Abstract
Purpose
To develop multimodality imaging techniques for measuring epidermal growth factor receptor (EGFR) as a therapy-relevant and metastasis-associated molecular marker in triple-negative mammary adenocarcinoma metastases.
Materials and Methods
An orthotopic bone metastasis EGFR-positive, triple-negative breast cancer (TNBC) model in rats was used for bioluminescence imaging, SPECT/CT, PET/CT, and MRI with quantitative analysis of transcripts (n = 22 rats). Receptor-specific MRI of EGFR expression in vivo was performed by acquiring spin-echo T1-weighted images after sequential administration of a pair of anti-EGFR antigen binding fragments, F(ab’)2, conjugated to either horseradish peroxidase or glucose oxidase, which have complementing activities, as well as paramagnetic (gadolinium[III]-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid, or Gd-5HT-DOTAGA) or positron-emitting (gallium 68-5HT-DOTAGA) substrates for MRI and PET/CT imaging, respectively. EGFR expression was confirmed by quantitative reverse transcriptase polymerase chain reaction and immunohistochemical analyses to compare with image findings.
Results
After surgical intraarterial delivery of TNBC cells, rats developed tumors that diverged into either rapidly growing osteolytic or slow-growing nonosteolytic tumors. Both tumor types showed receptor-specific initial MRI signal enhancement (contrast-to-noise ratio) that was three to six times higher than that of normal bone marrow (29.4 vs 4.9; P < .01). Micro PET/CT imaging of EGFR expression demonstrated a high level of heterogeneity with regional uptake of the tracer, which corresponded to region-of-interest MRI signal intensity elevation (121.1 vs 93.3; P < .001). Analysis of metastases with corroboration of imaging results showed high levels of EGFR protein and messenger RNA, or mRNA, expression in the invasive tumor.
Conclusion
Convergence of multimodal molecular receptor imaging enabled comprehensive assessment of EGFR overexpression in an orthotopic model of TNBC metastasis.
Keywords: Animal Studies, Molecular Imaging-Cancer, MR-Contrast Agent, Radionuclide Studies, Skeletal-Appendicular, Metastases
Supplemental material is available for this article.
© RSNA, 2021
Keywords: Animal Studies, Molecular Imaging-Cancer, MR-Contrast Agent, Radionuclide Studies, Skeletal-Appendicular, Metastases
Summary
Multimodality imaging of triple-negative mammary adenocarcinoma metastases by using luminescence enabled differentiation between osteolytic and nonosteolytic phenotypes, whereas specific assessment of epidermal growth factor receptor overexpression was achieved by using MRI and PET/CT in an orthotopic rat model.
Key Points
■ Longitudinal luminescence imaging differentiated between experimental orthotopic bone metastases phenotypes in a triple-negative breast cancer model.
■ Enzymatic activity complementation provided receptor-specific expression imaging in metastases after the administration of binary targeted conjugates followed by imaging substrates.
■ Both paramagnetic and positron-emitting substrates may potentially be used for receptor imaging using a pair of targeted enzymes with complementing activities.
Introduction
Triple-negative breast cancer (TNBC) is a diverse group of cancers lacking well-defined therapeutic molecular targets. TNBC is generally associated with poorer outcomes compared with other breast cancer subtypes (1,2), and TNBCs carry various driver lesions in their genomes (3). The presence of epidermal growth factor receptor (EGFR) driver amplicons was shown to correlate with programmed cell death ligand 1 (PD-L1) expression in TNBC (3). Furthermore, EGFR overexpression in TNBC is clearly linked to malignancy and metastatic spread of breast cancer to the brain (4) and to bone (ie, approximately 50% of all cases of metastatic spread [5–7]). Whereas available clinical experience is insufficient, these findings suggest that monoclonal antibodies (mAbs) specific to immune checkpoint receptors produce high rates of clinical benefit in the case of metastatic TNBC, with objective response rates exceeding 40% in some cases (8,9). These responses to programmed cell death 1 (PD-1) and PD-L1 monotherapy could be a consequence of differences in the expression levels of PD-1 and PD-L1 in TNBC compared with non-TNBC tumors (10). It is expected that combination therapeutic strategies will be required to surpass response rates observed to date for monotherapies (11–13); such strategies may involve repurposing of existing targeting approaches.
In addition to genomic profiling, the ability to spatially detect and monitor EGFR expression in patients with breast cancer through noninvasive molecular imaging could be useful in screening patients to select candidates who could benefit from combination anti-EGFR and checkpoint inhibitor therapies. There has been a consistent effort to use imaging to visualize EGFR expression as a marker of TNBC (14). Related cancer imaging efforts included developing molecular imaging agents based on high-affinity humanized or fully human antibodies (15,16), their proteolytic fragments (17), and engineered affibodies (18–20). The proposed imaging modalities for in vivo EGFR imaging range from tomographic and clinically approved techniques (PET, SPECT, and MRI) (21,22) to fluorescence imaging for emerging applications in imaging-guided surgery (23–25).
Certain authors of this study previously investigated EGFR imaging in animal models by using a strategy of receptor targeting with binary system for imaging signal amplification (26,27). The latter strategy is based on tagging antireceptor antibodies with two enzymes with complementing activities. Anti-EGFR antibody-enzyme conjugates colocalize in the tissue because of the binding of antibody to tumor cells in a receptor-specific manner (28). In this study, we used horseradish peroxidase (HRP) and glucose oxidase (GOx). The resultant complementation of enzyme activities yields trapping of labeled substrates (eg, paramagnetic imaging enzymatic probes [29,30]) at the site of colocalization. In our study, we further explore a multimodality receptor imaging approach combined with corroboration by using tumor-specific transcripts analysis and histopathologic outcomes with bioluminescence and CT imaging–based phenotyping of orthotopic TNBC tumor metastases in detecting EGFR-specific PET and MRI signatures in a model of human breast adenocarcinoma bone metastases in rats.
Materials and Methods
Study Design
All surgical, interventional, and imaging procedures related to animal use were performed according to the guidelines approved by the Institute Animal Care and Use Committee. Rowett nude rats (male and female rats; age, 3–4 weeks) were acquired from Charles River Laboratories and were used for orthotopic intraarterial cell implantation when animals reached the age of 6–12 weeks.
This study included a total of five different in vivo experiments with a total of 22 rats, with some overlap of animals between experiments. In total, three rats underwent interventional protocols with fLuc+MBA-MB-231 and 19 rats underwent surgical protocols with either fLuc+MBA-MD-231 (n = 9) or MDA-MD-231 (n = 10). The five experiments were as follows: comparison of interventional versus surgical protocols, identification and characterization of osteolytic versus nonosteolytic fLuc+MBA-MB-231 tumors by bioluminescence imaging (BLI) (n = 8), assessment of EGFR expression in osteolytic tumors (fLuc+MBA-MB-231 tumors) imaged with SPECT/CT and histopathologic comparison (three from experiment 1), MRI in osteolytic and nonosteolytic tumors for EGFR using GOx and HRP conjugated to anti-EGFR F(ab’)2 with coinjection of gadolinium[III]-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (Gd-5HT-DOTAGA) (eight from experiment 1), and imaging of osteolytic MDA-MB-231 tumors for EGFR by using GOx and HRP-conjugated anti-EGFR F(ab’)2 with coinjection of Gd-5HT-DOTAGA for MRI and gallium 68 (68Ga)-5HT-DOTAGA for PET/CT imaging (n = 10; six experimental and four control rats). The overview of animal numbers and their use in experiments is shown in Table E3 (supplement).
Cell Lines and Protocols for Orthotopic Tumor Cell Delivery
For all in vivo experiments, fLuc-MDA-MB-231 and MDA-MB-231 were used. For in vitro experiments, fLuc-MDA-MB-231 and MDA-MB-468 were used. Additional information on the cell lines used in this study can be found in Appendix E1 (supplement). The interventional and surgical protocols for tumor cell delivery to the bone are in Appendix E2 (supplement).
Preparation of Imaging Agents
We used click chemistry technique based on inverse electron demand Diels-Alder cycloaddition (31,32) to improve the previously used conjugation approach (28). The click chemistry reaction was used to conjugate the anti-EGFR and antiepithelial cell adhesion molecule (EpCAM; anti-EpCAM) antigen binding fragments, F(ab’)2, to deglycosylated HRP or GOx without the need for further purification (Fig E1A [supplement]).
Anti-EGFR antibodies were conjugated with HRP and GOx. Details regarding the conjugation process and characterization are described in Appendix E3 (supplement). Conjugation of anti-EGFR antibodies with technetium 99m (99mTc) mertiatide–labeled anti-EGFR F(ab’)2) for SPECT/CT imaging is described in Appendix E4 (supplement). Appendix E5 (supplement) describes the synthesis of 5HT-DOTAGA and the conjugation of gadolinium to 5HT-DOTAGA (Gd-5HT-DOTAGA) for MRI. Appendix E6 (supplement) describes the synthesis of 68Ga-labeled 5HT-DOTAGA tracer for PET/CT imaging.
