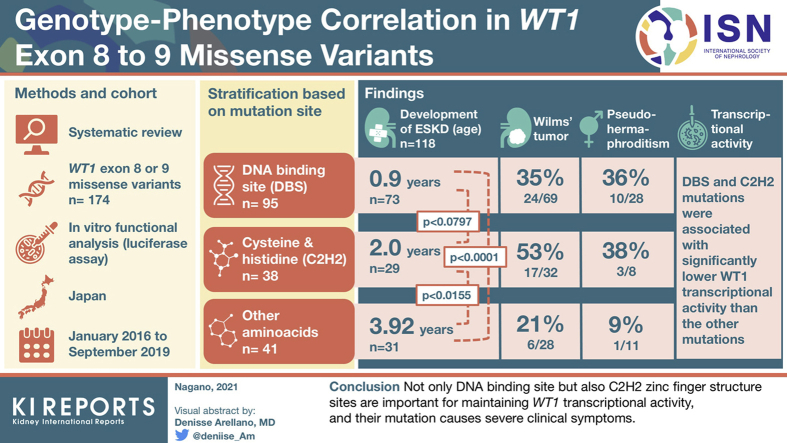
Abstract
Introduction
WT1 missense mutation in exon 8 or 9 causes infantile nephrotic syndrome with early progression to end-stage kidney disease (ESKD), Wilms tumor, and 46,XY female. However, some patients with missense mutations in exon 8 or 9 progress to ESKD in their teens or later. Therefore, we conducted a systematic review and functional analysis of WT1 transcriptional activity.
Methods
We conducted a systematic review of 174 cases with WT1 exon 8 or 9 missense variants from our cohort (n=13) and previous reports (n=161). Of these cases, mild and severe genotypes were selected for further in vitro functional analysis using luciferase assay.
Results
The median age of developing ESKD was 1.17 years. A comparative study was conducted among three WT1 genotype classes: mutations of the DNA-binding site (DBS group), mutations outside the DNA-binding site but at sites important for zinc finger structure formation by 2 cysteines and 2 histidines (C2H2 group), and mutations leading to other amino acid changes (Others group). The DBS group showed the severest phenotype and the C2H2 group was intermediate, whereas the Others group showed the mildest phenotype (developing ESKD at 0.90, 2.00, and 3.92 years, respectively, with significant differences). In vitro functional analysis showed dominant-negative effects for all variants; in addition, the DBS and C2H2 mutations were associated with significantly lower WT1 transcriptional activity than the other mutations.
Conclusion
Not only the DNA-binding site but also C2H2 zinc finger structure sites are important for maintaining WT1 transcriptional activity, and their mutation causes severe clinical symptoms.
Keywords: DNA binding, end-stage kidney disease, WT1, transcriptional activity
Graphical abstract
Wilms tumor is the most common renal tumor of childhood. A tumor suppressor gene responsible for this disease, WT1, was isolated from the short arm of chromosome 11, band p13, by positional cloning.1 The WT1 gene encodes the zinc finger DNA-binding protein WT1, which acts as a transcriptional activator or repressor depending on the cellular or chromosomal context.2 WT1 is required for normal formation of the genitourinary system and mesothelial tissues. Several human syndromes resulting from WT1 heterozygous mutations have been identified. WT1 heterozygous deletion is known as WAGR syndrome, which is a disorder affecting many body systems that is named for its main features: Wilms tumor, aniridia, genitourinary abnormalities, and intellectual disability.3 In addition, point mutations at the donor splice site on intron 9 of the WT1 gene cause Frasier syndrome, a rare disorder defined by male pseudohermaphroditism and progressive glomerulopathy.4,5 In contrast, heterozygous WT1 exon 8 to 9 missense point mutations lead to Denys-Drash syndrome (DDS), which is associated with the development of Wilms tumor, early-onset nephropathy, and male pseudohermaphroditism.6
A wealth of findings on the genotype-phenotype correlation of WT1-associated nephropathy has been reported, and a systematic review of mutations across the whole of the WT1 gene in 61 patients with steroid-resistant nephrotic syndrome was presented.7 In that report, the median age of developing ESKD in cases with DNA-binding site missense mutations (24 cases) was 2.5 years. In contrast, the corresponding age in cases with truncating mutations (9 cases) was 16.5 years. As such, patients with missense mutations in exons 8 to 9 show much more severe phenotypes than those with truncating mutations.7
It was previously reported7 that most of the WT1 missense mutations in exon 8 or 9 affect the DNA-binding site. However, recently, genetic testing has become widely available and led to the discovery of missense mutations in exon 8 to 9 outside of the region encoding the DNA-binding site of WT1.8 Such cases were found to exhibit a huge variety of phenotypes, from the early development of ESKD with DDS phenotypes to slow progression to ESKD. We have experienced patients with missense mutations in exon 9 of the WT1 gene, but showing relatively mild phenotypes. However, until now, no study has revealed the genotype-phenotype correlations concentrating only on missense mutations in exon 8 or 9. Therefore, we decided to conduct a systematic review and functional analysis of such variants associated with mild and severe phenotypes to investigate the mechanism determining disease severity. We also revealed the molecular mechanisms behind the dominant-negative effect of these mutations and correlations between WT1 transcriptional activity and phenotype for the first time. In addition, we attempted to show the DNA binding ability by computational analysis.
