Abstract
Introduction
Declining renal function results in the accumulation of solutes normally excreted by healthy kidneys. Data suggest that some of the protein-bound solutes mediate accelerated cardiovascular disease. Many of the poorly dialyzable protein-bound uremic retention solutes are products of gut bacterial metabolism.
Methods
We performed a blinded-randomized controlled trial comparing the changes in plasma concentrations of a panel of protein-bound solutes and microbiome structure in response to the once-weekly oral administration of 250 mg of vancomycin or placebo over a period of 12 weeks in a cohort of stable patients with end-stage kidney disease. We also examined the pattern of recovery of the solutes and gut microbiome over 12 weeks of placebo administration following vancomycin.
Results
We enrolled 15 subjects. Ten subjects provided sufficient plasma and stool samples to permit us to examine the effect of vancomycin on plasma solute levels. We showed that a weekly dose of vancomycin resulted in a reduction in the plasma concentration of 7 colon-derived solutes. We described a significant effect of vancomycin on the microbiome structure with a decrease in alpha diversity and change in beta diversity. Multiple taxa decreased with vancomycin including genera Clostridium and Bacteroides. We demonstrated microbiome recovery after stopping vancomycin. However, recovery in the solutes was highly variable between subjects.
Conclusions
We demonstrated that microbiome suppression using vancomycin resulted in changes in multiple gut-derived uremic solutes. Future studies are needed to address whether reduction in those uremic solutes results in improvement of cardiovascular outcomes in ESKD patients.
Keywords: antibiotic, indoxyl sulfate, microbiome, p-cresyl sulfate, uremic toxins
Graphical abstract
Declining renal function results in the accumulation of solutes normally excreted by healthy kidneys. Roughly one-half of these solutes are small water-soluble compounds, such as urea and creatinine, which would be readily filtered at the glomerulus and are easily removed by dialysis. About one-quarter are bound to protein, predominantly albumin, and so are poorly dialyzable.1, 2, 3 Observational studies4 and in vitro studies 5 suggest that some of the protein-bound solutes mediate accelerated cardiovascular disease in patients undergoing maintenance hemodialysis.6, 7, 8, 9 Most striking is the observed relationship between plasma levels of indoxyl sulfate (IS) and p-cresyl sulfate (PCS) and cardiovascular disease in patients with chronic kidney disease10, 11, 12 leading to their designation as uremic retention solutes (URS). Many of the poorly dialyzable, protein-bound uremic retention solutes are products of gut bacterial metabolism.13,14 This knowledge has led to studies of the effects of dietary and pharmacologic manipulation aimed at reducing the levels of URS.15, 16, 17, 18, 19
In a prior study, we observed that a single oral dose of 250 mg of vancomycin resulted in a significant decrease in the plasma concentrations of IS and PCS over a period of 1 week.20 Changes in several genera identified by 16S sequencing of the gut microbiome paralleled the changes in the concentrations of IS and PCS. Encouraged by the short-term effects of vancomycin, we performed a single-blinded randomized controlled trial comparing the changes in plasma concentrations of a panel of protein-bound solutes in response to the weekly oral administration of 250 mg of vancomycin or placebo over 12 weeks in a cohort of stable patients with end stage kidney disease (ESKD). Further, we examined the changes in the composition of the gut microbiome over 3 months during which vancomycin was administered and the stability of the microbiome composition during three-months of placebo. We examined the recovery pattern of the solutes and gut microbiome over 3 months of placebo administration after vancomycin.
Methods
Sample size and eligibility
Based on the intrasubject variation and change in mean solute level observed in our prior studies with PCS and IS20 and using the computer program G∗power v3.1.2 (http://www.gpower.hhu.de/en.html), we estimated that a test of the 1-tailed hypothesis that vancomycin administration would reduce the concentration of PCS by the amount seen in our prior study after 1, 2, or 3 months of administration with alpha = 0.05 and power = 0.80 would require 13 subjects. We anticipated a 15% to 20% dropout rate and thus, we enrolled 15 patients in the study. Patients were required to have dialysis access via either fistula or arteriovenous graft to reduce the likelihood of bloodstream infection which might necessitate antibiotics during the course of the study.
Sample collection, storage, and analysis
Fifteen subjects with established ESKD receiving thrice-weekly hemodialysis in the River Renal Dialysis Unit at Bellevue Hospital gave informed consent. They were randomly assigned, in a paired, single-blinded fashion, to receive either vancomycin or placebo weekly, followed by a crossover to the alternative treatment (Supplementary Figure S1A). During the study, subjects were interviewed regularly about the development of new symptoms, including diarrhea or fever, changes in diet, sick home contacts, and changes in medication use.
Blood
Predialysis samples were collected as baseline values on treatment day 0. Each subject in the antibiotic group ingested a capsule containing 250 mg vancomycin weekly, after dialysis, and subjects in the control group received a placebo capsule. Subsequent predialysis blood samples (5 ml) were obtained weekly at the end of weeks 1, 2, 3, 4, 8, and 12 before crossover to the alternate treatment. Blood samples were collected from the afferent dialysis line into ethylenediamine tetraacetic acid anticoagulant, kept on ice until they were centrifuged at 2500 rpm, and the plasma stored at –80o C. The concentration of a panel of solutes, including protein-bound anions, a cation (trimethylamine N-oxide [TMAO]), and several essential amino acids were measured by mass spectrometer high-performance liquid chromatography.21 The panel of solutes measured includes protein-bound anions, several amino acids, TMAO, and a cationic solute. This panel provided a range of solutes whose response to altering the gut microbiome was of interest.
Stool
Stool samples were collected from all subjects at home 2 days before starting treatment, and subsequent samples were obtained on the day preceding blood sampling in weeks 4, 8, and 12 of the treatment period. Stool samples were collected at home in 2 plastic vials, 1 prefilled with RNAlater (an RNA and DNA stabilization solution) and another empty. Samples were stored, with and without RNAlater, at –80o C.
