Abstract
The radiologic appearance of locally advanced lung cancer may be linked to molecular changes of the disease during treatment, but characteristics of this phenomenon are poorly understood. Radiomics, liquid biopsy of cell-free DNA (cfDNA), and next-generation sequencing of circulating tumor DNA (ctDNA) encode tumor-specific radiogenomic expression patterns that can be probed to study this problem. Preliminary findings are reported from a radiogenomic analysis of CT imaging, cfDNA, and ctDNA in 24 patients (median age, 64 years; range, 49–74 years) with stage III lung cancer undergoing chemoradiation on a prospective pilot study (NCT00921739) between September 2009 and September 2014. Unsupervised clustering of radiomic signatures resulted in two clusters that were associated with ctDNA TP53 mutations (P = .03) and changes in cfDNA concentration after 2 weeks of chemoradiation (P = .02). The radiomic features dissimilarity (hazard ratio [HR] = 0.56; P = .05), joint entropy (HR = 0.56; P = .04), sum entropy (HR = 0.53; P = .02), and normalized inverse difference (HR = 1.77; P = .05) were associated with overall survival. These results suggest heterogeneous and low-attenuating disease without a detectable ctDNA TP53 mutation was associated with early surges of cfDNA concentration in response to therapy and a generally better prognosis.
Keywords: CT-Quantitative, Radiation Therapy, Lung, Computer Applications-3D, Oncology, Tumor Response, Outcomes Analysis
Clinical trial registration no. NCT00921739
Supplemental material is available for this article.
© RSNA, 2021
Keywords: CT-Quantitative, Radiation Therapy, Lung, Computer Applications-3D, Oncology, Tumor Response, Outcomes Analysis
Summary
Radiogenomic expression patterns (combining features from CT imaging with cell-free DNA and circulating tumor DNA in serum) were identified in patients with stage III lung cancer and were associated with overall survival.
Key Points
■ Using 61 different radiomic features, two distinct radiomic signatures were identified in patients with lung cancer prior to undergoing chemoradiation therapy.
■ The presence of TP53 mutation within circulating tumor DNA (P = .03), as well as changes in cell-free DNA 2 weeks after chemoradiation (P = .02), were associated with the two identified radiomic signatures.
■ Heterogeneous and low-attenuating disease without a detectable circulating tumor DNA TP53 mutation was linked to early surges of cell-free DNA concentration in response to therapy and longer overall survival times.
Introduction
Locally advanced, inoperable lung cancer is a challenging disease. Despite aggressive treatment with chemoradiation, local cancer recurrence is observed in approximately 50% of patients (1), and the median survival rate is only 23 to 25 months (2). Imaging genomics, otherwise known as radiogenomics, is a promising technique which can potentially improve prognostication and help guide new treatment strategies. This emerging field aims to capture the molecular subtypes and genetic underpinnings of a disease based on associations between radiologic imaging and genomic sequencing.
Radiomics is a high-throughput computational technique in which mathematical features are derived from medical images. The analysis of cell-free DNA (cfDNA) is a complementary technique whereby traces of cfDNA are detectable in the bloodstream when apoptotic cells shed fragmented DNA. Using next-generation sequencing, cancer-specific mutations for genes implicated in the pathogenesis of lung cancer can be identified from a subset of cfDNA known as circulating tumor DNA (ctDNA). Both cfDNA and ctDNA, which are mechanistically and technologically different from tissue-based mutational assessment, are promising liquid biopsy-based markers of radiation response (3–7).
This brief report provides preliminary results demonstrating the feasibility of an integrated radiomic, cfDNA, and ctDNA analysis in patients with locally advanced lung cancer. Our method is based on a prospective pilot study of patients who underwent concurrent chemoradiation in which liquid biopsies were performed during treatment. We hypothesized that cell death due to chemoradiation therapy would lead to the release of measurable cfDNA into the bloodstream, which when combined with CT radiomics and next-generation sequencing, could provide patient-specific radiogenomic expression patterns associated with prognosis.