HRP and GOx Cross-Titration and MRI Enzyme-Linked Immunosorbent Assay
To determine the ratio of HRP:GOx EGFR-conjugated antibodies needed for optimal signal, an enzyme-linked immunosorbent assay was performed with varying concentrations of anti-EGFR F(ab’)2. Additional details on this assay are in Appendix E7 (supplement). Additionally, because one objective of this study was to assess the use of HRP and GOx-conjugated antibodies for MRI, an MRI enzyme-linked immunosorbent assay was performed to assess T1-weighted MRI signal (Appendix E8 [supplement]).
BLI Acquisition
Animals were anesthetized with isoflurane (1.75% in 30% oxygen and nitrogen mixture), and an intraperitoneal injection of a dose of 100 mg/kg D-luciferin in 0.75 mL of saline was performed. BLI (IVIS SpectrumCT; PerkinElmer) was performed on days 2, 6, 14, and 22 after implantation of TNBC 5 minutes after injection every 5 minutes for a total of 20 minutes to ensure the peak of luminescence was detected. The acquired 16-bit TIFF images were analyzed by using ImageJ (https://imagej.net/Welcome), and luminescent tumor area and integrated intensity (in relative light units) were measured within the boundaries of a region of interest (ROI) with margins determined by signal levels equivalent to twice the background levels.
MRI Acquisition
Two groups of animals (four with osteolytic tumors and four with nonosteolytic tumors) underwent imaging by using 3-T MRI (Philips Achieva) without contrast enhancement on the 30th day after cell implantation. The same animals were further imaged between the 30th and 60th day after implantation after the consecutive intravenous injection of F(ab’)2 conjugates and Gd-5HT-DOTAGA. Animals were maintained under gas anesthesia at 37°C throughout the experiments with respiratory monitoring. In experiments involving the injection of Gd-5HT-DOTAGA, a 26-gauge catheter capped with a needle port was placed in the tail vein for direct paramagnetic substrate administration.
A solenoid radiofrequency coil (50 mm × 50 mm) was used for MRI signal collection. Initially a multisection T2-weighted image was acquired by using a spin-echo pulse sequence (repetition time msec/echo time msec, 4000/40) with the following MRI parameters: 41 sections; thickness, 0.7 mm; intersection gap, 0; field of view, 44 mm × 44 mm; data acquisition matrix, 148 × 144 (reconstruction matrix, 384 × 384); and seven excitations. Contrast agent–only administration and imaging were performed as follows: a precontrast (ie, obtained before the administration of contrast material) image was acquired by using spin-echo pulse sequence (600/7.4) with the same geometric parameters as the T2-weighted image with fast-spin-echo factor of four; two or eight excitations. MRI contrast agent (0.15 mmol/kg Gd-5HT-DOTAGA in 0.5–0.8 mL Dulbecco phosphate-buffered saline) was intravenously injected by tail vein catheter, and T1-weighted images were acquired every 6 minutes for 90 minutes.
The next day the animals were administered 200 µg (dose, 1 mg/kg) of tumor cell–specific pair of anti-EGFR F(ab’)2 conjugated to HRP or anti-EGFR F(ab’)2 conjugated to GOx (schematically shown in Figure 1A) intravenously in 0.2 mL phosphate-buffered saline. Each of the injection mixtures (experimental or control mixture) contained a two-times lower amount of HRP conjugate than that of GOx (ie, a one-to-two weight ratio). Two types of control experiments were performed to test the specificity of the EGFR imaging: (a) anti-EGFR F(ab’)2 HRP conjugate in the mixture was replaced by anti-EpCAM F(ab’)2-HRP, and (b) animals were preinjected with a dose of cetuximab (an anti-EGFR mAb, used to block the HRP- and GOx-labeled antibodies from binding) at 5 mg/kg 10 minutes before the F(ab’)2 conjugates were administered and MRI was performed.
Figure 1:
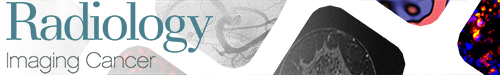
(A) A schema shows the main structural elements of F(ab’)2 conjugates with deglycosylated horseradish peroxidase (HRP) and glucose oxidase (GOx) synthesized by using metal-free click chemistry. (B) A schema of enzymatic activity complementation involving reaction between GOx conjugated to F(ab’)2 and glucose that provides hydrogen peroxide used by HRP in the presence of 5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid-chelated gadolinium (III) or gallium 68 (68Ga) (III).
Four hours after the intravenous injection of antibody conjugates, a precontrast image was acquired and then an intravenous injection of 0.15 mmol/kg Gd-5HT-DOTAGA was administered. Thereafter, MRI T1-weighted spin-echo images were acquired serially during a 1-hour period. All animals with enhancing nonosteolytic tumors at MRI were imaged after an 18-hour to 20-hour delay. Precontrast T2-weighted images were used for selecting ROIs for quantitative analysis.
A separate group of animals bearing osteolytic MDA-MB-231 tumors (six experimental rats and four control rats) was imaged by using 7.0-T MRI (ClinScan 70/30; Bruker) and PET/CT (see below) performed the next day by injecting the conjugates as described above followed by a 4-hour waiting period after which a dose of 0.2 mmol/kg Gd-5HT-DOTAGA was injected intravenously. The preinjection of free cetuximab was performed in a control group of animals as described above. Seven pre- and postcontrast MRI scans were acquired at 10-minute intervals by using spin-echo pulse sequence (repetition time, 600 msec; echo time, 6.6 msec; reconstruction matrix, 384 × 384; field of view, 45 mm × 45 mm).
SPECT Imaging
Animals were monitored for 26 days after implantation of cancer cells by using CT (NanoSPECT/CT; Bioscan). SPECT was subsequently performed when signs of osteolysis were identifiable on CT images by injecting 0.8 mCi (29.6 MBq in 0.25 mL of saline) of 99mTc mertiatide–labeled anti-EGFR F(ab’)2 in the tail vein and acquiring images 6 hours after injection (n = 3). CT and SPECT reconstruction, image fusion, and calculation of imaging volumes were performed by using InVivoScope 1.37 software (Bioscan).
PET Imaging
The 68Ga-labeled 5HT-DOTAGA diluted in saline and 0.25 mL (3–7 MBq per animal) were injected intravenously in the tail vein (three experimental and four control animals) at 4 hours after injection of the anti-EGFR F(ab’)2 HRP and anti-EGFR F(ab’)2-GOx conjugate mixture, at a dose of 200 µg (1 mg/kg). The PET images were acquired at 1 hour after tracer injection by using the Inveon PET/CT scanner (Siemens), at an acquisition time of 15 minutes. Images were analyzed and ROIs were drawn using Siemens Inveon Research Workplace 4.0 software.
MRI Analysis
Initial enhancement (10–12-minute peak), delayed enhancement (1 hour), and serially acquired T1-weighted spin-echo MRI signals (12 minutes to 20 hours) were analyzed as signal decay curves in the tumor area defined by an ROI drawn in each section based on precontrast brightening on T2-weighted spin-echo images. The mean ROI signal intensity was calculated using ImageJ (33). For each imaging section, contrast-to-noise ratios (CNRs) were calculated as a difference between signal-to-noise ratios in the tumor and in normal muscle by using the following equation: CNR = SNRt − SNRm = (SIt − SIm)/SDn, where SNR is signal-to-noise ratio, SNRt is the signal-to-noise ratio in the tumor, SNRm is the signal-to-noise ratio in normal muscle, SIt is the in-section mean ROI signal intensity of tumor, SIm is the in-section mean ROI signal intensity of the muscle (contralateral extremity), and SDn is the standard deviation of noise (determined by placing ROI outside the body imaging section).
PET and MRI Registration and Quantitative Analysis
MRI sections were chosen for analysis by the operator to provide the best matching to the corresponding PET/CT-fused image. Look-up-table-inverted spin-echo T1-weighted MRI scans and PET/CT images were registered using bone structure as anatomic landmarks using the TrakEM2 tool (Fiji) (34). PET/CT images were segmented using the Trainable Weka Segmentation machine learning tool (35) (ImageJ) using random forest classifier with binary manual annotations for classifier training (ie, “category 1” ROI with standardized uptake value > 0.2 vs “category 2” [all background]). The obtained PET/CT classified (segmented) image maps were processed as follows: threshold was adjusted to isolate ROIs, ROI smoothing was applied, and particle analysis was performed in ImageJ using PET/CT ROIs redirected to MRI grayscale images. Mean MRI signal intensity values measured per ROI were used for final analysis.
Transcript Analysis and Immunohistochemistry Analysis
Expression of EGFR, fLUC, EpCAM, CDH1 (E-cadherin), CTNNB1 (β-catenin), and VIM (vimentin) were assessed from MDA-MB-231 tumors. Further details on messenger RNA, or mRNA, extraction and quantitative reverse transcriptase (qRT) polymerase chain reaction (PCR) are described in Appendix E9 (supplement). Tissues were additionally assessed for the expression of EGFR and EpCAM using immunohistochemistry analysis. Protocols are described in Appendix E10 (supplement).