To test for the emergence of vancomycin resistant enterococci, stool samples (collected in empty vials) were plated on colistin nalidixic acid blood agar and incubated at 35 °C to 37 °C for 24 to 48 hours under room air conditions. Colonies suspicious for Enterococci were Gram stained. Those colonies showing Gram-positive cocci were subjected to identification using the VITEK MS (mass spectrometry, MALDI) (BioMerieux, France).22 Colonies identified as Enterococcus sp. were tested for vancomycin susceptibility by the disc diffusion method using disks containing 30 μg of vancomycin.
For microbiome analysis, DNA was extracted from fecal samples using the Mo Bio Laboratories (Carlsbad, CA) extraction kit following the manufacturer’s instructions. For amplicon library generation, the V4 region of the 16S rRNA gene was amplified with gene-specific primers, as described.23 The reverse amplification primers contained a 12-base pair Golay barcode for multiplexed sequencing runs.24 Amplicons were prepared in triplicate, and DNA concentrations were measured using QuantiT PicoGreen dsDNA Assay kit (Invitrogen, Carlsbad, CA) and pooled at equal DNA quantity. After samples were combined in subpools of up to 96 samples, excess primers were removed with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany). DNA concentrations in these subpools were quantified with the Qubit high-sensitivity dsDNA Assay (Invitrogen) and combined to an equal concentration. The 254 bp V4 region was sequenced using the Illumina MiSeq 2 × 150 bp platform at NYU Langone Health. Operational taxonomic units (OTUs) were picked using GreenGenes 13_8 for reference, using the DADA2 pipeline as part of the QIIME 2 pipeline.25
Data management and statistical analyses
All data were initially collected as Excel spreadsheets, comma-delimited datafiles, ∗.microbiome, or ∗.qua (QIIME2) datasets. They were imported into R version 3.5.3. and reformatted for statistical analysis. Serum levels were analyzed by a mixed model analysis in which cubic polynomial models were fitted with treatment and week as fixed factors and subject as a random factor. Microbiome data were assessed for alpha and beta diversity using the R Phyloseq library.26 The effect of treatment and subject on microbiome diversity was assessed by constrained correspondence analysis.
The data were rarefied using the rarefy_even_depth() function in the R Phyloseq library. Depths of 800, 1600, 3200, and 6400 were examined. A depth of 800 was selected for diversity analysis as showing the best balance between the stability of OTU count and sample dropout as recommended by Goodrich.27 For the recovery analysis, a depth of 480 was used.
We then compared the relative abundance of the top 10 most abundant genera (rarefied at 1000) before and at 4 weeks postantibiotics using paired t test. To identify the taxa whose contribution to the total microbiome increased or decreased significantly during treatment with vancomycin or following discontinuation of vancomycin, we performed an analysis using the R package DESeq2.28 DESeq2 is specifically designed to account for the specifics of high-throughput sequencing count data, such as nonnormality and a dependence of the variance on the mean, and the challenging analysis of a small number of samples. DESeq2 is a more powerful method than inferential methods that treat each gene separately, to describe differential expression of RNA-seq data. DESeq2 fits a negative binomial regression model to transformed count data for each OTU in the microbiome samples. All post hoc tests were performed controlling the family-wise error rate at 0.05 with the Tukey’s honestly significant difference, Bonferroni, or Dunn-Sidak methods. In the case of DESeq2, the false discovery rate was controlled at 0.1 using the Benjamini-Hochberg method. The DESeq2 analysis used the design “~ SubjectID + Treatment” where SubjectID is a unique subject identifier and Treatment is a variable with values of “Placebo” or “Vancomycin” as defined earlier. Time was not included as a variable. Wald statistics were used to assess statistical significance at a false discovery rate of 0.1. Adequacy of the fitting algorithm was assessed by visual inspection of the plotted dispersion estimates (using the function DESeq::plotDispEsts).
To test the correlation between the microbiota and solute levels, we examined the correlation between contemporaneous OTU counts and solute level in the 10 subjects. We identified significant correlations between the solute levels and the OTUs that mapped to bacterial genera we identified as changing significantly in our DESeq analysis. Benjamini-Hochberg false discovery rate of 0.1 was used.
Results
Subjects
We enrolled 15 subjects, 1 of whom exhibited a transient rash while taking vancomycin and was dropped from the study. Ten subjects provided sufficient plasma and stool samples to permit us to examine the effect of vancomycin on plasma solute levels (Table 1). No subjects received antibiotics or proton pump inhibitors during the study.
Table 1.
Demographics and other participant data
| Gender (male/female) | 9/1 |
| Mean age (years ± SD) | 56.6 ± 10.1 |
| Ethnicity (White/Black/Asian/Hispanic/>1 or other) | 1/2/2/4/1 |
| Dialysis shift (MWF/TTS)∗ | 5/5 |
| Time on HD (mean years ± SD) | 7.4 ± 7.7 |
| Length of HD session (mean hours ± SD) | 3.6 ± 0.4 |
| Heparin with HD (yes/no/unknown) | 6/3/1 |
| Cause of ESRD (DM, HTN, DM, and HTN, unknown)∗∗ | 4,4, 1, 1 |
| Urine production (yes/no) | 6/4 |
| Diagnosis of HTN (yes/no) | 9/1 |
| DM type II (yes/no) | 4/6 |
| Coronary artery disease (yes/no) | 2/8 |
| Diet (nonrestricted/low sodium and/or low potassium) | 6/4 |
| In the past month, have you experienced a change in bowel habits? (yes/no) | 2/8 |
DM, diabetes mellitus; HD, hemodialysis; HTN, hypertension; MWF, Monday, Wednesday, Friday; TTS, Tuesday, Thursday, Saturday.
