Abstract
The question associated with efficacy and longevity of SARS-CoV-2 protection post-vaccination is paramount. The cPass surrogate virus neutralization test (sVNT) has gained popularity globally as a dual application assay for: 1. Accurate SARS-CoV-2 population surveillance (seroprevalence) analysis and 2. Revealing the presence of antibodies that block and effectively neutralize the interaction between the SARS-CoV-2 receptor binding domain and the host cell ACE2 receptor in recovered or vaccinated individuals. This study describes an approach for accurate quantification of neutralizing antibodies using the cPass sVNT with an automated workflow on the Tecan EVO and Dynex Agility platforms that is applicable to other liquid handling systems. This methodology was used to assess the stability of SARS-CoV-2 neutralizing antibodies between freeze/thaw and refrigerated sample storage conditions. Furthermore, a subset of twenty-five samples from SARS-CoV-2 infected/recovered individuals revealed a 600-fold difference in the neutralizing antibody response where low titers were represented in about half of the samples. Finally, pre- and post-vaccination samples were tested for neutralizing antibodies using the qualitative and semi-quantitative cPass sVNT protocols revealing undetectable or relatively low levels after the first vaccine dose and a decline in levels longitudinally over the months following the second dose. This wide range in neutralizing (blocking) antibodies from both natural infection and vaccination supports a differential immune response that may be attributed to several physiological and genetic factors underlining the potential for measuring SARS-CoV-2 neutralizing antibody titer levels post-vaccination to help ensure robust and prolonged immunity.
1. Introduction
The cPass sVNT has proven to serve as a highly accurate serology assay that detects and measures the functional response of circulating antibodies that specifically “block or neutralize” the interaction of the SARS-CoV-2 receptor binding domain (RBD) to the host cell ACE2 receptor [1], [2], [3], [4], [5]. These dual detection/screening and functional properties uniquely positions the test for: 1. population surveillance (seroprevalence) [6], [7]; 2. vaccine development, associated clinical trials and post-vaccination follow-up testing [8], [9]; 3. convalescent donor plasma and drug screening [10], [11] and 4. longitudinal testing to track neutralizing antibody levels post-vaccination. Considering the broad applications and potential large-scale need for cPass sVNT, migration to an automated liquid handling platform is critical. However, differences between the cPass sVNT competition/inhibition test methodology and those of more traditional antigen-coated, ELISA-based SARS-CoV-2 assays create unique challenges [4], [12], [13], [14].
The current gold standard virus neutralization tests require live cells and virus in a BSL3 containment lab. These complex, manual assays span two to four days, require specialized equipment and highly trained technicians [15], [16], [17]. With the advent of SARS-CoV-2 global vaccination programs with vaccines of varying efficacies [18], [19], [20], it is important to correlate post-vaccination immune responses with the duration of protection against reinfection. Understanding this temporal component may be important in preventing future SARS-CoV-2 pandemics and outbreaks. Recent studies from SARS-CoV-2 infected individuals show antibody titers decline after recovery, distinct immunotypes between infected individuals, and weakened immune responses in older adults [21], [22], [23], [24], [25], [26]. This may warrant regular measurement of neutralizing antibody titers post-vaccination. Thus, a technology that permits the direct comparison of neutralizing antibody levels between samples while circumventing time-consuming and cost-prohibitive live cell neutralizing antibody assays like the plaque reducing neutralizing antibody test (PRNT) could be beneficial [17], [27], [28]. The cPass sVNT has been shown to give comparable data to live cell tests without the extensive processing and complexity in a simple 96-well plate-based assay that requires approximately 1.5 h to qualitatively interrogate up to 92 samples [1], [2], [3], [4], [29], [30], [31]. While the improved workflow with the sVNT is beneficial, further improvements can be made by transforming the assay to a fully automated and semi-quantitative test. This would facilitate at-scale, continued longitudinal assessment of neutralizing antibody titers in post-vaccination populations.
Accurate SARS-CoV-2 antibody testing also requires knowledge of neutralizing antibody stability in serum/plasma samples at 4 °C and −80 °C with multiple freeze/thaw cycles. Although the data are sparse, some work has been performed in the past with dengue, measles, mumps, and rubella as well as anticardiolipin immune response antibodies that give varying degrees of stability [32], [33], [34]. Considering the real-world variability in sample acquisition [35], [36], processing and storage [37], a better understanding concerning how those conditions impact assay results could improve downstream data accuracy and the conclusions drawn from this semi-quantitative test.
A methodology for automation and production of semi-quantitative data from the cPass sVNT is described. A set of SARS-CoV-2 positive and negative samples were qualitatively screened and delineated with this platform with selected samples processed using a novel, semi-quantitative protocol. Longitudinally collected samples from individuals both pre- and post-vaccination were tested using the qualitative and semi-quantitative cPass sVNT protocols. Finally, a subset of samples was assessed for stability under a number of freeze/thaw and refrigerated conditions to reveal the effect of storage on the measured antibody titers. Taken together the results pave a practical path forward to achieve accurate, high-throughput and semi-quantitative SARS-CoV-2 immune response testing in a post-COVID vaccination world.
2. Materials and methods
2.1. Serum samples from individuals previously infected and recovered from SARS-CoV-2
Seventy-four COVID-19 presumed positive samples collected for clinical diagnostic purposes were subsequently de-identified and released to the Kaiser Permanente (KP) Research Bank for secondary use. Fifty-seven of the presumed positive samples were used to compare the performance of cPass sVNT with two commercial IgG binding assays (see subsection “SARS-CoV-2 ELISA Tests” below). The remaining 17 samples were allocated to refrigerated and freeze/thaw stability studies. Twenty-nine serum samples collected prior to August 2019 (pre-pandemic) were randomly selected from the KP Research Bank Biorepository and subsequently deidentified. The SARS-CoV-2 positive serum specimens were collected from 3 to 15 weeks post PCR testing, processed in BD Vacutainer Serum Separation Transport Tubes (SST) according to the standard clinical protocol in CAP/CLIA certified Kaiser Permanente laboratories. These samples were stored for 3–15 days refrigerated prior to shipment to the KP Research Bank laboratory. All specimens except those used for stability testing, were introduced to one freeze/thaw cycle. Pre-pandemic specimens were stored at −80 °C prior to testing.
2.2. Serum samples from SARS-CoV-2 vaccinated individuals
The data in Fig. 5A and B were derived from de-identified samples from vaccinated individuals collected and tested by FourthWall Testing LLC and Cure-Hub LLC (https://www.cure-hub.com). Fig. 5C and D represent a longitudinal study from vaccinated individuals collected and tested by Cayman Chemical Company (https://www.caymanchem.com/). All individuals gave informed consent to use their samples for research purposes.
