Abstract
We previously conducted a genome-wide association study (GWAS) of attempted suicide within bipolar disorder, which implicated common variation in the 2p25 region primarily in males. The top association signal from our GWAS occurred in an intergenic region of 2p25 (p = 5.07×10−8) and was supported by two independent studies. In the current study, to better characterize the association of the 2p25 region with attempted suicide, we sequenced the entire 350kb 2p25 region in 476 bipolar suicide attempters and 473 bipolar non-attempters using targeted next-generation sequencing. This fine-mapping project identified 4,681 variants in the 2p25 region. We performed both gene-level and individual-variant tests on our sequencing results and identified 375 variants which were nominally significant (p < 0.05) and three common variants that were significantly associated with attempted suicide in males (corrected p = 0.035, odds ratio (OR) = 2.13). These three variants are in strong linkage disequilibrium with the top variant from our GWAS. Our top five variants are also predicted expression quantitative trait loci (eQTL) for three genes in the 2p25 region based on publicly available brain expression databases. Our sequencing and eQTL data implicate these three genes - SH3YL1, ACP1, and FAM150B - and three additional pathways - androgen receptor, Wnt signaling, and glutamatergic/GABAergic signaling - in the association of the 2p25 region with suicide. The current study provides additional support for an association of the 2p25 region with attempted suicide in males and identifies several candidate genes and pathways that warrant further investigation to understand their role in suicidal behavior.
Keywords: suicide, genetics, bipolar, sequencing, 2p25, sex-specific
Introduction
Genetic factors contribute to the risk for suicidal behavior, with current heritability estimates around 30–50% (Mann et al., 2009). Although this heritability is partly driven by psychiatric disorders, there is also an independent genetic component that may be specific to suicide (Brent and Mann, 2005). To elucidate this independent genetic factor, we previously conducted a genome-wide association study (GWAS) that implicated the 2p25 chromosomal region in the attempted suicide phenotype (Willour et al., 2012). The top finding from this GWAS (rs300774, p = 5.07 × 10−8) was driven mainly by the male-specific sample set (male p = 4.55 × 10−6, female p = 0.011). The associated region is intergenic and found in a large linkage disequilibrium (LD) block within 2p25. We also identified several other variants within the 2p25 region that were approaching genome-wide significance, suggesting that this region may play a role in the risk for suicidal behavior.
The 2p25 region includes the first 350kb of chromosome 2 and contains four genes, all of which are expressed in the brain (Hawrylycz et al., 2012): FAM110C, SH3YL1, ACP1, and FAM150B. FAM110C may have roles in cell spreading, migration, and proliferation (Hauge et al., 2009; Li et al., 2012). The SH3YL1 protein has been shown to interact with the N-terminus of androgen receptors to regulate gene expression (Blessing et al., 2015). FAM150B encodes a secreted factor that is expressed in neuroblastoma (Reshetnyak et al., 2015) and stimulates both leukocyte tyrosine kinase and anaplastic lymphoma kinase activation (Guan et al., 2015; Zhang et al., 2014). The final gene in the region, ACP1, is a protein tyrosine phosphatase whose expression is significantly upregulated in the brains of bipolar suicide completers (Higgs et al., 2006; Willour et al., 2012). The ACP1 protein has also been shown to influence the canonical Wnt signaling pathway in a manner that is the opposite of the effect of lithium, one of the major drugs shown to consistently reduce the risk for suicidal behavior (Gould et al., 2004; Taddei et al., 2002).
In the current study, we sequenced the entire 350kb 2p25 region in 476 bipolar suicide attempters and 473 bipolar non-attempters using a targeted next-generation sequencing approach. This next-generation sequencing approach allowed us to more deeply assess variation, both common and rare, and the LD structure within the 2p25 candidate region in order to fine map where the true association signal lies. We also used publicly available brain gene expression databases to assess the functionality of variants implicated through both our attempted suicide GWAS and the current next-generation sequencing study. The results of this study provide further support for a potential role of the 2p25 region in suicidal behavior and elucidate potential novel candidate genes and pathways for the male-specific association of the 2p25 region with suicidal behavior.
Materials and Methods
Sample Collection
The samples used for this study were a subset of our attempted suicide GWAS samples (Willour et al., 2012). These samples were from the National Institute of Mental Health Bipolar Initiative (https://www.nimhgenetics.org/), and their DNA was obtained from the Rutgers Cell and DNA Repository. The sample set included 476 bipolar suicide attempters (224 males and 252 females) and 476 bipolar non-attempters (224 males and 252 females). All subjects were unrelated and of European-American descent (Table 1). Subjects were all diagnosed with either bipolar I (BPI) or schizoaffective disorder, bipolar type (SABP). Suicide attempters responded positively to the question “Have you ever tried to kill yourself?” from the Diagnostic Interview for Genetic Studies (DIGS) (Nurnberger et al., 1994). While our GWAS examined all suicide attempters regardless of intent, the subset of attempters used in this study included only attempters that had definite or serious intent to die in their suicide attempt according to the DIGS questionnaire. We selected high intent attempters with the goal of increasing homogeneity and improving our chance to identify genetic contributors to suicide. All subjects provided Institutional Review Board-approved consent before participating in this study.
Table 1:
Sample Set Demographics
| Sex | Diagnosis | ||||||
|---|---|---|---|---|---|---|---|
| Male (%) | Female (%) | BP I | SABP | ||||
| Non-Attempters | 222 (46.93%) | 251 (53.07%) | 473 | 0 | 42.72 (18–88) | 473 EA | 473 |
| Suicide Attempters | 224 (47.06%) | 252 (52.94%) | 475 | 1 | 42.99 (19–82) | 476 EA | 476 |
BP I, bipolar I; SABP, schizoaffective disorder, bipolar type; EA, European-American
Subjects are all unrelated.
Age information was not obtained for one subject. No significant difference in age of attempters and non-attempters (p = 0.75).
All subjects have a European-American background as determined through a principal component analysis.
Three non-attempters were removed from the original sample set due to high levels of genotype mismatch with our genome-wide association study data. The final sample set totals are shown here.
