Dear Sir,
In this Journal, Sansone and colleagues measured effectiveness of BNT162b2 vaccine against the B.1.1.7 variant of SARS-CoV-2 among healthcare workers.1 India experienced a severe secod wave of SARS-CoV-2 infections during the months of April and May 2021. COVID-19 vaccination with BBV152 vaccine (Covaxin; Bharat Biotech) and ChAdOx1 nCoV-19 (Covishield, Serum Institute of India) was started in the country in January 2021, targeting healthcare workers in the first phase and later expanded to include adult population groups.2 Breakthrough infections following vaccination have been reported in India.3 , 4 Breakthrough infections could be due to emergence of newer mutant strains capable of escaping the host immune response.5 During March 2021, sequencing of more than 10,000 RT-PCR positive samples indicated circulation of viruses of B.1.1.7 (Alpha/UK variant), B.1.351 (beta/South African), P.1 (Gamma/Brazilian) lineage and Kappa/Delta Indian variants (B.1.617).6 During the course of second wave in India, Delta B.1.617.2 variant emerged as the major sub-lineage among variants that also included B1617.1, B.617.3 and B.1.1.7.3 Chennai was one of the worst affected cities in the second wave of COVID-19 in India, with nearly 6000 cases reported daily during the first three weeks of May 2021, despite a high seroprevalence of around 45% estimated during October -November 2020.7 Chennai has reported more than 520,000 COVID-19 cumulative cases and 7793 deaths since the beginning of the pandemic and vaccinated around 2 million people with at least one dose of COVID-19 vaccine.8 In this context, we described the prevalence of variants of concern (VOCs) among vaccinated and unvaccinated COVID-19 positive individuals in Chennai.
Newly diagnosed COVID-19 patients are triaged in screening centers established by the Greater Chennai Corporation. We selected three of the ten such triaging centers for the study with one center each from the northern, central and southern parts of Chennai to ensure representativeness. We consecutively enrolled consenting COVID-19 positive individuals visiting these centers who had taken at least one dose of COVID-19 vaccine 14 days prior to confirmation of the diagnosis. We also recruited unvaccinated COVID-19 cases attending the triage centers. We collected demographic details, clinical history, comorbidities, previous COVID-19 history and date of vaccination. Nasal and oro-pharyngeal (N/OP) swabs and blood samples were collected from the study participants. We tested the N/OP swab samples for the detection of E and RdRP gene using Real-time RT-PCR and only those with Ct<30 were included for preparation of RNA libraries Illumina Covidseq protocol (Illumina Inc, USA).9 Amplified and purified libraries were quantified using KAPA Library Quantification Kit (Kapa Biosystems, Roche Diagnostics Corporation, USA) and loaded on NextSeq 500/550 system after normalization. Bcl files generated were analysed after conversion to fastq using CLC genomics workbench version11.0 (CLC, QIAGEN, Germany). Reference-based mapping was performed to retrieve the sequence of the SARS-CoV-2 and a phylogenetic tree was generated using MEGA software version 7. Blood samples were tested for SARS-CoV-2 IgG antibodies against S1-RBD (Siemens, Munich, Germany).
The participants were followed up telephonically after four weeks to collect information about their symptoms, hospitalisation and treatment details, and clinical outcome. Patients with SpO2 < 94%, dyspnoea and requiring supplemental oxygen during hospitalization were considered as having moderate/severe illness and remaining as mild illness. Categorical variables were expressed as proportions and continuous variables as median and inter-quartile range (IQR). The study was approved by the Institutional Ethics Committee of ICMR-National Institute of Epidemiology, Chennai.
Of the 3790 COVID-19 cases who visited the triage centers between May 3 and May 7, 2021, 373 reported receiving at least one dose of vaccine 14 days prior to their COVID-19 diagnosis and the remaining 3417 were unvaccinated. We enrolled 354 (94.9%) of the 373 vaccinated (241 had taken one dose, (partially vaccinated) and 113 had taken two doses of COVID-19 vaccine (fully vaccinated)) and 185 (5.4%) of the 3417 unvaccinated individuals in the study. The median age of the individuals who were unvaccinated, received 1 and 2 doses were 47 years (IQR; 33–57), 53 years (IQR; 46–60) and 54 years (IQR: 42–64), respectively (Table 1 ). Most study participants were male and the proportion having comorbidities was not different in the three groups.
