Abstract
Glycosylation represents a critical quality attribute modulating a myriad of physiochemical properties and effector functions of biotherapeutics. Furthermore, a rising landscape of glycosylated biotherapeutics including biosimilars, biobetters, and fusion proteins harboring complicated and dynamic glycosylation profiles requires tailored analytical approaches capable of characterizing their heterogeneous nature. In this work, we perform in-depth evaluation of the glycosylation profiles of three glycoengineered variants of the widely used biotherapeutic erythropoietin. We analyzed these samples in parallel using a glycopeptide-centric liquid chromatography/mass spectrometry approach and high-resolution native mass spectrometry. Although for all of the studied variants the glycopeptide and native mass spectrometry data were in good qualitative agreement, we observed substantial quantitative differences arising from ionization deficiencies and unwanted neutral losses, in particular, for sialylated glycopeptides in the glycoproteomics approach. However, the latter provides direct information about glycosite localization. We conclude that the combined parallel use of native mass spectrometry and bottom-up glycoproteomics offers superior characterization of glycosylated biotherapeutics and thus provides a valuable attribute in the characterization of glycoengineered proteins and other complex biotherapeutics.
Introduction
Glycosylation of biologics is a critical quality attribute with over 80% of currently approved biologics being glycoproteins.1 Far from being a simple decoration, glycosylation influences the activity and pharmacokinetics of biotherapeutics.2,3 For instance, IgG core fucosylation is known to inhibit ADCC. For erythropoietin (EPO), which we study here, sialylation and branching of N-glycans strongly influence molecule half-life and hematopoietic activity.3−5 The advent of products with additional glycosylation sites and heterogeneous glycosylation profiles as well as rapidly emerging biosimilars and biobetters requires tailored analytical approaches for the characterization of biotherapeutic glycosylation at every stage of their lifecycle.3,6,7 Currently, the most common approaches for glycosylation analysis are based on released glycan analysis,8,9 where care has to be taken to avoid contaminations from copurified glycoproteins.10 However, with recent development in fragmentation11−14 and digestion techniques, the approaches based on intact glycopeptide analysis are gaining ground in the quality control of biotherapeutics as part of a multiattribute method.15 The multiattribute method replaces common conventional methods such as released N-glycan profiling, charge variants, and presence of clipping variants with a single mass spectrometry (MS)-based method.16
It is widely known that the glycosylation can negatively influence the ionization efficiency of modified peptides, complicating quantitative glycan analysis.17 Measuring intact glycoproteins under native conditions is considered to largely alleviate this ionization bias as the ionization efficiency is mainly driven by the protein backbone.18,19 For instance, in our previous work, direct comparison of IgG glycoprofiles obtained with native MS and those measured with standard glycomics approaches were in excellent quantitative agreement.18 In the present study, we pursue a face-to-face comparison of two analytical approaches: analysis of intact glycopeptides and native MS analysis in the characterization of EPO glycosylation.
EPO is used as treatment of anemia and is modified by three N-glycans and a single O-glycan. In contrast to antibodies which exhibit a limited repertoire of N-glycans, EPO exhibits highly heterogeneous N-glycans ranging from biantennary to poly-LacNAc-elongated and core-fucosylated tetra-antennary N-glycans.20,21 To probe the influence of various glycan structures on analytical method outcomes, we compare three EPO variants. The first is close to the clinically used variant with a well-characterized glycosylation profile. The second EPO sample exhibits partial occupancy (macro-heterogeneity) at the first N-glycosite. Third, we study an EPO variant lacking sialylation. Our results demonstrate a satisfying overall agreement between the methods in terms of identified glycan composition and also a divergence in quantitation of, in particular, sialylated glycoforms and site occupancy estimations.