Next-Generation Sequencing
We performed library preparation for the entire 350kb 2p25 region using the SureSelectXT Target Enrichment next-generation sequencing technology from Agilent (Santa Clara, CA). Sequencing was done using the Illumina (San Diego, CA) HiSeq 2000. The Burrows-Wheeler Aligner (BWA: v0.6.2) (Li and Durbin, 2009) was used to align 100bp paired-end reads to the hg19 reference human genome. SAMtools (v0.1.18) (Li et al., 2009) was used to create, sort, and index binary alignment (BAM) files of the sequencing data. Picard (http://picard.sourceforge.net:v1.88) was used to fix mispaired reads and remove duplicate reads. Reads with a mapping quality score <20 were removed using BAMtools (v2.2.3) (Barnett et al., 2011). Indels and single nucleotide polymorphisms (SNPs) were called using the Genome Analysis Tool Kit (GATK: v3.1.1) (McKenna et al., 2010). While the data in this study have an overall concordance rate of 99.75% with our GWAS variant calls, we removed three non-attempter subjects with high levels of genotype mismatch. Thus, our final sample for this study included 476 bipolar suicide attempters and 473 bipolar non-attempters (Table 1). Additional quality control filters are described in the Supplemental Methods.
Statistical Analyses
Individual-variant tests were performed using logistic regression with Firth’s correction method (Firth, 1993), which provides unbiased and robust results within small/sparse datasets, and were covariate-corrected for sex and the first three principal components for each subject from the attempted suicide GWAS (Willour et al., 2012). We did not correct for age, because our attempters and non-attempters did not differ significantly in their average age (p = 0.75; Table 1). Both common and rare individual variants were assessed in the entire sample set and in sex-specific subsets. Individual-variant tests were corrected for multiple testing using a permutation-based correction method in which the individual-variant tests were repeated 10,000 times with randomly swapped attempter and non-attempter labels for each subject. An individual-variant result was considered significant if the permutation-corrected p-value was less than 0.05. The permutation correction was applied for the overall and sex-specific analyses. The sequencing results were assessed using Haploview v4.2 (Barrett et al., 2005) to determine the LD structure of the 2p25 region. By recoding insertions and deletions as A/B alleles using PLINK (Purcell et al., 2007), we were able to include both SNPs and insertions/deletions in our Haploview analyses.
We also performed gene-level tests for the four known genes within the 2p25 region (FAM110C, SH3YL1, ACP1, FAM150B). Our goal was to assess the role of rare variation (minor allele frequency (MAF) < 0.05) in these genes. Two types of gene-level tests, gene-burden tests (Li and Leal, 2008) and sequence kernel association tests (SKAT) (Wu et al., 2011), were performed for both the entire sample set and sex-specific subsets. We ran these tests separately on coding and regulatory regions and on two separate groups of functional variants, which were defined bioinformatically (Purcell et al., 2014) (see Supplemental Methods).
Brain Expression Databases
We examined two publicly available brain gene expression databases to determine whether the top variants from our attempted suicide GWAS and the current sequencing study are expression quantitative trait loci (eQTLs). The first, BrainCloud (Collado-Torres et al., 2019) (http://braincloud.jhmi.edu), contains postmortem gene expression data for 551 brains (286 schizophrenia, 265 non-psychiatric controls). Of the 286 schizophrenia brains, ~19.6% of them were from individuals that died from suicide. RNA was isolated from both the dorsolateral prefrontal cortex and hippocampus, and gene expression analyses were performed using RNA-sequencing. Genotypes were determined using DNA isolated from cerebellar tissue and analyzed using HumanHap650Y_V3, Human 1M-Duo_V3, and Omni5 BeadChips (Illumina: San Diego, CA) (Jaffe et al., 2018). This genotype and gene expression data was then used to identify brain eQTLs.
The second publicly available dataset, Braineac (Trabzuni et al., 2011) (http://braineac.org), has expression data for 134 postmortem brains free of neurodegenerative disorders. Up to 10 regions were examined for each brain including the cerebellar cortex, frontal cortex, hippocampus, medulla, occipital cortex, putamen, substantia nigra, thalamus, temporal cortex, and intralobular white matter. Gene expression was determined using the Affymetrix (Thermo Fisher: Santa Clara, CA) Human Exon 1.0 ST arrays, and DNA was collected and genotyped using the Illumina Infinium Omni1-Quad BeadChip and Immunochip to allow for eQTL determination. For both BrainCloud and Braineac, the eQTL p-values reported are false discovery-rate corrected.
To further investigate the regulatory potential of our top variants, we used the atSNP database (http://atsnp.biostat.wisc.edu/search/). This database utilizes an in silico method to predict the effect that SNPs have on transcription factor binding based on known transcription factor motifs (Zuo et al., 2015).
Results
We identified 4,681 variants in the 2p25 region, 375 of which were nominally significant (p < 0.05; Table S1). Since the original goal of the gene-level tests was to assess rare variation, which is missed by GWAS, we performed gene-level analyses with functional rare variants (MAF < 0.05) on the four genes in the 2p25 region. However, none of the gene-level tests for rare variation produced significant evidence after correction for multiple testing (Table S2–S4).
We then decided to reassess the evidence for rare and common variation based on individual-variant testing. As seen in our GWAS, the most significant evidence came from the male samples. We identified 361 nominally significant male-specific variants (p<0.05; Table S5), and the top three male-specific variants, rs300802, rs300799, and rs300797, were study-wide significant for association with attempted suicide (p = 4.84 × 10−5, corrected p = 0.035, odds ratio (OR) = 2.13; Table 2). These variants are in an intergenic LD block between the FAM110C and SH3YL1 genes that also contains rs300774, our top GWAS variant. These three variants are all in strong LD with each other and rs300774 (r2 = 0.99) and fall between 5kb and 8kb from this variant. We also performed individual-variant tests for the overall and female-specific sample sets. We identified 375 nominally significant overall variants (p<0.05). The top overall variant was rs300802 (p = 2.20 × 10−4, corrected p = 0.15, OR = 1.59; Table S1), which was also one of the significant male-specific variants. We identified 41 nominally significant female-specific variants (p<0.05), and the top variant was a novel intergenic variant (p = 0.0062, corrected p = 0.99, OR = 0.064; Table S6).