Materials and Methods
Study Design
Our study design is summarized in Figure 1. All research was conducted in keeping with best clinical practice and regulatory compliance. Prospective data (images, plasma, patient outcomes) were generated from Pro00017361 (NCT00921739) approved by the Duke University Institutional Review Board. In addition, the Duke University Institutional Review Board approved a secondary retrospective analysis (radiomics, next-generation sequencing) of the original data with Pro00076119, which was completed under an approved waiver of consent, Health Insurance Portability and Protection Act Authorization, and decedent research notification. Next-generation sequencing was performed by Inivata, of which G.J. is an employee; all remaining authors had control of the data. NCT00921739 participants were previously studied with respect to radiation toxicity (8) and genomic analysis (9). The current article extends this prior work to radiogenomic analysis.
Figure 1:
Pilot study design. (A) Treatment and data acquisition. Prior to chemoradiation, CT images were acquired and phlebotomy was performed to obtain cell-free DNA (cfDNA) from baseline plasma. Patients were treated with a mean dose of 66 Gy with an accelerated fractionation scheme (six fractions/week) with concurrent cisplatin and etoposide. Additional plasma samples were obtained during weeks 2 and 5 of treatment, as well as 6 weeks after treatment. (B) Radiomic transcription and next-generation sequencing. High-throughput radiomic features were extracted from the gross disease within lung parenchyma on CT images, cfDNA concentration was quantified via spectrophotometry, and ctDNA was characterized via next-generation sequencing. (C) Radiogenomic expression panel. For each patient, 61 radiomic features were extracted from the CT image and 36 genes were analyzed for mutational changes. (D) Radiogenomic analysis. Unsupervised data clustering was used to quantify associations between radiomics and genomics, and Cox proportional hazards analysis was used to test the prognostic value of radiomic features.
Patients and Treatment
Between September 2009 and September 2014, 24 patients (Table 1) were enrolled into a phase 1 prospective clinical trial (NCT00921739) to receive cisplatin and etoposide with concurrent thoracic-accelerated radiation therapy for stage III lung cancer. CT images were acquired prior to chemoradiation, and phlebotomy was performed to obtain cfDNA from baseline plasma. Patients were given a mean dose of 66 Gy with an accelerated fractionation scheme (six fractions/week) and concurrent cisplatin and etoposide. Unless patients refused, additional plasma samples were obtained during weeks 2 and 5 of treatment and 6 weeks after treatment. The clinical endpoint was overall survival, patients were censored at the date of last follow-up, and the median follow-up time was calculated based on the reverse Kaplan-Meier method.
Table 1:
Participant and Tumor Characteristics

CT Imaging and Radiomic Feature Extraction
CT image acquisition was performed with either a GE Lightspeed CT scanner or a Siemens Biograph mCT scanner (0.8-mm in-plane resolution, 2.0–2.5-mm section thickness, 120 kVp). The gross tumor volume within the lung parenchyma was manually segmented by a radiation oncologist with 12 years of experience (M.N.C.). The image matrix size was resampled to 0.8-mm isotropic voxel spacing using a tricubic interpolation method, and the dynamic range was rebinned to 64 gray levels. Sixty-one mathematical features (Appendix E1 [supplement]) were extracted to quantify tumor-specific radiomic patterns. Features were encoded into the matrix
 |
where fi,j denotes the ith radiomic feature measured on the image of the jth patient. Each radiomic signature (column vectors of Ƒ) was designed to collectively capture tumor morphologic, intensity, and texture characteristics. Texture features were averaged over 13 directions to approximate rotational invariance. Columns of Ƒ were standardized to zero mean and unit variance. Feature extraction was performed in Matlab using custom software previously benchmarked on digital phantoms (10).
