Abstract
Purpose
To identify distinguishing CT radiomic features of pancreatic ductal adenocarcinoma (PDAC) and to investigate whether radiomic analysis with machine learning can distinguish between patients who have PDAC and those who do not.
Materials and Methods
This retrospective study included contrast material–enhanced CT images in 436 patients with PDAC and 479 healthy controls from 2012 to 2018 from Taiwan that were randomly divided for training and testing. Another 100 patients with PDAC (enriched for small PDACs) and 100 controls from Taiwan were identified for testing (from 2004 to 2011). An additional 182 patients with PDAC and 82 healthy controls from the United States were randomly divided for training and testing. Images were processed into patches. An XGBoost (https://xgboost.ai/) model was trained to classify patches as cancerous or noncancerous. Patients were classified as either having or not having PDAC on the basis of the proportion of patches classified as cancerous. For both patch-based and patient-based classification, the models were characterized as either a local model (trained on Taiwanese data only) or a generalized model (trained on both Taiwanese and U.S. data). Sensitivity, specificity, and accuracy were calculated for patch- and patient-based analysis for the models.
Results
The median tumor size was 2.8 cm (interquartile range, 2.0–4.0 cm) in the 536 Taiwanese patients with PDAC (mean age, 65 years ± 12 [standard deviation]; 289 men). Compared with normal pancreas, PDACs had lower values for radiomic features reflecting intensity and higher values for radiomic features reflecting heterogeneity. The performance metrics for the developed generalized model when tested on the Taiwanese and U.S. test data sets, respectively, were as follows: sensitivity, 94.7% (177 of 187) and 80.6% (29 of 36); specificity, 95.4% (187 of 196) and 100% (16 of 16); accuracy, 95.0% (364 of 383) and 86.5% (45 of 52); and area under the curve, 0.98 and 0.91.
Conclusion
Radiomic analysis with machine learning enabled accurate detection of PDAC at CT and could identify patients with PDAC.
Keywords: CT, Computer Aided Diagnosis (CAD), Pancreas, Computer Applications-Detection/Diagnosis
Supplemental material is available for this article.
© RSNA, 2021
An earlier incorrect version of this article appeared online. This article was corrected on July 30, 2021.
Keywords: CT, Computer Aided Diagnosis (CAD), Pancreas, Computer Applications-Detection/Diagnosis
Summary
Radiomic analysis of CT images with machine learning enabled accurate differentiation between patients with pancreatic cancer and healthy controls.
Key Points
■ Distinct radiomic features reflecting imaging intensity and heterogeneity were decreased and increased, respectively, in pancreatic ductal adenocarcinoma (PDAC).
■ In a radiomics machine learning model that was predominantly trained with Taiwanese training images, the model achieved 95.0% accuracy on Taiwanese test images and 86.5% on test images from the United States for differentiating between patients with and without PDAC; sensitivities of 96.4% and 90.9% were obtained for the two test data sets, respectively, for PDACs smaller than 2 cm.
■ In the Taiwanese test data set, the radiomics model correctly identified all PDACs missed by radiologists.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most lethal cancer and projected to become the second leading cause of cancer death in the United States by 2030 (1). CT is the major imaging modality for detection and diagnosis of PDAC, but small PDACs are often obscure. Approximately 40% of tumors that are smaller than 2 cm are missed at CT (2). Furthermore, the diagnostic performance of CT is interpreter dependent and thus influenced by disparities in radiologist availability and expertise. As patient survival sharply declines as a tumor grows larger than 2 cm (3), methods to improve the diagnostic performance of CT are urgently needed.
Radiomics is a method that extracts quantitative information on density, shape, and texture in medical images (4). Analysis of radiomic features by machine learning algorithms that can build predictive models through learning from examples have been shown to discover patterns and rules indiscernible to the naked eye (4,5). A recent single-institution study showed that CT radiomic features could distinguish PDAC from normal pancreas (6), but the generalizability of the results was not assessed.
Ascertaining the radiomic characteristics of PDAC may enable detection of small or occult PDACs and supplement radiologist interpretation. Understanding the generalizability of radiomics-based image analysis is essential before deployment in real clinical practice. We aimed to identify the distinguishing CT radiomic features of PDAC, investigate whether radiomic analysis with machine learning could differentiate between PDAC and noncancerous pancreas and supplement radiologist interpretation, and assess the generalizability of the model across institutions and populations.
Materials and Methods
This retrospective study was approved by the institute’s Research Ethics Committee (approval no. 201710050RINA), which waived individual patient consent. This study complied with the Health Insurance Portability and Accountability Act.