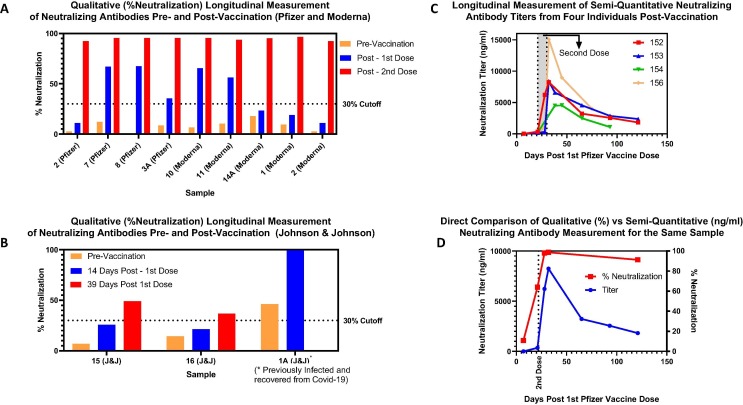
Fig. 5.
Longitudinal analysis of neutralizing antibodies pre and post-vaccination with cPass sVNT. A. Qualitative (% Neutralization) assessment of neutralizing antibodies with Pfizer and Moderna vaccines. Samples were tested before (pre-vaccination), or within two to three weeks after each dose (post-1st or 2nd dose). B. Qualitative (% Neutralization) analysis of neutralizing antibodies with Johnson & Johnson vaccine. Samples were tested before (pre-vaccination), two weeks after and 39 days after vaccination. C. Semi-quantitative testing of neutralizing antibody titers (Fig. 3) post-vaccination. Samples were collected from the same individuals over multiple time points post-vaccination and assessed semi-quantitatively. D. Direct comparison between qualitative (% neutralization) and semi-quantitative (ng/ml titers) post-vaccination. Samples were collected from subject 152 (Fig. 5C) using the cPass sVNT qualitative and semi-quantitative (Fig. 3) protocols with semi-quantitative data plotted on the left Y-axis and qualitative data on the right Y-axis.
2.3. Facility
Experimental work with samples from COVID-19 infected/recovered individuals was performed in the Kaiser Permanente Research Bank Biorepository – ISO 2001:2015 certified and College of American Pathologists (CAP) accredited (CAP ID # 9511943) BSL2 facility. The Biorepository adheres to industry best practices for processing, long-term storage, retrieval and distribution of specimens. The data generated from vaccinated individuals was derived from FourthWall Testing LLC, Cayman Chemical Company and Cure-Hub LLC.
2.4. SARS-CoV-2 ELISA tests
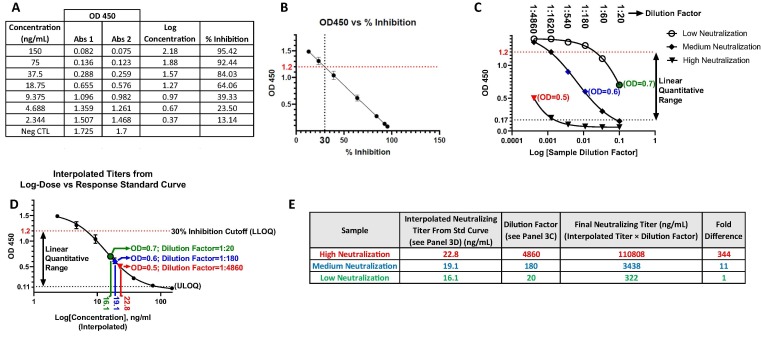
The SARS-CoV2 cPass Surrogate Virus Neutralizing Test (sVNT) (GenScript sVNT (Piscataway, NJ) #L00847) utilizes the recombinant receptor binding domain (RBD) of the SARS-CoV-2 spike protein to detect antibodies that specifically block the RBD from binding to hACE2 receptor [4]. A neutralizing antibody standard curve was designed to validate the kit and semi-quantitatively assess neutralizing antibody titers in diluted samples. Monoclonal Neutralizing Antibody (MAB) (GenScript - #A02051) was diluted in negative control matrix (SARS-CoV-2 negative serum diluted 1:10 in kit-specific dilution buffer) to a concentration of 300 ng/mL. This stock solution was then serially diluted 1:2 in negative matrix control (ie: a pool of SARS-CoV-2 negative serum diluted 1:10 in the kit-provided sample dilution buffer) for standards 2 through 7. Since each dilution is mixed with an equivalent volume of RBD conjugated horseradish peroxidase (RBD-HRP), the final, starting concentration for the MAB standard curve was 150 ng/ml (Fig. 3A). Negative matrix control alone was used for background wells as well as the kit supplied negative control. Plasma or serum samples were diluted 1:10 in kit-specific sample dilution buffer and positive control diluted 1:10 in negative matrix control and sample dilution buffer. Standards, samples and controls were tested and analyzed in a semi-automated approach according to the kit instructions. For semi-quantitative analysis, samples were diluted 1:10 and then serially 1:3 for five additional dilutions in sample dilution buffer. All six dilutions from each sample were then tested and interpolated against the MAB standard curve to determine a titer value that after accounting for the dilution factor, gave a final neutralizing titer per sample (Fig. 3).
Fig. 3.
Workflow for semi-quantitative analysis of SARS-CoV-2 immune response antibodies. A. Generate OD450 data for standard curve and diluted samples using the semi-quantitative protocol (section 2.5). Tabulate the OD450 values from the monoclonal neutralizing antibody (A02051) standard curve with the associated concentrations and calculated % inhibition values. B. Determine the OD450 corresponding to 30% inhibition cutoff. Plot OD450 versus % inhibition and fit to a linear regression model to interpolate the OD450 that gives 30% inhibition (cutoff between positive and negative samples) to define the lower limit of quantification (LLOQ) of the standard curve (Fig. 3D). C. Serially dilute samples to reveal OD450 values within the linear quantitative range (ie: between the LLOQ and ULOQ (Fig. 3D). Dilute individual samples 1:20 (ie: 1:10 in sample dilution buffer followed by 1:2 after mixing with RBD-HRP) and then serially 1:3 in sample dilution buffer for up to five additional dilutions to produce dilutions within the linear quantitative range (points in red, blue and green for examples of samples with low, medium and high neutralizing titers respectively). D. Plot the standard curve and interpolate the neutralizing antibody titers per sample. Plot the standard curve of OD450 versus Log Concentration and define the linear quantitative range used to interpolate the OD450 values from the unknown sample dilutions (Fig. 3C) to achieve semi-quantitative titers (red, blue and green). The LLOQ is defined in Fig. 3B and the upper limit of quantification (ULOQ) is determined from either the 95% confidence interval within the standard curve lower plateau [38] or by using a signal to noise of three (ie: 3 multiplied by the lowest OD450 between the most concentrated point on the standard curve or the positive control) [39]. E. Calculate the final Neutralizing Titer. Calculate the product of the interpolated titers from the standard curve (Fig. 3D) and the sample dilution factors required to achieve the linear range (Fig. 3C) as shown for samples with high (red) medium (blue) and low (green) neutralization titers. All steps were performed with the Tecan EVO automation system and have also been validated with the Dynex Agility. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The Platelia SARS-CoV-2 Total Ab (Bio-Rad (Hercules, California) # 12014591) is a total antibody test that measures the interaction between immune response antibodies with soluble, peroxidase conjugated nucleocapsid protein (PCNP) and immobilized nucleocapsid protein (INP) bound to the assay plate. The presence of immune response antibodies in the SARS-CoV-2 samples will form a stable complex between PCNP and INP giving a signal after washing the plate. The identical 86 samples (29 negative and 57 presumed positive) previously tested with cPass sVNT were analyzed according to the kit instructions.