Table 1.
Demographic characteristics, VOCs prevalence and clinical outcome in the COVID-19 vaccinated and Unvaccinated.
| Characteristics | Vaccinated for both doses(N = 113)n (% of total) | Vaccinated for one dose(N = 241)n (% of total) | Unvaccinated(N = 185)n (% of total) |
|---|---|---|---|
| Age (Years) | |||
| Median (Interquartile range) | 54 (42–64) | 53 (46–60) | 47 (33–57) |
| Gender | |||
| Male | 66 (58.4) | 149 (61.8) | 109 (58.9) |
| Female | 44 (38.9) | 87 (36.1) | 74 (40.0) |
| Other | 3 (2.7) | 5 (2.1) | 2 (1.1) |
| Comorbidities | |||
| Yes | 50 (44.6) | 110 (46.0) | 71 (39.0) |
| No | 62 (55.4) | 129 (54.0) | 111 (61.0) |
| missing | 1 | 2 | 3 |
| Type of Vaccine | |||
| Covaxin | 31 (27.4) | 80 (33.2) | - |
| Covishield | 80 (70.8) | 160 (66.4) | - |
| Do not Know | 2 (1.8) | 1 (0.4) | - |
| Variants of concern | |||
| B.1.617.2 | 84 (74.3) | 164 (68.1) | 134 (72.4) |
| B.1.617.1 | 1 (0.9) | 6 (2.5) | 4 (2.2) |
| AY.1 | 1 (0.9) | 2 (0.8) | 2 (1.1) |
| B.1.1.7 | 1 (0.9) | 2 (0.8) | 5 (2.7) |
| B.1.351 | 0 (0.0) | 2 (0.8) | 3 (1.6) |
| B.1.351.3 | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| B.1.1 | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| B.1.530 | 0 (0.0) | 0 (0.0) | 1 (0.5) |
| Could not be retrieved | 26 (23.0) | 65 (27.0) | 34 (18.4) |
| Presence of IgG at the time of sample collection | |||
| Yes | 96 (85.0) | 154 (63.9) | 27 (14.6) |
| No | 9 (8.0) | 64 (26.6) | 118 (63.8) |
| Not done | 8 (7.0) | 23 (9.5) | 40 (21.6) |
| Symptoms during the course of illness | N = 104 | N = 224 | N = 176 |
| Yes | 92 (88.5) | 212 (94.6) | 166 (94.3) |
| No | 12 (11.5) | 12 (5.4) | 10 (5.7) |
| Severity of illness | N = 104 | N = 224 | N = 176 |
| Moderate/Severe illness | 7 (6.7)* | 46 (20.5) | 34 (19.3) |
| Mild illness | 97 (93.3) | 178 (79.5) | 142 (80.7) |
| Clinical outcome | N = 104 | N = 224 | N = 176 |
| Alive | 104 (100.0) | 221 (98.7) | 169 (96.0) |
| Died | 0 (0.0)⁎⁎ | 3 (1.3)⁎⁎⁎ | 7 (4.0) |
p = 0.003 for the proportions with severe disease among fully vaccinated and unvaccinated individuals.
p value (1-tail) = 0.018 for the proportions of deaths among fully vaccinated and unvaccinated individuals.
p value (1-tail) = 0.046 for the proportions of deaths among partially vaccinated and unvaccinated individuals.