Study Design
The numbers of patients and images included are summarized in Figure 1. Patients with histologically or cytologically confirmed PDAC were identified from the cancer registry of National Taiwan University Hospital, a tertiary referral center in Taiwan. Contrast material–enhanced portal venous CT images in 436 patients with confirmed PDAC between January 1, 2012, and December 31, 2018, and 479 patients undergoing CT during the same period with a negative or unremarkable pancreas in the radiologist report were randomly split into the local training (262 patients with PDAC and 287 controls) and validation set (87 patients with PDAC and 96 controls) and a test set (local test 1: 87 patients with PDAC, 96 controls). Images of another 100 patients with PDAC and 100 individuals with normal pancreas imaged between October 1, 2004, and December 31, 2011, from the same institution were retrieved to form local test 2, with small PDACs (< 2 cm) preferentially selected. As early PDACs are often asymptomatic and may manifest as incidental findings at CT examinations performed for nonpancreatic diseases or conditions, the indications for CT were not considered in selecting the controls. Images in the controls were reviewed to confirm the absence of abnormalities during manual labeling of the pancreas by one of the two radiologists participating in this study (see below) and verified as not having PDAC by referencing the cancer registry of the institute (PDAC diagnosis updated to December 31, 2019, mortality from PDAC updated to September 30, 2020).
Figure 1:
Flow diagram of data sets.
Overall, the local data sets included 27 235 CT images (8392 PDACs and 18 843 controls) in 1115 patients (536 patients with PDAC and 579 controls). Formal radiologist reports of the images in the patients with PDAC in the local test sets, which were interpreted by attending radiologists in this academic tertiary center with a large volume of patients with PDAC, were retrieved from electronic health records to ascertain radiologist performance (details on image selection, assessment of radiologist report, and CT equipment and protocol in Appendix E1 [supplement]). A total of 475 of the 732 patients in the training and validation set and 290 of 383 in the local test sets had been used for training and testing in a previous study on distinguishing PDAC from normal pancreas with deep learning (7); however, this prior study did not assess radiomic features.
The external data set consisted of 12 769 CT images (5715 PDACs and 7054 controls) obtained in 264 patients (182 with PDAC and 82 controls) from the United States (Medical Segmentation Decathlon Dataset [8,9] and The Cancer Imaging Archive [10–12]) (Fig 1, details in Appendix E1 [supplement]).
CT Acquisition
CT examinations were performed using one of six CT scanners (Brilliance iCT 256, Philips Healthcare; Sensation 64 and SOMATOM Definition AS+, Siemens Healthcare; Aquilion ONE, Toshiba; Revolution CT and LightSpeed VCT, GE Healthcare) with 100, 120, or 130 kV, automatic milliampere control, and without extra noise reduction processes. The section thickness ranged between 0.7 mm and 1.5 mm, and the image size was 512 × 512 pixels. Portal venous phase images were obtained at 70–80 seconds after intravenous administration of contrast medium (Omnipaque-350, GE Healthcare; 1.5 mL per kilogram of body weight, with an upper limit of 150 mL). All images were reconstructed into 5-mm sections.
Segmentation and Image Preprocessing
The pancreases on CT images were manually segmented and labeled as regions of interest (ROIs). In patients with PDAC, the tumor was also segmented and labeled as ROI. The images were manually segmented by one of two experienced abdominal radiologists (P.T.C. and K.L.L.) who had 5 years and 20 years of experience, respectively. The images were subsequently cropped into patches for further analysis (detailed methods in Appendix E1 [supplement]).
Extraction of Radiomic Features and Model Training
We followed the Image Biomarker Standardization Initiative guideline in conducting the radiomic analysis (13). Because the tumor or pancreas was processed into patches, we excluded 13 features that were either shape-based or confounded by the volume of the analyzed object and extracted the remaining 88 nonfiltered radiomic features using PyRadiomics (version 2.2.0; https://pypi.org/project/pyradiomics/2.2.0/) (14) (details in Appendix E1 [supplement]).
For classifying patches as cancerous or noncancerous (patch-based analysis), features of all patches in the training set were input into a machine learning model (XGBoost) (15). A local model was trained with just Taiwanese training and validation data. To assess whether generalizability across populations could be improved by adding a modest number of U.S. images to the training data, a generalized model was also trained with local training and validation sets and external images, derived by randomly splitting the external data set into external training (110 PDAC and 50 controls) and validation data sets (36 PDAC and 16 controls) and an external test set (external test set 2, 36 PDAC and 16 controls) (Fig 1). The score for classifying a patient as with or without PDAC (patient-based analysis) was defined as the proportion of patches in the ROI that were predicted as cancerous in the patch-based analysis (methods of cutoff selection in Appendix E1 [supplement]).
For training the local and generalized patch-based models, features were sequentially added into reduced models in order of descending gain value, a measure of improvement in model performance conferred by individual features, until the retrained reduced model yielded comparable performance to the full model including all the features in the training and validation sets (ie, equal area under the receiver operating characteristic curve [AUC], rounded to the second decimal, in both patch- and patient-based analyses). As models with fewer variables are less susceptible to overfitting (16), the reduced models were used for further analyses and testing. The t-distributed stochastic neighbor embedding (t-SNE) (17) analysis was used to visualize how well the features selected in both models distinguished PDAC from normal pancreas.