Results and Discussion
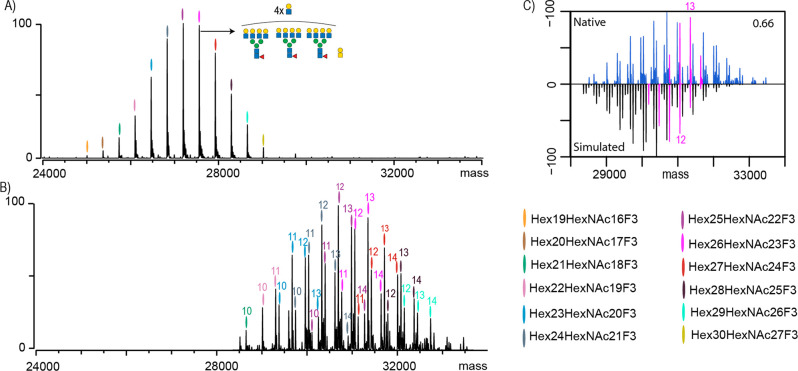
We first pursued characterization of variant 1, the EPO with the “standard” glycosylation profile (denoted as EPO-1). Specifically, all N-glycosylation sites are fully occupied and decorated with complex type of N-glycans capped with sialic acids. Based on our extensive experience with EPO analysis demonstrating incomplete sialylation as the main source of heterogeneity, we first analyzed sialidase-treated EPO22,23 (Figure 1A). In the native MS spectrum of desialylated EPO-1, we observed 12 glycan compositions starting from Hex19HexNAc16F3. From this composition, sequential additions of HexHexNAc can be observed culminating toward Hex30HexNAc27F3. The most abundant composition, as depicted in Figure 1A, corresponds most likely to an EPO-1 carrying three tetra-antennary N-glycans further decorated with four LacNAc extensions and one core O-glycan. We used these annotations to guide our annotations of the sialylated EPO-1 (Figure 1B). It became apparent that each glycan composition observed in the sialidase-treated EPO-1 appears as four distinct peaks in the non-sialidase-treated native mass spectra carrying anywhere between 10 and 14 sialic acid residues. This differential sialylation lowered our sensitivity as each peak observed in the spectra of sialidase-treated material was split into at least four separate peaks, leading to a diminished detection of just nine unique HexHexNAcF compositions as opposed to the 12 we could map for the sialidase-treated EPO-1. Although it has been reported by us and others that EPO may also carry sulfated, acetylated, and/or bisected glycans, we saw no evidence for any of those on the EPOs from the batches measured in this work.22,24 Next, EPO-1 glycopeptides were generated by GluC digestion, which enables coverage of all glycosylation sites, followed by liquid chromatography/mass spectrometry (LC-MS) analysis.25 Using our previously described algorithm,22,26 we used GluC bottom-up data to simulate a native MS spectrum. When we then compared the measured and simulated native MS spectra, we obtained a low similarity score of 0.66 (Figure 1C). Spectra simulated using the data from the GluC digest were found to be shifted toward lower masses when compared with the native mass spectra. For instance, we highlighted in pink the Hex26HexNAc23F3 (Figure 1C) composition which in the native spectra has the highest abundance as a species carrying 13 Sia residues. In contrast, species bearing 11 Sia residues were calculated to be the most abundant based on the GluC-digest glycoproteomics data. This is in line with the known observations that in bottom-up analysis sialylation can negatively affect the ionization efficiency of glycopeptides.17 In addition, the sialic acid residues can also be lost during the ionization process.17 Moreover, in glycopeptide analysis, the attached glycan can be significantly bigger than the peptide backbone to which it is attached. In the case of EPO, which has three N-glycosylation sites (N24, N38, and N83), there is a significant difference in the masses of the detected GluC glycopeptides. For instance, a fucosylated triantennary N-glycan carrying three sialic acids has a mass of 3007.06 Da (74% of the total intact glycopeptide mass), whereas the mass of the N24-bearing peptide (AENITTGCAE) corresponds to only 1065.43 Da. Thus, it is not surprising that the ionization efficiency for this glycopeptide would mainly be driven by the glycan moiety. An increase in sialic acid content would likely have an even more detrimental effect on ionization efficiency. Additionally, different glycosites can have a drastic difference in the peptide backbone size. For instance, for EPO, the peptide-bearing site N83 (VLRGQALLVNSSQPWEPLQLHVDKAVSGLRSLTTLLRALGAQKE) is approximately 4 times the size of the previously mentioned N24. If we now contrast N24 carrying one of the smaller glycan forms detected in our study, with N83 carrying a slightly bigger glycan, fucosylated tetra-antennary N-glycan with extra LacNAc repeat and four sialic acid residues, we obtain a total Mw of 8891.16 Da, which is more than double the size of previously mentioned N24 glycopeptide. If we were to perform the same exercise at the intact protein level (EPO carrying three N-glycans described for N24 and an O-glycan versus EPO carrying three N-glycans described for N83 and an O-glycan), the difference in mass between these two glycoproteoforms is only 8% of the total mass. The insights described above also hold true for O-glycopeptides. For instance, in our previous studies, we have compared O-glycosite occupancy in N-deglycosylated EPO, as determined by native MS and glycopeptide analysis and found values of 90 and 55%, respectively.23,27 Thus, glycan heterogeneity is less likely to exhibit a significant influence on the ionization efficiency of intact glycoproteins measured under nondenaturing conditions. On the downside of native MS, intact glycoprotein measurement only provides insight into the totality of glycans attached to the protein and lacks site specific information, whereas the glycoproteomics approach provides detailed characterization of modified sites as well as their glycan compositions. Hence, the ideal approach is a combination of both approaches, whereby the glycoproteomics provides detailed characterization of site heterogeneity and native MS serves to validate the quantitative results.