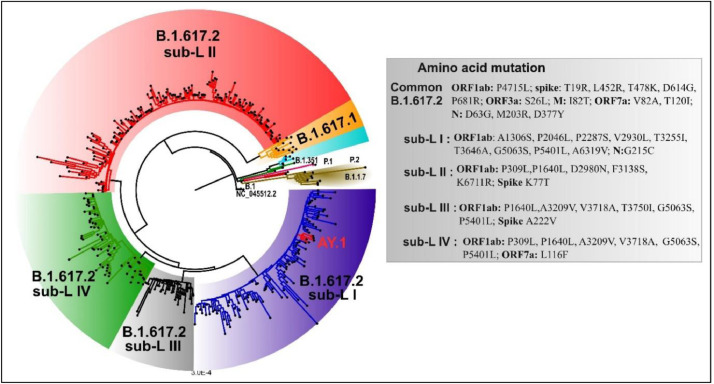
We could retrieve genomic sequences from 414 of the 539 samples. Median RT-PCR cycle threshold values were similar in the unvaccinated (22.4, IQR; 11.9–26.3) partially (22.5, IQR; 19.4–26.9) and fully (23.1, IQR; 18.3–26.4) vaccinated groups. B.1.617.2 (Delta variant) was the predominant VOC: 72.4% (134/185) in unvaccinated, 68.1% (164/241) in partially and 74.3% (84/113) in fully vaccinated groups (Table 1). AY.1 (Delta plus variant) was isolated in five study participants. Of the five patients with AY.1 infection, one required hospitalization for oxygen support and rest had mild disease. The proportion of other VOCs was low (Table 1). A neighbor joining tree was generated using Tamura-3-parameter model and a boot strap of 1000 replication cycle ( Fig. 1). Phylogenetic tree revealed the presence four distinct sub-clusters in the delta variant, similar to observed seen in the larger dataset (data unpublished).
Fig. 1.
A neighbor-joining tree was generated using a Tamura 3-parameter model with gamma distribution and a bootstrap replication of 1000 cycles. Common mutations observed in the B.1.617.2 lineage are mentioned in the figure. Further additional mutations were observed in B.1.617.2 cluster based on which four sub clusters were designated. The sub-clusters are highlighted in different colors: sub-cluster I: blue color; sub-cluster II: red; sub-cluster III: grey; sub-cluster IV: green color. The additional lineages that were found are marked on the nodes: B.1.351: green; red: P.1 and brown: P.2 and B.1.617.1 is highlighted in orange color. NC_0.45512.2 (Wuhan Hu-1) is the start of the root. The figure is edited in Figtree v1.4.4 and Inkscape (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Among the fully vaccinated, majority 85% (n = 96) had IgG antibody against SARS-CoV-2 S1-RBD whereas 63.9% (n = 154) partially vaccinated and 14.6% (n = 27) in the unvaccinated group were seropositive. We followed up 504 (93.5%) of the 539 participants and found that majority were symptomatic (88.5% in the fully vaccinated, 94.6% in the partially vaccinated and 94.3% in the unvaccinated). The proportion of patients with moderate/severe illness was significantly lower in the fully vaccinated group (7/104. 6.7%) than in the unvaccinated (34/176, 19.3%) group (p = 0.003). No deaths were reported in the fully vaccinated group, whereas 3 partially vaccinated group and seven unvaccinated COVID-19 patients died. The proportion of COVID-19 deaths was significantly lower in the partially vaccinated (1.3%, p value (1-tail) = 0.046) and fully vaccinated (0%, p value (1-tail) = 0.018) than the unvaccinated (4.0%).
The study findings indicate that the prevalence of B.1.617.2 was not different between the vaccinated and unvaccinated groups. Delta variant was the dominant circulating strain and one of the primary drivers for the second wave of SARS-CoV-2 in India.10 Studies have documented reduction in neutralization titres among Covishield and Covaxin recipients after infection with delta variant.11 , 12 This might be the reason for the breakthrough infections observed in the fully vaccinated individuals. However, the proportion of patients progressing to severe illness and mortality was lower in the vaccinated group.
Our study has certain limitations. We recruited majority of the vaccinated individuals visiting the triaging center but only 5% of the unvaccinated individuals could be recruited due to logistics challenges. We could not follow up around 5% of the study participants.