MWF: Monday, Wednesday, Friday. TTS: Tuesday, Thursday, Saturday,
DM: Diabetes Mellitus, HTN: Hypertension
Changes in solute levels
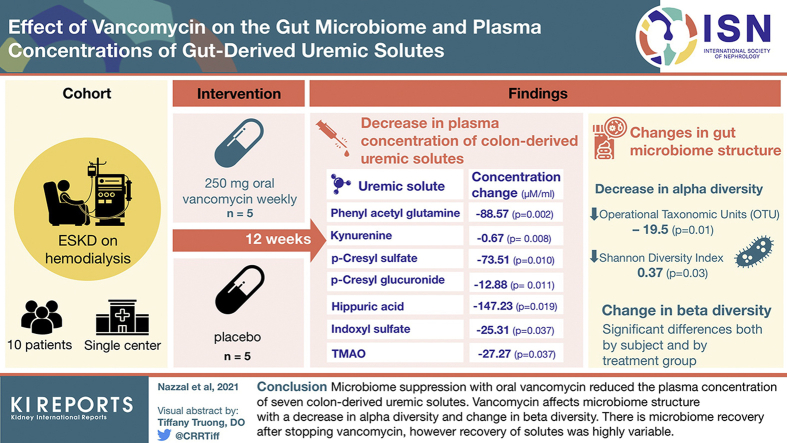
Five subjects (A1, A2, A4, A5, and A8) received vancomycin as the initial treatment, and 5 subjects (Z1, Z3, Z4, Z5, and Z6) initially received placebo. There was no “washout period” following vancomycin, thus for our analysis of the vancomycin effect, we compared changes in solute levels in all 10 subjects while receiving vancomycin to the changes seen in the 5 subjects while receiving placebo as the initial treatment. (Figure 1, Supplementary Figure S2). Over the first 4 weeks of vancomycin administration, concentrations of phenyl acetyl glutamine, kynurenine, p-cresyl sulfate, p-cresyl glucuronide, hippuric acid, indoxyl sulfate, and TMAO all decreased significantly (Figure 1a, Table 2). These 7 solutes are known to be at least partially derived from the gut microbiome.14 Four of these solutes—IS, PCS, phenyl sulfate, and hippurate—have been reported to be absent from the blood or stool of germ-free mice.29 Two of them, IS and PCS, are classified as colon-derived uremic solutes because their concentrations were reduced or absent from the plasma of ESKD patients who had undergone a prior total colectomy.14,30 Among the 7 solutes that significantly declined in concentration during vancomycin administration, there were positive correlations between a subject’s initial plasma level and the magnitude of that subject’s decline. The greater the initial concentration in a subject, the greater the decrease seen with vancomycin. Significant correlations were observed for hippuric acid (Spearman R = 0.83, P < 0.001), kynurenine (R = 0.82, P = 0.02), phenylacetylglutamine (R = 0.84, P < 0.001), and p-cresyl glucuronide (R = 0.77, P < 0.001). Similar but nonsignificant correlations were seen with the other three solutes.
Figure 1.
Changes in concentration of solutes from 10 subjects (126 samples) over the 12-week treatment periods. Changes in μmols/ml are shown on the right axis and changes in units of standard deviation on the left axis. Differences in the response to placebo and to oral weekly vancomycin were compared by likelihood ratio tests of mixed models. Red lines depict individual responses. Black lines and shaded areas depict fitted mixed models and 95% confidence limits. (a) Solutes for which the response to vancomycin significantly differed from that to placebo. (b) Solutes for which the response to vancomycin did not differ from that to placebo. The magnitude of individual subjects’ decline in solute concentration during vancomycin administration was statistically significantly correlated with their initial plasma concentration
Table 2.
Summary of statistical analysis of changes in solute concentrations
| Solute | Vancomycin effecta | Change in level ,Week 0–4 (μM/ml) |
|||
|---|---|---|---|---|---|
| Placebo |
Vancomycin |
||||
| Change | Pb | Change | Pb | ||
| Phenyl acetyl Glutamine | <0.001 | 16.41 | 0.683 | –88.57 | 0.002 |
| Kynurenine | 0.016 | 0.15 | 0.668 | –0.67 | 0.008 |
| p-cresyl sulphate | <0.001 | 0.64 | 0.98 | –73.51 | 0.010 |
| p-cresyl glucuronide | <0.001 | 1.83 | 0.792 | –12.88 | 0.011 |
| Hippuric acid | <0.001 | 72.73 | 0.406 | –147.23 | 0.019 |
| Indoxyl sulphate | <0.001 | 9.71 | 0.558 | –25.31 | 0.033 |
| TMAO | <0.001 | 12.38 | 0.498 | –27.27 | 0.037 |
| Indole-3-acetic acid | 0.093 | 1.06 | 0.699 | 4.00 | 0.041 |
| CMPF | 0.803 | 1.72 | 0.651 | 4.34 | 0.113 |
| Phenyl sulphate | 0.066 | 2.88 | 0.869 | 18.29 | 0.136 |
| Phenylalanine | 0.064 | 10.07 | 0.387 | –9.29 | 0.259 |
| Tyrosine | 0.339 | 8.01 | 0.363 | –4.73 | 0.447 |
| Kynurenic acid | 0.204 | 0.22 | 0.302 | –0.08 | 0.593 |
| Phenyl glucuronide | 0.452 | 0.19 | 0.852 | 0.36 | 0.627 |
| Tryptophan | 0.865 | 2.89 | 0.410 | 1.12 | 0.651 |
CMPF: 3-carboxy-4-methyl-5-propyl-2-furanpropionate; TMAO, trimethylamine N-oxide.
Rows are sorted by P values for change in vancomycin level (μM/ml) between 0 and 4 weeks. Solutes showing a significant decline in concentration with vancomycin are highlighted in green. The changes are estimated model means from the mixed polynomial models depicted in Figure 1.
Likelihood ratio test of mixed models (subjects as random variable) with fixed cubic polynomial terms compared with a similar model incorporating additive and interactive treatment effect terms. Shaded area shows solutes for which this comparison was statistically significant.
Tukey honest significant difference post hoc comparisons based on mixed model.