Table 2:
Top Individual-Variant Results
| Chromosome Locationa,b | Sample Set | Gene | dbSNP142 | Position | P-Value | Corrected P-Value | Odds Ratioc | Attempter MAF | Non-Attempter MAF |
|---|---|---|---|---|---|---|---|---|---|
| chr2:104,762 (A/G) | Male | NULL | rs300802 | intergenic | 4.84E-05 | 0.035 | 2.13 | 0.22 | 0.12 |
| chr2:105,514 (A/G) | Male | NULL | rs300799 | intergenic | 4.84E-05 | 0.035 | 2.13 | 0.22 | 0.12 |
| chr2:107,140 (A/G) | Male | NULL | rs300797 | intergenic | 4.84E-05 | 0.035 | 2.13 | 0.22 | 0.12 |
| chr2:108,411 (T/A) | Male | NULL | rs300793 | intergenic | 7.45E-05 | 0.053 | 2.08 | 0.22 | 0.13 |
| chr2:111,964 (A/C) | Male | NULL | rs300778 | intergenic | 7.45E-05 | 0.053 | 2.08 | 0.22 | 0.13 |
| chr2:119,346 (G/C) | Male | NULL | rs378456 | intergenic | 8.33E-05 | 0.058 | 2.07 | 0.22 | 0.13 |
| chr2:118,383 (C/A) | Male | NULL | rs1829220 | intergenic | 8.50E-05 | 0.059 | 2.07 | 0.22 | 0.13 |
| chr2:118,385 (G/A) | Male | NULL | rs2015419 | intergenic | 8.50E-05 | 0.059 | 2.07 | 0.22 | 0.13 |
| chr2:117,174 (G/A) | Male | NULL | rs167282 | intergenic | 9.50E-05 | 0.067 | 2.06 | 0.22 | 0.13 |
| chr2:138,363 (G/A) | Male | NULL | rs300713 | intergenic | 9.74E-05 | 0.069 | 2.08 | 0.22 | 0.12 |
MAF, minor allele frequency
Chromosome location was determined using the UCSC Genome Browser (hg19).
Variants are listed as (major allele/minor allele).
Odds ratios shown are for the minor allele.
The additional data from this sequencing study allowed us to characterize the LD structure of the 2p25 region more fully (Figure 1). Interestingly, almost all of our top common variants (97/100 top variants in entire sample set; 94/100 top variants in male-specific sample set) occurred within an 80kb LD block in 2p25 (hg19 – chr2: 100,000–180,000) that also contains our top GWAS variants. A bioinformatic assessment of the region using the UCSC Genome Browser revealed many potential regulatory elements across the region, including 132 predicted microRNAs, five predicted genes, DNase hypersensitivity sites, transcription factor binding sites, and peaks of conservation (Figure S1). Based on the regulatory potential of the region, we hypothesized that our top variants may be eQTLs. We investigated this possibility using two publicly available databases: BrainCloud and Braineac, both postmortem brain gene expression databases that include genotype and eQTL data. We specifically focused on the top five variants from the current study, including the three significant male-specific variants, and the top five variants from our attempted suicide GWAS. Two of the top variants overlapped between the two studies, so this resulted in a list of eight variants. All eight variants were eQTLs for SH3YL1 and ACP1 according to both databases (Table 3). Additionally, these variants were eQTLs for FAM150B according to Braineac (Table 3). The eQTL status of our top variants was further supported by the atSNP database (Zuo et al., 2015), which predicted that our top variants each affect between 42 and 109 transcription factor binding sites (Table S7).
Figure 1: Linkage disequilibrium (LD) structure of the 2p25 region.
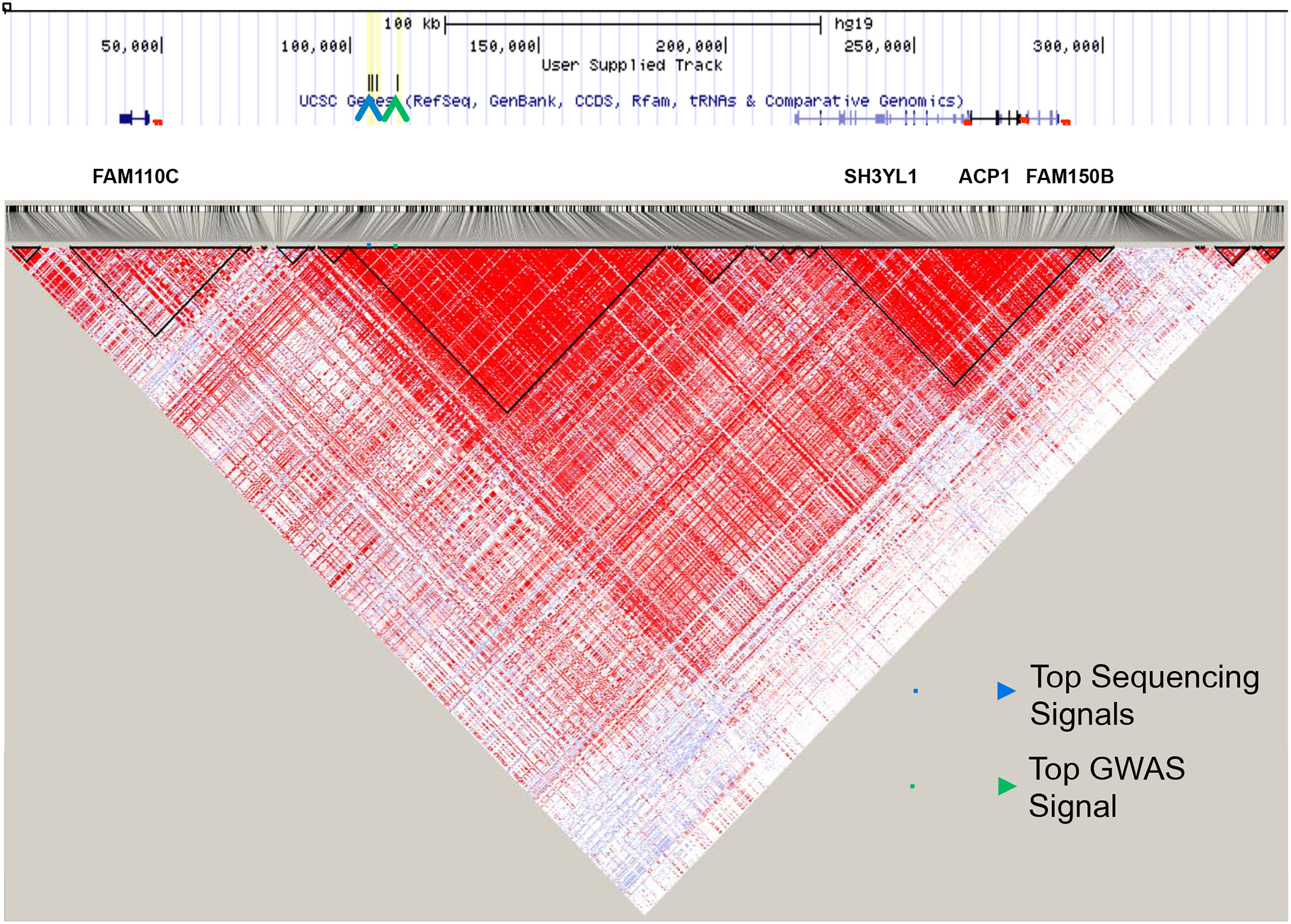
While our genome-wide association study (GWAS) indicated only two major LD blocks in this region, additional sequencing data from the current study allowed us to better characterize the LD structure of this region. We identified three major LD blocks and a number of small LD blocks across the region. Our three study-wide significant variants from the current study are indicated by the blue arrow, and the top GWAS variant (rs300774) is indicated by the green arrow.