Genomic Expression from cfDNA and ctDNA
Spectrophotometry was used to quantify the concentration of cfDNA within the collected plasma samples as previously described (9). Changes in cfDNA concentration during treatment were quantified based on logarithmic differences relative to baseline concentration
 |
where D(t) is the logarithmic difference in cfDNA concentration at time, t, [cfDNA]t is the concentration of cfDNA at time, t, and [cfDNA]0 is the baseline cfDNA concentration.
Based on the plasma ctDNA, cancer-specific mutations for genes implicated in the pathogenesis of lung cancer (Appendix E2 [supplement]) were characterized using the InvisionSeq next-generation sequencing platform by an enhanced tagged and targeted-amplicon sequencing (eTAm-Seq) technique (7,11). Gene mutations with a threshold detection rate of 20% or greater were tested for potential radiogenomic association.
Association between Radiomic Features and Genomic Expression
Unsupervised data clustering was implemented to investigate potential molecular underpinnings of the radiomic signal. The probability density of Ƒ was analyzed using a Langevin annealing technique (12) to determine the number of nondegenerate clusters, k. Patient partitioning was then derived using a k-means clustering approach in which patients were stratified into k groups based solely on intrinsic similarities of their radiomic signatures. Clustering solutions were naively compared with matched ctDNA eTAm-Seq mutation data and cfDNA concentration data with the Fisher exact test and t test, respectively. P ≤ .05 was considered to indicate a statistically significant difference.
Dimensionality Reduction and Feature Selection
We implemented the following unsupervised feature selection approach to interrogate the principal radiomic features driving the clustering process. Biclustering was applied, where columns and rows of Ƒ were clustered simultaneously. For each row-wise feature cluster, the top three features representing the largest median difference between column-wise patient clusters were encoded into the matrix
 |
where r is the number of row-wise feature clusters. The operation Ƒ→Ƒr produces a low-dimensional representation of Equation (1). Rows of Ƒr represent a subset of radiomic features that both maximize the separation of patients into clusters and minimize the redundancy of those clusters by selecting across different values of r.
Association between Radiomic Features and Clinical Outcome
Univariate Cox proportional hazards analysis was used to test the association between the selected radiomic features of Ƒr and overall survival. Our motivating assumption was that features that were highly discriminant across clusters were intrinsically associated with disparate outcomes. To test this hypothesis, univariate analyses were considered to reduce the likelihood of overfitting multivariate models. Statistical analysis was performed in Matlab using the coxphfit function.
Results
Association of Radiomic Signatures with Genomic Expression
As shown in Figure 2A, unsupervised clustering revealed k = 2 groups of patients who shared similar radiomic feature patterns (the k = 3 solution is provided in Fig E1 [supplement]). Comparison of radiomic clusters to corresponding eTAm-Seq data demonstrated a significant association with TP53 mutations in ctDNA (P = .03, Fisher exact test), which was present in 11 of 24 patients. Mutations were detected in 14 additional genes as shown in Figure 2A, but these genes demonstrated low detection rates (one to four occurrences) and were excluded from statistical analysis. Mutations in the remaining 21 genes were not detected in any of the patients. Radiomic cluster was not associated with latent variables, including CT manufacturer or any demographic information reported in Table 1.
Figure 2:
Associations between CT radiomics, cell-free DNA (ctDNA) gene expression, and intratreatment changes in cfDNA concentration. (A) The radiomic feature space, Ƒ, according to Equation (1), where column vectors denote tumor-specific radiomic signatures extracted from CT images, and row vectors denote different radiomic features. Two patient clusters were identified based on similarity of radiomic signatures. Comparison to matched eTAm-Seq gene expression data (below) demonstrated a significant association between radiomic signatures and ctDNA TP53 (P = .03, Fisher exact test). (B) Patient-specific changes in cfDNA concentration during treatment (baseline, n = 24; 2 weeks, n = 24; 5 weeks, n = 22; after treatment, n = 20), color-coded by radiomic cluster. (C) Clustered radiomic signatures were associated with changes in cfDNA concentration after 2 weeks of chemoradiation (P = .02, t test). The central line denotes the median value; the edges of the box represent the 25th and 75th percentiles; the whiskers are the most extreme data points. eTAm-Seq = enhanced tagged/targeted amplicon sequencing.