Statistical Analysis
Standardized uptake values in PET experiments (both mean and maximum standardized uptake value) were used to calculate tumor:muscle ratios, which were compared between PET/CT groups as well as with corresponding MRI ROI within the same groups using the nonparametric Mann-Whitney-Wilcoxon test. The comparison of mean luminescence and peak MRI CNRs in osteolytic versus nonosteolytic tumors was performed using the same Mann-Whitney-Wilcoxon test. Differences were considered significant if two-tailed P < .05. MRI signal-decay data were plotted to obtain time decay constant values using the comparison of mono- and biexponential fitting. For each model the better model was chosen when F test results were significant, and a simpler model (ie, the monoexponential) was chosen in the case of P > .05.
Results
Synthesis and Testing of F(ab’)2-HRP and F(ab’)2-GOx System in TNBC Cells
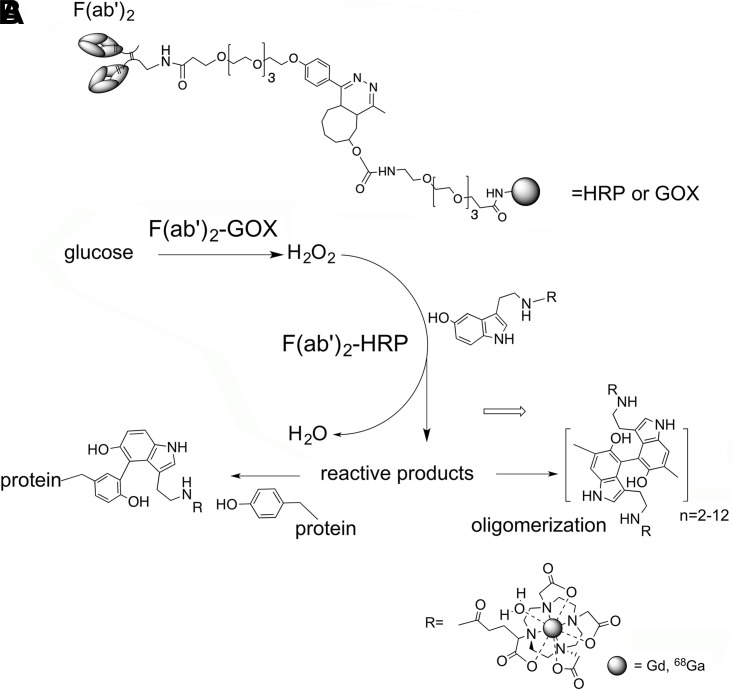
The half-maximum effective concentrations were 0.18 µg/mL to 0.19 µg/mL for both anti-EGFR F(ab’)2-HRP and anti-EGFR F(ab’)2-GOx conjugates as determined by cross-titration assay (Fig 2A). In the case of anti-EpCAM conjugates, we observed a much lower signal with the half-maximum effective concentration in the microgram range generated by either conjugate in the presence of the complementing enzyme activity (Fig E1B [supplement]).
Figure 2:
(A–C) Epidermal growth factor receptor (EGFR) expression by triple-negative breast cancer cells. Enzyme-linked immunosorbent assay and comparison with MRI signal change measured in MDA-MB-231 and MDA-MB-468 cells. (A) Results of cross titration of anti-EGFR-F(ab’)2–horseradish peroxidase (HRP) and anti-EGFR-F(ab’)2–glucose oxidase (GOX) conjugates on live firefly luciferase (fLuc)+MDA-MB-231 cells. The amounts of conjugates added per well are shown along the y-axis and the x-axis. (B) Plot shows dependence of EGFR-specific contrast-to-noise ratio (CNR) determined by using T1-weighted MRI pulse sequence (red and blue solid lines, left y-axis) and enzyme activity complementation (specific-to-nonspecific absorbance ratio A/Ao, red and blue dashed lines, right y-axis) as a function of log anti-EGFR-HRP conjugate concentration. The concentration of anti-EGFR-GOX conjugate was kept constant at 800 ng/mL; the concentration of paramagnetic substrate gadolinium(III)-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid) was 0.1 mM in 1% fetal calf serum, Hanks balanced salts solution. Blue indicates MDA-MB-468 cells (marked as 468), red indicates fLuc+MDA-MB-231 cells (marked as 231). (C) Bar graph shows specificity of conjugates. The fLuc+MDA-MB-231 cell-binding assay was performed by using technetium 99m mertiatide–labeled anti-EGFR F(ab’)2 with control samples containing a 10-fold molar excess of unlabeled conjugates.
To establish the concentration range corresponding to the optimized enzyme-linked immunosorbent assay signal, we performed cross-titration of conjugates on the surface of fLuc+MDA-MB-231 cells (Fig 2A). The optimal complementation was achieved at the weight ratio of 0.7:1.0 (anti-EGFR F(ab’)2-HRP: anti-EGFR F(ab’)2-GOx). We have previously observed the internalization of 70% to 75% of all bound anti-EGFR conjugates over the time course of 0.5 hours to 3 hours in various cell lines overexpressing EGFR (ie, between 25% and 30% of the conjugates carrying enzymes were located on the surface of the cells [27,28]).
To compare T1-weighted contrast-assisted MRI and enzyme-linked immunosorbent assay with regard to their ability to generate EGFR-specific signal we chose another TNBC line, MDA-MB-468, as a positive control (Fig 2B). MDA-MB-468 cells express high levels of EGFR (approximately 1.3 × 106 EGFR per cell [36]) as a result of wild-type EGFR allele amplification (37,38). Both approaches showed higher signal associated with high levels of EGFR expression by MDA-MB-468 versus MDA-MB-231 cells, which express approximately two-times lower numbers of EGFR on the cell surface (39). The MRI approach appeared more sensitive with 0.3 µg/mL anti-EGFR F(ab’)2-HRP versus 1.25 µg/mL (colorimetric enzyme-linked immunosorbent assay), resulting in significant differences in MRI signal intensity between the two cell lines (P < .05). However, the conjugate concentration window was wider in the case of colorimetric enzyme-linked immunosorbent assay and exceeded two orders of magnitude (Fig 2B). Both enzyme conjugates of anti-EGFR F(ab’)2 showed a high level of specific binding and uptake (Fig 2C), which exceeded 92% as was demonstrated by using an excess of unlabeled antibody fragment conjugates.
In Vitro Characterization of TNBC Cells
To determine the sensitivity of fLuc+MDA-MB-231 detection based on cell-specific bioluminescence, we serially diluted cells and analyzed the lysates in the presence of D-luciferin and adenosine triphosphate. This assay showed that luminescence (log counts/sec) was linearly dependent on log of cell number in the range spanning at least three orders of magnitude (100–100 000 cells), and the count rates were acceptable for tracking the increase of cell numbers by a factor of five with sufficient precision (Fig E1C [supplement]). The comparison by qRT PCR of five key gene expression levels and fLuc marker (Table E2 [supplement]) showed that EGFR levels were similar in both cells and EpCAM was slightly elevated in fLuc+MDA-MB-231, which was consistent with cell enzyme-linked immunosorbent assay results (Fig E1B [supplement]). The expression of VIM and CTNNB1, the markers of epithelial-mesenchymal transition, were both upregulated compared with the nonfLuc-expressing MDA-MB-231 cells (Table E2 [supplement]).
In Vivo Metastasis Model and Image-guided Differentiation between Tumor Phenotypes
The in vivo animal model of bone metastases used in this study was established by delivery of luciferase-expressing TNBC to the knee joint area in athymic rats. We tested both an interventional and a surgical (40) approach for TNBC cell delivery. Compared with the interventional procedure, the surgical approach was found to be superior regarding the tumor-take rate, resulting in reliable bone metastasis formation (Fig E2 [supplement]). All animals described underwent the surgical procedure.
Rats that underwent surgical protocols and that were injected with fLuc-MDA-MB-231 developed tumors that exhibited two distinct phenotypes. In half of the animals (n = 4), an exponential increase in both mean luminescence intensity (BLI counts, doubling time of 4.7 days) and tumor size (BLI area, doubling time of 16 days) occurred in the knee joint area over time (Fig 3A, Fig E3 [supplement]). The remaining animals with implanted cells (n = 4) showed BLI signal of fLuc+MDA-MD-231 cells stabilizing over time instead of increasing exponentially (Fig 3A). We further used CT of the lower extremities to assess whether the differences in the longitudinal changes of BLI signal corresponded to the expansion of the tumor beyond the boundaries of the bone as a result of osteolysis. Examination of CT reconstructions of radiographic images obtained on day 20–22 after tumor cell implantation established that all animals with stable and low BLI signal in the knee exhibited no signs of bone loss (Fig 3B); however, during the same time period the area of BLI-positive tumor increased approximately sixfold. Tumors in animals with exponentially increasing BLI signal intensity demonstrated at least 100-fold higher mean BLI intensity than in the BLI stable group (Fig 3A–3C). In these animals, high BLI signal in the area of the knee corresponded to the extensive osteolysis that involved the femoral and, sometimes, the tibial bones (Fig 3C). Based on this CT finding, animals were assigned to either an osteolytic or nonosteolytic group.