Model Testing and Statistical Analysis
Both the local and generalized models were tested with both local test sets. The local model was also tested with both external test sets (external data set 1 consisted of all 264 patients from the U.S. data set, and external data set 2 consisted of only 52 patients from the U.S. data set). Because the generalized model was trained on data from external data set 1, this model was only tested on external data set 2. Model performance was assessed with receiver operating characteristic curves and associated AUCs and compared between the local and generalized models using the DeLong test implemented with the pROC package in R (18,19). Sensitivity, specificity, and accuracy were calculated for patch- and patient-based analyses, with 95% CIs calculated on the basis of binomial distributions. Because sensitivity for detection is the major unmet clinical need, the sensitivity of patient-based analysis was compared with that of radiologists in the local test sets. As all controls were selected on the basis of absence of any pancreatic abnormality in the radiologist report, the specificity of radiologists could not be determined. Fisher exact test and Mann-Whitney U test were used for comparing categorical variables and continuous variables, respectively, with P values less than .05 considered as statistically significant.
Results
Patient Characteristics
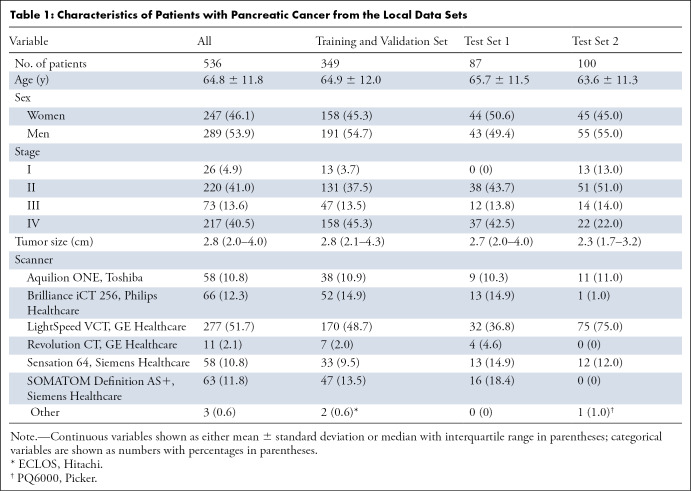
Patient characteristics were comparable between the training and validation sets and local test sets (Table 1). Median tumor size was 2.8 cm (interquartile range, 2.0–4.0 cm) in patients with PDAC (mean age, 65 years ± 12; 289 men). Local test set 2 had slightly smaller tumors compared with the training and validation sets.
Table 1:
Characteristics of Patients with Pancreatic Cancer from the Local Data Sets
Model Training
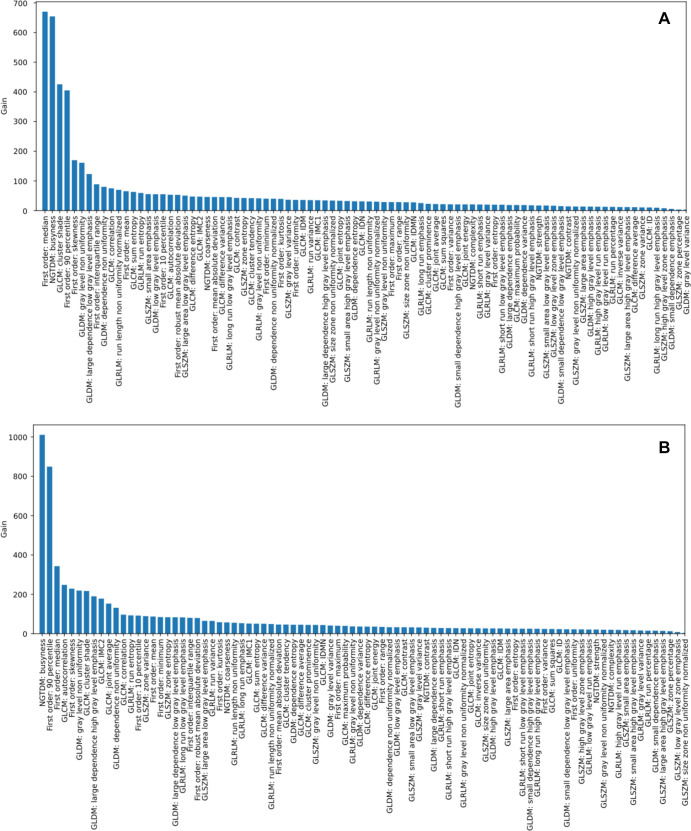
The local training and validation data set consisted of 34 164 cancerous patches and 100 955 noncancerous patches, and the combined training and validation data set (both internal and external data set patches) consisted of 41 088 cancerous patches and 137 955 noncancerous patches. Figure 2 shows the importance of features in distinguishing cancerous and noncancerous patches in the training and validation sets. A 14-feature model and a 20-feature model were selected as the local and the generalized models for subsequent testing (Tables E1, E2; Fig E1 [supplement]).
