Summary
The “omnigenic” model of the genetic architecture of complex traits proposed two categories of causal genes, core and peripheral. Core genes are hypothesized to directly regulate disease and may serve as therapeutic targets. Using a cell-type and timepoint-specific gene co-expression network for mineralizing osteoblasts, we identify a co-expression module enriched for genes implicated by bone mineral density (BMD) GWAS, correlated with in vitro osteoblast mineralization, and associated with skeletal phenotypes in human monogenic disease and mouse knockouts. Four genes from this module (B4GALNT3, CADM1, DOCK9, and GPR133) are located within BMD GWAS loci with colocalizing expression quantitative trait loci (eQTL) and exhibit altered BMD in mouse knockouts, suggesting that they are causal genetic drivers of BMD in humans. Our network-based approach identifies a “core” module for BMD and provides a resource for expanding our understanding of the genetics of bone mass.
Keywords: osteoporosis, co-expression network, GWAS, bone mineral density, core genes
Introduction
Osteoporosis is a disease characterized by low bone mineral density (BMD) and an increased risk of fracture (Black and Rosen, 2016). Worldwide, it is one of the most common diseases, affecting over 200 million individuals and causing 8.9 million fractures annually (Johnell and Kanis, 2006). Although osteoporosis is a multifactorial disease influenced by both environmental and genetic variation, fracture-related traits are influenced, in large part, by genetics (h2>0.5) (Ralston and de Crombrugghe, 2006; Ralston and Uitterlinden, 2010; Zheng et al., 2011). Over the last decade large-scale genome wide association studies (GWASs) have begun to dissect the genetics basis of bone traits with a primary focus on BMD (Hsu et al., 2020; Sabik and Farber, 2016) . These studies have been tremendously successful, identifying over 1100 independent BMD associations (Estrada et al., 2012; Kemp et al., 2017; Morris et al., 2018). However, despite the wealth of genetic signals, the genes and mechanisms through which these associations impact bone remain largely unknown (Hsu et al., 2020; Sabik and Farber, 2016).
Recently, the “omnigenic model” was proposed as a framework for understanding the genetic architecture of complex traits, such as BMD (Boyle et al., 2017a; Liu et al., 2019). The model posits that all genes expressed in disease-relevant cell-types have the potential to contribute to disease variation. One of the key concepts of the omnigenic model is the classification of causal genes as either “core” or “peripheral”. Core genes are predicted to directly modulate traits; whereas, peripheral genes are expected to impact traits via their effects on networks of core genes (Liu et al., 2019). The distinction between core and peripheral genes is logical given the evidence demonstrating that the contributions of genes to a disease or phenotype are not equal. As an example, RUNX2 is a transcription factor and master regulator of osteoblast activity and bone formation that initiates a transcriptional program absolutely required for the formation of a mineralized skeleton (Komori, 2009). In contrast, hundreds of genes have been identified participating in myriad pathways whose absence has subtle, often context-dependent (such as age and sex), effects on bone (Estrada et al., 2012; Kemp et al., 2017; Morris et al., 2019). Furthermore, we know the same distinction lies in biological processes, some of which play an intimate role in the regulation of a trait, while others play minor accessory roles. Thus, the identification of causal genes from GWAS data and the labeling of such genes as core or peripheral has the potential to highlight previously undiscovered key regulatory genes for specific trait-related biological processes, which may be more ideal therapeutic targets.
There are two main challenges in the identification of core genes. The first is how to precisely define them (Boyle et al., 2017b; Cox, 2017; Liu et al., 2019; Wray et al., 2018). In the omnigenic model, a gene is defined as a “core” gene “if and only if the gene product (protein, or RNA for a noncoding gene) has a direct effect—not mediated through regulation of another gene—on cellular and organismal processes leading to a change in the expected value of a particular phenotype” (Boyle et al., 2017a; Liu et al., 2019). This statistical definition is convenient for explaining the omnigenic model, but difficult to utilize for the identification of core genes in practice. It is also very strict; e.g. is RUNX2, as described above, a core gene for BMD? Instead we propose to use a set of biologically motivated criteria to distinguish genes with core-like properties from those that are likely peripheral by leveraging known pathways and processes that are essential to a disease-associated trait. For example, we would expect the expression of genes with core-like properties operating in pathways of critical importance in the regulation of BMD to be correlated with BMD and their severe perturbation to have a substantial impact on BMD (e.g., monogenic disease genes).
The second challenge is designing a strategy to identify genes with core-like properties, since GWAS alone is incapable of determining whether a locus is driven by a core or peripheral gene. One of the primary tenets of the omnigenic model is that peripheral genes account for a substantial component of the heritability of a trait because their effects are amplified by interactions with networks of co-expressed core genes (Liu et al., 2019). If one expects core genes to be co-expressed then integrating the results of GWAS with co-expression networks, which reflect the transcriptional programs associated with the trait of interest, is a logical approach to identify modules of genes with core-like properties. A number of studies have already successfully used co-expression networks to inform GWAS, however this approach has not been used in the context of the omnigenic model (Civelek and Lusis, 2014; Eising et al., 2016; Kogelman et al., 2014; Mäkinen et al., 2014; Rau et al., 2017).
Here, we combine weighted gene co-expression network analysis (WGCNA) and BMD GWAS data to identify genes that are causal genetic drivers of BMD with core-like properties. Our approach used a co-expression network for mature, mineralizing osteoblasts which we hypothesized would allow us to identify core genes specific for the process of mineralization. We first identified network modules enriched for genes implicated by GWAS and partitioned BMD heritability and then used the following biologically motivated filters to identify modules enriched for genes with core-like properties (i.e. “core” modules): (1) correlation with in vitro mineralization (a process of fundamental importance to BMD), (2) enrichment for genes that, when knocked-out in mice, alter BMD, and (3) enrichment for monogenic skeletal disease genes. Our analysis identified a single module (referred to as the “purple” module) fulfilling all the proposed criteria of a core module. As would be expected of a core module for mineralization, the purple module was enriched for genes with well-known roles in osteoblast activity and bone formation. Furthermore, we identified two submodules of genes within the purple module that followed distinct patterns of expression across osteoblast differentiation, the early and the late differentiation submodule (EDS and LDS). We found that the LDS, relative to the EDS, was more enriched for genes with core-like properties. Supporting the hypothesis that many LDS genes are causal genetic drivers, we observed that lead BMD SNPs located in GWAS loci harboring an LDS gene were more likely to overlap active regulatory elements in osteoblasts. Further characterization of the LDS identified four genes (B4GALNT3, CADM1, DOCK9, and GPR133) located within BMD GWAS loci that had colocalizing human eQTL and altered BMD in mouse knockout studies. We anticipate that this integrative approach will aid in the search for genes with core-like properties and pathways underlying BMD and risk of fracture.
Results
I. Construction of a co-expression network reflecting transcriptional programs in mineralizing osteoblasts
The goal of this work was to use a cell- and stage-specific co-expression network to identify genes with core-like properties that are causal for BMD GWAS associations. We chose to focus on generating a co-expression network using transcriptomic data from a single cell-type at a single-time point during differentiation: mature, mineralizing osteoblasts. We hypothesized this would allow us to focus on genes with core-like properties in the context of mineralization, a process critical in the regulation of BMD. We began by using WGCNA to construct a co-expression network using transcriptomic profiles generated from mineralizing primary calvarial osteoblasts from 42 strains of Collaborative Cross (CC) mice (Churchill et al., 2004). The CC is a panel of genetically diverse recombinant inbred strains. The resulting network consisted of 65 modules of transcripts, with an average of 292 transcripts representing 266 unique genes per module (Figure 1 and Table S2). Each co-expression module was distinguished by its assigned color (e.g., the purple module).
Figure 1. Weighted gene co-expression network generated using transcriptomic profiles from mineralizing osteoblasts.
The network, built using expression profiles of 42 unique strains of Collaborative Cross mice, was composed of 65 modules of co-expressed genes, many of which were enriched for specific biological processes relevant to osteoblasts. Padj = significance of enrichment of module genes in GO category and n = number of genes in the module. See also Table S2 and S3.