Methods
Study Participants
After obtaining informed consent from the patients and/or their guardians, we obtained clinical data and blood samples from individuals with severe proteinuria in Japan. Study approval was obtained from the Institutional Review Board of Kobe University Graduate School of Medicine (IRB approval number 301). Patients were enrolled between January 2016 and September 2019. We performed targeted next-generation sequencing in 303 families and detected WT1 exon 8 or 9 missense variants in 13 cases. All of these 13 patients were included in this study. We previously reported on some of them as part of the data in comprehensive gene screening for proteinuric patients.8 For variant description, the reference sequence NM_024426 was used. All of the identified variants were evaluated to determine their pathogenicity in accordance with the American College of Medical Genetics and Genomics guidelines9 for clinical sequence interpretation.
Systematic Review
A systematic search for patients with exon 8 or 9 missense mutations in WT1 was performed using the Human Gene Mutation Database (HGMD) Professional (https://portal.biobase-international.com/hgmd/pro/start.php) V2019.4. This identified 161 previously reported cases with clinical data. We calculated the median age at which ESKD developed and extracted information about phenotypes, including extrarenal symptoms and onset of Wilms tumor.
Functional Analysis of WT1 Transcriptional Activity
Plasmid construction
WT1 reporter plasmid was generated using the pNL1.2 vector (Promega, Madison, WI), which contained a WT1 response element that drives transcription of the luciferase reporter gene (Supplementary Figure S1). Chromatin immunoprecipitation followed by sequencing was performed to identify genome-wide WT1-binding sites, as previously described.10,11 Referring to these sequences, the WT1-binding sites (5′-GTGTGGGAG-3′) and minimal promoter sequence developed by Promega were selected for cloning. Single-stranded oligonucleotides were first annealed to form double-stranded oligonucleotides and then cloned into the pNL1.2 vector using In-Fusion cloning methods with In-Fusion HD Cloning Kit (Takara Bio Inc., Kusatsu, Japan), in accordance with the manufacturer’s instructions. The WT1 reporter plasmid contained a response sequence and a minimal promoter containing a TATA box necessary to initiate transcription of the gene. Then, the reporter gene itself was expressed in cultured cells, and binding of the target protein to the response element was shown to promote its transcription.
A plasmid based on cDNA of the ORF of the human WT1 gene (Sino Biological, Beijing, China) was used as an effector vector and was named WT1 expression vector. We introduced mutations into the WT1 expression vector by site-directed mutagenesis using PrimeSTAR Mutagenesis Basal Kit (Takara Bio Inc.), in accordance with the manufacturer’s instructions. The primers used are shown in Supplementary Table S1.
Luciferase Assay
Transfection of plasmids into HEK293T cells was performed in triplicate in 96-well plates using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) with plasmid DNA (94 ng of promoter vector, 1 ng of effector vector, and 5 ng of pGL4.53 vector [Promega] as an internal control per well). Cells were harvested 24 hours after transfection. The luciferase assay was performed with a dual luciferase reporter assay system (Promega) using EnSpire. The results are shown as the ratio of firefly luciferase activity to Renilla activity.
For promoter assays, we chose 3 mutations from each group: p.Arg467Trp, p.Arg439His, and p.Gln442His from the DBS group; p.Cys458Arg, p.His478Arg, and p.Cys433Tyr from the C2H2 group; and p.Pro455Ser, p.Arg463Gln, and p.Phe437Cys from the Others group. Luciferase reporter plasmids containing the WT1 response element were used to test whether mutations of WT1 affect its transcription. HEK293T cells were cotransfected with these reporter constructs plus a vector expressing WT1 wild type or mutant. Furthermore, to examine the dominant-negative pattern, vector expressing WT1 wild type only or cotransfected vectors expressing the wild type and mutant were added in equal amounts to express WT1 protein. Specifically, when we performed the luciferase assay to examine the dominant-negative pattern, we used a total of 1 ng of effector vector. For wild type, we used 0.5 ng of wild-type WT1 expression vector and 0.5 ng of empty vector. For mutants, we used 0.5 ng of wild-type WT1 expression vector and 0.5 ng of mutant WT1 expression vector. Subsequently, we measured the WT1 transcriptional activity.