The Lumit™ Dx SARS-CoV-2 Immunoassay (Promega (Fitchburg, Wisconsin) # VB1080) is a total antibody test that measures the interaction between immune response antibodies with two soluble, luminescent subunits each conjugated to an immunogenic SARS-CoV-2 antigen. The presence of immune response antibodies in the SARS-CoV-2 samples will form a stable complex between the luminescent subunits giving a signal without the requirement for washing the plate. The identical 86 samples used for the Platelia SARS-CoV-2 Total Ab test (29 negative and 57 presumed positive) previously tested with cPass sVNT were analyzed according to the kit instructions.
2.5. Automation for cPass sVNT
The samples were tested using Tecan EVO200 liquid handler with 8 Liquid Handling Arm (LiHa), 96 channels Multichannel arm (MCA) and robotic manipulator arm (ROMA), integrated 1D/2D Ziath scanner and Tecan M200 Infinite multimode detector. Two Tecan EVO scripts have been developed for each respective method: 1) Batch testing for qualitative analysis and 2) Sample of interest serial dilutions for semi-quantitative testing. Serial dilutions were prepared using 8 channels arm without changing the tips from high to low dilution. 96 MCA was used to decrease processing time and ensure simultaneous introduction of all test samples, controls and dilution series to various reagents or reaction components. The kit components and test sample have various liquid properties and pipetting volumes. Customized liquid classes have been developed/tested to accommodate various liquid properties, minimize droplet formation, ensure proper mixing, decrease dead volumes and minimize sample losses due to the tip retention. Integrated Tecan M200 Infinite multimode detector was used for on-deck dark ambient incubation and for absorbance detection. During all steps of the testing, sample dilution and test plates were moved using the Robotic Manipulator inside the instrument enclosure. To support completely unsupervised testing, photosensitive reagents (TMB) should be kept in a light protective trough or pipet the reagent directly from the container covered by perforated foil. Instead of manual test plate patting after wash steps, maximizing the removal of residual buffer is recommended by adjusting the MCA aspiration high (Z max close to the plate wells bottom) without touching the well surface.
The Dynex Agility was programmed to run the cPass sVNT with scripts produced for both qualitative (delineation between positive and negative) and quantitative (determination of neutralization antibody titers) sample analysis. Since the Dynex systems use a single channel pipette for sample transfer and mixing solutions and an eight-channel wash station, scripting to ensure consistent well-to-well incubation times was the major challenge. Minimizing incubation lag times and accounting for any extended lag times through software programming ensured high quality data.
For both the Tecan and Dynex systems, the cPass sVNT kit instructions for use were modified to a fully room temperature protocol to: a) eliminate pipetting lag times and intraplate data drift and b) produce a more streamlined protocol that is more adaptable to automation platforms that may not be equipped with temperature-controlled incubation. The incubation times were modified as follows:
-
1.
Neutralization Reaction: changed from 30 min at 37°to 45 min at room temperature.
-
2.
Incubation of neutralization reaction mixture in the ACE2-coated assay plate: altered from 15 min at 37° to between 20 and 25 min at room temperature (depends on lab temperature).
-
3.
TMB substrate reaction: altered from 15 min at 25° to between 18 and 25 min at room temperature (depends on lab temperature).
Also, to accommodate the dilution effect on TMB from residual wash buffer giving a reduced OD450 signal, the volume of TMB was adjusted to from 100 µL to 200 µL for the Tecan and 125 µL for the Dynex systems.
2.6. Data and statistical analysis
GraphPad Prism software was used for all graphical representations of the data as well as statistical analysis.
3. Results
3.1. Streamlining automation of the cPass sVNT: Transitioning the assay to ambient temperature incubations to achieve a walk-away testing solution
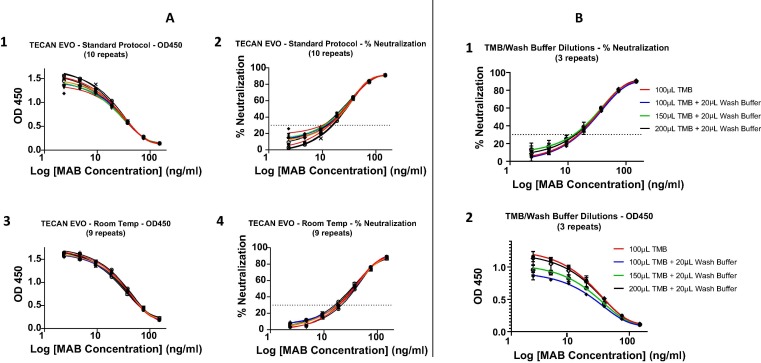
The product insert for the cPass sVNT calls for 37° and ambient temperature incubations at specific steps in the protocol. Using the Tecan automation platform, an optimized ambient temperature procedure was contrasted with the standard protocol giving comparable results (ie: within 20% CV for the second and third dilutions within the linear range) for both OD450 (Fig. 1 A1 and A3) and % neutralization (Fig. 1A2 and A4). However, the ambient, automated protocol requires a manual manipulation of the plate to remove excess wash buffer by patting the inverted plate against paper towel. The residual wash buffer remaining in the wells after washing was about 20 µL which significantly diluted the 100 µL of TMB required for the standard protocol leading to a decreased OD450. The effect of 20 µL residual wash buffer was tested in the presence of 100uL, 150uL and 200uL TMB against a control without residual wash buffer revealing a significant difference on OD450 (Fig. 1B2) but virtually none on % neutralization (Fig. 1B1). Similar data were obtained using the Dynex Agility system which had a smaller volume of residual wash buffer (about 7 µL) thus requiring 125 µL of TMB to offset the dilution effect of the wash buffer.
Fig. 1.
Optimization of cPass sVNT for liquid handler automation. A. Ambient temperature incubations. Ten replicates of a standard curve using the kit insert (KI) “Standard Protocol” were compared to nine replicates from an optimized ambient temperature “Room Temp” procedure (see section 2.5) using a well-characterized neutralizing monoclonal antibody (MAB) (GenScript #A02051) blocking the RBD-ACE2 interaction. The MAB was diluted serially by a factor of 1:2 from 150 ng/mL down each column of the plates to assess and compare intra-plate variability. The data between the two procedures were compared by plotting both the raw (OD450) values (A1 and A3) and the transformed % neutralization results (A2 and A4) vs Log of Ab Concentration. Within the linear range (ie: the second and third MAB dilutions) the difference between the standard (manual) and Tecan (automated) protocols was within 20% CV. B. Effect and compensation of wash buffer dilution of TMB. Mixtures of 20 µL residual wash buffer with the KI recommended 100 µL as well as 150 µL and 200 µL TMB were compared with the KI protocol of no residual wash buffer with 100 µL TMB. The data between the two procedures were compared by plotting both the raw (OD450) values (B2) and the transformed % neutralization results (B1) vs Log of Ab Concentration. Using a 2-way, multiple comparisons ANOVA test, a statistically significant difference was revealed within the linear range between the baseline (100 µL TMB) and the other conditions that included wash buffer with the exception of “20 µL wash buffer + 200 µL TMB” where there was no statistically significant difference.