Table 3:
eQTL Information for the Top Variants
| Variant | Studya | BrainCloud Hippocampus eQTL Gene | BrainCloud Hippocampus eQTL P-valueb,c | BrainCloud DLPFC eQTL Gene | BrainCloud DLPFC eQTL P-Valueb | Braineac eQTL Gene | Braineac eQTL P-Valueb,d |
|---|---|---|---|---|---|---|---|
| rs300802 | Current | SH3YL1 | 4.70E-04 | SH3YL1 | 1 15E-03 | SH3YL1 | 7.20E-04 |
| ACP1 | 0.012 | ACP1 | 8.50E-03 | ||||
| FAM150B | 6.30E-03 | ||||||
| rs300799 | Current | SH3 YL1 | 8.17E-04 | SH3YL1 | 9.67E-04 | SH3YL1 | 7.50E-04 |
| ACP1 | 0.040 | ACP1 | 8.20E-03 | ||||
| FAM150B | 6.20E-03 | ||||||
| rs300797 | Both | SH3YL1 | 3.26E-04 | SH3YL1 | 7.39E-04 | SH3YL1 | 7.60E-04 |
| ACP1 | 0.027 | ACP1 | 8.20E-03 | ||||
| FAM150B | 6.20E-03 | ||||||
| rs300793 | Current | SH3YL1 | 2.85E-04 | SH3YL1 | 1 78E-03 | SH3YL1 | 7.60E-04 |
| ACP1 | 9.45E-03 | ACP1 | 8.20E-03 | ||||
| FAM150B | 6.20E-03 | ||||||
| rs300778 | Both | SH3YL1 | 9.42E-04 | SH3YL1 | 7.18E-04 | SH3YL1 | 7.60E-04 |
| ACP1 | 8.20E-03 | ||||||
| ACP1 | 0.018 | FAM150B | 6.20E-03 | ||||
| rs436446 | GWAS | SH3YL1 | 3.98E-04 | SH3YL1 | 3 18E-03 | SH3YL1 | 7.60E-04 |
| ACP1 | 0 014 | ACP1 | 8.20E-03 | ||||
| FAM150B | 6.20E-03 | ||||||
| rs300774 | GWAS | SH3 YL1 | 1.77E-04 | SH3YL1 | 2.00E-03 | SH3YL1 | 7.60E-04 |
| ACP1 | 8.20E-03 | ||||||
| ACP1 | 0.017 | FAM150B | 6.20E-03 | ||||
| rs300773 | GWAS | SH3YL1 | 4.80E-04 | SH3YL1 | 2.78E-03 | SH3YL1 | 7.60E-04 |
| ACP1 | 6.45E-03 | ACP1 | 8.20E-03 | ||||
| FAM150B | 6.20E-03 |
eQTL, expression quantitative trait loci; GWAS, genome-wide association study; DLPFC, dorsolateral prefrontal cortex
Indicates whether the variant was identified in the current sequencing study, our GWAS, or both.
The p-values provided by both BrainCloud and Braineac are false discovery rate-corrected. For BrainCloud, the most significant probe from each brain region is listed. For Braineac, the most significant combined p-value from all brain regions and probes tested is listed.
The p-value for SH3YL1 is the top one listed. The p-value for ACP1 is the bottom one listed.
The p-value for SH3YL1 is the top one listed. The p-value for ACP1 is the middle one listed. The p-value for FAM150B is the bottom one listed.
Discussion
Our goal in the current study was to better understand the link between the 2p25 region and attempted suicide by assessing both common and rare variation through a targeted next-generation sequencing study of the entire candidate region. We first assessed rare variation within our sequencing data, but this study provided no evidence for an association of rare variants with attempted suicide. However, we did find additional evidence for an association of common variation. Applying targeted next-generation sequencing to the 2p25 region allowed the identification of substantially greater complexity in its LD structure and offered the means to pinpoint, with far greater accuracy, the area driving our previous male-specific GWAS findings. Specifically, we identified three variants that were significantly associated with attempted suicide within our male-specific sample set.
Suicidal behavior itself also displays sex-specific differences. While females are about three times as likely to attempt suicide, males are almost four times as likely to complete suicide (Prevention, 2013). This difference can be attributed in part to more violent suicide attempts among males, including firearms and suffocation, while females are much more likely to attempt suicide by poisoning (Prevention, 2013). Additionally, the age group most at risk for suicide differs between men and women. The highest rates of suicide for males occur in the 75 years old or older age group, while women between 45 and 54 years old are most at risk (Prevention, 2013). Past genetic studies have also identified sex-related suicide risk loci within a number of different psychiatric disorders, including bipolar disorder (Niculescu et al., 2017).
Since our GWAS was published, two independent studies have implicated the 2p25 region in suicidal behavior in bipolar disorder (Pawlak et al., 2016) and schizophrenia (SCZ) (Li et al., 2017), specifically in males. Pawlak et al. (2016) examined 577 bipolar, 248 major depression, and 847 control patients. They tried to replicate findings from several previous studies of suicidal behavior, including rs300774 from our attempted suicide GWAS. Within the bipolar group, Pawlak et al. found that rs300774 was significantly associated with attempted suicide in the male-specific sample set when comparing either attempters to non-psychiatric controls (p = 0.0002) or attempters to non-attempters (p = 0.0013). This variant was not significantly associated with attempted suicide in the overall or female-specific sample sets for bipolar disorder or in any sample sets for major depression. In the second independent study, Li et al. (2017) examined 162 Caucasian individuals (74 suicide attempters, 88 non-attempters) with either SCZ or schizoaffective disorder (SAD). They attempted to replicate three of the variants from our attempted suicide GWAS: rs300774, rs10437629, and rs7296262. Of the three, only rs300774 was significantly associated with attempted suicide in their SCZ and SAD sample set (p = 0.012). Their result was driven by the male-specific sample set (male p = 0.046, female p = 0.20). Li et al. also performed an in silico functional analysis of rs300774 and found that it is a predicted eQTL for several genes, including ACP1 and FAM150B. Along with our work, these two studies provide evidence from independent sample sets and multiple disorders that the 2p25 region is associated with attempted suicide in males. This association across multiple disorders suggests that variation within the 2p25 region may contribute to the independent heritable factor that is specific to suicide. Interestingly, studies on bipolar suicide that have not stratified by sex have been unable to find any association in the 2p25 region (Mullins et al., 2019; Zai et al., 2015). This evidence points to a potential sex-specific association of the 2p25 region with attempted suicide.