Western Blotting
The protein samples from HEK293T cells transfected with the expression vector WT1 wild type or mutant were diluted with Laemmli sample buffer (Bio-Rad Laboratories, Inc., Hercules, CA) and boiled for 10 minutes. Samples were loaded into 4% to 20% Mini-PROTEAN TGX Precast Protein gels (Bio-Rad Laboratories, Inc.) and electrophoresed in Tris-glycine-SDS running buffer. Proteins were transferred to Immun-Blot PVDF Membrane (Bio-Rad Laboratories, Inc.), in accordance with standard protocols, after which membranes were blocked with Block Ace (KAC Co., Ltd., Kyoto, Japan). Membranes were incubated overnight at 4°C with 10× Block Ace and rabbit monoclonal antibodies against WT1 ab89901 (Abcam, Cambridge, UK) or β-actin (Cat. No. 4967; Cell Signaling Technology, Danvers, MA). Subsequently, blots were washed and incubated for 1 hour at room temperature with goat anti-rabbit IgG antibody and HRP-conjugated secondary antibody. Immunoreactive proteins were visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore) followed by luminescence detection with ChemiDoc MP (Bio-Rad).
Cell Lines
HEK293T cells were obtained from the Cell Bank, Riken BioResource Center (Tsukuba, Japan).
Statistical Analysis
All experiments were conducted at least 3 times. Statistical analysis was performed using standard statistical software (JMP Version 11 for Windows; SAS Institute, Cary, NC). Values are expressed as mean ± SD. One-way analysis of variance was used when multiple comparisons were made, whereas individual comparisons were performed by Bonferroni’s multiple comparison test. Kaplan-Meier curves were generated for the time taken to develop ESKD. We compared curves for the 2 groups using the log-rank test. P <0.05 was considered to be statistically significant.
Structural Analysis In Silico
Substructures of WT1 mutants p.Pro455Ser, p.Arg467Trp, p.Cys458Arg, and p.His478Arg were prepared using MOE software12 from the 3-dimensional structure of part of wild-type WT1 (aa. 392–510, including the zinc finger region), which was obtained from PDB (PDB 6B0O chain A). The transferable intermolecular potential with 3 points (TIP3P) water model13 and the AMBER03 force field were used for structural optimization of 500-ps molecular dynamics using GROMACS software,14 as in our previous study.15 Then, these partial structures were input into APBS software for analysis of the electrostatic surface potential of the DNA-binding region, as described previously.16
Results
Clinical Characteristics of Our Patient Cohort
Among the 13 patients with exon 8 or 9 missense variants, neither Wilms tumor nor 46,XY female was detected. The clinical characteristics and variant information are shown in Table 1. Extrarenal symptoms occurred in 3 cases, and 2 cases had progressed to ESKD. Chromosomal analysis was performed in 3 cases (No. 6, 10, and 13), all of whom were female. Regarding the variants, 2 were novel and, in 9 cases, de novo mutations were confirmed.
Table 1.
Characteristics of 13 cases
| Case no. | ID | Sex | Age at onset (y) | Age at ESKD (y) | Extra-renal symptom | Exon | Gene variants (NM_024426) | Group | HGMD V2019.4 |
Transmission | gnomAD | ACMG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Neph7 | F | 0.3 | 0.5 | — | 9 | c.1399C>T Arg467Trp |
DBS | Reported | De novo | ND | Pathogenic |
| 2 | Neph90 | M | 3 | — | — | 9 | c.1407C>A Asp469Glu |
Others | Reported | De novo | ND | Pathogenic |
| 3 | Neph92 | F | 0 | 0 | Massive placenta | 8 | c.1315C>T Arg439Cys |
DBS | Reported | ND | ND | Likely pathogenic |
| 4 | Neph95 | M | 3 | 11 | Cryptochidism | 9 | c.1399C>T Arg467Trp |