3.2. Key considerations to achieve high quality data for semi-quantitative analysis of SARS-CoV-2 immune response with automated liquid handling platforms
To avoid system induced data drift, it is of critical importance for cPass sVNT (and most competition-based ELISA tests) that incubation times are strictly adhered and care is taken to minimize well-to-well differences to avoid introducing system induced drift in the data. Thus, especially for single but also for multichannel pipetting robotic manifolds, pre-diluting the samples, standards and controls into a “sample/standards plate” will minimize lag times. Pre-diluted samples/standards can then be mixed with the 1:1000 diluted RBD-HRP substrate (see materials and methods) in a separate “neutralization reaction plate”. Furthermore, when transferring the neutralizing reactions to the ACE2-coated assay plate, the well-to-well transfer lag time should be factored into the plate wash protocol. The following key points should be considered for programming the pipetting and plate washing with an automated liquid handling platform cPass sVNT:
-
1.
Pre-dilute samples, standards and controls into a 96-well “Sample Plate”.
-
2.
Pre-dilute the RBD-HRP into a pipetting trough or 96-well “RBD-HRP Plate”.
-
3.
Mix the diluted RBD-HRP with the samples, standards and controls into a 96-well “Neutralization Reaction Plate” accounting for lag times between wells to ensure all wells are incubating for the same time (45 min at room temperature or 30 min at 37 °C) prior to transfer to the ACE2-coated assay plate.
-
4.
Wash the ACE2-coated assay plate such that the lag time in transferring the neutralization reaction mixtures to the ACE2-coated assay plate is factored to ensure each well has incubated for a specific time in the 20 to 25 min range at room temperature or 15 min at 37 °C before washing.
-
5.
Add TMB to the washed ACE2 coated assay plate such that all wells have been incubating for a specific time in the 18 to 25 min range at room temperature or 15 min at 25 °C in the dark before adding Stop Solution.
-
6.
Add Stop Solution accounting for pipetting lag times between wells to ensure all wells were incubating for the same time with TMB (see step 5 above).
Room temperature scripts were written and successfully tested for the Tecan EVO and Dynex Agility systems enabling both qualitative and semi-quantitative sample analysis using cPass sVNT in a complete hands-off, walkaway, automated solution. The cPass sVNT, MAB dilution series (see Materials and Methods subsection 2.4) can be applied to each column over a full plate to validate intra-plate precision at each dilution which is especially useful for validation of automated protocols.
3.3. Qualitative versus semi-quantitative analysis of samples using cPass sVNT
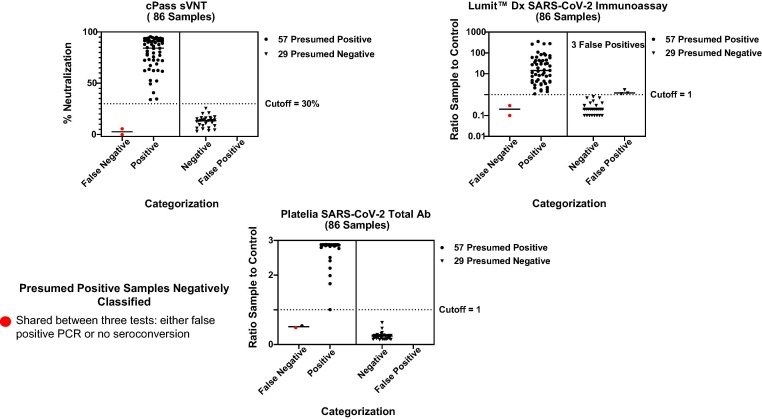
A set of 57 positive and 29 pre-pandemic negative samples were assessed with cPass sVNT, Platelia SARS-CoV-2 Total Ab and Lumit™ Dx SARS-CoV-2 Immunoassay tests (Fig. 2 – red dots were false negative samples shared between the tests). The associated data were tabulated to summarize specificity, sensitivity and accuracy (Table 1 ). In order to migrate the test from the qualitative (delineation between SARS-CoV-2 positive and negative samples using a 30% cutoff) to a semi-quantitative protocol for measurement of neutralization antibody titers, each sample may require dilution that is dependent on the individual immune response to infection or vaccination. This dilution step reveals OD450 values that fall within the linear quantitative dynamic range for interpolation against a standard curve (Fig. 3). The lower limit of quantification (LLOQ) of the standard curve (Fig. 3D) is defined by the OD450 that equates to the 30% neutralization cutoff derived by plotting the OD450 vs % inhibition (Fig. 3B). The upper limit of quantification (ULOQ) (Fig. 3D) for a 4PL fitted standard curve is commonly defined by the 95% confidence interval in the lower plateau [38]. Alternatively, the ULOQ can be set at a signal to noise of three [39]. For this test, the noise is the lowest measured OD from either the most concentrated standard curve dilution or the positive control and the LLOQ would be the OD450 of the noise multiplied by three.
Fig. 2.
Qualitative analysis of 86 SARS-CoV-2 positive and pre-pandemic samples using cPass sVNT, Platelia SARS-CoV-2 Total Ab and Lumit™ Dx SARS-CoV-2 Immunoassay tests. The identical set of 57 positive and 29 pre-pandemic samples were screened qualitatively using cPass sVNT, Platelia SARS-CoV-2 Total Ab and Lumit™ Dx SARS-CoV-2 Immunoassay tests. The same two positive samples were delineated negative (ie: false negative) for the cPass and Lumit assays and one of the false negative samples were in common with all three kits (red dots). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Combined data from the qualitative population surveillance study (Fig. 2) summarizing assay performance characteristics of cPass sVNT, Platelia SARS-CoV-2 Total Ab and Lumit™ Dx SARS-CoV-2 Immunoassay tests. Sensitivity, specificity, accuracy, positive and negative predictive values are shown. A prevalence of 10% was used for the calculations.
 |
A subset of the qualitatively assessed low (Fig. 4 A and C) and high (Fig. 4B and D) positive samples were serially diluted and interrogated for antibody response with cPass sVNT. The summarized results from 25 samples revealed large neutralizing antibody titer differences ranging up to about 600-fold (Table 2 – “Fold Difference to Sample 1 By Titer”). However, the differences between the samples were much smaller or non-existent when using the % neutralization data obtained through qualitative analysis (Table 2 – “Fold Difference to Sample 1 By % Neutralization”).