Figure 2:
Feature importance in differentiation between cancerous and noncancerous patches in the (A) local training and validation set and (B) combined (local and external) training and validation set.
Distinguishing Radiomic Features of PDAC
Eleven radiomic features were selected in both models (Fig 3, Table E3 [supplement]). A t-SNE based on these features separated cancerous and noncancerous patches (Fig 4), indicating the importance of these features in distinguishing PDAC from normal pancreas. Among the features, cancerous patches had lower values for three features reflecting image intensity (first order: median, first order: 90th percentile, and first order: mean) and higher values for three features reflecting heterogeneity (neighboring gray-tone difference matrix [NGTDM] busyness, gray-level dependence matrix [GLDM] gray-level nonuniformity, and GLDM dependence nonuniformity) compared with noncancerous patches (Table E4 [supplement]), consistent with the notion that PDACs typically manifest with heterogeneous hypoenhancement at CT (20,21).
Figure 3:
Radiomic features selected in the trained XGBoost models for patch-based analysis. Features are ranked in descending order of gain value. Features selected by both models are connected. GLCM = gray-level co-occurrence matrix, GLDM = gray-level dependence matrix, GLRLM = gray-level run-length matrix, GLSZM = gray-level size-zone matrix, IMC2 = information measure of correlation 2, NGTDM = neighboring gray-tone difference matrix.
Figure 4:

Separation of cancerous and noncancerous patches by t-distributed stochastic neighbor embedding (t-SNE). The t-SNE was based on the 11 features selected in both local and generalized models (perplexity = 30). The patches used for constructing the plot were selected by random sampling (sampling ratio, 0.1) with stratification by data set (local and external) and class (noncancerous and cancerous).
Model Performance in Local Test Sets
Model performance in the test sets is summarized in Tables 2 and 3. In local test set 1, the local model yielded 92.2% sensitivity (95% CI: 91.6, 92.7), 90.2% specificity (95% CI: 89.8, 90.5), and 90.6% accuracy (95% CI: 90.3, 90.9) in classifying patches (AUC 0.97 [95% CI: 0.97, 0.97]) (Fig E1E [supplement]) and 96.6% sensitivity (95% CI: 90.3, 99.3), 93.8% specificity (95% CI: 86.9, 97.7), and 95.1% accuracy (95% CI: 90.9, 97.7) (AUC 0.97 [95% CI: 0.94, 1.00]) in classifying patients (Fig E1F [supplement]).
Table 2:
Performance Metrics from Patch-based Models on Local and External Test Data sets
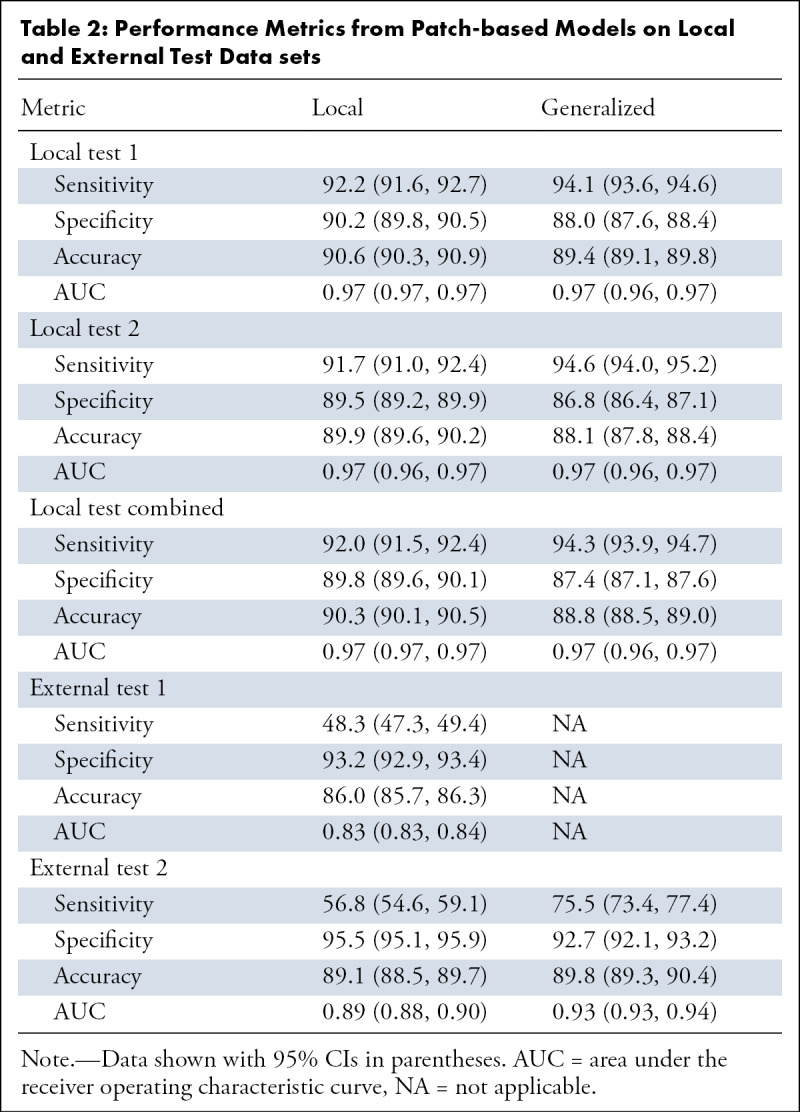
Table 3:
Performance Metrics from Patient-based Models on Local and External Test Data Sets
In local test set 2, the local model yielded 91.7% sensitivity (95% CI: 91.0, 92.4), 89.5% specificity (95% CI: 89.2, 89.9), and 89.9% accuracy (95% CI: 89.6, 90.2) in classifying patches (AUC 0.97 [95% CI: 0.96, 0.97]; Fig E1G [supplement]) and 91.0% sensitivity (95% CI: 83.6, 95.8), 90.0% specificity (95% CI: 82.4, 95.1), and 90.5% accuracy (95% CI: 85.6, 94.2) in classifying patients (AUC 0.94 [95% CI, 0.89–0.98]; Fig E1H [supplement]).