DBS | Reported | ND | ND | Likely pathogenic |
| 5 | Neph107 | F | 0.2 | 0.2 | — | 8 | c.1316G>A Arg439His |
DBS | Reported | De novo | ND | Pathogenic |
| 6 | Neph132 | F | 0.5 | 0.5 | — | 8 | c.1316G>T Arg439Leu |
DBS | Reported | De novo | ND | Pathogenic |
| 7 | Neph154 | M | 3.9 | — | Cryptochidism, hypospadias | 8 | c.1336C>T His446Tyr |
C2H2 | Reported | De novo | ND | Pathogenic |
| 8 | Neph171 | M | 9 | 29 | — | 9 | c.1363C>T Pro455Ser |
Others | Reported | De novo | ND | Pathogenic |
| 9 | Neph197 | F | 3 | 7 | — | 9 | c.1366T>A Phe456Ile |
Others | Novel | De novo | ND | Likely pathogenic |
| 10 | Neph208 | F | 0.4 | 0.4 | — | 8 | c.1349A>G His450Arg |
C2H2 | Reported | De novo | ND | Pathogenic |
| 11 | Neph243 | F | 0.4 | 0.4 | — | 9 | c.1399C>T Arg467Trp |
DBS | Reported | De novo | ND | Pathogenic |
| 12 | Neph248 | F | 0.3 | 0.3 | — | 9 | c.1400G>A Arg467Gln |
DBS | Reported | ND | ND | Likely pathogenic |
| 13 | Neph294 | F | 1 | 1.8 | — | 8 | c.1349A>C His450Pro |
C2H2 | Novel | ND | ND | Likely pathogenic |
ACMG, American College of Medical Genetics and Genomics; C2H2, zinc finger structure formed by 2 cysteines and 2 histidines; DBS, DNA-binding site; ESKD, end-stage kidney disease; F, female; gnomAD, Genome Aggregation Database; HGMD, Human Gene Mutation Database; M, male; ND, not determined
Systematic Review
We reviewed 174 cases in which WT1 exon 8 or 9 missense variants were detected, including our 13 cases. Characteristics of the 161 patients in previous reports are summarized in Supplementary Table S2. The age of onset was obtained in 168 cases, the median of which was 0.6 years. Extrarenal symptoms and Wilms tumor occurred in 17 of 75 (23%) cases and 47 of 129 (36%) cases for which such information was available, respectively. Among the 136 cases with data on kidney function, 118 cases had progressed to ESKD. It was revealed that the median age of developing ESKD was 1.17 years.
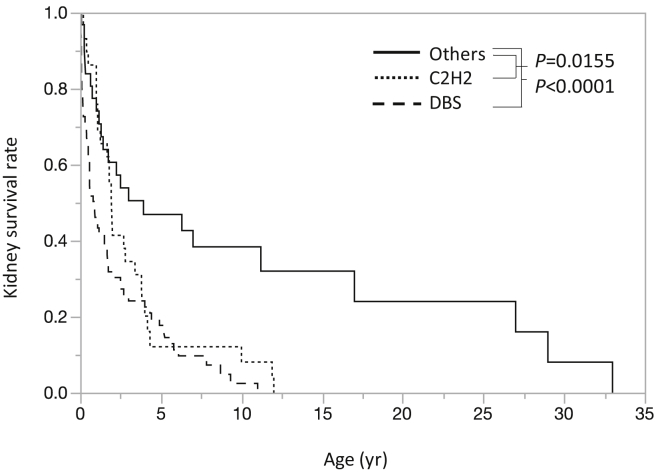
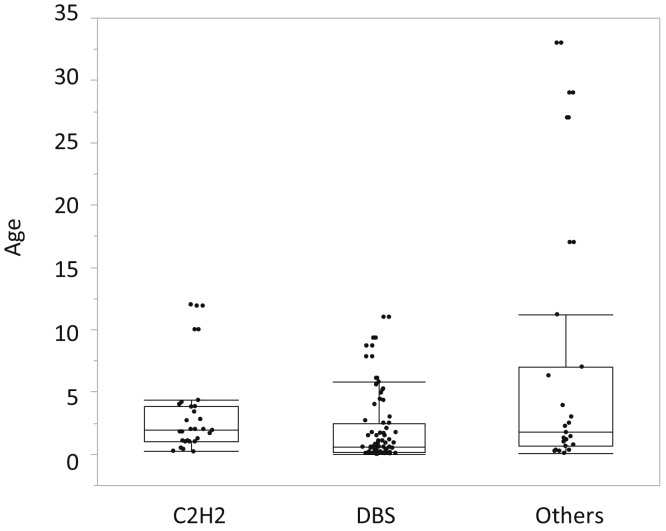
The patients were stratified based on the location of the mutation site: mutations in the DNA-binding site (DBS group, n=95), mutations in cysteine and histidine amino acids outside the DNA-binding site but important for zinc finger formation (C2H2 group, n=38), and mutations of other amino acids (Others group, n=41) (Figure 1). Kaplan-Meier analysis was performed on the age at development of ESKD. The results showed a significant difference between the DBS group (0.90 years, n=73) and the Others group (3.92 years, n=31) (P < 0.0001, log-rank test). There was also a significant difference between the C2H2 group (2.00 years, n=29) and the Others group (3.92 years, n=31) (P = 0.0155, log-rank test) (Figure 2). In contrast, there was only a tendency for a significant difference between the DBS group and the C2H2 group (P = 0.0797). When the ages at development of ESKD were compared, mild cases were more common in the Others group (Figure 3). Figure 3 shows the difference in the age of onset of ESKD only for the patients who developed ESKD. Because the Others group included older children who had not developed ESKD and are not shown in Figure 3, Kaplan-Meier analysis showed that the condition in the C2H2 group was significantly more severe than that in the Others group (Figure 2). Regarding the occurrence of Wilms tumor, there were 24 of 69 cases (35%) in the DBS group, 17 of 32 (53%) in the C2H2 group, and 6 of 28 (21%) in the Others group. Pseudohermaphroditism was detected in 10 of 28 individuals (36%) in the DBS group, 3 of 8 (38%) in the C2H2 group, and 1 of 11 (9%) in the Others group.