Figure 3:
(A–D) Multimodality imaging characterization of osteolytic and nonosteolytic fLuc-MDA-MB-231 tumors. (A) Graph shows that bioluminescence imaging (BLI) in tumors shows differences between osteolytic phenotype (high mean bioluminescence signal) and nonosteolytic phenotype (low bioluminescence signal) at 2 and 22 days after implantation. Mean data are ± standard deviation (four rats per group). (B) BLI scan (top) and CT image (bottom) in lower extremities, nonosteolytic tumor phenotype. (C) BLI scan (top) and CT image (bottom) in lower extremities, osteolytic tumor phenotype; the bone subjected to osteolysis is shown (arrow). (D) A representative SPECT/CT image of epidermal growth factor receptor (EGFR)–positive tumor (arrow) in the knee area of the animal shown in C by using technetium 99–labeled anti-EGFR-F(ab’)2 fragment (fused SPECT/CT image, maximum intensity pixel [MIP]). A = axial image projection, C = coronal, fLuc = firefly luciferase, RLU = relative light unit, S = sagittal.
Imaging of Tumor Marker Expression and Histologic Corroboration
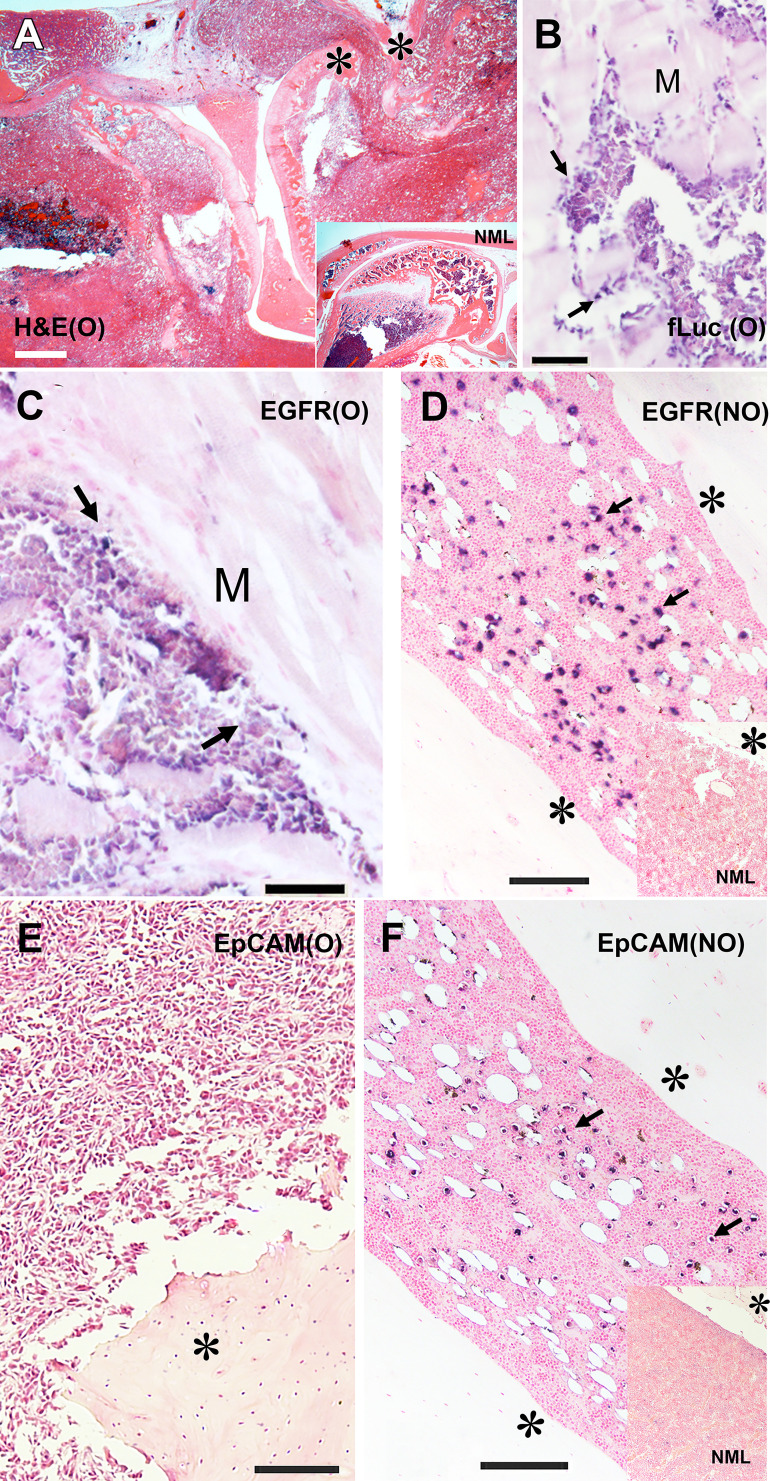
Tumor progression and osteolysis in fLuc+MDA-MB-231 tumors allowed for the assessment of whether the developed lesions were exhibiting EGFR expression in vivo. We performed SPECT/CT imaging of the animals (n = 3) with verified osteolytic phenotype after injecting 99mTc mertiatide–labeled anti-EGFR F(ab’)2, which was mapped to the area of the knee that exhibited femoral and tibial osteolysis (Fig 3D). These SPECT/CT and BLI imaging results obtained in osteolytic tumors were corroborated with histologic analysis of normal knee bone and osteolytic tumor structure (Fig 4A). Osteolytic tumors showed expansion of the tumor mass beyond the boundaries of the knee bones as assessed by histologic examination. The cells comprising the tumor front in the muscle tissue expressed EGFR during the invasion of the tumor (Fig 4B). Immunohistochemistry analysis also showed that the majority of tumor cells invading the muscle were positive for fLuc expression, confirming that the invading cells were the MDA-MB-231 type (Fig 4C). In nonosteolytic bone tumors that were positive for luciferase at BLI, immunohistochemistry analysis showed large areas of EGFR-positive tumor cell dissemination across the bone marrow, which contained multiple cystlike structures (Fig 4D). Using parallel section processing we also observed EpCAM-positive cells (Fig 4E), whereas no EpCAM-positive cells were detected in osteolytic tumors (Fig 4F).
Figure 4:
(A–F) Histologic and immunohistochemistry analysis of osteolytic (O) and nonosteolytic (NO) firefly luciferase (fLuc)+MDA-MB-231 bone metastasis models. (A) Hematoxylin-eosin (H&E) staining of decalcified knee area section of osteolytic tumor phenotype. Inset shows hematoxylin-eosin staining of a normal (NML) rat knee bone. (B) Anti–firefly luciferase immunohistochemistry staining of muscle (M) invading osteolytic tumor front (blue). (C) Anti-epidermal growth factor receptor (EGFR) immunohistochemistry staining of invading osteolytic tumor front (blue). (D) Nonosteolytic tumor dissemination in the bone marrow, anti-EGFR immunohistochemistry staining (blue). Inset, normal bone marrow section stained with anti-EGFR antibody (control animal). (E) Osteolytic tumor section immunohistochemistry staining with anti-epithelial cell adhesion molecule (EpCAM). (F) Nonosteolytic tumor section immunohistochemistry staining with anti-EpCAM antibody (blue). Inset, normal bone marrow section stained with anti-EpCAM antibody (control animal). * bone. Arrows indicate antibody-positive cells. Counterstaining (Fast Nuclear Red) bar in A, 400 µm; counterstaining bars in B–E, 200 µm.
MRI in Bone Tumor Metastasis Model
We performed MRI at 3 T with and without contrast enhancement in selected animals 20–30 days after the tumor cells were surgically delivered to the bone. To achieve MRI of receptor-specific signal, we used Gd-5HT-DOTAGA (Fig E4 [supplement]). The presence of glucose in the extracellular medium provided a source of hydrogen peroxide from catalytic activity of GOx conjugated to the anti-EGFR F(ab’)2. Hydrogen peroxide is then consumed by HRP, and quick reduction of oxidized HRP is enabled by the Gd-5HT-DOTAGA-reducing substrate (Fig 1B).
Before performing in vivo imaging of rats harboring bone tumors, we performed in vitro testing of Gd-5HT-DOTAGA at various concentrations by adding both GOx and HRP conjugates and measuring the change of water proton relaxation times. In vitro testing showed a time-dependent 1.6- to 1.9-fold increase of molar relaxivity of gadolinium (r1) measured at 0.47 T in the absence of albumin and a 2.5-fold increase of r1 in the presence of both albumin and conjugates (Fig E4B [supplement]). In the absence of conjugates, no appreciable increase of r1 was observed.