Analysis with the generalized model yielded comparable performance with the local model in both local test sets (Table 2, Fig E1E–H [supplement]). Analysis of the two local test sets combined yielded similar results as when separately analyzed, with comparable performance between the two models (Table 2; Fig 5A, 5B).
Figure 5:
Receiver operating characteristic curves of the local model and generalized model in the combined local test set (A patch-based and B patient-based) and external test set 2 (C patch-based [P < .001] and D patient-based [P = .08]). AUC = area under the receiver operating characteristic curve.
Model Performance on External Test Sets
When tested on external test set 1, the local model yielded 48.3% sensitivity (95% CI: 47.3, 49.4), 93.2% specificity (95% CI: 92.9, 93.4), and 86.0% accuracy (95% CI: 85.7, 86.3), with an AUC of 0.83 (95% CI: 0.83, 0.84) in classifying patches (Fig E1I [supplement]); the model yielded 65.9% sensitivity (95% CI: 58.6, 72.8), 96.3% specificity (95% CI: 89.7, 99.2), and 75.4% accuracy (95% CI: 69.7, 80.5) with an AUC of 0.76 (95% CI: 0.71, 0.82) for classifying patients (Fig E1J [supplement]).
On external test set 2, the local model yielded 56.8% sensitivity (95% CI: 54.6, 59.1), 95.5% specificity (95% CI: 95.1, 95.9), and 89.1% accuracy (95% CI: 88.5%, 89.7%) in classifying patches with an AUC of 0.89 (95% CI: 0.88, 0.90) (Fig 5C); the model yielded 72.2% sensitivity (95% CI: 54.8, 85.8), 93.8% specificity (95% CI: 69.8, 99.8), and 78.8% accuracy (95% CI: 65.3, 88.9) with an AUC of 0.85 (95% CI: 0.74, 0.95) in classifying patients (Fig 5D).
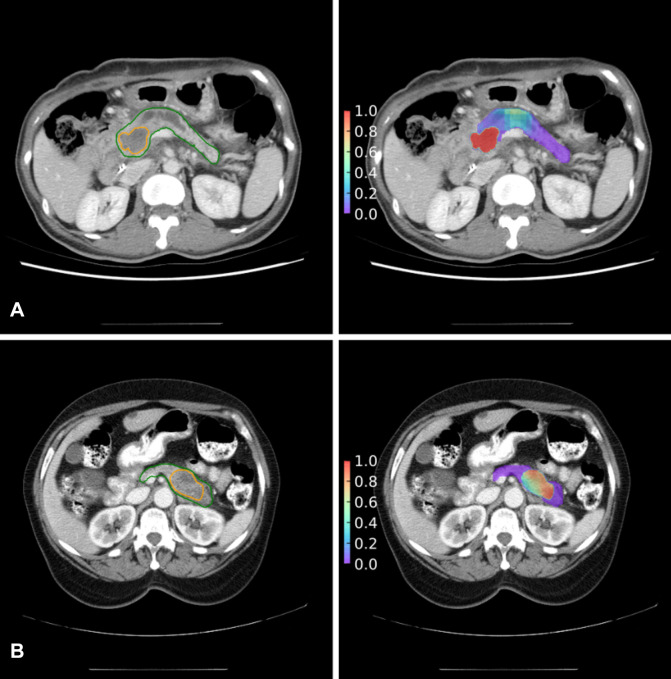
The generalized model outperformed the local model in classifying patches (AUC, 0.93 [95% CI: 0.93, 0.94]; P < .001) (Fig 5C) but performed similarly to the local model for classifying patients (AUC, 0.91 [95% CI: 0.83, 1.00]; P = .08; Fig 5D). Figure 6 shows correspondence in tumor location between model prediction and radiologist’s labeling.