Figure 1.
Analysis of EPO-1 by native MS and bottom-up glycoproteomics. (A) Native MS spectrum of desialylated EPO-1, where each peak is color-coded and represents a unique Hexx+3HexNAcxF3 composition. One of the most abundant peaks is annotated with its most likely glycan composition. (B) Native MS spectrum of non-sialidase-treated EPO-1 where the numbers above the color codes indicate the cumulative number of sialic acid residues attached to the EPO glycans. Upsized spectrum is available as Figure S1. (C) Comparison and cross-correlation of the native MS data (blue) of non-sialidase-treated EPO-1 with a simulated intact mass spectrum based on the GluC-digest glycoproteomics data. Highlighted in pink are peaks belonging to a Hex26HexNAc23F3 composition carrying between 9 and 14 sialic acids.
Characterization of Partially Glycosylated Erythropoietin
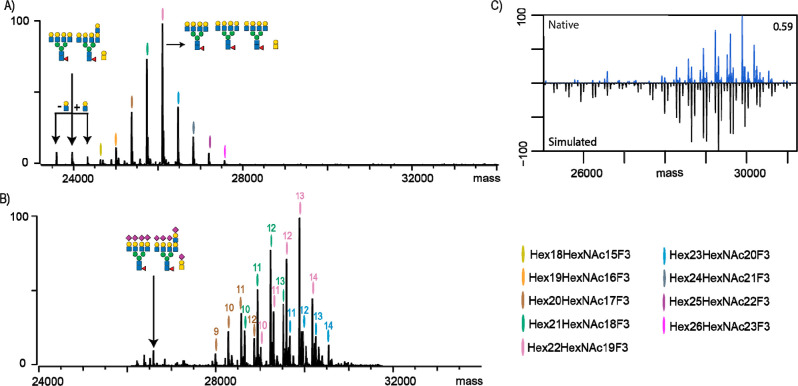
A major challenge in the analysis of glycoproteins is partial occupancy of N-glycosylation sites (macro-heterogeneity). It is crucial to determine the extent of site occupancy due to potential impacts on protein function. In glycoproteomics approaches, which often employ enrichment of glycopeptides prior to MS analysis, this information is typically lost. When the sample of interest is a purified glycoprotein, such an enrichment step can be omitted, but data obtained can still be biased due to the even more substantial differences in ionization efficiency of peptides versus glycopeptides.17 A more common approach is to use deglycosylation in heavy water, which leaves a 2.988 Da signature on the deamidated asparagine, enabling an accurate readout of the N-glycosylation stoichiometry.28 For these reasons, we sought to next characterize an EPO variant exhibiting partial N-glycan occupancy (EPO-2) at the first (N24) glycosylation site as measured by our multiattribute method.25 We, again, performed native MS analysis of desialylated EPO-2 (Figure 2A). We identified 13 glycan compositions where the most abundant one corresponded to EPO-2 carrying four tetra-antennary N-glycans and an O-glycan. Notably, the most abundant species is smaller than the most abundant species observed in the native MS analysis of EPO-1, which carried four extra LacNAc repeats. Additionally, we observed peaks corresponding to EPO-2 lacking one of the N-glycans. Presence of partially glycosylated EPO-2 was confirmed by the native MS examination of sialylated EPO-2 (Figure 2B). We compared the results of the GluC-digested EPO-2 glycopeptides with our native MS approach and obtained a similarity score of 0.59 (Figure 2C). This is slightly lower than the fully glycosylated EPO-1 characterized in Figure 1. However, the cause of lower similarity remains the same (i.e., a shift to lower sialylation states in the simulated spectra). Additionally, bottom-up analysis also revealed that a significant proportion of EPO-2 lacks one N-glycan, which was in contrast to the native MS data that revealed only trace amounts of partially glycosylated EPO. A closer examination of the glycopeptide data revealed that the most abundant signal at N24 stems from the unoccupied peptide (∼15% unoccupied vs ∼3% as measured by native MS). The difference between the bottom-up and native MS can again be explained by the striking differences in the size of measured peptide/glycopeptide (1065.43 vs 4072.49 Da, respectively). Additionally, considering the presence of negatively charged sialic acids, it is likely that this has a detrimental effect on bottom-up quantitation and results in underestimation of site occupancy.