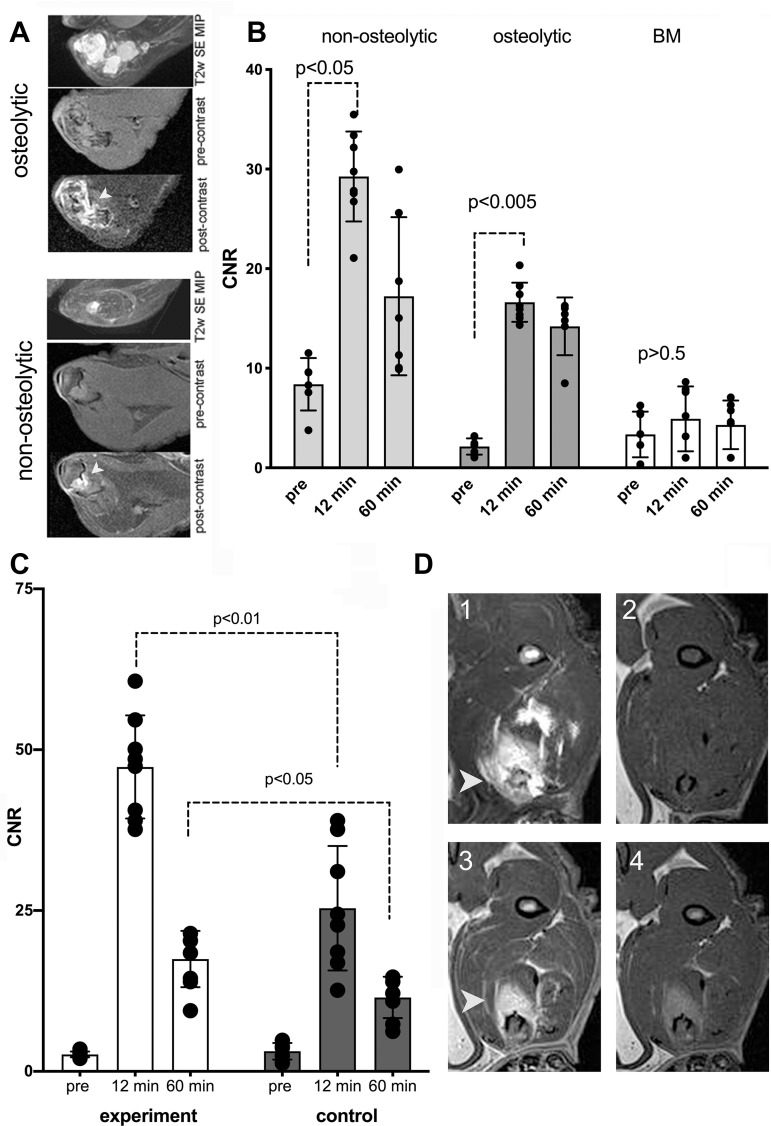
We then performed coinjection of HRP and GOx conjugates of anti-EGFR F(ab’)2 in rats harboring both phenotypes of fluc+MDA-MB-231 bone tumors by using the optimized ratio of HRP and GOx activity in the conjugates, which was determined by testing in cell culture. Consecutive intravenous injections of anti-EGFR conjugates and Gd-5HT-DOTAGA were separated by a 4-hour interval (ie, a period exceeding the half-life of conjugates in the bloodstream by approximately a factor of two [28]). Consequently, an increase of mean MRI signal was measured in the ROIs outlined by using T2-weighted enhancement at precontrast spin-echo MRI (Fig 5A). Unlike in normal bone marrow, in both osteolytic and nonosteolytic tumors the contrast enhancement was highly significant approximately 12 minutes after intravenous injection of 0.15 mmol of gadolinium per kilogram of Gd-5HT-DOTAGA (ie, at the measured peak of signal increase) (Fig 5B). The initial enhancement was followed by a period of MRI signal decay over time (Fig 5B, Table 1).
Figure 5:
(A–D) MRI scans and graphs show osteolytic and nonosteolytic bone tumors with gadolinium(III)-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid) (Gd-5HT-DOTAGA). (A) Comparative morphologic structure of representative osteolytic and nonosteolytic bone firefly luciferase+MDA-MB-231 tumors shown as T2-weighted (T2w) spin-echo (SE) maximum intensity pixel (MIP) projection images and T1-weighted spin-echo pulse sequence 3.0-T MRI scans acquired in-section pre-Gd-5HT-DOTAGA (pre-contrast) and after Gd-5HT-DOTAGA (post-contrast) (0.15 mmol/kg) administration. The animals were preinjected with a mixture of anti-epidermal growth factor receptor (EGFR)-F(ab’)2-horseradish peroxidase (HRP) and anti-EGFR-F(ab’)2-glucose oxidase (GOx) conjugates before Gd-5HT-DOTAGA injection. (B) Dynamic contrast enhancement in tumors (at 3.0-T MRI) expressed as contrast-to-noise (CNR) ratio. CNR of bone tumor images in animals preinjected with anti-EGFR conjugates before (pre) and 12 and 60 minutes after injection of Gd-5HT-DOTAGA. CNR in animals with CT-confirmed osteolysis and nonosteolytic tumors are shown compared with normal bone marrow (BM) enhancement. Mean data are ± standard deviation (four tumors per group, two to three MRI sections per animal). (C) CNR of tumors in experimental and control groups (three tumors per group, two to three MRI sections per animal); MRI was performed at 7.0 T. Control group animals were preinjected with a blocking dose of cetuximab (5 mg/kg) before injection of a mixture of anti-EGFR-F(ab’)2-HRP and anti-EGFR-F(ab’)2-GOx conjugates. Significant differences were observed at 12 minutes (first T1-weighted spin-echo pulse sequence acquisition) and at 60 minutes after injection of Gd-5HT-DOTAGA (0.2 mmol/kg). (D) Representative 7.0-T MRI scans of invasive tumor: (1) T2-weighted spin-echo maximum intensity pixel projection, (2) pre-Gd-5HT-DOTAGA injection T1-weighted spin echo, (3) 12 minutes after Gd-5HT-DOTAGA injection T1-weighted spin echo, (4) 60 minutes after Gd-5HT-DOTAGA injection T1-weighted spin echo. Arrowheads point to the tumor in panels A and D.
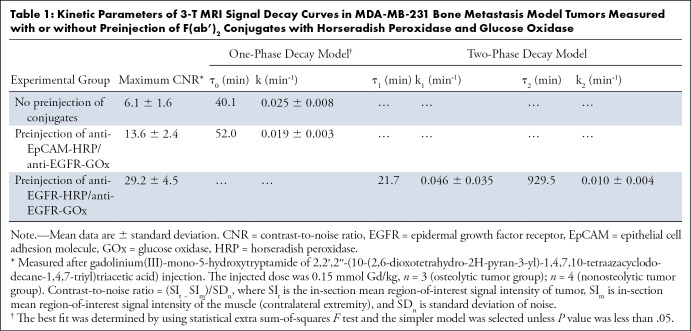
Table 1:
Kinetic Parameters of 3-T MRI Signal Decay Curves in MDA-MB-231 Bone Metastasis Model Tumors Measured with or without Preinjection of F(ab’)2 Conjugates with Horseradish Peroxidase and Glucose Oxidase
We further analyzed the differential MRI signal decay in vivo by using dynamic T1-weighted signal acquisition. The comparison of signal decay kinetics performed by data fitting of MRI signal change over time suggested monoexponential signal decay under conditions when either no anti-EGFR F(ab’)2 conjugates were injected or if one of the conjugates was replaced by a conjugate of anti-EpCAM F(ab’)2. In the case of the peak of EpCAM F(ab’)2 conjugate the enhancement measured as mean tumor CNR was still two times higher than the value of CNR at the peak when Gd-5HT-DOTAGA was injected alone (Table 1). The largest CNR value change (ie, 4.7 times higher than nonspecific CNR at the peak) and biexponential decay of CNR was observed after intravenous injection of both anti-EGFR conjugates and Gd-5HT-DOTAGA. The mean values of CNR in nonosteolytic tumors were higher overall than in the osteolytic group (29 vs 17; P < .01) in CNR between the groups because early MRI signal-to-noise ratio in nonosteolytic tumors was approximately two times higher than in osteolytic tumors (Fig 5B). The blocking study performed by using a preinjection of free cetuximab showed that the CNR for EGFR was reduced when blocked (mean CNR, 24 vs 48; P < .01); the difference in enhancement levels decreased over 1 hour but remained significant (mean CNR, 12 vs 19; P < .05; Fig 5C, 5D).
Multimodality Imaging in the Same Group of Animals
A separate group of animals (n = 10; six experimental and four control rats) was selected for sequential PET and MRI acquisitions. These rats were surgically implanted with MDA-MB-231 cells. The injection of anti-EGFR conjugates was followed by the corresponding imaging substrates (ie, 68Ga-labeled 5HT-DOTAGA for PET imaging and Gd-5HT-DOTAGA for MRI on two consecutive days [Fig 6).
Figure 6:
(A–D) Multimodality micro-PET/CT and 7.0-T MRI of osteolytic bone metastasis using anti–epidermal growth factor receptor (EGFR) conjugates injection followed by imaging substrate gallium 68-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid)-(5HT-DOTAGA) and gadolinium(III)-5HT-DOTAGA, respectively, and associated graphs. (A) Three-dimensional CT reconstruction map displaying osteolysis as a consequence of MDA-MB-231 tumor growth, with imaging planes numbered. (B) PET/CT images (left) after registering to corresponding MRI sections (right) acquired at plane positions shown in A. MRI scans were used as reference to target PET/CT images for image registration purposes by using a set of landmarks (Fig E5 [supplement]). Numbers correspond to numbered imaging planes in A. * Femur. (C) PET tumor-to-muscle mean standardized uptake value (SUV) ratios (open circles) and maximum SUV ratios (filled circles) measured in total tumor volume in control group that underwent blocking anti-EGFR antibody preinjection (black circles), experimental group (blue circles), and tissue-infiltrating bone tumor fraction in experimental groups (red circles) (three tumors per group). * Statistically significant difference (Mann-Whitney nonparametric test, P < .05). (D) MRI signal intensity measured by using TW-segmentation of PET/CT images for region of interest analysis (SUV > 0.2) identified as tumor (n = 15) versus background signal intensity measured in the muscle (n = 10). Data shown as mean ± standard deviation were analyzed by using Mann-Whitney nonparametric test (P < .001). AU = arbitrary units, TW = trainable Weka.