To confirm that modules of transcripts produced by the co-expression analysis represented transcriptional programs reflecting specific biological processes, we assessed whether modules were enriched for genes associated with specific gene ontology (GO) terms (van Dam et al., 2018). Most network modules were enriched for general biological processes, such as the immune response (Padj = 6.6 x 10−36) in the blue module, mRNA metabolism (Padj = 7.8 x 10−9) in the darkolivegreen module, and chromatin remodeling (Padj = 1.9 x 10−4) in the grey60 module (Figure 1 and Table S3). However, as would be expected, there were a subset of modules enriched for genes involved in the activity of osteoblasts. For example, the cyan module was enriched for members of the Wnt signaling pathway (a key regulator of osteoblast activity) (Padj = 2.3 x 10−4), the turquoise module was enriched for genes encoding extracellular matrix proteins (Padj = 3.5 x 10−25) (such as genes encoding for collagens (Padj = 0.4 x 10−10)), and the purple module was enriched for genes involved in skeletal system development (Padj = 2.3 x 10−10) and osteoblast differentiation (Padj = 2.0 x 10−6) (Figure 1 and Table S2). Given that network modules represented distinct biological processes, including those involved in mineralization and osteoblast activity, we were confident it would provide a platform for identifying core genes related to mineralization that potentially underlie BMD GWAS associations.
II. Identification of co-expression modules enriched for genes implicated by GWAS
To identify modules of co-expressed genes informative for GWAS, we first determined if any of the 65 modules were enriched for genes that overlapped GWAS associations. Using data from the two largest GWASs performed at the time, one study of Dual Energy X-Absorptiometry (DEXA) derived areal BMD measures at the lumbar spine and femoral neck (Estrada et al., 2012) (“Estrada et al. GWAS”; N=32,961) and one study of ultrasound determined heel estimated BMD (eBMD) (Kemp et al., 2017) (“Kemp et al. GWAS”, N=142,487), we developed a list of 789 human genes (NEstrada = 179, NKemp = 701, (91 shared genes)) intersecting BMD GWAS loci. A total of 723 (92%) of these had mouse homologs in the network (Table S4). Of the 65 modules in the network, 13 were enriched for mouse homologs of human genes implicated by BMD GWAS (Fisher’s exact test, Padj < 0.05) (Table S5 and Figure 2A). Additionally, we performed stratified LD score regression by calculating the BMD heritability partitioned by SNPs surrounding genes in each module using the Kemp et al. GWAS (Finucane et al., 2015; Kemp et al., 2017). We found 16 modules enriched for partitioned BMD heritability, including nine of the 13 enriched for BMD GWAS implicated genes (Figure 2B and Table S5).
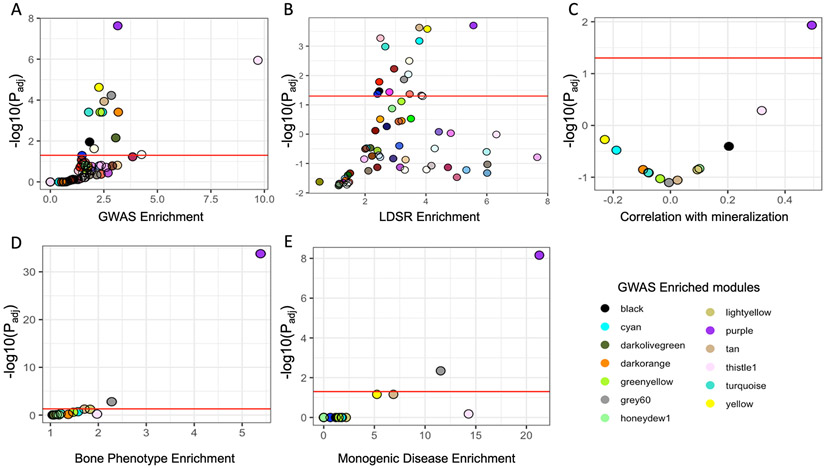
Figure 2. The purple module is enriched for genes with core-like properties.
(A) Module enrichments for genes overlapping a BMD GWAS association. (B) Enrichments for partitioned BMD heritability for each module determined using stratified LD score regression. (C) Correlation between each module eigengene and in vitro mineralization. (D) Module enrichments for genes that, when knocked out, produced a bone phenotype and (E) human monogenic bone disease genes. Red line in each panel represents Padj < 0.05. See also Figures S1 and S2.
III. The purple module is enriched for genes with core-like properties
Next, we focused on identifying which of the 13 modules identified above were enriched for genes with core-like properties. To accomplish this, we selected modules using biologically motivated criteria which likely reflected the properties of core genes. First, we compared the 13 module eigengenes with in vitro mineralization using osteogenic cultures from the same 42 CC strains used in the construction of the co-expression network (Figure S1). Only one, the purple module, had a pattern of expression that was significantly correlated with mineralization (r = 0.49, Padj = 0.012), suggesting the purple module was enriched for genes with a direct role in mineralization (Figure 2C and Figure S2).
Core genes have been broadly defined as those that directly influence a disease-relevant biological processes (Boyle et al., 2017a; Liu et al., 2019). Thus, severe perturbation of a core gene is more likely to result in a significant impact on a phenotype, as in the case of a mouse knockout or human monogenic disease. We identified all gene knockouts that produced a bone phenotype, defined as either a change in BMD, bone mineral content (BMC), abnormal bone morphology, or abnormal bone cell activity, by utilizing mouse knockout phenotype data from several databases (Bolser, 2004; Dyment et al., 2016; Freudenthal et al., 2016; Koscielny et al., 2014) (Table S6). Of the 13 modules enriched for BMD GWAS genes, two were enriched for genes whose deficiency impacted bone in mice (Figure 2D). The purple module was the most significantly enriched (OR=5.4, Padj = 1.6 x 10−34). We also compiled a list of 35 known drivers of monogenic bone diseases associated with osteoblast dysfunction, including osteogenesis imperfecta, hyperostosis, and osteosclerosis (Table S5) (Boudin and Van Hul, 2017; Johnson, 2016; Marini and Brandi, 2010; Robinson and Rauch, 2019; Rocha-Braz and Ferraz-de-Souza, 2016). Again, the purple module, containing 11 of 35 (31.4%) monogenic disease genes, was the most significantly enriched (OR = 21.3, Padj = 6.9 x 10−9) (Figure 2E). Together, these independent lines of evidence suggested the purple module was enriched for genes with core-like properties.
IV. New BMD GWAS associations further support the purple module as a core gene module
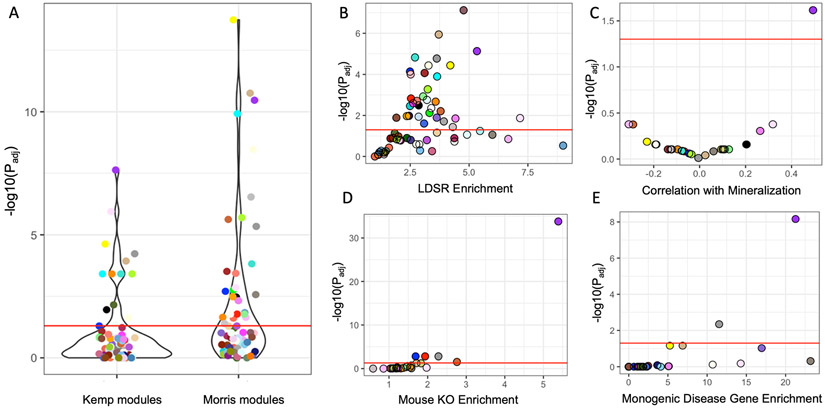
While we were analyzing the Kemp et al. GWAS data, an extension of this study, with a significantly increased eBMD sample size, was published (“Morris et al. GWAS”) (Morris et al., 2019). The Estrada et al. (N=32,961) and Kemp et al. (N=142,487) GWASs identified 56 and 307 conditionally independent associations, respectively (Estrada et al., 2012; Kemp et al., 2017). In comparison, the Morris et al. GWAS (N=426,824) identified 1103 eBMD associations; an increase of over 3.5-fold (Morris et al., 2019). The associations identified in the Morris et al. GWAS overlapped 1581 genes, as compared to 789 in the Estrada et al. and Kemp et al. GWASs (Table S3). Assuming the genetic architecture of BMD is consistent with the omnigenic model, we expected the inclusion of the Morris et al. GWAS data to increase the number of modules enriched for GWAS implicated genes. Consistent with this hypothesis, the number of modules enriched for GWAS-implicated genes doubled (NKemp = 13, NMorris = 26) using the Morris et al. GWAS (Figure 3A and Table S5). As observed in the first analysis, most (18/26, 69%) of the new modules enriched for GWAS-implicated genes were also enriched for partitioned BMD heritability (Table S5 and Figure 3C). These new modules were enriched for genes involved in general biological processes such as RNA splicing (brown module, Padj = 4.0 x 10−11), cell junctions (floralwhite module, Padj = 6.2 x 10−3), cell motor activity (orange, Padj = 6.6 x 10−3), the cell cycle (lightgreen, Padj = 3.2 x 10−4), ER to Golgi trafficking (salmon, Padj = 1.8 x 10−2), and the glycolytic process (red, Padj = 1.1 x 10−13), and not processes specific to osteoblast activity and/or mineralization (Table S3).