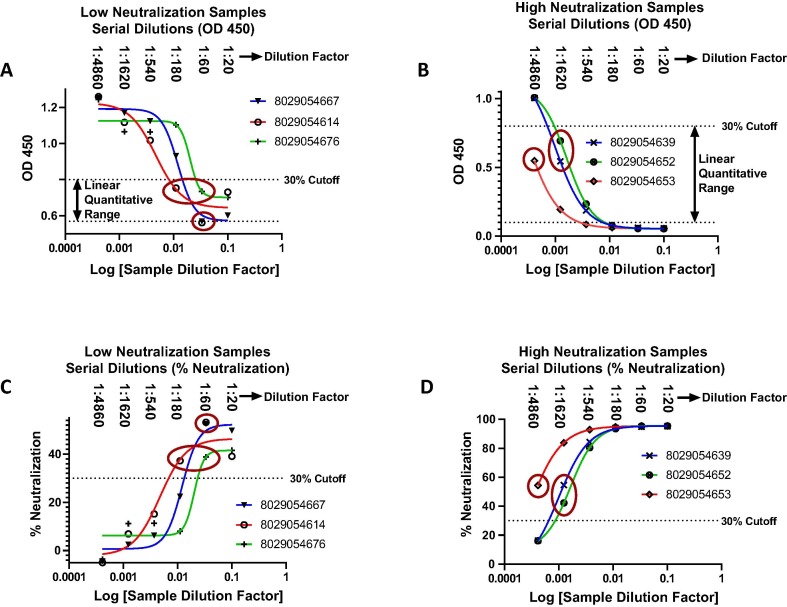
Fig. 4.
Semi-quantitative analysis of serum from SARS-CoV-2 infected and recovered individuals. Graphical representation of serially diluted SARS-CoV-2 positive samples with high and low % neutralization. An optimal dilution series starting from a 1:10 dilution of serum in sample buffer that is mixed equally with RBD-HRP (for a final dilution of 1:20) followed by up to five, serial 1:3 dilutions (ie: 1:20, 1:60, 1:180, 1:540, 1:1620, 1:4860) permits quantification of samples with both high and low neutralizing antibody titers assuring at least one dilution within the linear quantitative range. Graphical representation of OD450 versus log dilution factor for three low (A) and high (B) neutralizing antibody titers. Also represented are the plots of the converted % neutralization versus log dilution factor for the same low (C) and high (D) neutralization samples. Circled points are within the linear range for standard curve interpolation (Fig. 3D). The % recovery between two points within the linear range was examined for samples 8029054639 and 8029054652 (Fig. 4B) and were found to be within 40% of expected as predicted by the inhibitory effect of serum matrix on antibody-antigen interactions [48], [49].
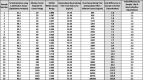
Table 2.
Standard curve interpolated titers of 25 SARS-CoV-2 positive samples with high and low % neutralization. Neutralizing antibody titers from OD450 values taken from the linear range of diluted samples (as exemplified in Fig. 4 – circled points in each panel) interpolated from the monoclonal neutralization antibody (GenScript: #A02051) standard curve (as shown in Fig. 3D). The titers are then multiplied by the associated sample dilution factor (described in Fig. 3E) used for standard curve interpolation to give the “Final Neutralizing Titer” from which the “Fold Difference to Sample 1 By Titer” can be calculated. Also represented are the “% Neutralization Using 1:20 Dilution Factor” values used for qualitative analysis (first column) and the associated “Fold Difference to Sample 1 By % Neutralization” (last column).
 |
3.4. Application of the cPass sVNT qualitative and semi-quantitative protocols for longitudinal assessment of SARS-CoV-2 neutralizing antibodies post-vaccination
Samples from individuals who received either the Pfizer, Moderna or Johnson & Johnson vaccines were acquired both pre and post-vaccination for longitudinal detection and/or quantification of neutralizing antibodies with cPass sVNT. Three separate sets of samples were tested either qualitatively (% neutralization) from 1:20 diluted samples (Fig. 5A and 5B) or semi-quantitatively (ng/ml titers) (Fig. 5C and D). Although the Moderna and Pfizer vaccines gave a high % neutralization after the second dose, several samples were below the 30% cut-off and therefore negative for neutralizing antibodies after receiving the first dose.
Of the three people who received the Johnson & Johnson vaccine, one was previously infected and recovered from COVID-19 who had detectable levels of neutralizing antibodies prior to vaccination and very high levels 14 days post-vaccination (Fig. 5B). Whereas the remaining two people were negative up to 2 weeks after vaccination and exhibited neutralizing antibody levels just above the 30% cut-off 39 days post-vaccination (Fig. 5B).
Four subjects receiving the Pfizer vaccine were tested semi-quantitatively with cPass sVNT giving low or undetectable levels of neutralizing antibodies up until the second dose where levels rose rapidly and peaked within days following the second dose (Fig. 5C). At peak concentration, an approximate 3-fold difference in titer was observed between individuals (compare samples 154 and 156) and all samples exhibited a sharp initial decline in neutralizing antibodies that began to tail off approximately 30 days after the second dose. For sample 152, the % neutralization was qualitatively assessed using the standard 1:20 sample dilution at each time point to reveal a marked difference between the neutralizing antibody levels measured qualitatively versus quantitatively (Fig. 5D).
3.5. Stability of SARS-CoV-2 antibodies at 4 °C and from multiple freeze/thaw cycles
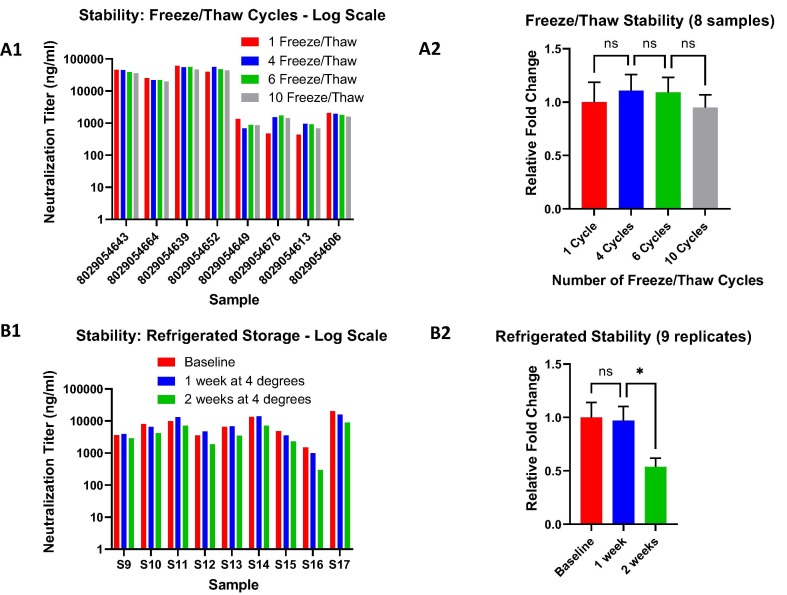
With the combined semi-quantitative format and automated platform for cPass sVNT, stability testing was straightforward and fast. A set of 17 samples were semi-quantitatively tested according to the protocol described in Fig. 3 for various refrigerated (9 samples) and freeze/thaw (8 samples) storage conditions to assess stability of measured antibodies. The data reveal a high degree of stability for circulating neutralizing antibodies with excellent tolerance to up to ten freeze/thaw cycles (Fig. 6 A1 and A2) and refrigerated storage up to one week with a two-fold decrease after two weeks (Fig. 6B1 and B2).