Virtual imaging planes were selected by performing three-dimensional CT reconstructions of the bone. Those planes corresponded to osteolytic and nonosteolytic areas of the tumor metastasis model, and they were further matched to individual MRI sections (Fig 6A, 6B). The analysis of tumor-to-muscle standardized uptake value ratios calculated across PET images showed that the ratios for whole tumors at PET/CT ranged between 1.2 and 5.8, and their muscle-invading margins EGFR-overexpressing tumors showed higher ratios ranging between 11.1 and 22.3; however, these differences were not statistically significant (Mann-Whitney test, P = .1) (Fig 6C). Furthermore, transaxial sections of the reconstructed image volumes were matched by applying PET/CT to MRI nonrigid image registration guided by anatomic landmarks, which were identifiable in both image data sets (ie, femur, fibula, and tibia; Fig 6B). Image registration enabled analysis of MRI by using ROI derived from PET/CT images. These ROIs were selected by applying classified image segmentation of PET/CT fused images with thresholding at standardized uptake value of greater than 0.2, and the measurements of mean MRI signal intensities associated with each individual ROI (Fig E5 [supplement]) were compared with muscle (background) MRI signal intensities (Fig 6D). The analysis of multiple ROIs showed that the areas with standardized uptake value of greater than 0.2 on PET images corresponded to the areas of MRI sections that exhibited elevated T1-weighted MRI signal intensity (P < .001) (Fig 6D) if compared with tumor-free muscle ROIs, which did not show enhancement on both PET and MRI.
To determine if the observed PET signal and matched MRI enhancement corresponded to overexpression of EGFR, we further performed partial molecular characterization of tumor transcripts by using qRT PCR analysis. The cDNA samples were synthesized after sampling the total tumor and highly enhancing areas that were identified at PET imaging (Table 2). The qRT PCR data showed an increase in the levels of EGFR expression in the invading and PET-enhanced tumors compared with proximally mapped tumor mass and the parent MDA-MB-231 cells (P < .001). These data also showed that in bone metastases there was a decrease in expression of both cancer-associated EpCAM and the markers of epithelial-to-mesenchymal transition in breast cancer cells (ie, β-catenin and vimentin; 41).
Table 2:
Relative Gene Transcription Levels in Experimental MDA-MB-231 Tumor Bone Metastases
Discussion
Tumor metastases frequently have a phenotype different from that of the primary tumor; molecular imaging may be a technique that can be used to differentially image these two tumor populations (42). Because assumptions made on the basis of primary tumor phenotype are potentially flawed, the direct characterization of metastases may result in more precise cancer treatment options. Our main research goal was to explore whether EGFR expression in TNBC cells could be tracked after tumor cells are delivered to the bone using high-spatial-resolution and/or high sensitivity imaging modalities. The secondary goal was to determine whether imaging signatures during tumor development could be used to predict whether tumor cells would undergo phenotype change from nonosteolytic to osteolytic. The major advantage of the orthotopic model used in our study (40) was the feasibility of longitudinal assessment of metastatic disease (43) because reduced observation time from disseminated disease is typical for other commonly used models of cancer metastases. Compared with models requiring local orthotopic tumor cell injections, which are usually performed in the tibia (44), the effects secondary to local bone destruction and resulting from drilling a hole in the tibia were not encountered because we used an intra-arterial approach for tumor cell inoculation in our study.
Toward the research goals of our study, we tested a multimodality imaging approach combining sensitivity of in vivo bioluminescent detection of highly aggressive MDA-MB-231 breast cancer variant (45) with high-resolution tomographic characterization of the whole tumor volume by enzyme-mediated imaging signal amplification strategy (26,29). The approach used in this work was previously designed and tested for imaging signal amplification (29). The approach is based on two antibody fragments linked to peroxidase (HRP in this study) and GOx, in which both enzymes have complementing activities. When the conjugates colocalize on the surface of the same cell, hydrogen peroxide generated by GOx is rapidly scavenged by peroxidase, which undergoes oxidation. Peroxidases are reduced to catalytically active state by synthetic substrates containing tyramide (29) or 5HT (46,47) moieties as well as macrocyclic chelates of paramagnetic or positron-emitting metal cations such as Gd-DOTAGA (Figs 1–E4 [supplement]). If these substrates are introduced by intravenous injection, the short-lived radicals generated by peroxidase-mediated catalysis in vivo may either undergo oligomerization or may bind directly to nearby proteins (Fig 1). In both cases, the paramagnetic products of peroxidase reaction are retained locally and have higher molar relaxivity than the initial substrates, which translates into high MRI signal. Radioactive reducing substrates are converted into products that are retained because of stable binding within the site of conjugate colocalization. In the case of radioisotope-based reducing substrates, the signal amplification effect is the consequence of multiple enzymatic catalytic cycles involving these substrates.
We previously demonstrated the feasibility of EGFR imaging in orthotopic rat models of glioblastoma by using enzyme-linked F(ab’)2 of matuzumab (EMD72000 mAb derived from mAb 425 [48] that has high affinity to a truncated EGFR variant [EGFRvIII] [27,28]). In the current work we took advantage of the high affinity and specificity of cetuximab (mAb C225) to the wild-type EGFR epitopes since the dissociation constant of cetuximab and its fragments is lower than that of EMD72000 mAb by at least two orders of magnitude (49,50), thus improving the affinity of the conjugates and their receptor association rates, which are both important for in vivo imaging. We also used the novel highly soluble chelate 5HT-DOTAGA, which functions as a reducing imaging substrate of peroxidases (47). In its native state Gd-5HT-DOTAGA was eliminated faster from both experimental tumor metastases (target) and muscle (nontarget) tissues in the absence of anti-EGFR F(ab’)2 conjugates in monoexponential fashion, unlike previously reported imaging substrates (28). This simplified the interpretation of experimental results because the retention of Gd-5HT-DOTAGA products with the average time constant t2 greater than 10 t1 was observed only in the case of injection of a combination of EGFR-specific MRI signal–amplifying anti-EGFR F(ab’)2 HRP and GOx conjugate. The prolonged retention of enzymatic reaction products in the tumors suggested specificity of enzyme activity complementation in the case of osteolytic and nonosteolytic MDA-MB-231 tumors. Anti-EpCAM conjugate imaging data were consistent with downregulation of this tumor marker, which was identified by qRT PCR analysis in osteolytic MDA-MB-231 tumors. The observed decrease in E-cadherin mRNA levels, in addition to downregulation of vimentin and upregulation of EGFR (Table 2) were consistent with markers of well-differentiated basal-like invasive phenotype of both fLuc+MDA-MB-231 and tissue-invading MDA-MB-231 wild-type tumor metastases (51,52).
The cations of gadolinium and gallium are trivalent and form very stable chelates with tetraazacyclododecanetetraacetic acid (53). Consequently, MRI signal amplifying strategy with enzymatic activity complementation may be repurposed for 68Ga PET imaging, which has certain advantages over MRI for imaging peroxidase activity in vivo because of greater sensitivity of detection (54). In the case of multimodality MRI and PET/CT imaging studies performed by using Gd-5HT-DOTAGA and 68Ga-labeled 5HT-DOTAGA tracer in the same groups of animals (Fig 6C, 6D) the consistency of imaging findings between PET and MRI studies was of prime importance.
This study had several limitations. The first was inherent in high-field-strength imaging of experimental animals; whereas 7-T MRI provided higher CNR primarily because of low noise (Fig 5) and ensured high image resolution, MRI signal gains because of peroxidase-generated products binding are lower at 7 T compared with 3 T. The benefits of gadolinium chelate binding to proteins resulting from longitudinal relaxivity increase are lower at higher magnetic fields (55). The second limitation was because of the overall low tumor-associated radioactivity by using PET/CT relative to MRI signal intensity increase. The likely explanation is in the nature of enzymatic reaction, which is at the core of signal amplification effect. The level of reaction products formation depends on local concentration of the substrate (ie, 68Ga-labeled 5HT-DOTAGA in the case of PET). Because PET tracers have high specific radioactivity and the injected physical amounts of the substrate were low, the local concentration of the reducing substrate in tumors was insufficient for achieving high signal-to-background ratios. Whereas qRT PCR corroboration confirmed the specificity of EGFR imaging by PET (ie, the correspondence between standardized uptake value and EGFR expression levels; Table 2) and high standardized uptake value regions generally overlapped with high MRI signal ROIs, there were no apparent benefits in comparison to alternative pretargeting PET imaging techniques (56). The third limitation was in the potential nonspecific toxicity of antibody-conjugated GOx. Although the enzyme has an acceptable systemic toxicity profile (57), receptor-mediated internalization of GOx in tumor and normal cells can lead to toxic effects (58,59).