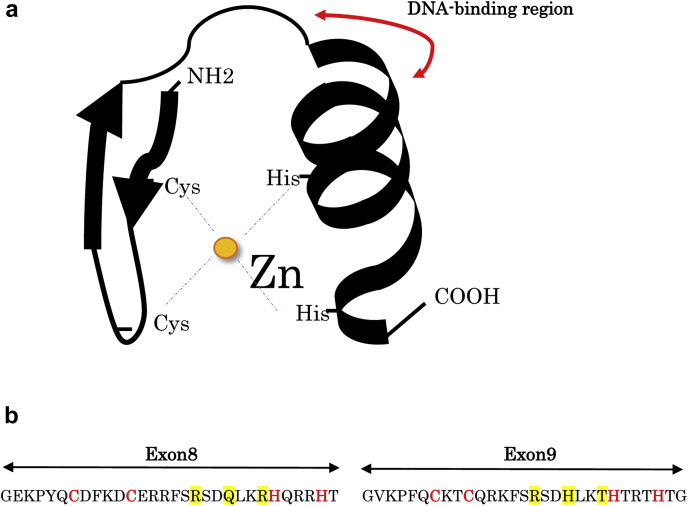
Figure 1.
(a) Schema of a C2H2 zinc finger motif: C2H2-type zinc finger contains a short beta hairpin and an alpha helix (beta/beta/alpha structure), where a single zinc atom is held in place by Cys(2)His(2) (C2H2) residues in a tetrahedral array. (b) Amino acid sequences of exons 8 and 9: the sequence-recognition amino acids at the protein-DNA interface in exons 8 and 9 (in yellow). The Cys2-His2 structural amino acids that coordinate the zinc ions and hydrophobic core are shown in red letters. C2H2, zinc finger structure formed by 2 cysteines and 2 histidines.
Figure 2.
Kaplan-Meier survival analysis of renal prognosis. Dashes indicate patients with mutations of the DNA-binding site (n=73). The median age for developing ESKD among these patients was 0.90 years. Dots indicate patients with mutations in the region encoding a zinc finger (n=29). The median age for developing ESKD among these patients was 2.00 years. Solid line indicates patients with mutations of other amino acids (n=31). The median age for developing ESKD among these patients was 3.92 years. DBS, DNA-binding site; ESKD, end-stage kidney disease.
Figure 3.
Difference in the age of onset of ESKD among DBS, C2H2, and Others groups. Some of those in the Others group developed ESKD in their late teens. DBS group: median age 0.60 (0.17–2.59) years; C2H2 group: median age 1.96 (1.02–3.83) years; Others group: median age 1.75 (0.64–7.00) years. C2H2: zinc finger structure formed by 2 cysteines and 2 histidines; DBS, DNA-binding site.
In Vitro Analysis
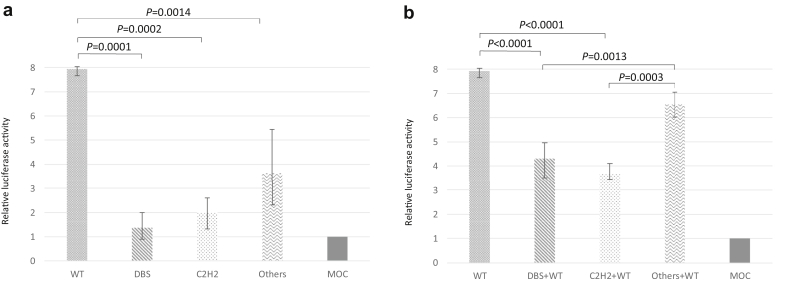
We hypothesized that the analyzed variants may affect the DNA-binding function of the WT1 protein. The measurement of luciferase activity revealed that WT1 mutant proteins were associated with significantly decreased promoter activation compared with the level for the WT1 wild-type protein (Figure 4a). When the WT1 mutant (DBS or C2H2 group) vector was cotransfected with the WT1 wild-type vector, in order to mimic heterozygosity, we observed that the variant inhibited the activation induced by the WT1 wild type, even though the amount of transfected wild-type vector was the same (Figure 4b). These findings indicate that the variants act via dominant-negative effects. In addition, the transcriptional activities of the WT1 mutants in the DBS and C2H2 groups were significantly lower than those in the Others group, which reflected the clinical severity of those genotypes. The transcriptional activities of all mutants are shown in Supplementary Figure S2. Furthermore, immunoblotting analysis indicated that, after transfection, mutants were expressed at the same level as the wild-type WT1 protein (Supplementary Figure S3), which means that a haploinsufficiency effect caused by the variants was unlikely.