Figure 6:
Correspondence in tumor location between model prediction (right panels) and radiologists’ labeling (left panels). Tumors are from the (A) local test set and (B) external test set. Areas encircled in the left panels are the cancer (orange) and pancreas (green) labeled by radiologists. Heatmaps (right panels) are constructed with the probabilities of cancer predicted by the generalized model.
Comparison with Radiologist Reports
The local and generalized models achieved sensitivities of 93.6% (95% CI: 89.1, 96.6) and 94.7% (95% CI: 90.4%, 97.4%), respectively, which was similar to radiologist sensitivity (92.1% [95% CI: 87.2, 95.6]; P = .59 and P = .33, respectively) (Table 2). Analyses with the local model, generalized model, and radiologist reports missed 12, 10, and 14 PDAC cases, respectively (Tables E5, E6 [supplement]). All PDAC cases missed by radiologists were correctly classified by radiomic analysis with either the local or generalized model, including five PDACs smaller than 2 cm. All the PDACs missed by radiomics-based analysis, except one without radiologist report, were correctly classified by radiologists.
Sensitivity according to Tumor Size and Stage
In both local test sets, the sensitivity of radiomics-based analysis for detecting PDAC was comparable to radiologists in all categories of primary tumor size and tumor stage (Tables 4, 5). For both local test sets combined, the sensitivities of radiomics-based analysis for PDACs smaller than 2 cm were 90.9% (95% CI: 80.0%, 97.0%) with the local model and 96.4% (95% CI: 87.5, 99.6) with the generalized model. The sensitivities for the resectable PDACs were 92.3% (95% CI: 64.0, 99.8) for stage I and 92.1% (95% CI: 84.5, 96.8) for stage II with the local model and 92.3% (95% CI: 64.0, 99.8) for stage I and 94.4% (95% CI: 87.4, 98.2) for stage II with the generalized model. In external test set 2, both models yielded a sensitivity of 90.9% (95% CI: 58.7, 99.8) for PDACs smaller than 2 cm. Information on cancer stage was not provided in the external test set and therefore was not assessed.
Table 4:
Sensitivity of Local and Generalized Model in Various Test Data Sets Stratified by Tumor Size
Table 5:
Sensitivity of the Local and Generalized Models in Various Data Sets Stratified by Tumor Stage
Discussion
This study aimed to investigate whether radiomic analysis with machine learning could differentiate between PDAC and noncancerous pancreas to supplement radiologist interpretation and assess its generalizability. We identified a panel of distinguishing radiomic features of PDAC (low first order: mean, median, and 90th percentile; high NGTDM: busyness; GLDM: gray-level nonuniformity and dependence nonuniformity), corresponding with the typical CT findings of PDACs as hypoattenuating heterogeneous masses (20,21). Radiomic analysis with a model trained with predominantly Taiwanese images could differentiate between patients with PDAC and controls in Taiwanese (accuracy, 95.0%) and U.S. (accuracy, 86.5%) test images, with 96.9% and 90.9% sensitivity, respectively, for PDACs smaller than 2 cm. Notably, radiomics-based analysis achieved comparable sensitivity to radiologists of a referral center and correctly detected all PDACs missed by radiologists, supporting the potential of supplementing radiologist interpretation.
Early detection is the most effective strategy to improve the prognosis of PDAC (3,22,23). However, 80% of PDACs have irregular contours and inconspicuous borders with surrounding tissue at CT. Therefore, small PDACs are often obscure and may evade detection (24), especially when the CT scan is ordered for nonpancreatic diseases and conditions. Hence, a CT scan, regardless of the indications for scanning, should be carefully evaluated with respect to the pancreas to reduce the probability of missed PDAC. A radiomics model could help reduce missed PDAC, which prompted our development of the models.
A recent study also used radiomics to differentiate between PDAC and normal pancreas and reported 100% sensitivity, 98.5% specificity, and 99.2% accuracy in test images from the same institution (6), but the generalizability was not assessed with external images. That study used the whole pancreas including the tumor in calculating the radiomic features; therefore, most of the obtained features represented mixtures of the tumor and nontumorous pancreas. By contrast, in this study, radiomic features were separately extracted from the tumor and nontumorous pancreas, which were then used to identify radiomic features that distinguish PDAC from normal pancreas across different patient populations and institutions.
Our results support that a well-trained local radiomic model may be optimized to achieve adequate generalizability to another population by adding a modest training data set from the latter population, obviating de novo training with a large amount of data from the latter population, which is often difficult to obtain (25). In this study, adding a comparatively small number of U.S. images to Taiwanese training data did not improve model performance in Taiwanese test sets, but in U.S. test set models, sensitivity increased by nearly 20% and 10% in patch- and patient-based analyses, respectively. Multiple factors could underlie the differences between the Taiwanese and U.S. data sets, such as differences in scanners or imaging protocols and parameters. Furthermore, disparity in pancreatic fat content might also cause differences in pancreatic radiomic profiles. Fatty pancreas, a condition manifesting as decreased intensity of the pancreas at CT and associated with obesity and metabolic syndrome, is more prevalent in the United States compared with Taiwan (prevalence 27.8% [26] vs 12.9% [27]).