The new sequencing data provided by the current study allowed us to more finely map the LD structure of the 2p25 region (Figure 1). Our attempted suicide GWAS indicated that the top association signal fell within a large LD block (~200kb) containing SH3YL1, ACP1, and FAM150B. However, following the incorporation of our new sequencing data, we found that there are actually several smaller LD blocks within the large LD block implicated in our GWAS. Among these is an 80kb LD block that contains almost all of our top common variants (97/100 top variants in entire sample set; 94/100 top variants in male-specific sample set) and our top 30 GWAS variants. This 80kb LD block encompasses an intergenic region between FAM110C and SH3YL1. Because this is an intergenic region, we performed a bioinformatic assessment of the region using the UCSC Genome Browser and identified many potential regulatory elements, including 132 predicted microRNAs, five predicted genes, DNase hypersensitivity sites, transcription factor binding sites, and peaks of conservation (Figure S1). Preliminary experiments that involved targeted genome editing indicate that this region has regulatory potential (data not shown). The clustering of our top variants allowed us to narrow our candidate region down from the large LD block implicated in our attempted suicide GWAS to this 80kb intergenic LD block. Additionally, because our top candidate region is intergenic with many predicted regulatory elements, we hypothesized that regulatory variation, not coding variation, within the 2p25 region influences the risk for attempted suicide in males.
Based on the regulatory potential of the region, we predicted that our top variants may be eQTLs that exert their effects on suicide risk through effects on gene expression. This is supported by the atSNP database (Zuo et al., 2015), which predicted that our top variants affect transcription factor binding (Table S7). Further, the genes that these variants are predicted to regulate may help explain the sex specificity of the 2p25 association (Figure 2). To investigate this, we used two separate gene expression databases examining postmortem brains: BrainCloud (Collado-Torres et al., 2019) (http://braincloud.jhmi.edu) and Braineac (Trabzuni et al., 2011) (http://braineac.org). Both databases indicated that the top variants from our attempted suicide GWAS and the current sequencing study are brain eQTLs for the SH3YL1 gene in the 2p25 region. The SH3YL1 protein interacts with androgen receptors (AR) to regulate gene expression of a number of AR-modulated genes (Blessing et al., 2015). Androgens are the major male sex hormones and have been previously implicated in suicidal behavior. Levels of testosterone, a type of androgen, have been associated with suicide attempts in men and women with bipolar disorder (Sher et al., 2012, 2014). Higher prenatal androgen loads and masculinized brain organization have been linked to higher rates of suicide completion in men (Lenz et al., 2016). Finally, androgens and the androgen receptor have been implicated in the regulation of aggressive and impulsive behaviors (Carré et al., 2017; Giammanco et al., 2005; Zuloaga et al., 2008). These behaviors have been linked to an increased risk for suicidal behavior (Brent and Mann, 2005; Stack, 2014), and males complete suicide at higher rates than women (Prevention, 2013). Thus, one hypothesis for the male-specific association of the 2p25 region with attempted suicide is that our variants affect expression of SH3YL1, which, in turn, affects expression of AR-regulated genes such as those regulating impulsive aggression, a known risk factor for suicidal behavior (Brent and Mann, 2005, 2006; Dumais et al., 2005; Mann et al., 2009). Because AR-regulated genes influence male-specific biological processes, this hypothesis could also explain why our variants contribute to the risk for suicide in a male-specific fashion.
Figure 2: Three hypotheses for how the male-specific 2p25 variants may contribute to the risk for attempted suicide.
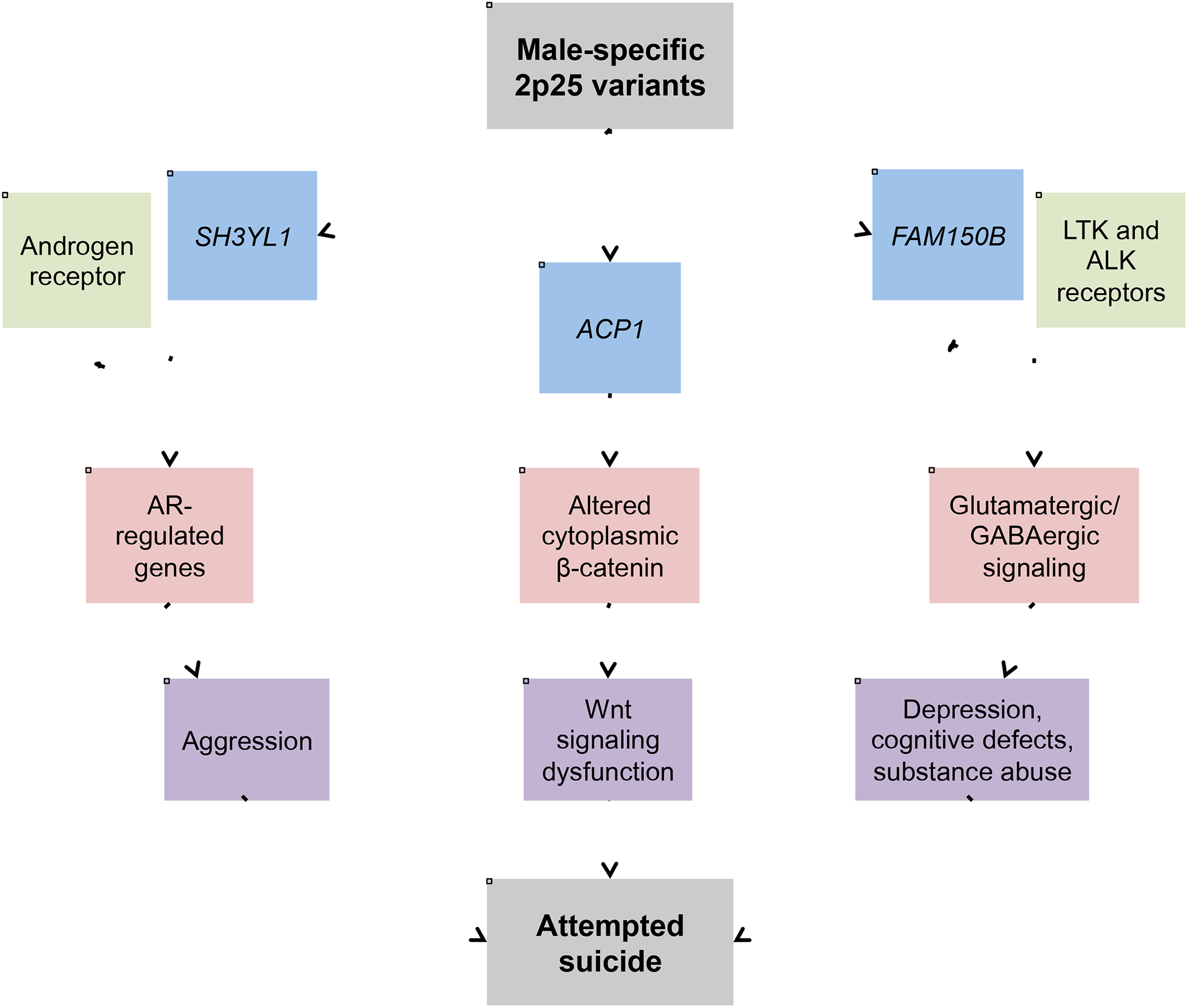
The implicated genes are boxed in blue with their known interacting partners in green. Shown in red are the pathways that would be affected by changes in expression of the implicated genes. Finally, the downstream effects shown in purple are the known risk factors for suicidal behavior. AR = androgen receptor; LTK = leukocyte tyrosine kinase; ALK = anaplastic lymphoma kinase.