Figure 2.
Characterization of partially glycosylated EPO-2 by native MS and bottom-up glycoproteomics. (A) Native MS spectrum of desialylated EPO-2, where each peak is color-coded and represents unique Hexx+3HexNAcxF3 compositions. The most abundant peak and peaks lacking one N-glycan are annotated, together with their likely glycan compositions. (B) Native MS spectrum of non-sialidase-treated EPO-2, where the numbers above the color codes indicate the cumulative number of sialic acid residues attached to EPO glycans. The peaks corresponding to the partially glycosylated EPO are annotated with the corresponding glycan compositions. (C) Comparison and cross-correlation of native MS data with the simulated intact mass spectrum based on the GluC-digest glycoproteomics data for non-sialidase-treated EPO-2.
Sialylation as the Main Driver of the Observed Discrepancies between Native MS and Glycoproteomics
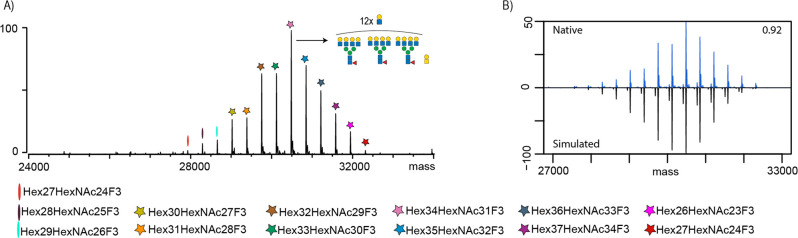
Next, we examined whether specific modifications, such as sialic acid moieties or the underlying N-glycan branching/LacNAc heterogeneity, are the main drivers of the observed discrepancy between the measured and simulated intact protein MS results. To this end, we investigated another EPO variant (termed EPO-3) that lacked sialic acids. Briefly, EPO-3 was a glycoengineered variant, whereby the proportion of poly-LacNAc extensions was enhanced (see Methods and Materials), and no sialic acid moieties were incorporated. This EPO-3 has an increased abundance of LacNAc structures, more than 10 mol/mol, compared to that of EPO-1. The native MS of EPO-3 is depicted in Figure 3A. From this relatively simple spectrum, we could assign 13 glycan compositions, confirming that the most abundant glycoproteoform had a significantly higher number of LacNAc repeats when compared to the EPO-1 and EPO-2 variants. We compared, as before, the native MS spectra of the EPO-3 variant with predicted spectra based on GluC glycoproteomics results. The increased similarity score for the two spectra (0.92) suggests that it is primarily the presence and amount of sialic acid residues rather than the N-glycan itself that are the key elements that induce the quantitative discrepancy between the native MS and glycoproteomics data. In the latter data for EPO-3, we could observe a few peaks originating from nonconfident identifications of sialylated glycopeptides, which were not readily observable in the native MS data. Of note, differences in ionization efficiency could also be caused by our selection of GluC as a protease of choice. Studying the effects of other proteases would be warranted. Of note, during the peer review period of this study, Miller et al.29 published the characterization of intact trimeric SARS-CoV-2 spike protein by charge detection mass spectrometry. When comparing their results to the glycopeptide studies utilizing trypsin, chymotrypsin, or alpha lytic protease, it was revealed that average glycan masses obtained by their intact approach are up to 47% larger than those reported by glycoproteomics studies. Taken together, this provides further credence to the claims put forward in this work.