For 8 solutes—indole-3-acetic acid, 3-carboxy-4-methyl-5-propyl-2-furanpropionate, phenyl sulfate, phenylalanine, tyrosine, kynurenic acid, phenyl glucuronide, and tryptophan, the response to vancomycin was not significantly different from the response to placebo (Figure 1b, Table 2). Of these solutes, only phenyl sulfate is microbiome-derived.29
Changes in Microbiome during vancomycin administration
Forty-three samples were tested for enterococci; all were positive for vancomycin-sensitive enterococci. No resistant strains were isolated before or after vancomycin therapy.
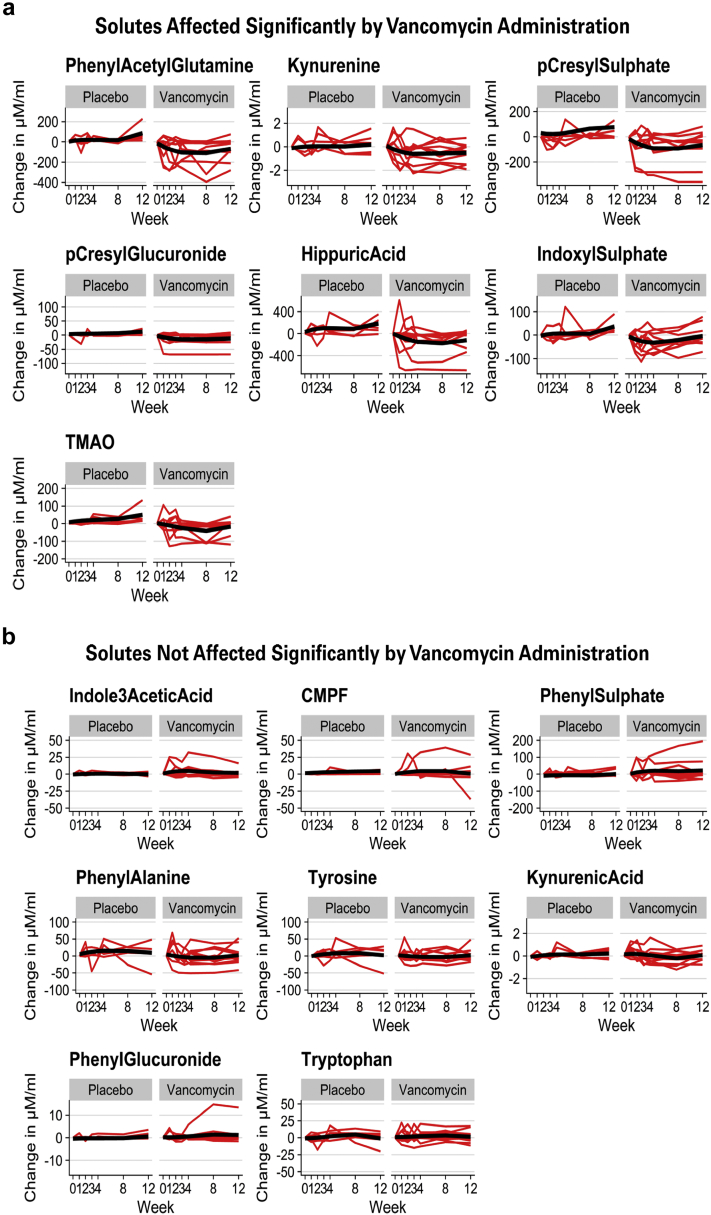
We next evaluated the effect of vancomycin on the microbiome structure. A reflection of the within-sample diversity, α-diversity relates to both the number of species in a sample and the quantitative distribution of those species. We computed the observed number of OTUs and the Shannon Diversity Index from the rarefied microbiome data. We analyzed each measure as the dependent variable in a mixed model with treatment week as a fixed and subject as a random effect (Figure 2a). The size of each index was compared between that seen while receiving placebo and that seen at each week as well as for the entire period. After adjustment for multiple comparisons, vancomycin was found to have reduced both indices significantly (mean reduction in OTUs while receiving vancomycin was –19.5, (95% confidence interval [CI]: –35.7 to –3.4, P = 0.01), and mean decline in Shannon Index was 0.37 (95% CI = –.71 to –0.03, P = 0.03).
Figure 2.
Microbiome diversity in treatment and placebo groups from 10 subjects (48 samples). (a) Alpha diversity. Total operational taxonomic units and Shannon indices versus weeks from starting vancomycin. The blue lines are mixed model estimates of the means for the placebo and the Vancomycin periods. Count data were rarefied to a depth of 800. (b) Beta diversity. Constrained correspondence analysis (CCA) of microbiome samples from subjects receiving weekly placebo or vancomycin over the course of the study. Count data were rarified to a depth of 800.
β-diversity captures the degree to which taxonomic membership or structure is shared between microbial communities. Because microbiome data are complex and multidimensional, we performed constrained correspondence analyses, a dimension reduction method, on the rarefied counts using a model corresponding to the experimental design with components for subject effect, treatment effect, and treatment-by-subject interaction. Total chi-squared for the OTU table was 13.51 with 19 degrees of freedom, and the modeled constraints accounted for 59% of the total chi-squared (7.98 with 19 degrees of freedom). Each of the three factors in the model was statistically significant at P < 0.001, indicating differences of beta diversity by subject and treatment group. Together the first two axes explain 11.2% of overall differences in beta diversity (Figure 2b). The plot shows clear clustering by subject (the subject effect) and consistent shifts in position while subjects are on antibiotics, indicating an effect of the antibiotic on beta diversity.