Figure 4.
Luciferase analysis. Luciferase analysis in HEK293T cells. HEK293T cells were transfected with the WT1 reporter plasmid, WT1 expression vector, and control vector. Twenty-four hours after transfection, cells were subjected to dual luciferase analysis. Luciferase reporter activities were calculated as firefly/Renilla and are normalized to the activities of the WT1 cDNA empty vector. Mock (MOC) shows the activity of the WT1 cDNA empty vector. We used 3 mutations (p.Arg467Trp, p.Arg439His, and p.Gln442His) as the DBS group, 3 (p.Cys458Arg, p.His478Arg, and p.Cys433Tyr) as the C2H2 group, and 3 (p.Pro455Ser, p.Arg463Gln, and p.Phe437Cys) as the Others group. (a) Promoter activities were significantly decreased in WT1 mutants compared with that in the WT1 wild type. (b) Promoter activities were significantly inhibited when both WT1 wild-type (WT) and mutant (DBS or C2H2 group) vectors were cotransfected. Promoter activity was higher on cotransfection of vectors encoding WT protein and proteins from the Others group than on cotransfection of vectors encoding other mutations (DSB or C2H2 group) and encoding the WT protein. When the same amount of effector vector was used for transfection, the promoter activity was decreased by cotransfection of the mutant protein compared with the wild-type activity. These results suggest that the mutant protein suppressed the activity of the wild-type protein. C2H2: zinc finger structure formed by 2 cysteines and 2 histidines; DBS, DNA-binding site.
Electrostatic Surface Potential Analysis
The electrostatic surface potential of the DNA-binding region was analyzed to determine the alteration of DNA-binding ability because of the amino acid substitutions (Supplementary Figure S4). The negative charge of the DNA-binding region was slightly increased by these amino acid substitutions. This increase would weaken the DNA-binding ability because DNA is also characterized by a negative charge. The findings also showed that the DNA-binding areas of the mutants were slightly decreased compared with those of the wild type, leading to a reduction of DNA binding ability.
Discussion
In this study, we determined the genotype-phenotype correlations and transcriptional activity–phenotype correlations for missense mutations in exons 8 and 9 of the WT1 gene for the first time. This is also the first report clearly showing evidence of dominant-negative effects by these heterozygous missense mutations.
Various combinations of renal and other findings associated with WT1 pathogenic variants were previously designated to reflect certain syndromes such as WAGR syndrome, Frasier syndrome, and DDS. In addition, truncating pathogenic variants have been shown to be associated with later-onset glomerulopathy, genital anomalies, and the risk of bilateral Wilms tumor.17 Missense variants affecting nucleotides encoding amino acid residues in the DNA-binding region in exons 8 and 9 of the WT1 gene are associated with early-onset rapidly progressive steroid-resistant nephrotic syndrome, pseudohermaphroditism, and Wilms tumor, which is known as DDS.7 WT1 gene variants also cause various types of glomerulonephritis globally.18,19 Especially in Japan, the most common disease-causing gene among patients with severe proteinuria is WT1.8 Among such patients, we noticed that some cases with missense variants in exons 8 to 9 did not conform to previous subgroups. We therefore focused on the cases with these variants.
Our results showed that there were patients with severe phenotypes other than those with mutations in the DNA-binding site, most of whom had C2H2 mutations affecting regions important for maintaining the structure of the DNA-binding site of the WT1 protein (Supplementary Figure S2). Therefore, we performed a systematic review and found for the first time that the condition of WT1 patients with C2H2 mutations was as severe as that of patients with DNA-binding site mutations. The findings clearly revealed that C2H2 mutations can also lead to a severe phenotype. The C2H2 zinc finger structure is stabilized by coordination of a zinc atom between 2 conserved cysteine residues at 1 end of a β-sheet and 2 conserved histidine residues at the C-terminus of an α-helix.20 The cysteine-histidine pair is conserved, as is the hydrophobic core that forms the α-helix.21 Therefore, we hypothesized that mutations in cysteine and histidine could cause major conformational changes. In this systematic review, patients with mutations in cysteine and histidine were found to have a more severe condition than those with mutations in other amino acids. The findings from in vitro analysis of transcriptional activity and conformation analysis were consistent with these results. In silico analysis successfully classified severe cases and mild cases using features of the molecular structure. These results were in accordance with the promoter assay findings. Previous reports of genotype-phenotype correlations have shown severe phenotypes in cases with DNA-binding site mutations.7 However, we found that it is not sufficient to determine whether or not a mutation is located at the region encoding the DNA-binding site to predict whether a severe phenotype could develop, as mutations at the C2H2 site could also lead to a severe phenotype. Our results suggested that conformational changes caused by mutations in regions such as C2H2 may affect WT1’s function. Changes in the volume and hydrophobicity of amino acids within a protein may disrupt its structural stability and cause dysfunction, resulting in disease.22 Even some cases in the Others group showed severe symptoms, so there might be other factors that influence disease severity. Further analysis is needed to clarify the other factors that determine the clinical phenotype.