In conclusion, receptor-specific imaging signal detection achieved by enzyme-mediated amplification of paramagnetic and PET probes indicated persistent EGFR expression in experimental metastases, particularly at the invasive edge of the lesions, which showed striking differences in time-dependent evolution of bioluminescence signal, suggesting potential for noninvasive imaging of the transition from nonosteolytic to osteolytic phenotype. The analysis of MRI signal alone and of PET/CT image registration with MRI showed that tumor-associated high standardized uptake value areas corresponded to the elevated MRI signal mapped to the same ROIs and was in line with EGFR mRNA expression levels. Our imaging-based observations were independently corroborated by tissue histopathologic examination and qRT PCR of major transcripts, which suggests that analysis of noninvasive multimodality imaging signatures of key surface molecules may have a substantial role in developing treatment combination options for patients with metastatic cancer.
Acknowledgments
Acknowledgments
We appreciate the effort of Manuel Geisthoff (Department of Nuclear Medicine, University Hospital Erlangen) for PET and 7.0-T MRI experiments, and of Dr Jean-Marc Gauguet (Department of Radiology, UMASS Medical School) during the earlier phase of this study.
Current address: Beth Israel Lahey Health, Tufts University School of Medicine, Burlington, Mass.
This work was supported in part by grants From the National Institutes of Health: R01DK095728 (A.B.), R01EB000858 (A.B.). T.B. supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) within the Priority Programme µBone (BA 4027/10–1), Collaborative Research Center 1181 “Checkpoints for Resolution of Inflammation” (CRC 1181, Project Z02) and CRC TRR 305 “Striking a moving target: From mechanisms of metastatic organ colonization to novel systemic therapies” (Project Z01).
Disclosures of Conflicts of Interest: T.B. disclosed no relevant relationships. S.G. disclosed no relevant relationships. S.Z. disclosed no relevant relationships. L.S. disclosed no relevant relationships. A.L. disclosed no relevant relationships. M.M. disclosed no relevant relationships. S.M. disclosed no relevant relationships. O.P. disclosed no relevant relationships. P.C. Activities related to the present article: institution receives grants/grant pending from the National Institutes of Health. Activities not related to the present article: disclosed money to author from Collagen Medical, Janssen for consultancies; disclosed grants/grants pending from National Institutes of Health, Celgene, Takeda, Janssen, Pliant Therapeutics, Indalo Therapeutics, Pfizer; disclosed patent for Factor 1A; disclosed stock/stock options for Reveal Pharmaceuticals, Collagen Medical. Other relationships: disclosed no relevant relationships. A.B. disclosed no relevant relationships.
Abbreviations:
- BLI
- bioluminescence imaging
- CNR
- contrast-to-noise ratio
- EGFR
- epidermal growth factor receptor
- EpCAM
- epithelial cell adhesion molecule
- GOx
- glucose oxidase
- HRP
- horseradish peroxidase
- mAb
- monoclonal antibody
- PCR
- polymerase chain reaction
- PD-1
- programmed cell death 1
- PD-L1
- programmed cell death ligand 1
- qRT PCR
- quantitative reverse transcriptase PCR
- ROI
- region of interest
- TNBC
- triple-negative breast cancer
References
- 1.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol 2008; 26(15):2568–2581. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121(7):2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett MT, Lenkiewicz E, Malasi S, et al. The association of genomic lesions and PD-1/PD-L1 expression in resected triple-negative breast cancers. Breast Cancer Res 2018; 20(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks DG, Short SM, Prescott NL, et al. Breast cancers with brain metastases are more likely to be estrogen receptor negative, express the basal cytokeratin CK5/6, and overexpress HER2 or EGFR. Am J Surg Pathol 2006; 30(9):1097–1104. [DOI] [PubMed] [Google Scholar]
- 5.Coleman RE, Rubens RD. The clinical course of bone metastases from breast cancer. Br J Cancer 1987; 55(1):61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley J, Nickerson NK, Nam S, Allen KT, et al. EGFR signaling in breast cancer: bad to the bone. Semin Cell Dev Biol 2010; 21(9):951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts MS, Anstine LJ, Finke VS, et al. KLF4 defines the efficacy of the epidermal growth factor receptor inhibitor, erlotinib, in triple-negative breast cancer cells by repressing the EGFR gene. Breast Cancer Res 2020; 22(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santa-Maria CA, Kato T, Park JH, et al. A pilot study of durvalumab and tremelimumab and immunogenomic dynamics in metastatic breast cancer. Oncotarget 2018; 9(27):18985–18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al Sayed AD, Elshenawy MA, Tulbah A, Al-Tweigeri T, Ghebeh H. Complete Response of Chemo-Refractory Metastatic Metaplastic Breast Cancer to Paclitaxel-Immunotherapy Combination. Am J Case Rep 2019; 20(1630):1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatalica Z, Snyder C, Maney T, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev 2014; 23(12):2965–2970. [DOI] [PubMed] [Google Scholar]
- 11.Force J, Leal JHS, McArthur HL. Checkpoint Blockade Strategies in the Treatment of Breast Cancer: Where We Are and Where We Are Heading. Curr Treat Options Oncol 2019; 20(4):35. [DOI] [PubMed] [Google Scholar]
- 12.Zimmer AS, Nichols E, Cimino-Mathews A, et al. A phase I study of the PD-L1 inhibitor, durvalumab, in combination with a PARP inhibitor, olaparib, and a VEGFR1-3 inhibitor, cediranib, in recurrent women’s cancers with biomarker analyses. J Immunother Cancer 2019; 7(1):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva JL, Cardoso Nunes NC, Izetti P, de Mesquita GG, de Melo AC. Triple negative breast cancer: A thorough review of biomarkers. Crit Rev Oncol Hematol 2020; 145102855. [DOI] [PubMed] [Google Scholar]
- 14.Schmadeka R, Harmon BE, Singh M. Triple-negative breast carcinoma: current and emerging concepts. Am J Clin Pathol 2014; 141(4):462–477. [DOI] [PubMed] [Google Scholar]
- 15.Koyama Y, Barrett T, Hama Y, Ravizzini G, Choyke PL, Kobayashi H. In vivo molecular imaging to diagnose and subtype tumors through receptor-targeted optically labeled monoclonal antibodies. Neoplasia 2007; 9(12):1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadri K, Ren Q, Zhang K, et al. PET imaging of EGFR expression in nude mice bearing MDA-MB-468, a human breast adenocarcinoma. Nucl Med Commun 2011; 32(7):563–569. [DOI] [PubMed] [Google Scholar]
- 17.Turker NS, Heidari P, Kucherlapati R, Kucherlapati M, Mahmood U. An EGFR targeted PET imaging probe for the detection of colonic adenocarcinomas in the setting of colitis. Theranostics 2014; 4(9):893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolmachev V, Friedman M, Sandström M, et al. Affibody molecules for epidermal growth factor receptor targeting in vivo: aspects of dimerization and labeling chemistry. J Nucl Med 2009; 50(2):274–283. [DOI] [PubMed] [Google Scholar]
- 19.Miao Z, Ren G, Liu H, Qi S, Wu S, Cheng Z. PET of EGFR expression with an 18F-labeled affibody molecule. J Nucl Med 2012; 53(7):1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samkoe KS, Tichauer KM, Gunn JR, Wells WA, Hasan T, Pogue BW. Quantitative in vivo immunohistochemistry of epidermal growth factor receptor using a receptor concentration imaging approach. Cancer Res 2014; 74(24):7465–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiao J, Xue S, Pu F, et al. Molecular imaging of EGFR/HER2 cancer biomarkers by protein MRI contrast agents. J Biol Inorg Chem 2014; 19(2):259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corcoran EB, Hanson RN. Imaging EGFR and HER2 by PET and SPECT: a review. Med Res Rev 2014; 34(3):596–643. [DOI] [PubMed] [Google Scholar]
- 23.Antaris AL, Chen H, Cheng K, et al. A small-molecule dye for NIR-II imaging. Nat Mater 2016; 15(2):235–242. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Ballou B, Schmidt BF, et al. Affibody-targeted fluorogen activating protein for in vivo tumor imaging. Chem Commun (Camb) 2017; 53(12):2001–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Souza AL, Marra K, Gunn J, et al. Fluorescent Affibody Molecule Administered In Vivo at a Microdose Level Labels EGFR Expressing Glioma Tumor Regions. Mol Imaging Biol 2017; 19(1):41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogdanov A Jr, Kang HW, Querol M, Pretorius PH, Yudina A. Synthesis and testing of a binary catalytic system for imaging of signal amplification in vivo. Bioconjug Chem 2007; 18(4):1123–1130. [DOI] [PubMed] [Google Scholar]
- 27.Shazeeb MS, Sotak CH, DeLeo M 3rd, Bogdanov A Jr. Targeted signal-amplifying enzymes enhance MRI of EGFR expression in an orthotopic model of human glioma. Cancer Res 2011; 71(6):2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shazeeb MS, Gupta S, Bogdanov A Jr. MR signal amplification for imaging of the mutant EGF receptor in orthotopic human glioma model. Mol Imaging Biol 2013; 15(6):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdanov A Jr, Matuszewski L, Bremer C, Petrovsky A, Weissleder R. Oligomerization of paramagnetic substrates result in signal amplification and can be used for MR imaging of molecular targets. Mol Imaging 2002; 1(1):16–23. [DOI] [PubMed] [Google Scholar]