In total, 90 phlebotomy samples were analyzed with spectrophotometry, and the resulting changes in cfDNA concentration are reported in Figure 2B. Comparison between radiomic clusters and intratreatment measurements of cfDNA concentration revealed a relationship between radiomic features and changes in cfDNA concentration after 2 weeks of chemoradiation (+89.8% ± 109.4 vs −23.1% ± 106.4; P = .02, t test; Fig 2C). Additional cfDNA concentration data (absolute values, relative changes) are provided in Figure E2 (supplement).
Low-dimensional Interrogation of Radiomic Signatures
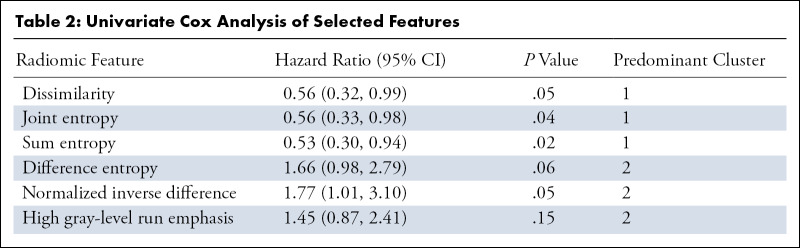
Reducing Ƒ→Ƒr was driven by r = 2 row-wise feature clusters. The subset of features representing the largest median difference between column-wise patient clusters is reported in Figure 3. The patient cluster associated with a detectable TP53 mutation and minimal cfDNA change (cluster 2) was enriched with difference entropy, normalized inverse difference (both measures of local homogeneity) (13), and high gray-level run emphasis (a measure of high density) (14). In contrast, dissimilarity, joint entropy, and sum entropy—all texture measures of local heterogeneity—were present in high abundance within the cluster associated with undetectable TP53 and early therapeutic changes in cfDNA concentration (cluster 1).
Figure 3:
Low-dimensional radiomic feature patterns. The reduced radiomic feature space, Ƒr, following the unsupervised dimensionality reduction and feature selection operation, Ƒ→Ƒr, according to Equation (3). Column vectors of Ƒr denote low-dimensional radiomic signatures, and row vectors of Ƒr represent a subset of radiomic features from the complete feature space, Ƒ, that maximize the separation of patients into clusters and minimize the redundancy within those clusters. GLCOM = gray level co-occurrence matrix.
Association of Radiomic Signatures with Clinical Outcome
The median clinical follow-up time was 60 months (80% CI: 40, 61), and median overall survival was 28 months (80% CI: 20, 52). Univariate Cox analysis results of the selected features from Ƒr are reported in Table 2. Dissimilarity, joint entropy, sum entropy, and normalized inverse difference were all significantly associated with overall survival. Collectively, these results suggest that patients with longer overall survival presented with heterogeneous and low-attenuating disease at CT, in the absence of a detectable TP53 mutation in ctDNA. Although the average survival time of cluster 1 was longer compared with that of cluster 2, this result was not statistically significant (35.8 months ± 22.1 vs 24.2 months ± 16.3; P = .16, t test).
Table 2:
Univariate Cox Analysis of Selected Features
Discussion
Although changes in cfDNA during chemoradiation have not yet been fully characterized, preliminary results of this pilot study demonstrate its potential as a companion diagnostic to radiomics. Quantitative features derived from imaging, cfDNA, and ctDNA may enable new treatment strategies for advanced lung cancer, including escalation of prescription doses and modification of radiation target volumes. They may also serve as surrogate endpoints and enable health surveillance for patients at high risk of recurrence.