Figure 3.
EPO-3 exhibits a high extent of poly-LacNAc elongation. (A) Native MS spectrum (non-sialidase-treated) of EPO-3 with glycan compositions color-coded and legend provided below the spectra. The most abundant peak is depicted with the likely glycan composition carrying 12 LacNAc repeats. (B) Comparison of native and simulated mass spectrum based on the GluC-digest glycoproteomics data for EPO-3.
Conclusion
Here, we compared face-to-face two emerging mass spectrometry approaches used in the characterization of biotherapeutics, namely, native mass spectrometry and bottom-up glycoproteomics. Both approaches provide complementary data and were found to be in good agreement in terms of identified glycan compositions associated with each of the three EPO variants analyzed in this work. Additionally, we demonstrated an added benefit of the glycoproteomics workflow to confidently localize the identified glycan compositions to each of the EPO glycosites, something which cannot be readily extracted from the native MS data. On the other hand, interplay between co-occurring glycoforms can only be probed at an intact glycoprotein level.30 We also observed a significant divergence between these two approaches in regards to quantitative characterization of EPO glycosylation profiles where we demonstrated that the increased size of glycopeptides and, especially, the presence of negatively charged sialic acids can have a detrimental effect on the ionization efficiency of glycopeptides, whereas it had minimal effect on the measurements of intact glycoproteins under native conditions. In conclusion, while glycoproteomics analysis provides excellent qualitative and site-specific characterization of glycopeptides, care has to be taken when dealing with large multiply sialylated N-glycans or partially occupied glycosites. These are best characterized at the intact glycoprotein level under native conditions.
Methods and Materials
EPO samples used in this study were prepared from NeoRecormon (EPO beta) drug substance material from Roche Diagnostics GmbH (Penzberg, Germany). Endoproteinase GluC, guanidinium hydrochloride (Gua-HCl), tris(hydroxymethyl)aminomethane (Tris), 1,4-dithiothreitol (DTT), and NAP-5 gel filtration columns were from Sigma-Aldrich/Merck KGaA (Darmstadt, Germany). Ammonium hydrogen carbonate, acetonitrile (ACN), formic acid (FA), and iodoacetic acid (IAA) were obtained from Fisher Scientific International Inc. (Pittsburgh, PA, USA).
Preparation of EPO Samples
Samples with modified glycan profile EPO-1 and EPO-2 were obtained as previously described.25 Briefly, EPO drug substance was separated by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Grace VYDAC C4 column using 0.1% TFA in purified water as solvent A and increasing volumetric ratios of 0.1% TFA in ACN as solvent B. Fractions from early (EPO-1) and late (EPO-2) elution time points were separated, collected, and further purified by a repeated RP-HPLC. Fractions containing EPO-1 and EPO-2 were collected again subjected to anion exchange chromatography at 2 to 8 °C on a eiethylaminoethyl (DEAE) sepharose fast flow anion exchange chromatography resin (GE Healthcare). For washing, 10 mM sodium/potassium phosphate, pH 7.5, and 30 mM sodium acetate, pH 4.5, were used alternately. Elution was performed with a 10 mM sodium/potassium phosphate, 80 mM NaCl, pH 7.5. The eluate was collected, sterile filtrated, aliquoted, and stored at ≤−60 °C. For preparation of EPO-3, representative EPO drug substance material was incubated with neuraminidase. Next, desialylated EPO was incubated with recombinant human β-1,3-N-acetylglucosaminyltransferase 2 in a buffer containing uridine diphosphate N-acetylglucosamine. Following a buffer exchange step, the resulting EPO was incubated with β-1,4-galactosyltransferase in a buffer containing UDP-galactose. Afterward, EPO material was purified using cation exchange chromatography. In a final enzyme treatment, EPO was incubated with two N- and O-glycan-specific β-galactoside α-2,3-sialyltransferases in a buffer containing cytidine-5′-monophospho-N-acetylneuraminic acid. Anion exchange chromatography was applied to purify the resulting EPO material. Subsequently, concentration, diafiltration, and conductivity correction were performed to yield the EPO drug substance buffer composition.