Both the BrainCloud and Braineac databases also indicated that our top variants are brain eQTLs for ACP1. ACP1 is a protein tyrosine phosphatase that is expressed in the brain and has been shown to have higher overall expression in the brains of males than females (Higgs et al., 2006). ACP1 has also been shown to have increased expression in the postmortem brains of bipolar suicide completers even after controlling for sex (Willour et al., 2012), and variation in this gene has been associated with suicide (Campos et al., 2017). Interestingly, rs300774, our top GWAS finding, may influence the expression of ACP1 in peripheral blood (Li et al., 2017) and lithium responsiveness in bipolar patients (Szczepankiewicz et al., 2018). Overexpression of ACP1, as seen in bipolar suicide completers, may inhibit the Wnt signaling pathway by causing decreased cytoplasmic β-catenin, while lithium, the major drug used to treat bipolar disorder and suicidal behavior, has the opposite effect (Gould et al., 2004; Taddei et al., 2002). Wnt signaling dysfunction has also been previously implicated in suicidal behavior (Flory et al., 2017; Ren et al., 2013). This evidence supports a second hypothesis in which our variants lead to altered expression of ACP1, an effect potentially more pronounced in males as they have higher baseline expression of ACP1 than females.
Our top variants were also predicted by Braineac to be brain eQTLs for FAM150B. BrainCloud did not identify these variants as eQTLs for FAM150B, potentially due to the different brain regions examined in each database (see Materials and Methods). FAM150B encodes a secreted factor that is expressed in neuroblastoma (Reshetnyak et al., 2015). It acts as a ligand to stimulate both leukocyte tyrosine kinase (LTK) and anaplastic lymphoma kinase (ALK) receptors (Guan et al., 2015; Zhang et al., 2014). While not much is known about the FAM150B gene itself, the LTK and ALK receptors have been implicated in brain-related functions. Both are expressed in neural tissues and have been shown to regulate neurogenesis (Weiss et al., 2012). There is evidence from mouse studies that the ALK receptor may affect glutamatergic and GABAergic signaling (Mangieri et al., 2017; Schweitzer et al., 2016). Additionally, mouse studies have implicated the ALK receptor in depression (Bilsland et al., 2008), cognition (Weiss et al., 2012), and substance use disorders (Mangieri et al., 2017; Schweitzer et al., 2016), all of which are affected by glutamatergic and GABAergic signaling (Cohen Kadosh et al., 2015; Filip and Frankowska, 2008; Kim and Yoon, 2017; Sequeira et al., 2009) and have been previously implicated in suicide risk. In the human brain, the ALK gene has higher baseline expression in males than females (Higgs et al., 2006). This evidence supports a third hypothesis, where the 2p25 variants affect expression of FAM150B, which affects the activity of ALK and LTK receptors. This may lead to alteration of glutamatergic and GABAergic signaling and an increased risk for suicide, an effect potentially more pronounced in males as they have higher baseline expression of ALK. For both eQTL databases, it is important to note that all of our top variants are in very high LD (r2 > 0.99). Future functional work will be needed to identify the causal variant(s) for the observed changes in expression.
There are several limitations to our study. The first is that our sample size was only large enough to detect a genotypic relative risk ≥ 1.70 with 80% power for individual-variant tests looking at variants with a MAF of 0.25 (calculated using Quanto v1.2.4; http://biostats.usc.edu/Quanto.html). For the gene-level tests, we could significantly detect a genotypic relative risk ≥ 2.12 with 80% power and a MAF of 0.05. Another limitation to our study is that several genomic regions within the 2p25 region are highly repetitive and inherently difficult to sequence. Thus, while we were able to cover 94.6% of our target region with at least 10X coverage (Table S8), we may have missed potential regulatory elements within these repetitive regions due to poor sequencing quality. A final limitation is that the two gene expression databases used, BrainCloud and Braineac, do not assess eQTL status in bipolar disorder. BrainCloud contains data from schizophrenia and non-psychiatric controls, and Braineac includes data from individuals free of neurodegenerative disorders. There are no published brain gene expression studies that focus on male-specific risk factors for suicide in the context of bipolar disorder for direct comparison. Future gene expression studies using bipolar disorder males with and without suicidal behavior will be required to confirm the potential role that our variants and the 2p25 genes may play in suicide risk. Additionally, further whole-genome sequencing studies in larger sample sets of both males and females will be necessary to confirm the sex-specific association of common and/or rare variation in the 2p25 region with attempted suicide in bipolar disorder.