Fig. 6.
Stability testing of immune response antibody activity. A. Freeze/Thaw cycles. Up to 10 freeze/thaw cycles were assessed for samples with high and low neutralization antibody titers (A1) with no statistically significant difference between them as measured with a one way ANOVA (multiple comparisons) test on the combined, normalized sample data (A2). B. Refrigerated storage. Samples stored for one and two weeks at 4°were tested (B1). Although, there was a significant difference between the baseline/1 week and the 2 week refrigerated cycles the decrease in titers were only about two-fold (B2). For both refrigerated and frozen stability, all samples were tested semi-quantitatively as described in Fig. 3. ns: non-significant difference; * statistically significant (p-value = 0.0451).
4. Discussion
4.1. Automation of the cPass sVNT on high throughput liquid handling platforms
An ambient temperature protocol with compensated incubation times was optimized for cPass sVNT and found to be comparable to the standard methodology (Fig. 1A) which is useful for the many single channel liquid pipetting platforms that cannot support the temperature-controlled incubations. Furthermore, since plate inversion and patting to remove residual wash buffer prior to addition of TMB is challenging on robotics liquid handlers, pipetting 100 µL additional TMB for a total of 200 µL was found to rescue the dilution effect on OD450 from residual wash buffer with the Tecan system (Fig. 1B2). For the Dynex Agility, a total volume of 125 µL TMB was required (data not shown). However, the inhibitory effect of wash buffer on the % neutralization was minimal at all antibody dilutions (Fig. 1B1). The scripts for qualitative population surveillance (seroprevalence) and semi-quantitative analysis of immune response neutralizing antibody titers using cPass sVNT were produced for both the Tecan EVO and Dynex Agility.
4.2. Application of cPass sVNT for seroprevalence analysis of SARS-CoV-2 samples
SARS-CoV-2 positive and pre-pandemic negative samples were qualitatively screened to delineate positive samples ranging from low (31%) to high (96%) neutralization in addition to a pre-pandemic negative group (Fig. 2). These data substantiate the high sensitivity, specificity and accuracy of the cPass sVNT test (Table 1) versus other commercial serology assays with previous studies [2], [3], [4], [29], [30], [31], [40].
5. Utility of cPass sVNT for semi-quantitative analysis of neutralization antibodies associated with SARS-CoV-2 infection, vaccination and treatment
In simple terms, vaccination should illicit a robust and general immune response that stimulates T-cell, memory B-cell and frontline neutralization antibodies that together ensure long term protection from the disease for several months or years [18], [19], [41]. However, for SARS-CoV-2, recent studies have shown that the immune response from infected/recovered individuals may be more complicated with declining immune response antibodies and differential T and B-cell levels post-infection particularly in the aging population [21], [22], [23], [24], [25], [26], [42], [43]. Furthermore, a recent study has shown that neutralizing antibodies are highly predictive of immune protection from SARS-CoV-2 infection [44]. Thus, periodic assessment of neutralizing antibody titers post-vaccination may be warranted to determine cutoff titer levels triggering booster vaccinations for ongoing protection.
Since individual neutralizing antibody response to infection and vaccination can vary widely [8], [9], [45], [46], [47], the dilution of samples is required to ensure the data fall within the linear quantitative range of the assay (Fig. 3). However, it is important to note that some variability in the data will be inherent to the effect of matrix dilution on the antibody activity [48], [49]. This was exemplified by examining the neutralizing antibody titers derived from multiple dilutions within the linear range of the same samples revealing up to a 40% difference between the dilution-factor corrected concentrations (samples 8029054639 and 8029054652 in Fig. 4B (data not shown)). However, the larger % differences between the dilution-factor corrected concentrations that exceeded 20% were more associated with comparing sample dilutions that were near the ULOQ and LLOQ where variability increases. This underlines the “semi” quantitative nature of serology assays requiring different sample dilutions to achieve data points within the linear quantitative range of the standard curve whereby variability can arise from both matrix dilution and standard curve interpolation. This underlines the importance of using samples that are adequately diluted (to minimize matrix effect) to give a data point that is well within the linear range of the standard curve for more accurate and precise quantification.
A subset of 25 positive samples qualitatively delineated between 31% and 96% neutralization (Table 2 “% Neutralization using 1:20 Dilution Factor (Qualitative Analysis)”) were serially diluted 1:3 from an initial 1:20 dilution to produce six dilutions per sample (1:20; 1:60; 1:180; 1:540; 1:1620 and 1:4860). For each sample, a dilution within the linear quantitative range was chosen for interpolation against a standard curve according to Fig. 3. The data support a diverse immune response to SARS-CoV-2 infection with a broad range of neutralization titers ranging up to 600-fold (Table 2 – “Fold Difference to Sample 1 By Titer”) that is likely dependent on multiple factors including age, general health and environment [45].
For vaccination, the neutralizing antibody response was generally low to negative after the first dose of both the Pfizer and Moderna vaccines (Fig. 5A) underlining a cautionary message for those who have received only one dose of vaccine. This was also the case for the people receiving the Johnson and Johnson vaccine two-weeks post-vaccination with the exception of subject 1A who was previously infected and exhibited a very high response post-vaccination (Fig. 5B). Those individuals with a negative response should likely remain vigilant with protective measures and social distancing since they may not have front line neutralizing antibodies to block the virus from binding to cells and propagating infection [44], [50]. For all three vaccines, all subjects ultimately gave positive neutralizing antibody levels either after several weeks (Johson & Johnson – Fig. 5B) or within two weeks after the second dose (Pfizer and Moderna - Fig. 5A). However, there remains the question concerning how long immunity persists after vaccination and although the sample set was limited to four subjects, there was a consistent and rapid initial decline in neutralizing antibody titers after the second dose of Pfizer vaccine (Fig. 5C). Furthermore, the qualitative (% Neutralization) data did not reveal the downward quantitative trend in neutralizing antibody titers (ng/ml) as shown with sample 152 (Fig. 5D) which is a direct consequence to the sample dilutions required to acheive accurate, semi-quantitative titer analysis.
Given that cPass sVNT gives highly comparable results to the gold standard live cell neutralization tests [1], [3], [4], [29], these data support the use of the semi-quantitative protocol for cPass sVNT in comparing neutralizing antibody titers between SARS-CoV-2 vaccinated or infected/recovered individuals longitudinally. At the very least, samples can initially be measured two-weeks post-vaccination using the qualitative cPass sVNT protocol at a single 1:20 dilution factor to ensure the presence of neutralizing antibodies. As immunity wanes over time, subjects can potentially submit samples several months post-vaccination for semi-quantitative analysis (Fig. 3) to determine and more closely follow their titer levels.
5.1. Stability of SARS-CoV-2 immune response antibodies in serum samples
The stability of circulating antibodies from viral infections has remained a topical and important subject for sample testing globally [33], [34]. Selected samples quantified using cPass sVNT underwent successive freeze/thaw cycles (Fig. 6A) and refrigerated storage up to two weeks (Fig. 6B). Both refrigerated storage and freeze/thaw cycles showed little difference (well within a half log change) in antibody stability. However, between one and two weeks at 4° did give an approximate two-fold decrease that was statistically significant (Fig. 6B2). Since the collection and storage of samples at various locations does not permit storage under optimal laboratory conditions, these data provide evidence that even suboptimal storage conditions do not greatly affect antibody stability such that the samples may remain useful for downstream analysis.