Our radiomic analysis indicated that tumors appearing more homogeneous with higher attenuation on CT images were associated with detectable ctDNA TP53 mutations and stagnant changes in cfDNA concentration early in treatment. For example, difference entropy and normalized inverse difference are both by definition measures of local homogeneity where high values indicate a more homogeneous attenuation pattern on CT images. Furthermore, high gray-level run emphasis is proportional to CT attenuation, such that high values indicate a high-attenuating tumor. These features were all enriched in cluster 2. In contrast, dissimilarity, joint entropy, and sum entropy are all measures of local heterogeneity and were enriched in cluster 1. These radiomic characteristics imply that imaging is potentially capturing a larger, more difficult to treat tumor burden, which is consistent with previous studies (15–17).
The association between radiomic features and increased cfDNA concentration after 2 weeks of therapy in patients with undetectable ctDNA TP53 mutations is of potential interest. For example, cancer cell death as a result of chemoradiation may lead to increased cfDNA concentration in the plasma. This is consistent with a recent study showing that radiation therapy increases cfDNA concentration in non–small cell lung cancer (18). However, normal tissue cell death may contribute to stochastic fluctuations in cfDNA concentration, and decreased concentration may indicate cfDNA cleared from plasma at the time of phlebotomy in responding tumors.
Low specificity of cfDNA thus substantiates the need for more specific measures of ctDNA (9). Most notably, mutations in the DNA binding domain of TP53 suppress its ability to sense DNA damage, which reduces the probability of cell cycle arrest and apoptosis. TP53 mutated tumors have a more radioresistant phenotype that typically portends a poor prognosis. Our results are therefore biologically relevant and should be further interrogated with mechanistic preclinical experiments that probe the biologic underpinnings of radiomic signals.
The main limitation of this analysis was the small sample size due to the prospective nature of our data. Because of this, we designed our study using conservative methods. We implemented a fully unsupervised feature selection process to reduce the dimension of the data without any observation of dependent variables that may otherwise overfit predictors. We also limited our evaluation of prognosis to hypothesis-driven univariate analysis. Future work will focus on cluster generalization, which requires more samples to achieve stable results (19,20).
In conclusion, our radiogenomics data suggest that heterogeneous and low-attenuating tumors at CT without a detectable ctDNA TP53 mutation may be associated with early therapeutic response. This is indicated by changes in plasma cfDNA concentration after 2 weeks of chemoradiation and a generally better prognosis. These findings provide hypothesis-generating data and illustrate the importance of larger clinical trials to investigate the combined prognostic effect of radiomics, cfDNA, and ctDNA in locally advanced lung cancers.
Clinical Trial Information: All research was conducted in keeping with best clinical practice and regulatory compliance. Prospective data (ie, CT images, blood samples, and patient outcomes) were generated from Pro00017361 (NCT00921739) approved by the Duke University Health System Institutional Review Board (DUHS IRB). Additionally, the DUHS IRB approved a secondary retrospective analysis (ie, radiomic analysis and next-generation sequencing) of the original data with Pro00076119, which was completed under an approved waiver of consent, HIPAA authorization, and decedent research notification.
Authors declared no funding for this work.
Disclosures of Conflicts of Interest: K.J.L. disclosed no relevant relationships. M.N.C. disclosed no relevant relationships. J.G. disclosed no relevant relationships. C.D.J. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Premera Blue Cross. Other relationships: disclosed no relevant relationships. J.W. disclosed no relevant relationships. Y.C. disclosed no relevant relationships. C.W. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Duke University Medical Center. Other relationships: disclosed no relevant relationships. A.H. disclosed no relevant relationships. E.X. disclosed no relevant relationships. G.J. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Inivata; has stock options issued as employee of Inivata. Other relationships: disclosed no relevant relationships. C.R.K. disclosed no relevant relationships. F.F.Y. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: employed by Duke University Hospital; institution receives grant from Varian Medical Systems that is used for other work not related to this study. Other relationships: disclosed no relevant relationships.