Figure 3. The purple module was the only core module after increasing the number of analyzed GWAS associations by 3.5-fold.
(A) A greater number of modules (N = 65) were identified as enriched for GWAS implicated genes in the Morris et al. GWAS versus the Kemp et al. GWAS. (B) Module enrichments for partitioned BMD heritability for each module determined using stratified LD score regression. (C) Correlation between each module eigengene and in vitro mineralization. Related to figure S2. (D) Module enrichments for genes that, when knocked out, produced a bone phenotype and (E) human monogenic bone disease genes. Red line in each panel represents Padj < 0.05. The most highly connected genes in the purple module also have higher alizarin red correlations and are more likely to be implicated by BMD GWAS. See also Figures S1 and S2.
Similar to the analysis of the Kemp et al. data, the purple module was among the most enriched for GWAS implicated genes (OR = 2.67, Padj = 3.4 x 10−11) (Figures 3A) and BMD heritability captured (OR = 5.8, Padj = 4.7 x 10−6) (Figures 3B). Using the Estrada et al. and Kemp et al. GWAS, the purple module contained 45 genes implicated by GWAS (OR = 3.15, Padj = 2.3 x 10−8) (5.7% of GWAS-implicated genes; 8.9% of purple module genes) and explained 26% +/− 4% of the SNP-heritability (hg2) in the study. Using the Morris et al. GWAS, the number of purple module genes implicated by GWAS increased to 77 (OR = 2.7, Padj = 3.4 x 1011) (4.9% of GWAS-implicated genes; 15.2% of purple module genes) explaining 25% +/− 4% of the hg2. Additionally, the purple module was still the only one correlated with in vitro mineralization (Figure 3C), the most significantly enriched for genes eliciting a bone phenotype when knocked-out in mice (Figure 3D), and human monogenic bone disease genes (Figure 3E). These data indicate that even with a significant increase in the number of GWAS-implicated genes included in the analysis, the purple module is the only one enriched for genes with core like properties.
V. The purple module contains genes belonging to one of two distinct transcriptional programs across osteoblast differentiation
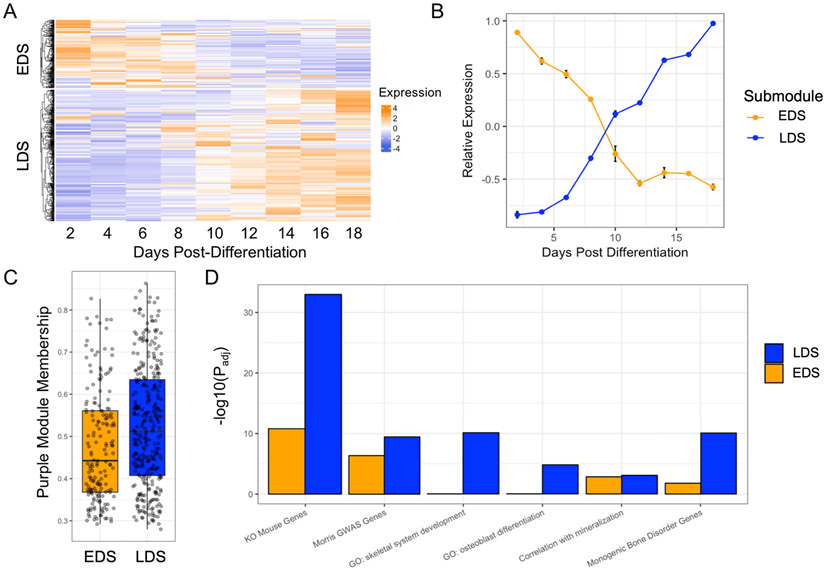
The purple module was enriched for GO categories important for the function of osteoblasts. Consistent with this observation, it contained many genes known to play key roles in osteoblast differentiation and mineralization, including Runx2 (Komori, 2009), Sp7 (Yoshida et al., 2012), Sost (Atkins et al., 2011; Semënov et al., 2005), Bglap (Nakamura et al., 2009), Alpl (Golub and Boesze-Battaglia, 2007), among many others (Table S7). Thus, to further investigate the purple module, we evaluated the expression of its genes with regards to osteoblast differentiation. To do this, we utilized transcriptomic profiles collected from purified osteoblasts at multiple time points across differentiation (GSE54461). Using k-means clustering, we found that the genes within the purple module clearly partitioned into two distinct transcriptional profiles with regards to differentiation (Figure 4A,B). We have termed these groups the Early Differentiation Submodule (EDS; high expression early and low expression late) (N=192 transcripts; 175 unique genes) and the Late Differentiation Submodule (LDS; low expression early and high expression late) (N=423 transcripts; 323 unique genes).
Figure 4. The purple module consists of genes representing two distinct transcriptional profiles across osteoblast differentiation, one of which, the late differentiation submodule (LDS), is more enriched for genes with properties consistent with core genes for mineralization.
(A) Purple module genes show two distinct patterns of expression across differentiation, (B) Genes in cluster 1 (or the early differentiation submodule; EDS; N=192 transcripts; 175 unique genes) are expressed high early in osteoblast differentiation. Genes in cluster 2 (or the late differentiation submodule; LDS; N=423 transcripts; 323 unique genes) are expressed high late in osteoblast differentiation (relative expression = average z-score of expression of genes in each submodule; error bars = standard deviation of relative expression across three replicates). (C) LDS genes have a significantly higher purple module membership score (P = 3.0 x 10−4), see also Figure S4. (D) The LDS is more significantly enriched than the EDS for genes implicated by BMD GWAS in humans, associated with GO terms for bone development, for genes that when knocked out, produce a bone phenotype, and for genes involved in monogenic bone disorders. See also Figure S4.
We assessed whether there were differences between the EDS and the LDS with regard to network parameters and their enrichment for functional annotations seen in the purple module. We first looked at intramodular connectivity, measured by module membership (correlation between the expression of each gene and the module eigengene). On average, LDS genes had higher module membership scores than EDS genes (P = 3.0 x 10−4) (Figure 4C), suggesting they may play more critical roles in the context of overall module behavior. Additionally, the LDS was more significantly enriched for genes implicated by GWAS (OR = 3.0, Padj = 5.2 x 10−10), osteoblast relevant GO terms (e.g. “ossification“ (Padj = 3.24x10−14), skeletal development” (Padj = 9.6 x 10−11), “osteoblast differentiation” (Padj = 1.4 x 10−4), and “biomineral tissue development” (Padj = 4.1x10−6), genes that when knocked-out result in a bone phenotype (OR = 7.3, Padj = 1.1 x 10−33) and monogenic bone disease genes (OR = 33.2, Padj = 8.4 x 10−11) (Figure 4D). As one would expect based on their higher expression later in differentiation, many of the most well-known regulators of mineralization, such as Phospho1 (Roberts et al., 2007), Bglap (Neve et al., 2013), Fam20c (Liu et al., 2018), Mepe (Cho and Ryoo, 2008), Phex (Quarles and Darryl Quarles, 2003), to name a few, were members of the LDS (Table S6). These observations, together with the fact that LDS genes are expressed at high levels during late differentiation, coincident with when the osteoblasts are actively mineralizing, suggest that LDS contains genes with core-like properties specific for the process of mineralization. For all downstream analyses we focused on the LDS.