This study lends further support to the notion that radiomic analysis with machine learning holds promise for developing computer-aided detection and diagnosis tools. XGBoost is a machine learning algorithm that has consistently outperformed other algorithms for classification (15). During model training, XGBoost adds decision trees sequentially such that a latter tree improves classification for the samples misclassified by the preceding tree (ie, each tree minimizes the errors of the previous tree), and the final model aggregates the predictions made by all trees. XGBoost also quantifies improvement in model performance conferred by each feature (ie, gain value) and thereby guides feature selection.
While the major unmet clinical need is the sensitivity for detection, given the relatively lower incidence of PDAC compared with more common malignancies (eg, lung and colorectal cancers), a computer-aided diagnostic tool for PDAC must also have high specificity to be applicable in real clinical practice. The finding that both sensitivity and specificity increased in the patient-based analysis compared with those in the patch-based analysis reaffirmed the notion that the two-level analytical approach is a solution to the dilemma of requiring high sensitivity and specificity simultaneously (7). A trade-off between sensitivity and specificity is inevitable if the presence and/or absence of PDAC is determined by a single analysis. Instead, our patch-level analysis examined the tumor at multiple fine-grained subregions, rather than once only with the whole-pancreas approach. Because the patches were overlapping, each subregion was analyzed multiple times, each with different surrounding subregions, to increase the sensitivity for PDAC. It is worth noting that the narrow 95% CIs in patch-based analysis could be attributed to the large number of patches and thus should not be overinterpreted. To prevent inflated type I error (ie, false-positive findings and reduced specificity) from multiple examinations at the patch level, diagnosing PDAC at the patient level required the proportion of patches predicted as cancerous to exceed a threshold.
This study had limitations. Radiologist reports were not available in the U.S. data set, precluding comparison between radiomic analysis and radiologist interpretation. Second, due to barriers limiting sharing of patient data and paucity of open data sets (25), the PDAC cases in both Taiwanese and U.S. data sets were from a single institution. However, those institutions were major PDAC referral centers in Taiwan and the United States and thus should have provided a representative sample of PDAC in respective populations. We seek to increase the volume and diversity of training and testing data sets in future study. Last, this proof-of-concept study aimed to investigate whether radiomic analysis might differentiate between PDAC and normal pancreas; therefore, the specificity of the trained model in patients with suspected PDAC whose images may have included various conditions that mimic or coincide with PDAC was not assessed in this study. As different cutoffs may be needed in different target populations and/or conditions, the models derived from this study should be further evaluated in patients suspected of having PDAC to ascertain its performance and the associated optimal cutoffs for various conditions.
In conclusion, this study identified the key radiomic features that distinguish PDAC from normal pancreas and demonstrated that radiomic analysis with a machine learning model could enable accurate detection and diagnosis of PDAC at CT and supplement radiologist interpretation. Further research to validate these findings and explore its potential clinical applications is warranted.
Acknowledgments
Acknowledgments
The authors thank Nico Wu for assistance in data management and preprocessing and Tinghui Wu and Pochuan Wang for computational technical support.
P.T.C and D.C contributed equally to this work.
W.C.L. and W.W. are co-senior authors who contributed equally to this work.
Supported by the Ministry of Science and Technology, Taiwan (grants MOST 107-2634-F-002-014-, MOST 107-2634-F-002-016-, MOST 108-2634-F-002-011-, MOST 108-2634-F-002-013-, MOST 109-2634-F-002-028-, and MOST 110-2634-F-002-030-), MOST All Vista Healthcare Sub-center, and National Center for Theoretical Sciences Mathematics Division.