- 30.Chen JW, Pham W, Weissleder R, Bogdanov A Jr. Human myeloperoxidase: A potential target for molecular MR imaging in atherosclerosis. Magn Reson Med 2004; 52(5):1021–1028. [DOI] [PubMed] [Google Scholar]
- 31.Blackman ML, Royzen M, Fox JM. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc 2008; 130(41):13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeglis BM, Mohindra P, Weissmann GI, et al. Modular strategy for the construction of radiometalated antibodies for positron emission tomography based on inverse electron demand Diels-Alder click chemistry. Bioconjug Chem 2011; 22(10):2048–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arganda-Carreras I, Kaynig V, Rueden C, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 2017; 33(15):2424–2426. [DOI] [PubMed] [Google Scholar]
- 36.Filmus J, Pollak MN, Cailleau R, Buick RN. MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun 1985; 128(2):898–905. [DOI] [PubMed] [Google Scholar]
- 37.El Guerrab A, Bamdad M, Kwiatkowski F, Bignon YJ, Penault-Llorca F, Aubel C. Anti-EGFR monoclonal antibodies and EGFR tyrosine kinase inhibitors as combination therapy for triple-negative breast cancer. Oncotarget 2016; 7(45):73618–73637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subik K, Lee JF, Baxter L, et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl) 2010; 4(35):4.[Published correction appears in Breast Cancer (Auckl) 2018;12:1178223418806626.]. [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzpatrick SL, LaChance MP, Schultz GS. Characterization of epidermal growth factor receptor and action on human breast cancer cells in culture. Cancer Res 1984; 44(8):3442–3447. [PubMed] [Google Scholar]
- 40.Bäuerle T, Adwan H, Kiessling F, Hilbig H, Armbruster FP, Berger MR. Characterization of a rat model with site-specific bone metastasis induced by MDA-MB-231 breast cancer cells and its application to the effects of an antibody against bone sialoprotein. Int J Cancer 2005; 115(2):177–186. [DOI] [PubMed] [Google Scholar]
- 41.Gilles C, Polette M, Mestdagt M, et al. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res 2003; 63(10):2658–2664. [PubMed] [Google Scholar]
- 42.Curtit E, Nerich V, Mansi L, et al. Discordances in estrogen receptor status, progesterone receptor status, and HER2 status between primary breast cancer and metastasis. Oncologist 2013; 18(6):667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bretschi M, Fränzle A, Merz M, et al. Assessing treatment response of osteolytic lesions by manual volumetry, automatic segmentation, and RECIST in experimental bone metastases. Acad Radiol 2014; 21(9):1177–1184. [DOI] [PubMed] [Google Scholar]
- 44.Kjønniksen I, Winderen M, Bruland O, Fodstad O. Validity and usefulness of human tumor models established by intratibial cell inoculation in nude rats. Cancer Res 1994; 54(7):1715–1719. [PubMed] [Google Scholar]
- 45.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res 2005; 7(4):R444–R454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wadghiri YZ, Hoang DM, Leporati A, et al. High-resolution Imaging of Myeloperoxidase Activity Sensors in Human Cerebrovascular Disease. Sci Rep 2018; 8(1):7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Rodríguez A, Shuvaev S, Rotile N, et al. Peroxidase Sensitive Amplifiable Probe for Molecular Magnetic Resonance Imaging of Pulmonary Inflammation. ACS Sens 2019; 4(9):2412–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamat V, Donaldson JM, Kari C, et al. Enhanced EGFR inhibition and distinct epitope recognition by EGFR antagonistic mAbs C225 and 425. Cancer Biol Ther 2008; 7(5):726–733. [DOI] [PubMed] [Google Scholar]
- 49.Schmiedel J, Blaukat A, Li S, Knöchel T, Ferguson KM. Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer Cell 2008; 13(4):365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel D, Guo X, Ng S, et al. IgG isotype, glycosylation, and EGFR expression determine the induction of antibody-dependent cellular cytotoxicity in vitro by cetuximab. Hum Antibodies 2010; 19(4):89–99. [DOI] [PubMed] [Google Scholar]
- 51.Gamallo C, Palacios J, Suarez A, et al. Correlation of E-cadherin expression with differentiation grade and histological type in breast carcinoma. Am J Pathol 1993; 142(4):987–993. [PMC free article] [PubMed] [Google Scholar]
- 52.Thike AA, Iqbal J, Cheok PY, Tse GM, Tan PH. Ductal carcinoma in situ associated with triple negative invasive breast cancer: evidence for a precursor-product relationship. J Clin Pathol 2013; 66(8):665–670. [DOI] [PubMed] [Google Scholar]
- 53.Kubícek V, Havlícková J, Kotek J, et al. Gallium(III) complexes of DOTA and DOTA-monoamide: kinetic and thermodynamic studies. Inorg Chem 2010; 49(23):10960–10969. [DOI] [PubMed] [Google Scholar]
- 54.Wang C, Keliher E, Zeller MWG, et al. An activatable PET imaging radioprobe is a dynamic reporter of myeloperoxidase activity in vivo. Proc Natl Acad Sci U S A 2019; 116(24):11966–11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caravan P, Farrar CT, Frullano L, Uppal R. Influence of molecular parameters and increasing magnetic field strength on relaxivity of gadolinium- and manganese-based T1 contrast agents. Contrast Media Mol Imaging 2009; 4(2):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reiner T, Lewis JS, Zeglis BM. Harnessing the bioorthogonal inverse electron demand Diels-Alder cycloaddition for pretargeted PET imaging. J Vis Exp 2015;(96):e52335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samoszuk M, Ehrlich D, Ramzi E. Preclinical safety studies of glucose oxidase. J Pharmacol Exp Ther 1993; 266(3):1643–1648. [PubMed] [Google Scholar]
- 58.Samoszuk MK, Wimley WC, Nguyen V. Eradication of interleukin 5-transfected J558L plasmacytomas in mice by hydrogen peroxide-generating Stealth liposomes. Cancer Res 1996; 56(1):87–90. [PubMed] [Google Scholar]
- 59.Christofidou-Solomidou M, Pietra GG, Solomides CC, et al. Immunotargeting of glucose oxidase to endothelium in vivo causes oxidative vascular injury in the lungs. Am J Physiol Lung Cell Mol Physiol 2000; 278(4):L794–L805. [DOI] [PubMed] [Google Scholar]


![(A–D) Multimodality imaging characterization of osteolytic and nonosteolytic fLuc-MDA-MB-231 tumors. (A) Graph shows that bioluminescence imaging (BLI) in tumors shows differences between osteolytic phenotype (high mean bioluminescence signal) and nonosteolytic phenotype (low bioluminescence signal) at 2 and 22 days after implantation. Mean data are ± standard deviation (four rats per group). (B) BLI scan (top) and CT image (bottom) in lower extremities, nonosteolytic tumor phenotype. (C) BLI scan (top) and CT image (bottom) in lower extremities, osteolytic tumor phenotype; the bone subjected to osteolysis is shown (arrow). (D) A representative SPECT/CT image of epidermal growth factor receptor (EGFR)–positive tumor (arrow) in the knee area of the animal shown in C by using technetium 99–labeled anti-EGFR-F(ab’)2 fragment (fused SPECT/CT image, maximum intensity pixel [MIP]). A = axial image projection, C = coronal, fLuc = firefly luciferase, RLU = relative light unit, S = sagittal.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1b9b/8344354/c10e327dca9d/rycan.2021200069.fig3.jpg)
![(A–D) Multimodality micro-PET/CT and 7.0-T MRI of osteolytic bone metastasis using anti–epidermal growth factor receptor (EGFR) conjugates injection followed by imaging substrate gallium 68-mono-5-hydroxytryptamide of 2,2′,2′′-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid)-(5HT-DOTAGA) and gadolinium(III)-5HT-DOTAGA, respectively, and associated graphs. (A) Three-dimensional CT reconstruction map displaying osteolysis as a consequence of MDA-MB-231 tumor growth, with imaging planes numbered. (B) PET/CT images (left) after registering to corresponding MRI sections (right) acquired at plane positions shown in A. MRI scans were used as reference to target PET/CT images for image registration purposes by using a set of landmarks (Fig E5 [supplement]). Numbers correspond to numbered imaging planes in A. * Femur. (C) PET tumor-to-muscle mean standardized uptake value (SUV) ratios (open circles) and maximum SUV ratios (filled circles) measured in total tumor volume in control group that underwent blocking anti-EGFR antibody preinjection (black circles), experimental group (blue circles), and tissue-infiltrating bone tumor fraction in experimental groups (red circles) (three tumors per group). * Statistically significant difference (Mann-Whitney nonparametric test, P < .05). (D) MRI signal intensity measured by using TW-segmentation of PET/CT images for region of interest analysis (SUV > 0.2) identified as tumor (n = 15) versus background signal intensity measured in the muscle (n = 10). Data shown as mean ± standard deviation were analyzed by using Mann-Whitney nonparametric test (P < .001). AU = arbitrary units, TW = trainable Weka.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/1b9b/8344354/bc6e2e93684e/rycan.2021200069.fig6.jpg)