VI. LDS genes CADM1, B4GALNT3, DOCK9, and GPR133 are novel genetic determinants of BMD
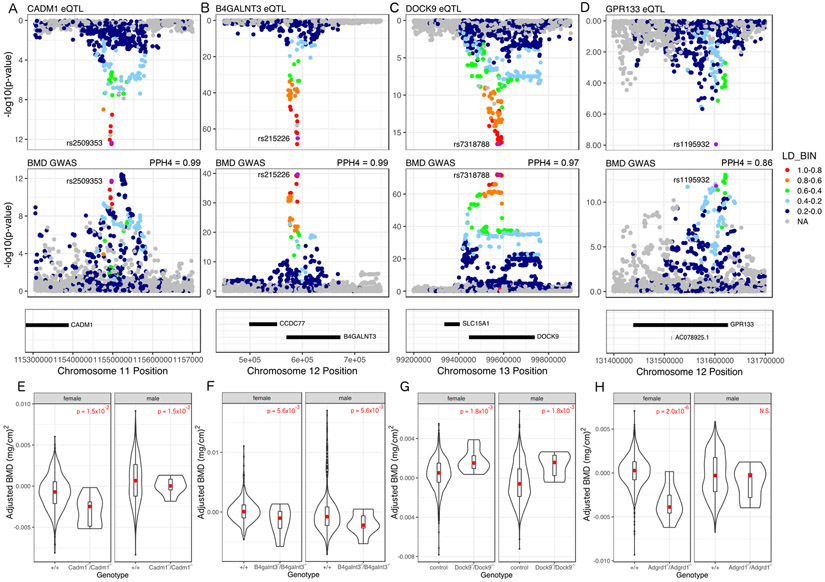
The overarching goal of this study was to identify causal genes from a module enriched for genes with core-like properties underlying BMD GWAS associations. As described above, 48 (14.9%) LDS genes overlapped an eBMD GWAS association from the Morris et al. study and these SNPs were also enriched in regions of the osteoblast genome marked as promoter and enhancers and depleted in repressed regions, supporting their potential role as causal genes for BMD (Table S7 and Figure S3). To further identify those with strong evidence of being causal, we utilized expression quantitative trait locus (eQTL) data from the Gene Tissue Expression (GTEx) project to identify local eQTL colocalizing with BMD associations (GTEx Consortium et al., 2017). We also used total body BMD data on LDS gene knockouts collected as part of the International Mouse Phenotyping Consortium (IMPC) (Koscielny et al., 2014). Together, these data allowed us to directly link BMD associated variants to LDS genes and LDS genes to pathways regulating BMD. We performed a colocalization analysis for each eQTL/BMD association pair for all 48 genes in 48 GTEx tissues and identified 12 LDS genes with colocalizing eQTL (PP4>0.7) (Table S6 and Figures 5A, B, C, and D). We also queried each of 12 LDS genes with a colocalizing eQTL and found that BMD had been measured by the IMPC in 5 mutants. Of these, four genes (Cadm1, B4galnt3, Dock9, and Gpr133) had significantly altered total body BMD (Padj < 0.05) (Table S1 and Figures 5E, F, G and H). For Cadm1 and Dock9 the direction of effect inferred from the eQTL/BMD association matched the direction of the effect observed in the mouse knockout; however, for B4galnt3 and Gpr133 the directions did not match (Table S1).
Figure 5. CADM1, B4GALNT3, DOCK9, and GPR133 (aka Adgrd1) are genetic regulators of BMD.
(A-D) All four genes have an eQTL in at least one tissue in the GTEx database that colocalizes with a proximal BMD GWAS association. (E-H) Knockout mice from the KOMP for each gene exhibit altered BMD (E) N control male = 816, N control female = 778, N KO male = 7 N KO female = 7; (F) N control male = 445, N control female = 459, N KO male = 8, N KO female = 8; (G) N control male = 1692, N control female = 1729, N KO male = 8, N KO female = 8; (H) N control male = 1477, N control female = 1466, N KO male = 7, N KO female = 7, see also Figures S3 and S4 and Table S1.
Lastly, we evaluated network parameters of Cadm1, B4galnt3, Dock9 and Adgrd1. Given prior work that has established a connection between high network connectivity and functional importance to disease, we evaluated the inter-module connectivity of the genes in the LDS (Carlson et al., 2006; Farber, 2010; Langfelder et al., 2013). We observed that Cadm1 and B4galnt3 were ranked in the top 20 based on LDS connectivity (Table S7). In fact, Cadm1 was the 2nd most highly connected gene. Together, the four genes had, on average, higher module membership than the average LDS gene (0.72 vs. 0.52; P = 0.002). In support of the importance of connectivity in the LDS, we observed that more highly connected LDS genes were more likely (P=0.008) to overlap a BMD GWAS locus (Figure S4A) and there was a strong positive correlation between connectivity and in vitro mineralization for all LDS genes (r = 0.71, P< 2.2 x 10−16) (Figure S4B). These data suggest that connectivity is an important feature of the LDS and a strong proxy for biological importance. Furthermore, these data strongly support CADM1, B4GALNT3, DOCK9 and GPR133 as genetic drivers of BMD in humans.
Discussion
Osteoporosis is an increasingly common disease associated with reduced BMD and negative health outcomes, namely fracture (Black and Rosen, 2016). Despite its significant genetic component, we do not fully understand the genes and mechanisms that influence osteoporosis and its determinants, such as BMD. Moreover, current therapeutics for osteoporosis have been associated with rare side effects, leading to decreased compliance (Kolata, 2016). Identification of the causal genes with core-like properties that regulate BMD will help us to further understand the etiology of osteoporosis and lead to the development of novel therapeutics. In this study, we identified the LDS, a co-expression submodule enriched for genes with core-like properties influencing BMD, by integrating a cell- and timepoint-specific co-expression network with the results of BMD GWAS. We then used this information to identify four LDS genes that overlapped GWAS loci, had colocalizing eQTL, and altered BMD in knockouts, suggesting they are causal for their respective BMD GWAS association.
In this work, we hypothesized that the genes underlying BMD GWAS associations are not created equal with respect to “biological importance” or membership in pathways with direct impacts on bone mass. Substantial prior evidence supports this prediction (International Schizophrenia Consortium et al., 2009; Manolio et al., 2009)and it is one of the primary tenets of the omnigenic model (Boyle et al., 2017a; Liu et al., 2019). Identification of genes whose activity or function is more proximal to BMD is important for a number of reasons. First, the identification of genes with core-like properties has the potential to identify critical new players in pathways known to directly impact bone and to uncover new processes essential to skeletal growth and maintenance. Second, it provides a way to prioritize hundreds of BMD GWAS loci for further investigation. Third, based on their central role in the regulation of BMD, it is logical to use the concept of a core gene as a way to prioritize gene discovery in the context of selecting promising new therapeutic targets for evaluation.
The omnigenic model uses a strict statistical definition to define core genes and many have debated the utility of this designation(Boyle et al., 2017a, 2017b; Cox, 2017; Wray et al., 2018). Some have argued that focusing on core genes underestimates the complexity of complex traits, attributing biologically nuanced diseases to a small set of genes 13. Others have argued that the focus should not be on thoroughly defining core genes, but instead on identifying the underlying biological pathways and mechanisms 14. In practice, it is likely that the designation of core genes follows a spectrum rather than a discrete classification. If so, then it should be possible to rank genes based on their continuous “core” attributes, which would be analogous to ranking genes based on their proximity to a disease or phenotype. In essence, that is what we have done in the current study with the goal of identifying genes on the end of the “core” attribute distribution for mineralization. Importantly, it is not likely that all genes in the LDS are causal genetic drivers or, if they are causal, it is possible that several will have few core attributes. However, based on our analysis and results, it is likely that many are causal genes that participate in “core” pathways and processes that directly impact mineralization, bone formation, and BMD.