LC-MS Multiattribute Monitoring Method
First, 250 μg of each EPO sample was denatured with 0.4 M Tris and 8 M Gua-HCl, pH 8.5. Next, samples were reduced with 21 mM DTT at 50 °C for 60 min and then alkylated with 50 mM IAA at room temperature (RT) for 30 min in the dark. Prior to Glu-C digestion, samples were buffer-exchanged to 50 mM aqueous ammonium hydrogen carbonate, pH 7.8, with NAP-5 columns. Samples were then digested with GluC at 25 °C for 16–18 h at a 1:20 enzyme/protein ratio.
Five micrograms of digested EPO was loaded on a Waters Corp. (Milford, MA, USA) ACQUITY ultraperformance liquid chromatography system and C8 ethylene bridged hybrid columns (2.1 mm × 150 mm, 1.7 μm, 130 Å) coupled to Q-TOF Synapt G2 HDMS by Waters, Q-TOF by Bruker Corporation (Billerica, MA, USA), or Orbitrap Velos or Fusion from Thermo Fisher Scientific. The column was kept at 65 °C and ran as a 65 min gradient method with 0.1% FA in H2O (solvent A) and 0.1% FA in ACN (solvent B) applied at a flow rate of 300 μL/min (0–30 min: 1–20% B, 30–60 min: 20–5% B, 60–65 min: 35–60% B, 65–75 min: 80–99% B, 75–80 min: 1% B). MS measurements were performed in positive ion mode, and data were acquired in the 500–2000 m/z range. Extracted ion chromatograms were generated for m/z values of expected glycopeptides; peaks were integrated, and the resulting areas were used to obtain relative glycan abundance at each site.
Native Mass Spectrometry
EPO-1–3 samples (20 μg each) were buffer exchanged into a 150 mM aqueous ammonium acetate (pH 7.5) by ultrafiltration (vivaspin500 10 kDa cutoff, Sartorius, Stedim Biotech, Germany) at 10,000g. The concentration was adjusted to 5 μM, and 4 μL was used for native MS analysis. Part of the EPO-1 and EPO-2 samples were additionally treated with 0.02 U of sialidase (Roche, IN, USA) at RT overnight. Samples were analyzed on a modified Exactive Plus Orbitrap instrument with an extended mass range (Thermo Fisher Scientific, Bremen) acquiring an m/z range of 500–10,000. Voltage offsets on the transport multipoles and ion lenses were manually tuned for optimal transmission of intact protein. Nitrogen was used in the HCD cell at a gas pressure of 6 × 10–10 bar. MS parameters were as follows: spray voltage 1.2–1.3 V, source fragmentation and collision energy were varied from 5 to 30 to achieve optimal desolvation, and resolution (at m/z 200) of 17,500. The instrument was mass calibrated in the 500–5000 m/z range using CsI clusters.31 Measured spectra were deconvoluted to zero-charge with Intact Mass software32 (Protein Metrics, CA, USA) using default settings, except for the mass range, which was adjusted based on the smallest and the largest identified glycoproteoforms to minimize the presence of artifacts.
For comparison of native and glycopeptide data, we performed an in silico data construction to simulate a deconvoluted intact MS spectrum based on the mass and relative abundances of (glyco)peptides mapped in the multiattribute method (Table S1). Simulated sample was compared with deconvoluted native MS spectra of each EPO and a similarity score based on Pearson correlation was calculated. Algorithm used for the analysis is publicly available at https://github.com/Yang0014/glycoNativeMS.22
Acknowledgments
We acknowledge support from The Netherlands Organization for Scientific Research (NWO) funding The Netherlands Proteomics Centre through the X-omics Road Map program (project 184.034.019). T.C. and A.J.R.H. acknowledge further support by the NWO TOP-Punt Grant 718.015.003, the Satin Grant 731.017.202, and the ENPPS.LIFT.019.001.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jasms.1c00060.
The authors declare the following competing financial interest(s): Alexander Buettner, Markus Haberger, and Dietmar Reusch are employees of Roche, a major manufacturer and distributor of erythropoietin.
