Abstract
Onchocerca lupi, Rodonaja 1967, is an emerging, zoonotic filarial nematode parasite that causes ocular disease in dogs, cats, wild canids, and humans. It is the causative agent of ocular onchocercosis in canines with increasing incidence in both North America and the Old World during the early twenty-first century. We report the complete mitochondrial genome of an O. lupi isolate from a dog from Arizona, southwestern USA, and its genetic differentiation from related Onchocerca species. The whole mitochondrial genome was obtained from whole genome sequencing of genomic DNA isolated from an adult worm. This mitogenome is 13,766 bp in size and contains 36 genes and a control region. This mitogenome provides a valuable resource for future studies involving epidemiological surveillance, population genetics, phylogeography, and comparative mitogenomics of this emerging pathogen and other parasitic nematodes.
Keywords: O. lupi, mitogenome, Bayesian phylogenetic analyses, Nematode, Onchocercosis, Filaria
Nematodes of the genus Onchocerca are found world-wide and infect a range of hosts such as humans, wild and domestic ungulates and carnivores, and comprise in excess of 30 valid species (Lefoulon et al. 2017). Onchocerca lupi is a tissue-dwelling zoonotic filarial parasite that causes ocular onchocercosis in dogs, cats, coyotes, wolves, and recently, humans (Roe et al. 2020). The putative biological vector of O. lupi are black flies (Diptera: Simuliidae), which transfer the infective third-stage larva to the mammal definitive host. Infection of the definitive host is usually associated with ocular disease, and may lead to blindness. Since 2010, there have been increasing reports of onchocercosis in domestic dogs and cats in North America and areas of Europe, North Africa and the Middle East (Labelle et al. 2011; Otranto et al. 2013; McLean et al. 2017), recent reports in coyotes (Roe et al. 2020), and zoonotic infections (Eberhard et al. 2013; Cantey et al. 2016). There is currently a knowledge gap regarding the genomics, population structure, and phylogeography of O. lupi. Previous research has utilized single mitochondrial genes (COI, ND5) for phylogenetic analyses providing low resolution relationships from global samples (Roe et al. 2020; Verocai et al. 2016); however, recent research of other zoonotic parasites identified phylogenetic relationships using whole mitochondrial sequencing that were indiscernible when examining short gene sequences (Kinkar et al. 2016; Laurimäe et al. 2018). Here, we report the complete mitochondrial genome of a single O. lupi nematode from the United States.
One adult nematode was collected by a local veterinarian from a privately-owned, infected dog from Flagstaff, AZ, USA (35°11′53.05″N, −111°39′4.57″W) in 2010. The specimen was pulverized by reciprocal shaking with steel beads at 30 Hz and DNA was extracted using a phenol-chloroform protocol (Doyle and Doyle 1990) modified with prolonged heated lysis and stored at −20 °C. Taxonomic identity was determined by Sanger sequencing using the previously published COI mitochondrial gene primers (Hassan et al. 2015). Genomic DNA was prepared for sequencing using previously published methods (Roe et al. 2016) and sequenced on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA). An O. lupi specimen was deposited at the Pathogen and Microbiome Institute (https://in.nau.edu/pmi/, Roxanne Nottingham, Roxanne.nottingham@nau.edu) under voucher number OL-202101.
Due to the presence of both nuclear parasite as well as host DNA, mitochondrial reads were extracted bioinformatically through alignment to Onchocerca volvulus reference mitogenome (AF015193; 13,747bp); reads that mapped to the O. volvulus mitochondrial genome were separated using SAMtools v1.9 (Li et al. 2009), assembled using SPAdes (Bankevich et al. 2012) to an average depth of 303X, and examined for circularity using Circlator (Hunt et al. 2015). The entire circularized O. lupi mitogenome is 13,766 bases (GenBank accession MW266120). The mitogenome consists of 12 protein-coding genes (PCG), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 noncoding region (NCR). One DNA strand serves as the template strand for all genes. All PCGs are syntenic with the O. volvulus mitogenome; however, total synteny differs by the placement of one tRNA-Lys. The nucleotide distribution of the mitogenome was biased toward A + T (68%) which is similar to other reported filarial nematodes (Keddie et al. 1998).
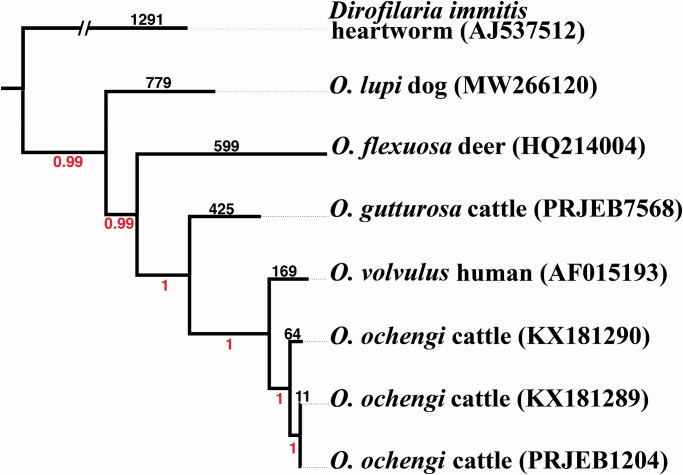
Bayesian phylogenetic analyses of seven Onchocerca samples were implemented with the program ExaBayes v1.5.1 (Aberer et al. 2014) using the concatenated, shared SNP loci produced by the SNP pipeline NASP (Sahl et al. 2016), which spanned 12,259 shared positions (89%) across four Onchocerca species with the canine heartworm, Dirofilaria immitis, serving as the outgroup (Figure 1) . This analysis revealed a total of 2,260 SNPs; 852 SNPs were parsimony-informative positions revealing O. lupi as the most basal species within the genus Onchocerca, considering the limited number of species with characterized mitogenomes. We report the completed mitogenome of O. lupi by the next-generation sequencing and molecular phylogenetic placement within the genus Onchocerca. This mitogenome will provide a novel, strong foundation for further studies involving phylogenetic relationships among Onchocerca species and observations of the origin and range expansion of O. lupi.