6. Conclusion
The cPass sVNT provides a highly accurate dual functional test for population surveillance and semi-quantitative analysis of neutralizing antibody titers post-infection and vaccination. The assay protocol was modified to permit ambient temperature incubations (Fig. 1A) with accommodation of residual wash buffer (Fig. 1B) permitting straightforward migration to automated liquid handing systems. Using automation supports accurate population screening for seroprevalence and contact tracing (Fig. 2 and Table 1). The application of cPass sVNT for semi-quantitative analysis of immune response antibodies was achieved by broadly diluting a subset of samples (Fig. 3, Fig. 4 ) to reveal a wide range (600 fold in our sample set) in neutralizing antibody titers between recovered individuals (Table 2 – “Fold Difference to Sample 1 By Titer”). Furthermore, the qualitative protocol permitted the longitudinal delineation of neutralizing antibody positive and negative samples both pre- and post-vaccination and the quantitative protocol revealed a decline in neutralizing antibody titers post-vaccination (Fig. 5). Finally, using assay automation and semi-quantitative analysis permitted stability testing of positive samples to elucidate the effect of various sample storage conditions on neutralizing antibody activity (Fig. 6).
7. Regulatory status
The cPass SARS-CoV-2 Neutralization Antibody Test is CE Marked for diagnostic use in European Union and authorized for emergency use by Health Sciences Authority in Singapore and the US Food and Drug Administration for qualitative delineation between positive and negative patient samples. The semi-quantitative and automation protocols have not yet been authorized by FDA, European Union or Singapore and are Research Use Only.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Lisa Wilson and FourthWall Testing LLC as well as Cure-Hub LLC for their generous contribution in longitudinal data from vaccinated individuals. This work was supported by the Kaiser Permanente Research Bank (KPRB) and the Kaiser Permanente NCAL TPMG Regional Laboratory, Medical Director Dr. Jeffrey Schapiro, Technical Director, Microbiology LaRonda S. Frazier and Lab Clinical R&D Scientist Ivy Yeung. We also wanted to acknowledge contribution of KPRB Lab Support Specialists, Shiyun Liang, Joe Nunoo and Shigeshi Yamamoto. We are grateful to Dynex for conducting the feasibility study of cPass sVNT on their Agility automated liquid handling systems.
References
- 1.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.-I.-C., Tiu C., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat Biotechnol. 2020:1–6. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 2.Tan S.S., Saw S., Chew K.L., Huak C.Y., Khoo C., Pajarillaga A., et al. Head-to-head evaluation on diagnostic accuracies of six SARS-CoV-2 serological assays. Pathology. 2020;52(7):770–777. doi: 10.1016/j.pathol.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perera RAPM, Ko R, Tsang OTY, Hui DSC, Kwan MYM, Brackman CJ, et al. Evaluation of a SARS-CoV-2 surrogate virus neutralization test for detection of antibody in human, canine, cat and hamster sera. Journal of Clinical Microbiology. 2020:JCM.02504-20. [DOI] [PMC free article] [PubMed]
- 4.Taylor SC, Hurst B, Charlton CL, Bailey A, Kanji JN, McCarthy MK, et al. A New SARS CoV-2 Dual Purpose Serology Test: Highly Accurate Infection Tracing and Neutralizing Antibody Response Detection. J Clin Microbiol. 2021. [DOI] [PMC free article] [PubMed]
- 5.Garritsen A., Scholzen A., van den Nieuwenhof D.W., Smits A.P., Datema E.S., van Galen L.S., et al. Two-tiered SARS-CoV-2 seroconversion screening in the Netherlands and stability of nucleocapsid, spike protein domain 1 and neutralizing antibodies. Infect Dis. 2021;1–15 doi: 10.1080/23744235.2021.1893378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olbrich L, Castelletti N, Schälte Y, Garí M, Pütz P, Bakuli A, et al. A Serology Strategy for Epidemiological Studies Based on the Comparison of the Performance of Seven Different Test Systems - The Representative COVID-19 Cohort Munich. medRxiv. 2021:2021.01.13.21249735.
- 7.Woon YL, Lee YL, Chong YM, Ayub NA, Krishnabahawan SL, Lau JFW, et al. Serology surveillance of SARS-CoV-2 antibodies among healthcare workers in COVID-19 designated facilities in Malaysia. The Lancet Regional Health-Western Pacific. 2021;9:100123. [DOI] [PMC free article] [PubMed]
- 8.Kanji JN, Bailey A, Fenton J, Ling SH, Rivera R, Plitt S, et al. Detection of SARS-CoV-2 antibodies formed in response to the BNT162b2 and mRNA-1237 mRNA vaccine by commercial antibody tests. medRxiv. 2021:2021.03.30.21254604. [DOI] [PMC free article] [PubMed]
- 9.Bradley T., Grundberg E., Selvarangan R., LeMaster C., Fraley E., Banerjee D., et al. Antibody Responses after a Single Dose of SARS-CoV-2 mRNA Vaccine. N Engl J Med. 2021;384(20):1959–1961. doi: 10.1056/NEJMc2102051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Rhein C, Scholz T, Henss L, Kronstein-Wiedemann R, Schwarz T, Rodionov RN, et al. Comparison of potency assays to assess SARS-CoV-2 neutralizing antibody capacity in COVID-19 convalescent plasma. Journal of virological methods. 2021;288:114031. [DOI] [PMC free article] [PubMed]
- 11.Bajpai M., Maheshwari A., Chabra K., Kale P., Gupta A., Gupta E., et al. Efficacy of Convalescent Plasma Therapy compared to Fresh Frozen Plasma in Severely ill COVID-19 Patients: A Pilot Randomized Controlled Trial. medRxiv. 2020 [Google Scholar]
- 12.Charlton CL, Kanji JN, Johal K, Bailey A, Plitt SS, MacDonald C, et al. Evaluation of Six Commercial Mid- to High-Volume Antibody and Six Point-of-Care Lateral Flow Assays for Detection of SARS-CoV-2 Antibodies. Journal of Clinical Microbiology. 2020;58:e01361-20. [DOI] [PMC free article] [PubMed]
- 13.Whitman J.D., Hiatt J., Mowery C.T., Shy B.R., Yu R., Yamamoto T.N., et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong F., Yuan L., Zheng Y.F., Chen W. Automatic liquid handling for life science: a critical review of the current state of the art. J Lab Autom. 2012;17(3):169–185. doi: 10.1177/2211068211435302. [DOI] [PubMed] [Google Scholar]
- 15.Control CfD, Prevention. Interim guidelines for COVID-19 antibody testing. Retrieved June. 2020;18:2020.