Supplementary Material
References
- Walsh G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. 10.1038/nbt.4305. [DOI] [PubMed] [Google Scholar]
- Hajba L.; Szekrényes Á.; Borza B.; Guttman A. On the glycosylation aspects of biosimilarity. Drug Discovery Today 2018, 23, 616. 10.1016/j.drudis.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Woen S.; Wang T.; Liau B.; Zhao S.; Chen C.; Yang Y.; Song Z.; Wormald M. R.; Yu C.; Rudd P. M. Challenges of glycosylation analysis and control: An integrated approach to producing optimal and consistent therapeutic drugs. Drug Discovery Today 2016, 21, 740–765. 10.1016/j.drudis.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Walsh G.; Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- Fukuda M. N.; Sasaki H.; Lopez L.; Fukuda M. Survival of Recombinant Erythropoietin in the Circulation: The Role of Carbohydrates. Blood 1989, 73, 84–89. 10.1182/blood.V73.1.84.84. [DOI] [PubMed] [Google Scholar]
- Hossler P.; Khattak S. F.; Li Z. J. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 2009, 19, 936–949. 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- Sha S.; Agarabi C.; Brorson K.; Lee D.-Y.; Yoon S. N-Glycosylation Design and Control of Therapeutic Monoclonal Antibodies. Trends Biotechnol. 2016, 34, 835–846. 10.1016/j.tibtech.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Planinc A.; Bones J.; Dejaegher B.; Van Antwerpen P.; Delporte C. Glycan characterization of biopharmaceuticals: Updates and perspectives. Anal. Chim. Acta 2016, 921, 13–27. 10.1016/j.aca.2016.03.049. [DOI] [PubMed] [Google Scholar]
- Saldova R.; Kilcoyne M.; Stöckmann H.; Millán Martín S.; Lewis A. M.; Tuite C. M. E.; Gerlach J. Q.; Le Berre M.; Borys M. C.; Li Z. J.; Abu-Absi N. R.; Leister K.; Joshi L.; Rudd P. M. Advances in analytical methodologies to guide bioprocess engineering for bio-therapeutics. Methods 2017, 116, 63–83. 10.1016/j.ymeth.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Lauc G.; Vučković F.; Bondt A.; Pezer M.; Wuhrer M. Trace N-glycans including sulphated species may originate from various plasma glycoproteins and not necessarily IgG. Nat. Commun. 2018, 9, 10–12. 10.1038/s41467-018-05173-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q.; Wang B.; Chen Z.; Urabe G.; Glover M. S.; Shi X.; Guo L. W.; Kent K. C.; Li L. Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glycoproteome Characterization. J. Am. Soc. Mass Spectrom. 2017, 28, 1751–1764. 10.1007/s13361-017-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T.; Zhu J.; Heck A. J. R. Simply Extending the Mass Range in Electron Transfer Higher Energy Collisional Dissociation Increases Confidence in N-Glycopeptide Identification. Anal. Chem. 2019, 91, 10401–10406. 10.1021/acs.analchem.9b02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J. Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides. Glycoconjugate J. 2016, 33, 261–272. 10.1007/s10719-016-9649-3. [DOI] [PubMed] [Google Scholar]
- Reiding K. R.; Bondt A.; Franc V.; Heck A. J. R. The benefits of hybrid fragmentation methods for glycoproteomics. TrAC, Trends Anal. Chem. 2018, 108, 260–268. 10.1016/j.trac.2018.09.007. [DOI] [Google Scholar]
- Rogers R. S.; Nightlinger N. S.; Livingston B.; Campbell P.; Bailey R.; Balland A. Development of a quantitative mass spectrometry multi-attribute method for characterization, quality control testing and disposition of biologics. MAbs. 2015, 7, 881–890. 10.1080/19420862.2015.1069454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogstad S.; Yan H.; Wang X.; Powers D.; Brorson K.; Damdinsuren B.; Lee S. Multi-Attribute Method for Quality Control of Therapeutic Proteins. Anal. Chem. 2019, 91, 14170–14177. 10.1021/acs.analchem.9b03808. [DOI] [PubMed] [Google Scholar]
