.D.M., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26(10):1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 29.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20(10):615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Seow, C. Graham, B. Merrick, et al., Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 5 (12) (2020) 1598-1607. [DOI] [PMC free article] [PubMed]
- 31.Sariol A., Perlman S. Lessons for COVID-19 immunity from other coronavirus infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B.o., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 33.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M., Mikolajek H., Malinauskas T., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 35.Bostancıklıoğlu M. SARS-CoV2 entry and spread in the lymphatic drainage system of the brain. Brain Behav. Immun. 2020;87:122. doi: 10.1016/j.bbi.2020.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S., Shen J., Fang S., et al. Genetic spectrum and distinct evolution patterns of SARS-CoV-2. Front. Microbiol. 2020;11:2390. doi: 10.3389/fmicb.2020.593548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padoan A., Sciacovelli L., Basso D., Negrini D., Zuin S., Cosma C., Faggian D., Matricardi P., Plebani M. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin. Chim. Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.H.Q. Yu, B.Q. Sun, Z.F. Fang, et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients, European Respiratory J. 56 (2) (2020). [DOI] [PMC free article] [PubMed]
- 39.Cao W.-C., Liu W., Zhang P.-H., Zhang F., Richardus J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007;357(11):1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 40.Wu L.-P., Wang N.-C., Chang Y.-H., Tian X.-Y., Na D.-Y., Zhang L.-Y., Zheng L., Lan T., Wang L.-F., Liang G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13(10):1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.https://www.gavi.org/vaccineswork/covid-19-vaccine-race, 2021.
- 42.Y. Liu, J. Liu, H. Xia, et al., Neutralizing activity of BNT162b2-elicited serum, New England J. Med. 384 (15) (2021) 1466-1468. [DOI] [PMC free article] [PubMed]
- 43.https://www.cidrap.umn.edu/news-perspective/2021/03/covid-19-antibodies-appear-ward-b117-better-b1351.
- 44.T. Bradley, E. Grundberg, R. Selvarangan, et al., Antibody Responses after a Single Dose of SARS-CoV-2 mRNA Vaccine, New England J. Med. (2021). [DOI] [PMC free article] [PubMed]
- 45.Saadat S., Tehrani Z.R., Logue J., et al. Binding and neutralization antibody titers after a single vaccine dose in health care workers previously infected with SARS-CoV-2. JAMA. 2021 doi: 10.1001/jama.2021.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krammer F., Srivastava K., Alshammary H., et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N. Engl. J. Med. 2021;384(14):1372–1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebinger J.E., Fert-Bober J., Printsev I., Wu M., Sun N., Prostko J.C., Frias E.C., Stewart J.L., Van Eyk J.E., Braun J.G., Cheng S., Sobhani K. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021;27(6):981–984. doi: 10.1038/s41591-021-01325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prendecki M., Clarke C., Brown J., Cox A., Gleeson S., Guckian M., Randell P., Pria A.D., Lightstone L., Xu X.-N., Barclay W., McAdoo S.P., Kelleher P., Willicombe M. Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine. The Lancet. 2021;397(10280):1178–1181. doi: 10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manisty C., Otter A.D., Treibel T.A., McKnight Á., Altmann D.M., Brooks T., Noursadeghi M., Boyton R.J., Semper A., Moon J.C. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. The Lancet. 2021;397(10279):1057–1058. doi: 10.1016/S0140-6736(21)00501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soiza R.L., Scicluna C., Thomson E.C. Efficacy and safety of COVID-19 vaccines in older people. Age Ageing. 2021;50(2):279–283. doi: 10.1093/ageing/afaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z., Schmidt F., Weisblum Y., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021:1–7. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anichini G., Terrosi C., Gandolfo C., Gori Savellini G., Fabrizi S., Miceli G.B., Cusi M.G. SARS-CoV-2 Antibody Response in Persons with Past Natural Infection. N. Engl. J. Med. 2021;385(1):90–92. doi: 10.1056/NEJMc2103825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H. Blain, E. Tuaillon, L. Gamon, et al., Spike Antibody Levels of Nursing Home Residents With or Without Prior COVID-19 3 Weeks After a Single BNT162b2 Vaccine Dose, JAMA, (2021). [DOI] [PMC free article] [PubMed]
- 54.https://www.news-medical.net/news/20210330/Study-finds-rapid-increase-in-IgG-and-IgA-antibody-levels-following-COVID-19-mRNA-vaccination.aspx.
- 55.J.C. Luna, A.V. Wisnewski, C.A. Redlich, Human IgG and IgA responses to COVID-19 mRNA vaccines, medRxiv, (2021). [DOI] [PMC free article] [PubMed]
- 56.Z. Wang, J.C. Lorenzi, F. Muecksch, et al., Enhanced SARS-CoV-2 neutralization by dimeric IgA, Sci. Trans. Med. 13 (577) (2021). [DOI] [PMC free article] [PubMed]
- 57.D. Sterlin, A. Mathian, M. Miyara, et al., IgA dominates the early neutralizing antibody response to SARS-CoV-2, Sci. Trans. Med. 13 (577) (2021). [DOI] [PMC free article] [PubMed]
- 58.Xiong N.a., Fu Y., Hu S., Xia M., Yang J. CCR10 and its ligands in regulation of epithelial immunity and diseases. Protein Cell. 2012;3(8):571–580. doi: 10.1007/s13238-012-2927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384(19):1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. The Lancet. 2021;397(10278):952–954. doi: 10.1016/S0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D. Skegg, P. Gluckman, G. Boulton, et al., Future scenarios for the COVID-19 pandemic, The Lancet 397 (10276) (2021) 777-778. [DOI] [PMC free article] [PubMed]