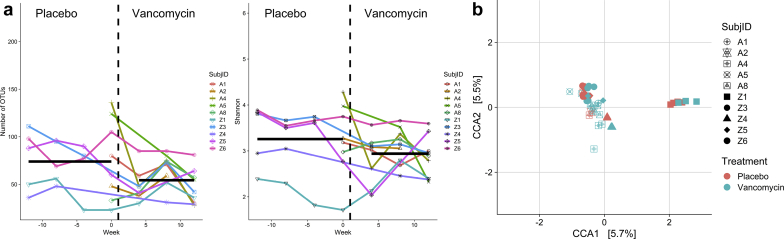
We then examined the effect of vancomycin treatment on the abundance of the topmost abundant genera (Supplementary Figure S3). Although we see changes in the average abundance of several genera, this difference was not statistically significant (Supplementary Table S1). The DESeq2 analysis was performed using all 36 available microbiome samples for the 7 patients who had contributed both week 0 (baseline) and week 4 (vancomycin) stool specimens. Placebo specimens included 16 stool samples from those participants who received placebo for 12 weeks before vancomycin as well as the baseline (week 0) samples of those participants who received vancomycin for 12 weeks before crossing over to placebo (Supplementary Figure 1B). Vancomycin specimens included 20 stool specimens taken while receiving vancomycin for 4, 8, or 12 weeks. We also identified taxa that changed in abundance while receiving vancomycin Using DESeq2 and a false discovery rate of 0.1, we identified 41 OTUs that declined significantly and 13 that increased significantly while receiving vancomycin (Figure 3). Multiple genera (Roseburia, Coprococcus, and Cuminococcus) from the family Lachnospiraceae significantly declined during vancomycin administration, whereas genus Bilophila increased significantly. We only detected genus enterococcus in 3 of the 10 subjects. RRA2 was the only subject where enterococcus was not detected at baseline and detected at 1 month after antibiotics. For the other 2 subjects (RRZ1 and RRZ2), it was detected only at baseline or while on placebo. There was no significant change in Enterococcus abundance by DESeq2 analysis.
Figure 3.
Taxa that increased or decreased in abundance while receiving vancomycin in 7 subjects (36 samples).
Correlation between bacterial taxa and solute levels
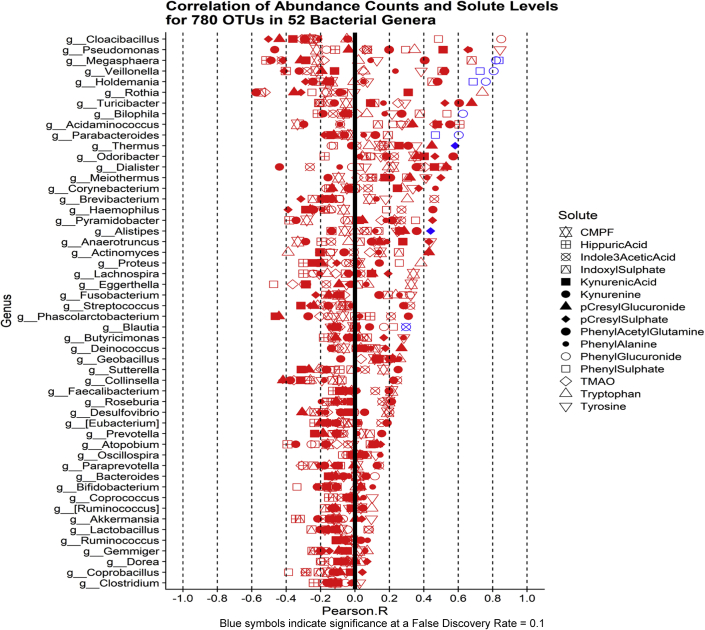
We examined, in all 10 patients (57 samples), the correlation between contemporaneous OTU count and solute levels for the 780 OTUs that mapped to the 54 bacterial genera we identified as changing significantly during the study. Among this set of 780 correlations using a Benjamini-Hochberg false discovery rate of 0.1, we identified 12 significant correlations between solute level and OTU count. Genera Megasphaera, Veillonella, Holdemenia, Thermus, and Parabacteroides were correlated with level of phenyl sulfate. Genera Megasphaera, Veillonella, Holdemania, Bilophila, and Parabacteroides were correlated with the level of phenyl glucuronide. Levels of PCS were correlated with genera Alistipes and Thermus, whereas indole 3 acetic acid is associated with Blautia counts (Figure 4).
Figure 4.
Correlation between contemporaneous operational taxonomic units count and solute levels in 10 subjects (57 stool and 57 serum samples). Significant correlations are shown in blue, nonsignificant correlations in red.
Microbiome and solute recovery after completion of vancomycin
We examined the recovery of the stool microbiome following discontinuation of vancomycin. Stool samples for this period were available for four subjects (A1, A4, A7, and A8).
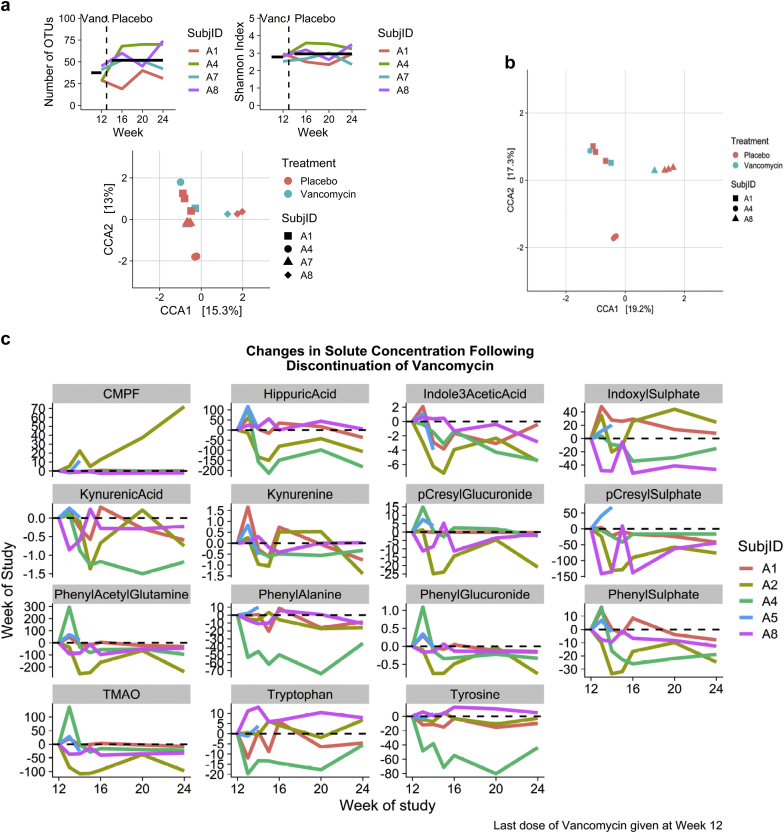
After rarefication to the minimum sample count, the mean number of OTUs at 12 weeks, the time of vancomycin discontinuation, was 35.8. At 16 weeks it had increased to 49.8, at 20 weeks to 51.5, at 24 weeks to 54.2. However, post hoc testing comparing the 16-, 20-, and 24-week averages to that at 12 weeks was nonsignificant. With respect to the Shannon index, the likelihood ratio test for the addition of week to the mixed model was also nonsignificant (Figure 5A).