Figure 1.
Bayesian phylogenetic relationships of Onchocerca lupi and 3 additional Onchocerca species based on concatenated SNPs that covered 89% total bases of the reference O. lupi mitogenome. Bayesian posterior probability values are shown below branches while SNP numbers are shown above corresponding branches. A“//” indicates a broken branch length that was shortened for visual purposes .
Funding Statement
This work was supported by the Pacific Southwest Center of Excellence in Vector-Borne Diseases under Grant no. [1004601].
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW266120.1 under the accession no. MW266120. The associated BioProject and Bio-Sample numbers are PRJNA733160 and SAMN19369283.
References
- Aberer AJ, Kobert K, Stamatakis A.. 2014. ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol Biol Evol. 31(10):2553–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantey PT, Weeks J, Edwards M, Rao S, Ostovar GA, Dehority W, Alzona M, Swoboda S, Christiaens B, Ballan W, et al. 2016. The emergence of zoonotic Onchocerca lupi infection in the United States – a case-series. Clin Infect Dis. 62(6):778–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Doyle J.. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15. [Google Scholar]
- Eberhard ML, Ostovar GA, Chundu K, Hobohm D, Feiz-Erfan I, Mathison BA, Bishop HS, Cantey PT.. 2013. Zoonotic Onchocerca lupi infection in a 22-month-old child in Arizona: first report in the United States and a review of the literature. Am J Trop Med Hyg. 88(3):601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HK, Bolcen S, Kubofcik J, Nutman TB, Eberhard ML, Middleton K, Wekesa JW, Ruedas G, Nelson KJ, Dubielzig R, et al. 2015. Isolation of Onchocerca lupi in dogs and black flies, California, USA. Emerg Infect Dis. 21(5):789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR.. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie EM, Higazi T, Unnasch TR.. 1998. The mitochondrial genome of Onchocerca volvulus: sequence, structure and phylogenetic analysis. Mol Biochem Parasitol. 95(1):111–127. [DOI] [PubMed] [Google Scholar]
- Kinkar L, Laurimäe T, Simsek S, Balkaya I, Casulli A, Manfredi MT, Ponce-Gordo F, Varcasia A, Lavikainen A, González LM, et al. 2016. High-resolution phylogeography of zoonotic tapeworm Echinococcus granulosus sensu stricto genotype G1 with an emphasis on its distribution in Turkey, Italy and Spain. Parasitology. 143(13):1790–1801. [DOI] [PubMed] [Google Scholar]
- Labelle AL, Daniels JB, Dix M, Labelle P.. 2011. Onchocerca lupi causing ocular disease in two cats. Vet Ophthalmol. 14(Suppl 1):105–110. [DOI] [PubMed] [Google Scholar]
- Laurimäe T, Kinkar L, Romig T, Omer RA, Casulli A, Umhang G, Gasser RB, Jabbar A, Sharbatkhori M, Mirhendi H, et al. 2018. The benefits of analysing complete mitochondrial genomes: deep insights into the phylogeny and population structure of Echinococcus granulosus sensu lato genotypes G6 and G7. Infect Genet Evol. 64:85–94. [DOI] [PubMed] [Google Scholar]
- Lefoulon E, Giannelli A, Makepeace BL, Mutafchiev Y, Townson S, Uni S, Verocai GG, Otranto D, Martin C.. 2017. Whence river blindness? The domestication of mammals and host-parasite co-evolution in the nematode genus Onchocerca. Int J Parasitol. 47(8):457–470. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean NJ, Newkirk K, Adema CM.. 2017. Canine ocular onchocerciasis: a retrospective review of the diagnosis, treatment, and outcome of 16 cases in New Mexico (2011–2015). Vet Ophthalmol. 20(4):349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otranto D, Dantas-Torres F, Giannelli A, Latrofa MS, Papadopoulos E, Cardoso L, Cortes H.. 2013. Zoonotic Onchocerca lupi infection in dogs, Greece and Portugal, 2011–2012. Emerg Infect Dis. 19(12):2000–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe CC, Horn KS, Driebe EM, Bowers J, Terriquez JA, Keim P, Engelthaler DM.. 2016. Whole genome SNP typing to investigate methicillin-resistant Staphylococcus aureus carriage in a health-care provider as the source of multiple surgical site infections. Hereditas. 153:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe CC, Yaglom H, Howard A, Urbanz J, Verocai GG, Andrews L, Harrison V, Barnes R, Lyons T, Bowers JR, et al. 2020. Coyotes as reservoirs for Onchocerca lupi, United States, 2015–2018. Emerg Infect Dis. 26(12):2989–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, et al. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genome. 2(8):e000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verocai GG, Conboy G, Lejeune M, Marron F, Hanna P, MacDonald E, Skorobohach B, Wilcock B, Kutz SJ, Gilleard JS, et al. 2016. Onchocerca lupi nematodes in dogs exported from the United States into Canada. Emerg Infect Dis. 22(8):1477–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MW266120.1 under the accession no. MW266120. The associated BioProject and Bio-Sample numbers are PRJNA733160 and SAMN19369283.