Functional analysis showed that the DNA-binding site mutations were associated with significantly lower transcriptional activity than the wild type, whereas other mutations also led to lower transcriptional activity. We also revealed that WT1 missense variants disturb WT1 transcription and proved the existence of dominant-negative effects for these variants for the first time. Only 1 previous report mentioned the dominant-negative effects of a WT1 variant, specifically, one associated with deletion of the whole of zinc finger 3. This is a truncating variant and not a missense variant,23 so this result does not correctly reflect the condition seen in DDS. In our study, we introduced a total of 9 missense variants in exon 8 or 9 of the WT1 gene, including 3 common DBS variants, and successfully showed dominant-negative effects for all variants. Thus, our experiments can support clinical phenotypes.
The mechanisms by which the mutations exert dominant-negative effects were unclear in this study, as also previously reported.23 In vitro assays previously showed that WT1 forms a homodimer, which requires its first 180 amino acids.23 Thus, the authors proposed that a dominant-negative mutant of the WT1 protein that cannot bind DNA may play a role in tumorigenesis and the development of DDS by binding to wild-type WT1 protein and reducing its transcriptional activity.23 WT1 not only binds directly to DNA as a transcription factor but also is said to be related to other transcription factors and to form coactivation and corepression complexes.24 Indeed, several other coactivators, corepressors, and transcription factors, including p53 (TRP53), have been shown to interact with WT1, modulating its target sites and activities, at least in cell lines.25 Therefore, further studies on these issues are needed, including on whether or not WT1 mutant proteins can form dimers.
Owing to the popularity of genetic testing and the recent increase in the number of WT1 mutation cases, new variants of unknown pathogenicity have been detected. Detection of pathogenic variants of the WT1 gene is clinically important to predict renal prognosis, detect extrarenal symptoms, and perform genetic counseling. Early diagnosis is needed because tumorigenesis and sex reversal are notable characters in cases with WT1 gene mutation. There is a need for a reliable system to determine whether the novel missense variants detected in patients are pathogenic. In this study, several WT1 missense cases were examined, and our findings suggested that our method for measuring WT1 transcriptional activity was useful for detecting pathogenicity.
This study had several limitations. First, the promoter assays and in silico analysis were performed for only limited genotypes. In future work, we will increase the number of cases and perform promoter assays for WT1 missense variants in regions other than exon 8 or 9. Second, the sensitivity of the promoter assay may not be high enough to detect the difference in effects on disease severity between variants in the C2H2 and Others groups. Third, because the sequence of the transcription factor binding site is short, the luciferase assay may have been nonspecific. Lastly, our study population was small and we were also limited by the retrospective nature of this work.
We determined the genotype-phenotype correlation for WT1-associated nephropathy with exon 8 or 9 mutation. Promoter assay and in silico analysis can be useful for examining the severity of this disease.
Disclosure
All the authors declared no competing interests.
Acknowledgments
The authors thank Edanz (https://en-author-services.edanz.com/ac) for editing the English text of a draft of this manuscript. This study was supported by a Grant-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (subject ID: 20K16926 to C.N.).