In this study, we have presented a targeted next-generation sequencing study of the 2p25 region in attempted suicide. We identified three male-specific variants that are significantly associated with the attempted suicide phenotype. We also found that our top variants are predicted brain eQTLs for SH3YL1, FAM150B, and ACP1, potentially affecting the expression of these genes. These effects on expression may then dysregulate pathways previously linked to suicidal behavior (Figure 2). Based on these findings, we generated several hypotheses that may help explain the association of the 2p25 region with attempted suicide in males. This study provides further support for a putative male-specific role of the 2p25 region in the attempted suicide phenotype and presents several new candidate genes and pathways for how this region may affect the risk for suicidal behavior.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
Declarations of interest: none
References
- Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT, 2011. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27(12), 1691–1692. 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2), 263–265. 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bilsland JG, Wheeldon A, Mead A, Znamenskiy P, Almond S, Waters KA, Thakur M, Beaumont V, Bonnert TP, Heavens R, Whiting P, McAllister G, Munoz-Sanjuan I, 2008. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 33(3), 685–700. 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- Blessing AM, Ganesan S, Rajapakshe K, Ying Sung Y, Reddy Bollu L, Shi Y, Cheung E, Coarfa C, Chang JT, McDonnell DP, Frigo DE, 2015. Identification of a Novel Coregulator, SH3YL1, That Interacts With the Androgen Receptor N-Terminus. Molecular Endocrinology 29(10), 1426–1439. 10.1210/me.2015-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent DA, Mann JJ, 2005. Family genetic studies, suicide, and suicidal behavior. Am. J. Med. Genet. C Semin. Med. Genet 133C(1), 13–24. 10.1002/ajmg.c.30042. [DOI] [PubMed] [Google Scholar]
- Brent DA, Mann JJ, 2006. Familial pathways to suicidal behavior--understanding and preventing suicide among adolescents. The New England Journal of Medicine 355(26), 2719–2721. 10.1056/NEJMp068195. [DOI] [PubMed] [Google Scholar]
- Campos SB, Brasil Rocha PM, Neves FS, Miranda DM, Correa H, 2017. ACP1 Gene Polymorphism Associated with Suicide Attempt Type in Bipolar Disorder Patients. Psychiatry Investigation 14(6), 909–910. 10.4306/pi.2017.14.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré JM, Geniole SN, Ortiz TL, Bird BM, Videto A, Bonin PL, 2017. Exogenous Testosterone Rapidly Increases Aggressive Behavior in Dominant and Impulsive Men. Biol. Psychiatry 82(4), 249–256. 10.1016/j.biopsych.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Cohen Kadosh K, Krause B, King AJ, Near J, Cohen Kadosh R, 2015. Linking GABA and glutamate levels to cognitive skill acquisition during development. Hum. Brain Mapp 36(11), 4334–4345. 10.1002/hbm.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Torres L, Burke EE, Peterson A, Shin J, Straub RE, Rajpurohit A, Semick SA, Ulrich WS, Price AJ, Valencia C, Tao R, Deep-Soboslay A, Hyde TM, Kleinman JE, Weinberger DR, Jaffe AE, Consortium B, 2019. Regional Heterogeneity in Gene Expression, Regulation, and Coherence in the Frontal Cortex and Hippocampus across Development and Schizophrenia. Neuron 103(2), 203–216.e208. 10.1016/j.neuron.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumais A, Lesage AD, Alda M, Rouleau G, Dumont M, Chawky N, Roy M, Mann JJ, Benkelfat C, Turecki G, 2005. Risk factors for suicide completion in major depression: a case-control study of impulsive and aggressive behaviors in men. The American Journal of Psychiatry 162(11), 2116–2124. 10.1176/appi.ajp.162.11.2116. [DOI] [PubMed] [Google Scholar]
- Filip M, Frankowska M, 2008. GABA(B) receptors in drug addiction. Pharmacol. Rep 60(6), 755–770. [PubMed] [Google Scholar]
- Firth D, 1993. Bias reduction of maximum likelihood estimates. Biometrika 80(1), 27–38. [Google Scholar]
- Flory JD, Donohue D, Muhie S, Yang R, Miller SA, Hammamieh R, Ryberg K, Yehuda R, 2017. Gene expression associated with suicide attempts in US veterans. Translational Psychiatry 7(9), e1226. 10.1038/tp.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammanco M, Tabacchi G, Giammanco S, Di Majo D, La Guardia M, 2005. Testosterone and aggressiveness. Med. Sci. Monit 11(4), RA136–145. [PubMed] [Google Scholar]
- Gould TD, Chen G, Manji HK, 2004. In vivo evidence in the brain for lithium inhibition of glycogen synthase kinase-3. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 29(1), 32–38. 10.1038/sj.npp.1300283. [DOI] [PubMed] [Google Scholar]
- Guan J, Umapathy G, Yamazaki Y, Wolfstetter G, Mendoza P, Pfeifer K, Mohammed A, Hugosson F, Zhang H, Hsu AW, Halenbeck R, Hallberg B, Palmer RH, 2015. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife 4, e09811. 10.7554/eLife.09811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauge H, Fjelland KE, Sioud M, Aasheim HC, 2009. Evidence for the involvement of FAM110C protein in cell spreading and migration. Cell. Signal 21(12), 1866–1873. 10.1016/j.cellsig.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, Abajian C, Beckmann CF, Bernard A, Bertagnolli D, Boe AF, Cartagena PM, Chakravarty MM, Chapin M, Chong J, Dalley RA, David Daly B, Dang C, Datta S, Dee N, Dolbeare TA, Faber V, Feng D, Fowler DR, Goldy J, Gregor BW, Haradon Z, Haynor DR, Hohmann JG, Horvath S, Howard RE, Jeromin A, Jochim JM, Kinnunen M, Lau C, Lazarz ET, Lee C, Lemon TA, Li L, Li Y, Morris JA, Overly CC, Parker PD, Parry SE, Reding M, Royall JJ, Schulkin J, Sequeira PA, Slaughterbeck CR, Smith SC, Sodt AJ, Sunkin SM, Swanson BE, Vawter MP, Williams D, Wohnoutka P, Zielke HR, Geschwind DH, Hof PR, Smith SM, Koch C, Grant SGN, Jones AR, 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489(7416), 391–399. 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs BW, Elashoff M, Richman S, Barci B, 2006. An online database for brain disease research. BMC Genomics 7, 70. 10.1186/1471-2164-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, Kam-Thong T, Xi HS, Quan J, Chen Q, Colantuoni C, Ulrich WS, Maher BJ, Deep-Soboslay A, BrainSeq C, Cross AJ, Brandon NJ, Leek JT, Hyde TM, Kleinman JE, Weinberger DR, 2018. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci 21(8), 1117–1125. 