Figure 5.
Recovery of microbiome and solutes after discontinuation of Vancomycin. (a) Alpha diversity in 3 subjects (12 samples). Change in operational taxonomic units (OTU) numbers and Shannon index after discontinuation of Vancomycin. (b) Beta diversity in 3 subjects (12 samples). Constrained correspondence analysis (CCA) of stool microbiomes. The model included effects for Treatment, Subject, and Subject by Treatment Interactions. (c) Solutes recovery data in each of the 5 individuals (41 samples). Data shown for weeks 12, the last week of vancomycin administration and for weeks 16, 20, and 24 thereafter.
Beta diversity in the recovery period was evaluated using constrained correspondence analysis. Together the first 2 axes accounted for 28.3% of differences in beta diversity, and the model accounted for 65% of the total chi-squared. Each of the factors in the model was statistically significant (Subject, P < 0.001, Treatment, P = 0.007, Subject by Treatment Interaction, P =0.001) with shifts across CCA1 and CCA2 illustrating the effect of stopping vancomycin (Figure 5B).
We then examined whether the levels of solutes increased after stopping vancomycin, indicating recovery of the taxa that produce precursors of uremic solutes (Figure 5c). Solutes increased in concentration in some subjects, for example, levels of IS in patient A1; however, no overall pattern of recovery was found (Supplementary Figure S4). Hippuric acid, indole-3-acetic acid, kynurenic acid levels, phenyl sulphate, and TMAO declined significantly after discontinuation of vancomycin (Figure 5C).
Discussion
Although maintenance hemodialysis, widely available in the United States and throughout the world, achieved dramatic improvement in the acute management of uremia, the early reports from Belding Scribner’s group31 indicated that accelerated cardiovascular disease was prevalent in patients undergoing maintenance hemodialysis. To date, accelerated cardiovascular disease is thought to be responsible for the nearly 50% 3-year mortality in patients with ESKD.32 Many modifications of dialysis techniques that reduce the plasma concentrations of blood urea and creatinine, assumed to be surrogates for uremic toxins, have failed to significantly reduce the poor cardiovascular outcomes.33 The report by the Eutox consortium in 2003 identified more than 100 compounds that were at a greater concentration in the plasma of patients with identified impairment of renal function.1,34 At least some of these toxins have been identified as gut-derived, and their levels have been associated with cardiovascular disease.7,14
Multiple therapies have been developed to reduce the levels of these toxins. Oral administration of microcrystalline charcoal directed at reducing the concentration of indole resulted in reduced plasma concentration of uremic solutes in only a small subgroup of patients with end-stage renal disease.16 Meijers et al. administered a prebiotic (oligofructose-enriched inulin) with the goal of altering the substrate available to gut microbes with a small effect.17,19 In PD patients, Gao et al. showed that p-inulin supplementation was associated with microbiome, microbial metabolic pathways, and plasma metabolome.35 Mishima et al. reported that the SGLT inhibitor, canagliflozin, an inhibitor of the renal glucose transporter SGLT 2, reduced the production of IS in a rodent renal failure model, an effect that might be attributable to partial inhibition of SGLT 1 in the bowel and delivery of more carbohydrate to the distal colon.18 To the best of our knowledge, we were the first to establish the effect of microbiome suppression using an antibiotic on the levels of uremic toxins in ESKD patients.
In this randomized controlled trial, we showed that a weekly dose of vancomycin resulted in a reduction in the plasma concentration of 7 colon-derived solutes associated with changes in the microbiome structure and multiple bacterial taxa. The observation that only microbiome-derived solutes exhibited a reduction in plasma concentration during vancomycin administration strongly suggests that the effect is attributable to changes in the composition or metabolic activity of some of the gut microbiome, rather than either an effect on gut motility or an effect on the renal excretion of solutes which might have been expected to affect all the solutes. The very great variation in plasma concentrations of IS and PCS among patients with roughly comparable degrees of reduced glomerular filtration, which we have previously noted20 is most consistent with variations in the gut microbiome composition or diet as important determinants of the plasma concentration of uremic solutes.
The decrease in the concentration of 7 solutes occurred quickly. It was maximal within the first month of vancomycin administration, consistent with the recognition that these solutes are dependent on microbial metabolism for their synthesis.29 It is notable that the vancomycin-induced decline in solute levels correlated with initial concentration consistent with the dependence of solute levels on gut microbiota.