Disclosures of Conflicts of Interest: P.T.C. disclosed no relevant relationships. D.C. Activities related to the present article: author’s institution has grant with the Ministry of Science and Technology, Taiwan, and author receives salary from this grant. Activities not related to the present article: author is one of the inventors of pending patent entitled Medical Image Analyzing System and Method Thereof. Other relationships: disclosed no relevant relationships. H.Y. disclosed no relevant relationships. K.L.L. disclosed no relevant relationships. S.Y.H. disclosed no relevant relationships. H.R. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: author is full-time employee of NVIDIA. Other relationships: disclosed no relevant relationships. M.S.W. disclosed no relevant relationships. W.C.L. Activities related to the present article: author’s institution has grant from the Ministry of Science and Technology; author’s institution received support for travel to meetings for the study or other purposes from the Ministry of Science and Technology. Activities not related to the present article: disclosed no relevant relationships. Other relationships: author’s institution has patent pending with National Taiwan University. W.W. Activities related to the present article: author’s institution has grant from the Ministry of Science and Technology (grant ID 110-2634-F-002-030). Activities not related to the present article: disclosed no relevant relationships. Other relationships: author’s institution has patent pending with National Taiwan University, for Medical Image Analyzing System and Method Thereof.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- GLDM
- gray-level dependence matrix
- NGTDM
- neighboring gray-tone difference matrix
- PDAC
- pancreatic ductal adenocarcinoma
- ROI
- region of interest
- t-SNE
- t-distributed stochastic neighbor embedding
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74(11):2913–2921. [DOI] [PubMed] [Google Scholar]
- 2.Dewitt J, Devereaux BM, Lehman GA, Sherman S, Imperiale TF. Comparison of endoscopic ultrasound and computed tomography for the preoperative evaluation of pancreatic cancer: a systematic review. Clin Gastroenterol Hepatol 2006;4(6):717–725.quiz 664. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal B, Correa AM, Ho L. Survival in pancreatic carcinoma based on tumor size. Pancreas 2008;36(1):e15–e20. [DOI] [PubMed] [Google Scholar]
- 4.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. RadioGraphics 2017;37(2):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu LC, Park S, Kawamoto S, et al. Utility of CT radiomics features in differentiation of pancreatic ductal adenocarcinoma from normal pancreatic tissue. AJR Am J Roentgenol 2019;213(2):349–357. [DOI] [PubMed] [Google Scholar]
- 7.Liu KL, Wu T, Chen PT, et al. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: a retrospective study with cross-racial external validation. Lancet Digit Health 2020;2(6):e303–e313. [DOI] [PubMed] [Google Scholar]
- 8.Medical Segmentation Decathlon. http://medicaldecathlon.com/index.html. Accessed July 29, 2020.
- 9.Simpson AL, Antonelli M, Bakas S, et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv:1902.09063 [cs.CV] [preprint] http://arxiv.org/abs/1902.09063. Posted February 25, 2019. Accessed July 29, 2020. [Google Scholar]
- 10.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pancreas-CT-The Cancer Imaging Archive (TCIA) Public Access - Cancer Imaging Archive Wiki. https://wiki.cancerimagingarchive.net/display/Public/Pancreas-CT. Accessed April 19, 2020.
- 12.Roth HR, Lu L, Farag A, et al. DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds.Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science, vol 9349.Cham, Switzerland:Springer,2015;556–564. [Google Scholar]
- 13.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv:1612.07003v11 [cs.CV] [preprint] http://arxiv.org/abs/1612.07003v11. Posted December 21, 2016. Accessed October 21, 2020. [Google Scholar]
- 14.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. In: KDD ‘16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, NY: Association for Computing Machinery, 2016; 785–794. [Google Scholar]
- 16.Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and generalizability in radiomics modeling: possible strategies in radiologic and statistical perspectives. Korean J Radiol 2019;20(7):1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Maaten L, Hinton G. Visualizing Data using t-SNE. J Mach Learn Res 2008;9(86):2579–2605.https://www.jmlr.org/papers/v9/vandermaaten08a.html. [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44(3):837–845. [PubMed] [Google Scholar]
- 19.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietryga JA, Morgan DE. Imaging preoperatively for pancreatic adenocarcinoma. J Gastrointest Oncol 2015;6(4):343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman MK, Gandhi NS. Imaging Evaluation of Pancreatic Cancer. Surg Clin North Am 2016;96(6):1235–1256. [DOI] [PubMed] [Google Scholar]
- 22.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 23.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371(22):2140–2141. [DOI] [PubMed] [Google Scholar]
- 24.Frampas E, David A, Regenet N, Touchefeu Y, Meyer J, Morla O. Pancreatic carcinoma: Key-points from diagnosis to treatment. Diagn Interv Imaging 2016;97(12):1207–1223. [DOI] [PubMed] [Google Scholar]
- 25.Liao WC, Simpson AL, Wang W. Convolutional neural network for the detection of pancreatic cancer on CT scans - Authors’ reply. Lancet Digit Health 2020;2(9):e454. [DOI] [PubMed] [Google Scholar]
- 26.Sepe PS, Ohri A, Sanaka S, et al. A prospective evaluation of fatty pancreas by using EUS. Gastrointest Endosc 2011;73(5):987–993. [DOI] [PubMed] [Google Scholar]
- 27.Wu WC, Wang CY. Association between non-alcoholic fatty pancreatic disease (NAFPD) and the metabolic syndrome: case-control retrospective study. Cardiovasc Diabetol 2013;12(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]





![Receiver operating characteristic curves of the local model and generalized model in the combined local test set (A patch-based and B patient-based) and external test set 2 (C patch-based [P < .001] and D patient-based [P = .08]). AUC = area under the receiver operating characteristic curve.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1376/8344348/be1fbdacf34c/rycan.2021210010fig5.jpg)