Abbreviations:
- cfDNA
- cell-free DNA
- ctDNA
- circulating tumor DNA
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Bradley JD, Paulus R, Komaki R, et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol 2015;16(2):187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17(4):223–238. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri AA, Binkley MS, Osmundson EC, Alizadeh AA, Diehn M. Predicting Radiotherapy Responses and Treatment Outcomes Through Analysis of Circulating Tumor DNA. Semin Radiat Oncol 2015;25(4):305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deig CR, Mendonca MS, Lautenschlaeger T. Blood-Based Nucleic Acid Biomarkers as a Potential Tool to Determine Radiation Therapy Response in Non-Small Cell Lung Cancer. Radiat Res 2017;187(3):333–338.28186469 [Google Scholar]
- 6.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20(5):548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4(136):136ra68. [DOI] [PubMed] [Google Scholar]
- 8.Kelsey CR, Das S, Gu L, Dunphy FR 3rd, Ready NE, Marks LB. Phase 1 Dose Escalation Study of Accelerated Radiation Therapy With Concurrent Chemotherapy for Locally Advanced Lung Cancer. Int J Radiat Oncol Biol Phys 2015;93(5):997–1004. [DOI] [PubMed] [Google Scholar]
- 9.Corradetti MN, Torok JA, Hatch AJ, et al. Dynamic Changes in Circulating Tumor DNA During Chemoradiation for Locally Advanced Lung Cancer. Adv Radiat Oncol 2019;4(4):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Lafata K, Wang C, et al. Digital phantoms for characterizing inconsistencies among radiomics extraction toolboxes. Biomed Phys Eng Express 2020;6(2):025016. [DOI] [PubMed] [Google Scholar]
- 11.Plagnol V, Woodhouse S, Howarth K, et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One 2018;13(3):e0193802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafata KJ, Zhou Z, Liu JG, Yin FF. Data clustering based on Langevin annealing with a self-consistent potential. Q Appl Math 2019;77(3):591–613. [Google Scholar]
- 13.Haralick R, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern 1973;SMC-3(6):610–621. [Google Scholar]
- 14.Tang X. Texture information in run-length matrices. IEEE Trans Image Process 1998;7(11):1602–1609. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Tang C, Hobbs BP, et al. Development and Validation of a Predictive Radiomics Model for Clinical Outcomes in Stage I Non-small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2018;102(4):1090–1097. [DOI] [PubMed] [Google Scholar]
- 16.Ye J, Chang J, Li ZQ, et al. (P097) tumor density, size, and histology in the outcome of stereotactic body radiation therapy for early-stage non-small-cell lung cancer: a single-institution experience. Oncology (Williston Park) 2015;29(4 Suppl 1):205053. [PubMed] [Google Scholar]
- 17.Lafata KJ, Hong JC, Geng R, et al. Association of pre-treatment radiomic features with lung cancer recurrence following stereotactic body radiation therapy. Phys Med Biol 2019;64(2):025007. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama SI, Nihei K, Karasawa K, et al. Radiotherapy increases plasma levels of tumoral cell-free DNA in non-small cell lung cancer patients [Published correction appears in Oncotarget 2018;9(34):23844.]. Oncotarget 2018;9(27):19368–19378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Cui Y, Sun X, et al. Unsupervised Clustering of Quantitative Image Phenotypes Reveals Breast Cancer Subtypes with Distinct Prognoses and Molecular Pathways. Clin Cancer Res 2017;23(13):3334–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itakura H, Achrol AS, Mitchell LA, et al. Magnetic resonance image features identify glioblastoma phenotypic subtypes with distinct molecular pathway activities. Sci Transl Med 2015;7(303):303ra138. [DOI] [PMC free article] [PubMed] [Google Scholar]