- 16.Yan Y, Chang L, Wang L. Laboratory testing of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 (2019‐nCoV): Current status, challenges, and countermeasures. Reviews in Medical Virology. 2020;30:e2106. [DOI] [PMC free article] [PubMed]
- 17.D'Cruz RJ, Currier AW, Sampson VB. Laboratory Testing Methods for Novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2). Front Cell Dev Biol. 2020;8:468-. [DOI] [PMC free article] [PubMed]
- 18.Hotez PJ, Nuzhath T, Callaghan T, Colwell B. COVID-19 Vaccine Decisions: Considering the Choices and Opportunities. Microbes and Infection. 2021:104811. [DOI] [PMC free article] [PubMed]
- 19.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 vaccines. Jama. 2021;325:1318-20. [DOI] [PubMed]
- 20.Mahase E. Covid-19: Where are we on vaccines and variants? : British Medical Journal Publishing Group; 2021.
- 21.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. [DOI] [PMC free article] [PubMed]
- 22.Ward H, Cooke G, Atchison C, Whitaker M, Elliott J, Moshe M, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020:2020.10.26.20219725.
- 23.Nikolich-Zugich J., Knox K.S., Rios C.T., Natt B., Bhattacharya D., Fain M.J. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. Geroscience. 2020;42(2):505–514. doi: 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marot S., Malet I., Leducq V., Zafilaza K., Sterlin D., Planas D., et al. Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat Commun. 2021;12:1–7. doi: 10.1038/s41467-021-21111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., et al. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Clinical performance of different SARS-CoV-2 IgG antibody tests. J Med Virol. 2020;92(10):2243–2247. doi: 10.1002/jmv.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timiryasova T.M., Bonaparte M.I., Luo P., Zedar R., Hu B.T., Hildreth S.W. Optimization and validation of a plaque reduction neutralization test for the detection of neutralizing antibodies to four serotypes of dengue virus used in support of dengue vaccine development. American J Tropical Med Hygiene. 2013;88:962–970. doi: 10.4269/ajtmh.12-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray M.J., McIntosh M., Atkinson C., Mahungu T., Wright E., Chatterton W., et al. Validation of a commercially available indirect assay for SARS-CoV-2 neutralising antibodies using a pseudotyped virus assay. J Infect. 2021 doi: 10.1016/j.jinf.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putcharoen O, Wacharapluesadee S, Chia WN, Paitoonpong L, Tan CW, Suwanpimolkul G, et al. Early detection of neutralizing antibodies against SARS-CoV-2 in COVID-19 patients in Thailand. PloS one. 2021;16:e0246864 [DOI] [PMC free article] [PubMed]
- 31.Fox-Lewis S., Whitcombe A., McGregor R., Carlton L., Hwang Y., Austin P., et al. A comparison of SARS-CoV-2 antibody assays evaluated in Auckland, New Zealand. New Zealand Med J (Online) 2020;133:127–131. [PubMed] [Google Scholar]
- 32.Ruangturakit S, Rojanasuphot S, Srijunggravanvong A, Duangchanda S. Storage stability of dengue IgM and IgG antibodies in whole blood and serum dried on filter paper strips detected by ELISA. Southeast Asian journal of tropical medicine and public health. 1994;25:560-. [PubMed]
- 33.Brey R.L., Cote S.A., McGlasson D.L., Triplett D.A., Barna L.K. Effects of Repeated Freeze-Thaw Cycles on Anticardiolipin Antibody Immunoreactivity. Am J Clin Pathol. 1994;102(5):586–588. doi: 10.1093/ajcp/102.5.586. [DOI] [PubMed] [Google Scholar]
- 34.Pinsky N.A., Huddleston J.M., Jacobson R.M., Wollan P.C., Poland G.A. Effect of multiple freeze-thaw cycles on detection of measles, mumps, and rubella virus antibodies. Clin Diagn Lab Immunol. 2003;10(1):19–21. doi: 10.1128/CDLI.10.1.19-21.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentine-Graves M, Hall E, Guest JL, Adam E, Valencia R, Shinn K, et al. At-home self-collection of saliva, oropharyngeal swabs and dried blood spots for SARS-CoV-2 diagnosis and serology: Post-collection acceptability of specimen collection process and patient confidence in specimens. PloS one. 2020;15:e0236775. [DOI] [PMC free article] [PubMed]
- 36.Gaugler S., Sottas P.-E., Blum K., Luginbühl M. Fully automated dried blood spot sample handling and extraction for serological testing of SARS-CoV-2 antibodies. Drug Test Anal. 2021;13(1):223–226. doi: 10.1002/dta.2946. [DOI] [PubMed] [Google Scholar]
- 37.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinn C.P., Semenova V.A., Elie C.M., Romero-Steiner S., Greene C., Li H., et al. Specific, sensitive, and quantitative enzyme-linked immunosorbent assay for human immunoglobulin G antibodies to anthrax toxin protective antigen. Emerg Infect Dis. 2002;8(10):1103–1110. doi: 10.3201/eid0810.020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little TA. Method validation essentials, limit of blank, limit of detection, and limit of quantitation. ADVANSTAR COMMUNICATIONS INC 131 W 1ST STREET, DULUTH, MN 55802 USA; 2015.
- 40.Bal A, Pozzetto B, Trabaud M-A, Escuret V, Rabilloud M, Langlois-Jacques C, et al. Evaluation of high-throughput SARS-CoV-2 serological assays in a longitudinal cohort of mild COVID-19 patients: sensitivity, specificity and association with virus neutralization test. medRxiv. 2020:2020.09.30.20194290. [DOI] [PMC free article] [PubMed]
- 41.O’Callaghan KP, Blatz AM, Offit PA. Developing a SARS-CoV-2 Vaccine at Warp Speed. JAMA. 2020;324:437-8. [DOI] [PubMed]
- 42.Saletti G, Gerlach T, Jansen JM, Molle A, Elbahesh H, Ludlow M, et al. Older adults lack SARS CoV-2 cross-reactive T lymphocytes directed to human coronaviruses OC43 and NL63. Scientific reports. 2020;10:1-10. [DOI] [PMC free article] [PubMed]
- 43.Bajaj V., Gadi N., Spihlman A.P., Wu S.C., Choi C.H., Moulton V.R. Aging, immunity, and COVID-19: how age influences the host immune response to coronavirus infections? Front Physiol. 2021;11:1793. doi: 10.3389/fphys.2020.571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 45.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182(3) doi: 10.1016/j.cell.2020.06.011. 744-53. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu J., Bennett P. Parallelism experiments to evaluate matrix effects, selectivity and sensitivity in ligand-binding assay method development: pros and cons. Bioanalysis. 2017;9(14):1107–1122. doi: 10.4155/bio-2017-0084. [DOI] [PubMed] [Google Scholar]
- 49.Thakur K, Sharma S, Prabhakar S, Gupta P, Anand A. Revisiting the dilution factor as vital parameter for sensitivity of ELISA assay in CSF and Plasma. Ann Neurosci. 2015;22:37-42. [DOI] [PMC free article] [PubMed]
- 50.Lucas C., Klein J., Sundaram M.E., Liu F., Wong P., Silva J., et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat Med. 2021 doi: 10.1038/s41591-021-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]