B.1.617.2 has the potential to infect both the vaccinated and unvaccinated individuals. However, the progression of illness seems to be prevented by vaccination. Therefore, non-pharmaceutical interventions must continue to slow down the transmission. Additionally, the pace and scale of vaccination has to be increased to mitigate the further waves of the pandemic. Systematic genomic surveillance must be carried out to monitor the emergence of newer variants and assess their capacity to evade infection/vaccine induced immunity.
Declaration of Competing Interest
None
Acknowledgements
We sincerely thank Dr Priya Abraham, Director, ICMR-NIV, Pune for encouragement and Ms Manisha Dudhmal, Mr. Yash Joshi, for their support in Genomic sequencing and analysis of sequences. We also thank Augustine D, Punitha, Karunakaran, C.Kanagasivam, P.Tamilselvi, R.Sivakumar, Namratha S Prabhu, MP Sarath Kumar, Arya Vinod, R.Sivakumar, Arun Prasath EB, S Sarath Kumar for data collection and laboratory processing of samples. We acknowledge the support from Greater Chennai Corporation health officials in field operations.
References
- 1.Sansone E., Tiraboschi M., Sala E., Albini E., Lombardo M., Castelli F., et al. Effectiveness of BNT162b2 vaccine against the B.1.1.7 variant of SARS-CoV-2 among healthcare workers in Brescia, Italy. J Infect. 2021;83(1):e17–e18. doi: 10.1016/j.jinf.2021.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ministry of Health and Family Welfare, Govt of India. Covid-19 vaccination. Frequently asked questions 2021. Available at https://www.mohfw.gov.in/covid_vaccination/vaccination/faqs.html#who-will-get-the-vaccine. Accessed on 11 August 2021
- 3.Singh U.B., Rophina M., Chaudhry R., Vigneshwar S., Bala K., Bhoyar R.C., et al. Variants of concern responsible for SARS-CoV-2 vaccine breakthrough infections from India 2021. OSF Pre-print Available at 10.31219/osf.io/fgd4x [DOI] [PMC free article] [PubMed]
- 4.Rana K., Mohindra R., Pinnaka L. Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med. 2021;385(2):e7. doi: 10.1056/NEJMc2107808. [DOI] [PubMed] [Google Scholar]
- 5.Prévost J., Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29(3):322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ministry of Health and Family Welfare, Govt of India. Genome sequencing by INSACOG shows variants of concern and a Novel variant in India_24 MAR 2021 2021. Available at https://pib.gov.in/PressReleaseIframePage.aspx?PRID=1707177. Accessed on 11 August 2021
- 7.Malani A., Ramachandran S., Tandel V., Parasa R., Sudharshini S., Prakash V. et al., SARS-CoV-2 seroprevalence in Tamil Nadu in October-November 2020. Available at MedRxiv2021:2021.02.03.21250949. 10.1101/2021.02.03.21250949, accessed on 11 August 2021
- 8.State Control Room. Directorate of public health and preventive medicine health and family welfare department, government of Tamil Nadu, Media Bulletin 2021. Available at: https://stopcorona.tn.gov.in/wp-content/uploads/2020/03/Media-Bulletin-15-06-21-COVID-19.pdf, accessed on 11 August 2021
- 9.Bhoyar R.C., Jain A., Sehgal P., Divakar M.K., Sharma D., Imran M., et al. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhar M.S., Marwal R., Radhakrishnan V.S., Ponnusamy K., Jolly B., Bhoyar R.C. et al., Genomic characterization and Epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Available at: MedRxiv2021:2021.06.02.21258076.,10.1101/2021.06.02.21258076, accessed on 11 August 2021. [DOI] [PMC free article] [PubMed]
- 11.Yadav P.D., Sapkal G.N., Ella R., Sahay R.R., Nyayanit D.A., Patil D.Y. et al., Neutralization against B.1.351 and B.1.617.2 with sera of COVID-19 recovered cases and vaccinees of BBV152. Available at: BioRxiv2021 :2021.06.05.447177. 10.1101/2021.06.05.447177, accessed on 11 August 2021.
- 12.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah Maaran M. et al., Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. Available at: BioRxiv2021 :2021.05.26.445838. 10.1101/2021.05.26.445838, accessed on 11 August 2021.