Footnotes
Figure S1. Schema of functional analysis
Figure S2. Luciferase analysis
Figure S3. Western blot analysis of WT1 proteins
Figure S4. Electrostatic surface potential of DNA-binding region
Table S1. Primers for cloning and mutagenesis
Table S2. Characteristics of 161 cases
Supplementary Material
Figure S1. Schema of functional analysis
Figure S2. Luciferase analysis
Figure S3. Western blot analysis of WT1 proteins
Figure S4. Electrostatic surface potential of DNA-binding region
Table S1. Primers for cloning and mutagenesis
Table S2. Characteristics of 161 cases
References
- 1.Rose E.A., Glaser T., Jones C. Complete physical map of the WAGR region of 11p13 localizes a candidate Wilms' tumor gene. Cell. 1990;60:495–508. doi: 10.1016/0092-8674(90)90600-j. [DOI] [PubMed] [Google Scholar]
- 2.Little M., Wells C. A clinical overview of WT1 gene mutations. Hum Mutat. 1997;9:209–225. doi: 10.1002/(SICI)1098-1004(1997)9:3<209::AID-HUMU2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Riccardi V.M., Sujansky E., Smith A.C. Chromosomal imbalance in the Aniridia-Wilms' tumor association: 11p interstitial deletion. Pediatrics. 1978;61:604–610. [PubMed] [Google Scholar]
- 4.Frasier S.D., Bashore R.A., Mosier H.D. Gonadoblastoma associated with pure gonadal dysgenesis in monozygous twins. J Pediatr. 1964;64:740–745. doi: 10.1016/s0022-3476(64)80622-3. [DOI] [PubMed] [Google Scholar]
- 5.Haning R.V., Jr., Chesney R.W., Moorthy A.V. A syndrome of chronic renal failure and XY gonadal dysgenesis in young phenotypic females without genital ambiguity. Am J Kidney Dis. 1985;6:40–48. doi: 10.1016/s0272-6386(85)80036-6. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier J., Bruening W., Kashtan C.E. Germline mutations in the Wilms' tumor suppressor gene are associated with abnormal urogenital development in Denys-Drash syndrome. Cell. 1991;67:437–447. doi: 10.1016/0092-8674(91)90194-4. [DOI] [PubMed] [Google Scholar]
- 7.Lipska B.S., Ranchin B., Iatropoulos P. Genotype-phenotype associations in WT1 glomerulopathy. Kidney Int. 2014;85:1169–1178. doi: 10.1038/ki.2013.519. [DOI] [PubMed] [Google Scholar]
- 8.Nagano C., Yamamura T., Horinouchi T. Comprehensive genetic diagnosis of Japanese patients with severe proteinuria. Sci Rep. 2020;10:270. doi: 10.1038/s41598-019-57149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motamedi F.J., Badro D.A., Clarkson M. WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat Commun. 2014;5:4444. doi: 10.1038/ncomms5444. [DOI] [PubMed] [Google Scholar]
- 11.Kann M., Ettou S., Jung Y.L. Genome-wide analysis of Wilms' tumor 1-controlled gene expression in podocytes reveals key regulatory mechanisms. J Am Soc Nephrol. 2015;26:2097–2104. doi: 10.1681/ASN.2014090940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takaoka Y., Takeuchi A., Sugano A. Establishment of the experimental procedure for prediction of conjugation capacity in mutant UGT1A1. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorgensen W.L., Chandrasekhar J., Madura J.D. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 14.Abraham M.J., Murtola T., Schulz R. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 15.Ohta M., Sugano A., Hatano N. Co-precipitation molecules hemopexin and transferrin may be key molecules for fibrillogenesis in TTR V30M amyloidogenesis. Transgenic Res. 2018;27:15–23. doi: 10.1007/s11248-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunadi, Miura K., Ohta M. Two novel mutations in the ED1 gene in Japanese families with X-linked hypohidrotic ectodermal dysplasia. Pediatr Res. 2009;65:453–457. doi: 10.1203/PDR.0b013e3181991229. [DOI] [PubMed] [Google Scholar]
- 17.Lehnhardt A., Karnatz C., Ahlenstiel-Grunow T. Clinical and molecular characterization of patients with heterozygous mutations in wilms tumor suppressor gene 1. Clin J Am Soc Nephrol. 2015;10:825–831. doi: 10.2215/CJN.10141014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadowski C.E., Lovric S., Ashraf S. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26:1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F., Zhang Y., Mao J. Spectrum of mutations in Chinese children with steroid-resistant nephrotic syndrome. Pediatr Nephrol. 2017;32:1181–1192. doi: 10.1007/s00467-017-3590-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackeh R., Marr A.K., Fadda A. C2H2-type zinc finger proteins: evolutionarily old and new partners of the nuclear hormone receptors. Nucl Recept Signal. 2018;15 doi: 10.1177/1550762918801071. 1550762918801071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Razin S.V., Borunova V.V., Maksimenko O.G. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry (Mosc) 2012;77:217–226. doi: 10.1134/S0006297912030017. [DOI] [PubMed] [Google Scholar]
- 22.Gaines J.C., Clark A.H., Regan L. Packing in protein cores. J Phys Condens Matter. 2017;29:293001. doi: 10.1088/1361-648X/aa75c2. [DOI] [PubMed] [Google Scholar]
- 23.Reddy J.C., Morris J.C., Wang J. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270:10878–10884. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- 24.Hastie N.D. Wilms' tumour 1 (WT1) in development, homeostasis and disease. Development. 2017;144:2862–2872. doi: 10.1242/dev.153163. [DOI] [PubMed] [Google Scholar]
- 25.Toska E., Roberts S.G. Mechanisms of transcriptional regulation by WT1 (Wilms' tumour 1) Biochem J. 2014;461:15–32. doi: 10.1042/BJ20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.