As we have previously demonstrated (Calabrese et al., 2017; Farber, 2010), there are a number of advantages to using co-expression networks to inform GWAS. First, it allowed us to group genes across the genome based on function and pathway membership and then identify groups of functionally similar genes that had core-like properties. Second, it allowed us to predict the function of potentially casual genetic drivers of BMD. Based on the strong GO enrichments and membership of genes with well-known roles in bone formation and mineralization, it is likely that many LDS genes, including those with no known function, impact mineralization in some manner. The idea of the LDS playing a central role in bone formation was further supported by the strong overlap observed between lead BMD GWAS SNPs for associations containing LDS genes regulatory elements in osteoblasts. Third, it begins to provide a systems-level context for causal genetic drivers. Once genes underlying GWAS loci are identified it is then important to begin to understand their role in complicated cellular networks, defining how a set of genetic variants may converge on multiple genes all involved in a particular process. We can use the LDS to begin to identify sets of variants that all work to influence genes which impact mineralization and the hierarchy of relationships between these genes.
This work extends our use of co-expression networks to inform GWAS. Previously, we used a network generated using cortical bone expression profiles from the Hybrid Mouse Diversity Panel to identify two “osteoblast” modules (enriched for genes involved in osteoblast differentiation and function) enriched for genes implicated by BMD GWAS. We used these modules to identify 35 genes potentially causal for GWAS loci, including two (MARK3 and SPTBN1) that we experimentally validated their involvement in BMD. Comparing the two modules to the LDS we observed a modest overlap (96 of 323 genes; 29.7%), even though they both demonstrated a strong “osteoblast” enrichment signature. While a number of differences (microarray vs. RNA-seq transcriptomic data, different mouse populations, etc.) confound the interpretation of the seemingly low overlap, it is likely due in large part to our use of osteoblast-specific network capturing the transcriptome at peak mineralization instead of the whole bone tissue representing a small number of osteoblasts.
We provided strong supporting evidence that four LDS genes (CADM1, B4GALNT3, DOCK9 and GPR133) are novel regulators of BMD and causal for their respective GWAS association. Prior to this study, none of these genes had been directly connected to the regulation of BMD. CADM1 (Cell Adhesion Molecule 1) is a ubiquitously expressed cell adhesion molecule involved in many biological processes, including cancer, spermatogenesis, and neuronal/mast/epithelial cell function(Cao et al., 2017; Wakayama and Iseki, 2009; Zhang et al., 2016) that had been implicated in osteoclast proliferation and activity(Nakamura et al., 2017) and as an osteoblast-specific marker in the context of osteosarcoma(Inoue et al., 2013; Mentink et al., 2013). B4GALNT3 (Beta-1,4-N-Acetyl- Galactosaminyltransferase 3) is a glycosyltransferase that transfers N-acetylgalactosaine (GalNAc) onto glucosyl residues, thus forming N,N-prime-diacetyllactoseadiamine (LacdiNAc), which serves as a terminal structure of cell surface N-glycans that contributes to cell signaling 51,52. B4GALNT3 is expressed in bone and associated with circulating levels of sclerostin 53-55. DOCK9 (Dedicator of Cytokinesis 9) is a guanine nucleotide-exchange factor (GEF) that activates Cdc42(Meller et al., 2002), which has been shown to regulate osteoclast differentiation and ossification 57,58. GPR133 (Adhesion G Protein-Coupled Receptor D1) is a G protein-coupled receptor that participates in cell-cell and cell-matrix interactions 59. Our results demonstrate the utility of the LDS in broadening our understanding of the molecular and genetic basis of BMD.
Our study is not without limitations. First, we used gene expression data from the mouse as a discovery platform, however this may limit the translational applications of the work due biological differences and missing homologs between mouse and human. Secondly, this was not a comprehensive study of the genetic effects driving osteoporosis, because we focused exclusively on the contributions of just one cell type, bone-forming osteoblasts. In future work, this approach could also be applied to other bone cell types. For example, one could use in vitro measures of osteoclast activity as a filter to identify groups of genes influencing bone resorption, and ultimately BMD. Finally, the eQTL comparisons made in this study were not derived from expression data in bone tissue, as bone tissue expression was not measured in the GTEx project. While we identified colocalizing eQTL in other tissues, these eQTL may be irrelevant to BMD or the direction of eQTL effects in non-bone tissues may not reflect the direction of effect in osteoblasts.
While we identified four novel regulators of bone mineral density, there is still much to be gleaned from the late differentiation submodule. The LDS is a promising resource for two key applications: (1) causal gene discovery and functional follow up and (2) studying the impact of genetic variation on biological networks and complex phenotypes. There are still many genes with no known connection to BMD in the LDS that are likely important to osteoblast biology and mineralization. Additionally, the LDS is not just a list of candidate genes; it also provides insight into the molecular hierarchy driving osteoblast differentiation and mineralization, which can demonstrate how genetic variation impacts biological networks. The network topology of the LDS can also be used to infer the causal relationships between genetic variants and the many genes that influence osteoblast activity. Moving forward, the LDS can serve as a platform for the identification of novel determinants of BMD and for furthering our understanding of the nuanced relationship between genetic variation, molecular phenotypes, and complex traits.
In summary, we have used an integrative, network-based method to identify core genes for the process of mineralization and BMD. While the definition of a core gene is still open to debate, we found the expected properties of core genes are effective lenses through which to contextualize GWAS associations. Integrating gene co-expression networks, GWAS data, in vitro and in vivo phenotypic data reflecting “core” properties, and eQTL information has led us to a more complete understanding of the biology and genetics of BMD.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Charles Farber (crf2s@virginia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The accession numbers for the RNA sequencing data reported in this paper are GEO: GSE134081.
Experimental Model and Subject Details
Primary cell cultures of neonatal murine osteoblasts were used to generate the RNA sequencing data and to conduct the in vitro mineralization experiments. Neonatal collaborative cross heads were received from the University of North Carolina. At UNC, neonatal (3-5 days) collaborative cross mice were euthanized by CO2, decapitated onto paper towels soaked in 70% ethanol, and placed in cold PBS on ice for overnight shipping. Once received, calvaria were dissected, paying special attention to brain and interparietal bone removal. Isolated calvaria were placed in 24 well plates containing 0.5 mL of digest solution (0.05% trypsin and 1.5 U/ml collagenase P) and incubated on a rocking platform at 37 degrees during six, fifteen-minute digestions in 0.5 mL of digestion solution. Fraction 1 is discarded and fractions 2-6 are collected. Fractions 2-6 are added to an equal volume of cold plating media (89 mL DMEM, 1 mL 100x Pen/Strep solution, and 10 mL Lot tested FBS). The resulting cells are filtered using a 70-100 mm cell strainer to remove clots, centrifuged at 1000 rpm for 5 minutes and re-suspended in 0.5 ml plating media. The resulting cells are plated in a T25 flask. 24 hours later, cells are washed with PBS, treated with trypsin, counted, and plated at a density of 1.5x105 cells per well in a 12-well plate, and allowed to grow to confluence for 48 hours. After 48 hours of growth, cells are switched to differentiation media (10 mL lot tested FCS, 1 mL 100x Pen/Strep solution, 283.8 uL ascorbic acid (0.1 M), 400 uL B-glycerol phosphate (1 M), and 88.3 mL alpha-MEM per 100 mL) and allowed to differentiate for 10 days and then either RNA is collected or mineral is quantified. The strain and sex of cells used are listed in Table S8.
Method Details
RNA sequencing in mineralizing osteoblasts:
On day 10, total RNA was extracted from the mineralized cultures using mirVana RNA isolation kit (ThermoFisher Scientific). RNA-Seq libraries were constructed from 200 ng of total RNA using Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold sample prep kits (Illumina, Carlsbad, CA). Constructed libraries contained RNAs >200 nt (both unpolyadenylated and polyadenylated) and were depleted of cytoplasmic and mitochondrial rRNAs. An average of 39.7 million 2 x 75 bp paired-end reads were generated for each sample on an Illumina NextSeq 500 (Illumina, Carlsbad, CA). FastQC was used to evaluate the quality of the reads, and all samples passed the QC stage (Bioinformatics, 2011). Reads were mapped to the eight collaborative cross founder transcriptomes based on build mm9 using Bowtie, and quantified using EMASE (Raghupathy et al., 2018). EMASE output transcript level expression estimates calculated by assigning multi-mapping reads across the genome using an expectation-maximization algorithm to allocate reads that differentiate between genes, then isoforms of a gene, and then alleles.