Our earlier study20 suggested that genera prominent in the production of tryptophanase were selectively affected by a single 250-mg dose of vancomycin administered orally. In the current study, bacterial taxa from the genera Clostridium and Bacteroides significantly decreased with vancomycin. These taxa are known to produce indole, the precursor of IS.36,37 The decline in multiple taxa is consistent with a broad effect of the vancomycin on the microbiome. We also found correlation between the levels of several solutes and bacterial taxa. To further explore the mechanistic role of microbiota in the generation of uremic toxins, metagenomic and metatranscriptomic analyses are needed. These analyses will allow identify taxa and pathways involved in URS production.38
We observed a decline in several genera of the Lachnospiraceae family and an increase in the genus Bilophila. Lachnospiraceae decline as a response to vancomycin treatment has been described in other studies.39 Because species in this family have been identified as important short-chain fatty acid producers,40,41 particularly butyric acid, it will be important to study the effects of this low-dose vancomycin on short-chain fatty acid production and the host physiology. Similarly, the functional significance in Bilophila increase need to be evaluated if vancomycin is to be used as a therapy for reduction of uremic toxins. Bilophila is an important organism involved in fat metabolism and has been shown to aggravate high fat diet–induced metabolic dysfunctions in mice.42
Our data show that there is microbiome recovery after stopping vancomycin. However, we could not demonstrate a clear relationship between microbiome recovery and an increase in solute levels in all subjects. Recovery in the solutes is highly variable from individual to individual, possibly due to differential acquisition of microbes providing the needed precursors; therefore, a much larger population is needed to study this question. The lack of uniform recovery of solutes derived from the microbiome in different subjects is consistent with the data by Isaac et al., who described drastic and consistent changes in the gut microbiome in response to daily vancomycin; however, the microbiota’s recovery rate was considerably different among subjects.43 The continuous decline in some solutes suggests long-lasting effects of the vancomycin on the microbiome in at least some subjects, as previously described.44
The lack of VRE emergence or significant side effects over the 3-month period suggests that the treatment might be safe for longer-term use. Data suggest that gut abundance of Enterococcus is associated with future development of Enterococcus infections.45,46 In our study, there was no significant change in Enterococcus abundance by DESeq2 analysis. Similarly, in the study by Basolo et al., there was no enrichment of Enterococcus genus with oral vancomycin.39 Oral vancomycin has been shown to significantly reduce the gut microbiome diversity and change its composition (Reijnders et al., 2016; Vrieze et al., 2014).39,47,48 In obese patients, it also reduced the conversion of primary to secondary bile acids and the production of SCFAs in the gut.47 Vancomycin also significantly affects host physiology by decreased bile acid dehydroxylation and peripheral insulin sensitivity in subjects with metabolic syndrome.48 However, these studies used a higher dose of daily vancomycin than what we used in our trial. Therefore, future studies need to evaluate the effects of the lower dose of vancomycin on bile acids, short chain fatty acids, and host physiology.
This study has several limitations. Although it is a randomized study, the sample size is small. Of the 15 subjects randomized, only 10 provided a complete set of plasma and fecal samples. In addition, the data for those subjects who crossed over from the vancomycin treatment to the placebo showed substantial carryover effects that precluded pooling their placebo period data with that of those who were initially given the placebo. We also did not account for residual renal function in our analysis; however, because of randomization, we do not expect it will have a significant effect on our final conclusions. Finally, although we screened for changes in dietary habits during the study, dietary intake, an important determinant of the microbiome and serum solutes, was not fixed during the trial.
Disclosures
The authors declared no competing interests.
Acknowledgments
Supported in part by the NYU CTSA grant UL1TR000038 from the National Center for the Advancement of Translational Science (NCATS), the Oxalosis and Hyperoxaluria Foundation-American Society of Nephrology Career Development Award (to L.N.).
Footnotes
Figure S1. Study design.
Figure S2. Changes in concentration of solutes from 10 subjects over the 12-week treatment periods.
Figure S3. Vancomycin-related changes in relative abundance of genera.
Figure S4. Indoxyl sulfate recovery after 12 weeks of vancomycin.
Table S1. Vancomycin-related changes in relative abundance of most abundant genera.
Supplementary Material
Figure S1. Study design.
Figure S2. Changes in concentration of solutes from 10 subjects over the 12-week treatment periods.
Figure S3. Vancomycin-related changes in relative abundance of genera.
Figure S4. Indoxyl sulfate recovery after 12 weeks of vancomycin.
Table S1. Vancomycin-related changes in relative abundance of most abundant genera.
References
- 1.Vanholder R., De Smet R., Glorieux G. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63:1934–1943. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 2.Duranton F., Cohen G., De Smet R. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23:1258–1270. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wikoff W.R., Nagle M.A., Kouznetsova V.L. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1) J Proteome Res. 2011;10:2842–2851. doi: 10.1021/pr200093w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu I.W., Hsu K.H., Lee C.C. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol Dial Transplant. 2011;26:938–947. doi: 10.1093/ndt/gfq580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gondouin B., Cerini C., Dou L. Indolic uremic solutes increase tissue factor production in endothelial cells by the aryl hydrocarbon receptor pathway. Kidney Int. 2013;84:733–744. doi: 10.1038/ki.2013.133. [DOI] [PubMed] [Google Scholar]