- Stavenhagen K.; Hinneburg H.; Thaysen-Andersen M.; Hartmann L.; Silva D. V. V.; Fuchser J.; Kaspar S.; Rapp E.; Seeberger P. H.; Kolarich D. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: An evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 2013, 48, 627–639. 10.1002/jms.3210. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Wang G.; Song T.; Lebrilla C. B.; Heck A. J. R. Resolving the micro-heterogeneity and structural integrity of monoclonal antibodies by hybrid mass spectrometric approaches. MAbs. 2017, 9, 638–645. 10.1080/19420862.2017.1290033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati S.; Yang Y.; Barendregt A.; Heck A. J. R. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc. 2014, 9, 967–976. 10.1038/nprot.2014.057. [DOI] [PubMed] [Google Scholar]
- Sasaki H.; Ochi N.; Dell A.; Fukuda M. Site-Specific Glycosylation of Human Recombinant Erythropoietin: Analysis of Glycopeptides or Peptides at Each Glycosylation Site by Fast Atom Bombardment Mass Spectrometry. Biochemistry 1988, 27, 8618–8626. 10.1021/bi00423a017. [DOI] [PubMed] [Google Scholar]
- Takegawa Y.; Ito H.; Keira T.; Deguchi K.; Nakagawa H.; Nishimura S. I. Profiling of N- and O-glycopeptides of erythropoietin by capillary zwitterionic type of hydrophilic interaction chromatography/electrospray ionization mass spectrometry. J. Sep. Sci. 2008, 31, 1585–1593. 10.1002/jssc.200700679. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Liu F.; Franc V.; Halim L. A.; Schellekens H.; Heck A. J. R. Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun. 2016, 7, 13397. 10.1038/ncomms13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T.; Tian W.; Yang Z.; Clausen H.; Heck A. J. R. Direct quality control of glycoengineered erythropoietin variants. Nat. Commun. 2018, 9, 3342. 10.1038/s41467-018-05536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. J.; Hua S.; Kim U.; Kim H. J.; Lee J.; Kim J.-H.; An H. J. Analytical detection and characterization of biopharmaceutical glycosylation by MS. Bioanalysis 2016, 8, 711–727. 10.4155/bio.16.20. [DOI] [PubMed] [Google Scholar]
- Buettner A.; Maier M.; Bonnington L.; Bulau P.; Reusch D. Multi-Attribute Monitoring of Complex Erythropoetin Beta Glycosylation by GluC Liquid Chromatography-Mass Spectrometry Peptide Mapping. Anal. Chem. 2020, 92, 7574–7580. 10.1021/acs.analchem.0c00124. [DOI] [PubMed] [Google Scholar]
- Lin Y.-H.; Zhu J.; Meijer S.; Franc V.; Heck A. J.R. Glycoproteogenomics: a frequent gene polymorphism affects the glycosylation pattern of the human serum fetuin/α-2-HS-glycoprotein. Mol. Cell. Proteomics 2019, 18, 1479–1490. 10.1074/mcp.RA119.001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T.; de Haan N.; Konstantinidi A.; Vakhrushev S. Y. Quantitative characterization of O-GalNAc glycosylation. Curr. Opin. Struct. Biol. 2021, 68, 135–141. 10.1016/j.sbi.2020.12.010. [DOI] [PubMed] [Google Scholar]
- Shajahan A.; Heiss C.; Ishihara M.; Azadi P. Glycomic and glycoproteomic analysis of glycoproteins—a tutorial. Anal. Bioanal. Chem. 2017, 409, 4483–4505. 10.1007/s00216-017-0406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. M.; Barnes L. F.; Raab S. A.; Draper B. E.; El-Baba T. J.; Lutomski C. A.; Robinson C. V.; Clemmer D. E.; Jarrold M. F. Heterogeneity of Glycan Processing on Trimeric SARS-CoV-2 Spike Protein Revealed by Charge Detection Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 3959. 10.1021/jacs.1c00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T.; Heck A. J. R.; Reiding K. R. Meta-heterogeneity: evaluating and describing the diversity in glycosylation between sites on the same glycoprotein. Mol. Cell. Proteomics. 2021, 20, 100010. 10.1074/mcp.R120.002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati S.; Yang Y.; Barendregt A.; Heck A. J. R. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc. 2014, 9, 967–976. 10.1038/nprot.2014.057. [DOI] [PubMed] [Google Scholar]
- Bern M.; Caval T.; Kil Y. J.; Tang W.; Becker C.; Carlson E.; Kletter D.; Sen K. I.; Galy N.; Hagemans D.; Franc V.; Heck A. J. R. Parsimonious Charge Deconvolution for Native Mass Spectrometry. J. Proteome Res. 2018, 17, 1216–1226. 10.1021/acs.jproteome.7b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.