In vitro mineralization measurement:
In order to identify the modules of coexpressed genes with patterns of expression correlated with mineralization, we measured in vitro mineralization in osteogenic cells from the calvaria of 42 strains of collaborative cross mice. After 10 days of differentiation and mineral production, cells are washed with PBS and treated with 10% NBF (1 mL per well) and incubated at room temperature for 15 minutes. The NBF is removed and cells are washed with H2O (1mL x 2). Next, wells are stained with alizarin red (0.5 mLs, 40 mM @ pH 5.6) for 20 minutes on a shake plate at 120 rpm. Alizarin red stain is then removed, and cells are washed 5 times with deionized H2O for 5 minutes on a shake plate at 180 rpm. Once rinsed, the mineralized wells are scanned, and .tiff images are retained to extract geometric parameters of the mineral deposits. After imaging, the wells are de-stained by incubation with 5% perchloric acid (1 mL) at room temperature for 5 minutes while shaking at 120 rpm. Eluent is collected and read at 405 nm. The levels of in vitro mineralization varied significantly across the population, with a 63-fold change from the highest to lowest mineralization samples (max_mmAR = 2.995993, min_mmAR = 0.04719). In this population, in vitro mineralization had a heritability of 47.8%(p=1.8x10−46), indicating that the between-strain variation is larger than the within strain variation and that there is a genetic contribution to the process of mineralization.
Quantification and Statistical Analysis
WGCNA network construction:
Estimated transcript count data was used as the basis for co-expression network construction from each of the 96 samples sequenced. We removed transcripts with less than an average tpm <= 0.3 tpm across all samples, resulting in 29,000 transcripts used to construct the network. We used a variance stabilizing transformation from the DESeq2 package that decouples the variance from the mean (Love et al., 2014). Next, we used PEER in order to remove latent confounding batch effects from our data (Stegle et al., 2012). As per PEER recommendations, we estimated PEER factors equal to one quarter of the number of samples (sample N = 96, PEER factors = 24) and included covariates in the calculation. We carried out the downstream analysis with the residual values from PEER transformation. Finally, we used quantile normalization to match the distribution of each of the samples in the analysis and averaged the expression across samples of the same strain, resulting in 42 expression profiles.
The resulting expression data was used to construct a signed, weighted gene co-expression network using the weighted gene co-expression network analysis (WGCNA) package (Langfelder and Horvath, 2008). There were no evident outliers from the hierarchical clustering analysis. The pickSoftThreshold() function from the wgcna package was used to determine the power used to calculate the network. The minimum power value that had an R2 >= 0.9 for the scale-free topology model fit was used, and the network was calculated using a power of 9. We then used the blockwiseModules() function to construct a signed network with a merge cut height of 0.15, and a minimum module size of 20 genes. Using WGCNA, we constructed a signed network composed of 65 modules of co-expressed genes, with an average of 292 transcripts and 266 unique genes per module.
Gene Ontology Analysis:
For those modules that were enriched for BMD GWAS genes, we conducted gene ontology analysis to identify the functional categories represented by each module. Using the ToppFun tool on the ToppGene site, we identified the significantly enriched categories for GO molecular functions, GO biological processes, GO cellular components, human and mouse phenotypes, and pathways (Chen et al., 2009). The adjusted p-values reported for these enrichments were Benjamini & Hochberg corrected FDR q-values, correcting for multiple testing across many gene ontology categories.
Creating BMD GWAS list:
In order to identify co-expression modules enriched for BMD GWAS genes, we identified all genes overlapping a BMD GWAS locus using the 2012 and 2017 BMD GWAS (Estrada et al., 2012; Kemp et al., 2017). For each BMD locus, a bin was defined by the furthest upstream and downstream SNPs with LD >= 0.7 as calculated from the European populations in the 1000 genomes phase III data identified using the LDLink LDProxy tool (Machiela and Chanock, 2015). Then, using the Genomic Ranges tool, we identified all genes from the GRCh37/hg19 Ensembl gene set overlapping a BMD GWAS bin (Hubbard et al., 2002; Lawrence et al., 2013). If no gene intersected a bin, we identified the nearest upstream and downstream genes from the bin. The Estrada GWAS resulted in 179 genes and the eBMD GWAS resulted in 701 genes, resulting in a list of 731 unique genes. We converted the list of human genes to mouse homologs.
BMD GWAS gene enrichment:
In order to identify modules of genes enriched for GWAS genes, we used a fisher’s exact test to measure the statistical significance of the representation of GWAS genes in each module. We then applied a Bonferroni correction to correct for testing the enrichment of all 65 modules, and applied a significance cutoff of 0.05 to the adjusted p-values, resulting in 13 modules of genes enriched for 2012 and 2017 GWAS genes, and 26 modules of genes enriched for 2012, 2017, and 2018 GWAS genes.
In vitro mineralization correlation with module expression:
Using the WGCNA package, the eigengene of each module was calculated, and the correlation between the eigengene and the in vitro mineralization phenotype was calculated using the cor() function in R. The p-values associated with the correlation between the module eigengenes and in vitro mineralization were corrected for multiple testing using a Bonferroni correction across 65 co-expression modules and a p-value cutoff of 0.05 was applied to the adjusted p-values.
LD Score Regression:
In order to evaluate the relevance of the BMD GWAS gene enriched modules we calculated the partitioned heritability of the SNPs in the regions surrounding the genes in each module. We used the LD score regression method, which takes gene lists as an input and returns the enrichment of the associated SNP set for heritability for the tested trait. For each set of modules we tested using this method, we corrected the enrichment p-values for multiple testing across 13 co-expression modules using a Bonferroni correction, and applied a p-value cutoff of 0.05 to the adjusted p-values. Additionally, we report the standard error of the heritability estimate where applicable.
Module enrichment for genes with associated bone phenotypes and monogenic bone disease:
In order to identify modules of co-expressed BMD GWAS genes that are enriched for genes with bone phenotype annotations, we curated a list of genes which produce a bone phenotype when knocked out. We used four databases of gene perturbations that result in bone phenotypes, including genes annotated with a bone phenotype in the Mouse Genome Informatics database (MGI), the Origins of Bone and Cartilage Disease (OBCD) database, the International Mouse Phenotyping Consortium (IMPC), and the Bonebase Database (Bolser, 2004; Dyment et al., 2016; Freudenthal et al., 2016; Koscielny et al., 2014). Specifically, we pulled BMD, altered bone morphology, altered bone cell activity, changes in ossification or mineralization, or association with a known bone disease from the MGI database. The OBCD database contained genes with changes in bone mineral content (BMC), bone volume fraction (BV/TV), and BMD of the femur and BMD of the vertebra. We mined the IMPC database for any genes with altered BMD, and we pulled all Bonebase genes with altered BV/TV in the femur or vertebra. This resulted in a list of 923 unique “bone” genes (Table S6).
We also curated a list of genes associated with monogenic bone disorders using a literature review, specifically focusing on genes that disrupt osteoblast function, leading to monogenic bone disorders (Boudin and Van Hul, 2017; Johnson, 2016; Marini and Brandi, 2010; Robinson and Rauch, 2019; Rocha-Braz and Ferraz-de-Souza, 2016) (Table S5). We used a fisher’s exact test to measure the statistical significance of the representation of genes with associated mouse knockout bone phenotypes and monogenic bone disease in each module. We then applied a Bonferroni correction to correct for testing the enrichment of all 13 or 26 modules and applied a significance cutoff of 0.05 to the adjusted p-values.
Clustering analysis in osteoblast differentiation gene expression data:
We investigated the expression profiles of all purple module genes in the context of differentiation. Using gene expression data from osteoblasts throughout differentiation (Series GSE54461), we used k-means clustering to identify differentiation-related transcriptional programs in the purple module. We tested k = 1:5, and found two robust clusters of genes within the purple module. Enrichment analysis of the two clusters in all function categories were conducted as described above.