- 6.Ravid J.D., Chitalia V.C. Molecular mechanisms underlying the cardiovascular toxicity of specific uremic solutes. Cells. 2020;9:2024. doi: 10.3390/cells9092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mair R.D., Sirich T.L., Meyer T.W. Uremic toxin clearance and cardiovascular toxicities. Toxins (Basel) 2018;10 doi: 10.3390/toxins10060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapp N., Evenepoel P., Stenvinkel P. Uremic toxins and vascular calcification-missing the forest for all the trees. Toxins (Basel) 2020;12 doi: 10.3390/toxins12100624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velasquez M.T., Centron P., Barrows I. Gut microbiota and cardiovascular uremic toxicities. Toxins (Basel) 2018;10 doi: 10.3390/toxins10070287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nigam S.K., Bush K.T. Uraemic syndrome of chronic kidney disease: altered remote sensing and signalling. Nat Rev Nephrol. 2019;15:301–316. doi: 10.1038/s41581-019-0111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C.H., Lai Y.H., Kuo C.H. Association between serum indoxyl sulfate levels and endothelial function in non-dialysis chronic kidney disease. Toxins (Basel) 2019;11 doi: 10.3390/toxins11100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opdebeeck B., Maudsley S., Azmi A. Indoxyl sulfate and p-cresyl sulfate promote vascular calcification and associate with glucose intolerance. J Am Soc Nephrol. 2019;30:751–766. doi: 10.1681/ASN.2018060609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evenepoel P., Meijers B.K., Bammens B.R. Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl. 2009:S12–S19. doi: 10.1038/ki.2009.402. [DOI] [PubMed] [Google Scholar]
- 14.Mair R.D., Sirich T.L., Plummer N.S. Characteristics of colon-derived uremic solutes. Clin J Am Soc Nephrol. 2018;13:1398–1404. doi: 10.2215/CJN.03150318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rastall R.A., Gibson G.R., Gill H.S. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol Ecol. 2005;52:145–152. doi: 10.1016/j.femsec.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Schulman G., Berl T., Beck G.J. Randomized placebo-controlled EPPIC trials of AST-120 in CKD. J Am Soc Nephrol. 2015;26:1732–1746. doi: 10.1681/ASN.2014010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijers B.K., De Preter V., Verbeke K. p-Cresyl sulfate serum concentrations in haemodialysis patients are reduced by the prebiotic oligofructose-enriched inulin. Nephrol Dial Transplant. 2010;25:219–224. doi: 10.1093/ndt/gfp414. [DOI] [PubMed] [Google Scholar]
- 18.Mishima E., Fukuda S., Kanemitsu Y. Canagliflozin reduces plasma uremic toxins and alters the intestinal microbiota composition in a chronic kidney disease mouse model. Am J Physiol Renal Physiol. 2018;315:F824–F833. doi: 10.1152/ajprenal.00314.2017. [DOI] [PubMed] [Google Scholar]
- 19.Sirich T.L., Plummer N.S., Gardner C.D. Effect of increasing dietary fiber on plasma levels of colon-derived solutes in hemodialysis patients. Clin J Am Soc Nephrol. 2014;9:1603–1610. doi: 10.2215/CJN.00490114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazzal L., Roberts J., Singh P. Microbiome perturbation by oral vancomycin reduces plasma concentration of two gut-derived uremic solutes, indoxyl sulfate and p-cresyl sulfate, in end-stage renal disease. Nephrol Dial Transplant. 2017;32:1809–1817. doi: 10.1093/ndt/gfx029. [DOI] [PubMed] [Google Scholar]
- 21.de Loor H., Poesen R., De Leger W. A liquid chromatography–tandem mass spectrometry method to measure a selected panel of uremic retention solutes derived from endogenous and colonic microbial metabolism. Anal Chim Acta. 2016;936:149–156. doi: 10.1016/j.aca.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 22.Febbraro F., Rodio D.M., Puggioni G. MALDI-TOF MS Versus VITEK((R))2: Comparison of systems for the identification of microorganisms responsible for bacteremia. Curr Microbiol. 2016;73:843–850. doi: 10.1007/s00284-016-1121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pebenito A.M., Liu M., Nazzal L. Development of a humanized murine model for the study of oxalobacter formigenes intestinal colonization. J Infect Dis. 2019;220:1848–1858. doi: 10.1093/infdis/jiz370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox L.M., Yamanishi S., Sohn J. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E., Rideout J.R., Dillon M.R. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich J.K., Di Rienzi S.C., Poole A.C. Conducting a microbiome study. Cell. 2014;158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishima E., Fukuda S., Mukawa C. Evaluation of the impact of gut microbiota on uremic solute accumulation by a CE-TOFMS-based metabolomics approach. Kidney Int. 2017;92:634–645. doi: 10.1016/j.kint.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Aronov P.A., Luo F.J., Plummer N.S. Colonic contribution to uremic solutes. J Am Soc Nephrol. 2011;22:1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindner A., Charra B., Sherrard D.J. Accelerated atherosclerosis in prolonged maintenance hemodialysis. N Engl J Med. 1974;290:697–701. doi: 10.1056/NEJM197403282901301. [DOI] [PubMed] [Google Scholar]
- 32.Saran R., Robinson B., Abbott K.C. US Renal Data System 2019 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis. 2020;75:A6–A7. doi: 10.1053/j.ajkd.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Eknoyan G., Beck G.J., Cheung A.K. Effect of dialysis dose and membrane flux in maintenance hemodialysis. N Engl J Med. 2002;347:2010–2019. doi: 10.1056/NEJMoa021583. [DOI] [PubMed] [Google Scholar]
- 34.Vanholder R., Baurmeister U., Brunet P. A bench to bedside view of uremic toxins. J Am Soc Nephrol. 2008;19:863–870. doi: 10.1681/ASN.2007121377. [DOI] [PubMed] [Google Scholar]
- 35.Gao B., Alonzo-Palma N., Brooks B. A pilot study on the effect of prebiotic on host-microbial co-metabolism in peritoneal dialysis patients. Kidney Int Rep. 2020;5:1309–1315. doi: 10.1016/j.ekir.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elsden S.R., Hilton M.G., Waller J.M. The end products of the metabolism of aromatic amino acids by Clostridia. Arch Microbiol. 1976;107:283–288. doi: 10.1007/BF00425340. [DOI] [PubMed] [Google Scholar]
- 37.Wang G., Huang S., Wang Y. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci. 2019;76:3917–3937. doi: 10.1007/s00018-019-03190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joossens M., Faust K., Gryp T. Gut microbiota dynamics and uraemic toxins: one size does not fit all. Gut. 2019;68:2257–2260. doi: 10.1136/gutjnl-2018-317561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basolo A., Hohenadel M., Ang Q.Y. Effects of underfeeding and oral vancomycin on gut microbiome and nutrient absorption in humans. Nat Med. 2020;26:589–598. doi: 10.1038/s41591-020-0801-z. [DOI] [PubMed] [Google Scholar]
- 40.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 41.Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 42.Natividad J.M., Lamas B., Pham H.P. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun. 2018;9:2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isaac S., Scher J.U., Djukovic A. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J Antimicrob Chemother. 2017;72:128–136. doi: 10.1093/jac/dkw383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magruder M., Sholi A.N., Gong C. Gut uropathogen abundance is a risk factor for development of bacteriuria and urinary tract infection. Nat Commun. 2019;10:5521. doi: 10.1038/s41467-019-13467-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taur Y., Xavier J.B., Lipuma L. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reijnders D., Goossens G.H., Hermes G.D. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 2016;24:63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Vrieze A., Out C., Fuentes S. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.