Epigenetic enrichment analysis for LDS BMD GWAS associations:
For BMD GWAS lead SNP (and proxies with LD >= 0.7) overlapping an LDS gene (n = 84), GenomicRanges (Lawrence et al., 2013) was used to calculate the proportion of lead SNPs overlapping regions marked by epigenetic modifications, including H3K4me1, H3K4me2, H3K4me3, H3K9me3, H3K27ac, H3K27me3, H3K26me3, H3K79me2 and H4K20me1, and histone H2AZ from the Roadmap Project (Chadwick, 2012). Using the GenomicRanges function findOverlaps(), we quantified the overlap between the LDS-associated lead SNPs and each epigenetic mark. To assess the enrichment of this overlap, we compared against 1000 sets of control SNPs (n = 84). We chose sets of control SNPs that were within +/− 20% of the mean distance from a transcription start site for the BMD GWAS lead SNPs, and within +/− 20% of the mean minor allele frequency of the BMD GWAS lead SNPs. P-values were calculated by taking the proportion of random sets of SNPs with a more extreme enrichment in the tail of the distribution with which we are comparing our experimental proportion. If the experimental proportion is more extreme than any measured random set, the p-value is reported as < 1x10−3. This same procedure was used to evaluate the tissue specificity for each mark. For each mark, the overlap with the LDS BMD SNP set and the 1000 random SNP sets were computed and the ratio between the proportion of overlapping LDS BMD SNPs and the mean proportion of overlapping random SNPs was computed. Higher ratios indicated greater enrichment of the LDS BMD SNPs over random SNPs with a given mark in a given tissue (Table S7).
Colocalization analysis:
For each gene in the LDS that overlapped a BMD GWAS association from the Morris et al. study, eQTL from all GTEx tissues were identified (GTEx Consortium et al., 2017; Morris et al., 2018). Using the coloc package, we assessed the potential for colocalization between the QTL for BMD and the proximal cis-eQTL (Giambartolomei et al., 2014). Two associations were considered to colocalize if the posterior probability of hypothesis four (PPH4), which is the probability of colocalization, is > 0.7. The RACER package was used to plot the two associations in a mirrorPlot (Sabik and Farber, 2018).
Mouse phenotype statistical comparisons:
Using the International Mouse Phenotyping Consortium (IMPC) database, we identified genes from the LDS that had eQTL that colocalized with BMD QTL and exhibited a difference in BMD when knocked out in mouse (Koscielny et al., 2014). Using the PhenStat package, we analyzed the differences between control and knockout animals using a mixed model framework (Kurbatova et al., 2015). The specific equation used for each analysis are in Table S1.
Network Topology Analysis:
A t-test was used to compare the module membership of the four causal genes and the remainder of the LDS genes and the connectivity of the LDS genes overlapping a BMD GWAS locus as opposed to those that do not. A linear model was used to assess the relationship between gene connectivity and gene correlation with in vitro mineralization.
Supplementary Material
Table S2: Network Summary file, containing module membership scores for all transcripts, related to Figure 1.
Table S3: Gene ontology results for all 64 modules, related to Figure 1.
Table S4: List of Estrada et al., Kemp et al., and Morris et al. BMD GWAS genes, related to Figure 2 and Figure 3.
Table S5: Module enrichment for BMD GWAS genes and partitioned heritability from Kemp et al. GWAS and Morris et al. GWAS, related to Figure 2 and Figure 3.
Table S6: Curated list of genes associated with bone phenotypes and monogenic bone disorders, related to Figure 2 and Figure 3.
Table S7: Summary of purple module genes, network properties, and clustering results, epigenetic enrichment analysis for LDS BMD GWAS associations, and colocalization test for LDS gene GTEx eQTL and BMD GWAS associations, related to Figure 4 and Figure S4.
Table S8: Sex and strain of biological samples, related to Figure 1, Figure S1, and STAR Methods Biological Samples.
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR064790 to C.L.A-B. and C.R.F. O.L.S. was supported by a Wagner Fellowship from the University of Virginia. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript (v7) were obtained from: the GTEx Portal on 01/11/2018. The International Mouse Phenotyping Consortium is partially funded by the NIH Knockout Mouse Programme (KOMP) project and the IMPC informatics and the data portal are supported by NIH grant U54 HG006370.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Atkins GJ, Rowe PS, Lim HP, Welldon KJ, Ormsby R, Wijenayaka AR, Zelenchuk L, Evdokiou A, and Findlay DM (2011). Sclerostin is a locally acting regulator of late-osteoblast/preosteocyte differentiation and regulates mineralization through a MEPE-ASARM-dependent mechanism. Journal of Bone and Mineral Research 26, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioinformatics B (2011). FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Babraham Institute. [Google Scholar]
- Black DM, and Rosen CJ (2016). Clinical Practice. Postmenopausal Osteoporosis. N. Engl. J. Med 374, 254–262. [DOI] [PubMed] [Google Scholar]
- Bolser D (2004). Mouse Genome Informatics (MGI, Mouse Genome Database, MGD). Dictionary of Bioinformatics and Computational Biology. [Google Scholar]
- Boudin E, and Van Hul W (2017). MECHANISMS IN ENDOCRINOLOGY: Genetics of human bone formation. Eur. J. Endocrinol 177, R69–R83. [DOI] [PubMed] [Google Scholar]
- Boyle EA, Li YI, and Pritchard JK (2017a). An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li Y, and Pritchard JK (2017b). The omnigenic model: Response from the authors. J. Psychiatry Brain Sci 2, S8. [Google Scholar]
- Calabrese GM, Mesner LD, Stains JP, Tommasini SM, Horowitz MC, Rosen CJ, and Farber CR (2017). Integrating GWAS and Co-expression Network Data Identifies Bone Mineral Density Genes SPTBN1 and MARK3 and an Osteoblast Functional Module. Cell Syst 4, 46–59.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Shi P, and Ge J-J (2017). miR-21 enhances cardiac fibrotic remodeling and fibroblast proliferation via CADM1/STAT3 pathway. BMC Cardiovasc. Disord 17, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MRJ, Zhang B, Fang Z, Mischel PS, Horvath S, and Nelson SF (2006). Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 7, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH (2012). The NIH Roadmap Epigenomics Program data resource. Epigenomics 4, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, and Ryoo H-M (2008). MEPE: A Mineralization Regulating Bone Matrix Protein. Journal of Korean Endocrine Society 23, 71. [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. (2004). The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet 36, 1133–1137. [DOI] [PubMed] [Google Scholar]
- Civelek M, and Lusis AJ (2014). Systems genetics approaches to understand complex traits. Nat. Rev. Genet 15, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ (2017). Comments on Pritchard Paper. Journal of Psychiatry and Brain Science 2, S5. [Google Scholar]
- van Dam S, Võsa U, van der Graaf A, Franke L, and de Magalhães JP (2018). Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform 19, 575–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment NA, Jiang X, Chen L, Hong S-H, Adams DJ, Ackert-Bicknell C, Shin D-G, and Rowe DW (2016). High-Throughput, Multi-Image Cryohistology of Mineralized Tissues. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eising E, Huisman SMH, Mahfouz A, Vijfhuizen LS, Anttila V, Winsvold BS, Kurth T, Ikram MA, Freilinger T, Kaprio J, et al. (2016). Gene co-expression analysis identifies brain regions and cell types involved in migraine pathophysiology: a GWAS-based study using the Allen Human Brain Atlas. Hum. Genet 135, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu Y-H, Duncan EL, Ntzani EE, Oei L, Albagha OME, Amin N, Kemp JP, et al. (2012). Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet 44, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR (2010). Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J. Bone Miner. Res 25, 2359–2367. [DOI] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenthal B, Logan J, Sanger Institute Mouse Pipelines, Croucher PI, Williams GR, and Bassett JHD (2016). Rapid phenotyping of knockout mice to identify genetic determinants of bone strength. J. Endocrinol 231, R31–R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, and Plagnol V (2014). Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EE, and Boesze-Battaglia K (2007). The role of alkaline phosphatase in mineralization. Curr. Opin. Orthop 18, 444. [Google Scholar]
- GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI, et al. (2017). Genetic effects on gene expression across human tissues. Nature 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-H, Farber CR, and Kiel DP (2020). Genetic determinants of bone mass and osteoporotic fracture. Principles of Bone Biology 1615–1630. [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. (2002). The Ensembl genome database project. Nucleic Acids Res. 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hagiyama M, Enoki E, Sakurai MA, Tan A, Wakayama T, Iseki S, Murakami Y, Fukuda K, Hamanishi C, et al. (2013). Cell adhesion molecule 1 is a new osteoblastic cell adhesion molecule and a diagnostic marker for osteosarcoma. Life Sci. 92, 91–99. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, and Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnell O, and Kanis JA (2006). An estimate of the worldwide prevalence and disability associated with osteoporotic fractures. Osteoporos. Int 17, 1726–1733. [DOI] [PubMed] [Google Scholar]
- Johnson ML (2016). How rare bone diseases have informed our knowledge of complex diseases. Bonekey Rep 5, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, Zheng J, Gregson CL, Grundberg E, Trajanoska K, et al. (2017). Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet 49, 1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogelman LJA, Cirera S, Zhernakova DV, Fredholm M, Franke L, and Kadarmideen HN (2014). Identification of co-expression gene networks, regulatory genes and pathways for obesity based on adipose tissue RNA Sequencing in a porcine model. BMC Med. Genomics 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolata G (2016). Fearing drugs’ rare side effects, millions take their chances with osteoporosis. NY Times; . [Google Scholar]
- Komori T (2009). Regulation of Osteoblast Differentiation by Runx2. Advances in Experimental Medicine and Biology 43–49. [DOI] [PubMed] [Google Scholar]
- Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen C-K, Easty R, Di Fenza A, et al. (2014). The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42, D802–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurbatova N, Mason JC, Morgan H, Meehan TF, and Karp NA (2015). PhenStat: A Tool Kit for Standardized Analysis of High Throughput Phenotypic Data. PLoS One 10, e0131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Mischel PS, and Horvath S (2013). When is hub gene selection better than standard meta-analysis? PLoS One 8, e61505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhang H, Jani P, Wang X, Lu Y, Li N, Xiao J, and Qin C (2018). FAM20C regulates osteoblast behaviors and intracellular signaling pathways in a cell-autonomous manner. Journal of Cellular Physiology 233, 3476–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li YI, and Pritchard JK (2019). Trans Effects on Gene Expression Can Drive Omnigenic Inheritance. Cell 177, 1022–1034.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, and Chanock SJ (2015). LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkinen V-P, Civelek M, Meng Q, Zhang B, Zhu J, Levian C, Huan T, Segrè AV, Ghosh S, Vivar J, et al. (2014). Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 10, e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, and Brandi ML (2010). Genetic determinants of osteoporosis: common bases to cardiovascular diseases? Int. J. Hypertens 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller N, Irani-Tehrani M, Kiosses WB, Del Pozo MA, and Schwartz MA (2002). Zizimin1, a novel Cdc42 activator, reveals a new GEF domain for Rho proteins. Nat. Cell Biol 4, 639–647. [DOI] [PubMed] [Google Scholar]
- Mentink A, Hulsman M, Groen N, Licht R, Dechering KJ, van der Stok J, Alves HA, Dhert WJ, van Someren EP, Reinders MJT, et al. (2013). Predicting the therapeutic efficacy of MSC in bone tissue engineering using the molecular marker CADM1. Biomaterials 34, 4592–4601. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai R, Vulpescu NA, Forgetta V, Kleinman A, Mohanty S, et al. (2018). An Atlas of Human and Murine Genetic Influences on Osteoporosis. [Google Scholar]
- Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, Vulpescu NA, Forgetta V, Kleinman A, Mohanty ST, et al. (2019). An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet 51, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Dohi Y, Akahane M, Ohgushi H, Nakajima H, Funaoka H, and Takakura Y (2009). Osteocalcin secretion as an early marker of in vitro osteogenic differentiation of rat mesenchymal stem cells. Tissue Eng. Part C Methods 15, 169–180. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Koyama T, Izawa N, Nomura S, Fujita T, Omata Y, Minami T, Matsumoto M, Nakamura M, Fujita-Jimbo E, et al. (2017). Negative feedback loop of bone resorption by NFATc1-dependent induction of Cadm1. PLoS One 12, e0175632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve A, Corrado A, and Cantatore FP (2013). Osteocalcin: Skeletal and extra-skeletal effects. Journal of Cellular Physiology 228, 1149–1153. [DOI] [PubMed] [Google Scholar]
- Quarles LD, and Darryl Quarles L (2003). FGF23, PHEX, and MEPE regulation of phosphate homeostasis and skeletal mineralization. American Journal of Physiology-Endocrinology and Metabolism 285, E1–E9. [DOI] [PubMed] [Google Scholar]
- Raghupathy N, Choi K, Vincent MJ, Beane GL, Sheppard KS, Munger SC, Korstanje R, Pardo-Manual de Villena F, and Churchill GA (2018). Hierarchical analysis of RNA-seq reads improves the accuracy of allele-specific expression. Bioinformatics 34, 2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston SH, and de Crombrugghe B (2006). Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 20, 2492–2506. [DOI] [PubMed] [Google Scholar]
- Ralston SH, and Uitterlinden AG (2010). Genetics of osteoporosis. Endocr. Rev 31, 629–662. [DOI] [PubMed] [Google Scholar]
- Rau CD, Romay MC, Tuteryan M, Wang JJ-C, Santolini M, Ren S, Karma A, Weiss JN, Wang Y, and Lusis AJ (2017). Systems Genetics Approach Identifies Gene Pathways and Adamts2 as Drivers of Isoproterenol-Induced Cardiac Hypertrophy and Cardiomyopathy in Mice. Cell Syst 4, 121–128.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Narisawa S, Harmey D, Millán JL, and Farquharson C (2007). Functional involvement of PHOSPHO1 in matrix vesicle-mediated skeletal mineralization. J. Bone Miner. Res 22, 617–627. [DOI] [PubMed] [Google Scholar]
- Robinson M-E, and Rauch F (2019). Mendelian bone fragility disorders. Bone. [DOI] [PubMed] [Google Scholar]
- Rocha-Braz MGM, and Ferraz-de-Souza B (2016). Genetics of osteoporosis: searching for candidate genes for bone fragility. Arch Endocrinol Metab 60, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabik OL, and Farber CR (2016). Using GWAS to identify novel therapeutic targets for osteoporosis. Transl. Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabik OL, and Farber CR (2018). RACER: A data visualization strategy for exploring multiple genetic associations. [Google Scholar]
- Semënov M, Tamai K, and He X (2005). SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J. Biol. Chem 280, 26770–26775. [DOI] [PubMed] [Google Scholar]
- Stegle O, Parts L, Piipari M, Winn J, and Durbin R (2012). Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc 7, 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T, and Iseki S (2009). Role of the spermatogenic–Sertoli cell interaction through cell adhesion molecule-1 (CADM1) in spermatogenesis. Anat. Sci. Int 84, 112–121. [DOI] [PubMed] [Google Scholar]
- Wray NR, Wijmenga C, Sullivan PF, Yang J, and Visscher PM (2018). Common Disease Is More Complex Than Implied by the Core Gene Omnigenic Model. Cell 173, 1573–1580. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Komori H, Maruyama Z, Miyazaki T, Kawasaki K, Furuichi T, Fukuyama R, Mori M, Yamana K, Nakamura K, et al. (2012). SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS One 7, e32364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xie H-Y, Ding S-M, Xing C-Y, Chen A, Lai M-C, Zhou L, and Zheng S-S (2016). CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb-E2F pathway in hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int 15, 289–296. [DOI] [PubMed] [Google Scholar]
- Zheng H-F, Spector TD, and Richards JB (2011). Insights into the genetics of osteoporosis from recent genome-wide association studies. Expert Rev. Mol. Med 13, e28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2: Network Summary file, containing module membership scores for all transcripts, related to Figure 1.
Table S3: Gene ontology results for all 64 modules, related to Figure 1.
Table S4: List of Estrada et al., Kemp et al., and Morris et al. BMD GWAS genes, related to Figure 2 and Figure 3.
Table S5: Module enrichment for BMD GWAS genes and partitioned heritability from Kemp et al. GWAS and Morris et al. GWAS, related to Figure 2 and Figure 3.
Table S6: Curated list of genes associated with bone phenotypes and monogenic bone disorders, related to Figure 2 and Figure 3.
Table S7: Summary of purple module genes, network properties, and clustering results, epigenetic enrichment analysis for LDS BMD GWAS associations, and colocalization test for LDS gene GTEx eQTL and BMD GWAS associations, related to Figure 4 and Figure S4.
Table S8: Sex and strain of biological samples, related to Figure 1, Figure S1, and STAR Methods Biological Samples.
Data Availability Statement
The accession numbers for the RNA sequencing data reported in this paper are GEO: GSE134081.