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Yoon BE, 2017. Altered GABAergic Signaling in Brain Disease at Various Stages of Life. Exp. Neurobiol 26(3), 122–131. 10.5607/en.2017.26.3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz B, Thiem D, Bouna-Pyrrou P, Mühle C, Stoessel C, Betz P, Kornhuber J, 2016. Low digit ratio (2D:4D) in male suicide victims. J. Neural Transm. (Vienna) 123(12), 1499–1503. 10.1007/s00702-016-1608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM, 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American Journal of Human Genetics 83(3), 311–321. 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Jang H, Puttabyatappa M, Jo M, Curry TE Jr., 2012. Ovarian FAM110C (family with sequence similarity 110C): induction during the periovulatory period and regulation of granulosa cell cycle kinetics in rats. Biology of Reproduction 86(6), 185. 10.1095/biolreprod.112.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14), 1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing, S., 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16), 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yoshikawa A, Meltzer HY, 2017. Replication of rs300774, a genetic biomarker near ACP1, associated with suicide attempts in patients with schizophrenia: Relation to brain cholesterol biosynthesis. J. Psychiatr. Res 94, 54–61. 10.1016/j.jpsychires.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Mangieri RA, Maier EY, Buske TR, Lasek AW, Morrisett RA, 2017. Anaplastic Lymphoma Kinase Is a Regulator of Alcohol Consumption and Excitatory Synaptic Plasticity in the Nucleus Accumbens Shell. Front. Pharmacol 8, 533. 10.3389/fphar.2017.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, Currier D, Dougherty DM, Haghighi F, Hodge SE, Kleinman J, Lehner T, McMahon F, Mościcki EK, Oquendo MA, Pandey GN, Pearson J, Stanley B, Terwilliger J, Wenzel A, 2009. Candidate endophenotypes for genetic studies of suicidal behavior. Biol. Psychiatry 65(7), 556–563. 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20(9), 1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins N, Bigdeli TB, Borglum AD, Coleman JRI, Demontis D, Mehta D, Power RA, Ripke S, Stahl EA, Starnawska A, Anjorin A, Psych MRC, Corvin A, Sanders AR, Forstner AJ, Reif A, Koller AC, Swiatkowska B, Baune BT, Muller-Myhsok B, Penninx B, Pato C, Zai C, Rujescu D, Hougaard DM, Quested D, Levinson DF, Binder EB, Byrne EM, Agerbo E Dr. Med.Sc, Streit F, Mayoral F, Bellivier F, Degenhardt F, Breen G, Morken G, Turecki G, Rouleau GA, Grabe HJ, Volzke H, Jones I, Giegling I, Agartz I, Melle I, Lawrence J M.R.C.Psych, Walters JTR, Strohmaier J, Shi J, Hauser J, Biernacka JM, Vincent JB, Kelsoe J, Strauss JS, Lissowska J, Pimm J M.R.C.Psych, Smoller JW, Guzman-Parra J, Berger K, Scott LJ, Jones LA, Azevedo MH, Trzaskowski M, Kogevinas M, Rietschel M, Boks M, Ising M, Grigoroiu-Serbanescu M, Hamshere ML, Leboyer M, Frye M, Nothen MM, Alda M, Preisig M, Nordentoft M, Boehnke M, O’Donovan MC, Owen MJ, Pato MT, Renteria ME, Budde M, Dipl P, Weissman MM, Wray NR, Bass N M.R.C.Psych, Craddock N, Smeland OB, Andreassen OA, Mors O, Gejman PV, Sklar P, McGrath P, Hoffmann P, McGuffin P, Lee PH, Mortensen PB, Kahn RS, Ophoff RA, Adolfsson R, Van der Auwera S, Djurovic S, Kloiber S, Heilmann-Heimbach S, Jamain S, Hamilton SP, McElroy SL, Lucae S, Cichon S, Schulze TG, Hansen T, Werge T, Air TM, Nimgaonkar V, Appadurai V, Cahn W, Milaneschi Y, Major Depressive Disorder Working Group of the Psychiatric Genomics, C., Bipolar Disorder Working Group of the Psychiatric Genomics, C., Schizophrenia Working Group of the Psychiatric Genomics, C., Fanous AH, Kendler KS, McQuillin A, Lewis CM, 2019. GWAS of Suicide Attempt in Psychiatric Disorders and Association With Major Depression Polygenic Risk Scores. Am. J. Psychiatry, appiajp201918080957. 10.1176/appi.ajp.2019.18080957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, Le-Niculescu H, Levey DF, Phalen PL, Dainton HL, Roseberry K, Niculescu EM, Niezer JO, Williams A, Graham DL, Jones TJ, Venugopal V, Ballew A, Yard M, Gelbart T, Kurian SM, Shekhar A, Schork NJ, Sandusky GE, Salomon DR, 2017. Precision medicine for suicidality: from universality to subtypes and personalization. Mol. Psychiatry 22(9), 1250–1273. 10.1038/mp.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI Jr., Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T, 1994. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Archives of General Psychiatry 51(11), 849–859; discussion 863–844. [DOI] [PubMed] [Google Scholar]
- Pawlak J, Dmitrzak-Weglarz M, Wilkosc M, Szczepankiewicz A, Leszczynska-Rodziewicz A, Zaremba D, Kapelski P, Rajewska-Rager A, Hauser J, 2016. Suicide behavior as a quantitative trait and its genetic background. J. Affect. Disord 206, 241–250. 10.1016/j.jad.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Prevention, Centers for Disease Control and, 2013. Web-based Injury Statistics Query and Reporting System (WISQARS). https://www.cdc.gov/injury/wisqars/index.html.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81(3), 559–575. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P, 2014. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506(7487), 185–190. 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Rizavi HS, Khan MA, Dwivedi Y, Pandey GN, 2013. Altered Wnt signalling in the teenage suicide brain: focus on glycogen synthase kinase-3beta and beta-catenin. The International Journal of Neuropsychopharmacology 16(5), 945–955. 10.1017/S1461145712001010. [DOI] [PubMed] [Google Scholar]
- Reshetnyak AV, Murray PB, Shi X, Mo ES, Mohanty J, Tome F, Bai H, Gunel M, Lax I, Schlessinger J, 2015. Augmentor alpha and beta (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proceedings of the National Academy of Sciences of the United States of America 112(52), 15862–15867. 10.1073/pnas.1520099112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer P, Cates-Gatto C, Varodayan FP, Nadav T, Roberto M, Lasek AW, Roberts AJ, 2016. Dependence-induced ethanol drinking and GABA neurotransmission are altered in Alk deficient mice. Neuropharmacology 107, 1–8. 10.1016/j.neuropharm.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G, 2009. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS One 4(8), e6585. 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher L, Grunebaum MF, Sullivan GM, Burke AK, Cooper TB, Mann JJ, Oquendo MA, 2012. Testosterone levels in suicide attempters with bipolar disorder. J. Psychiatr. Res 46(10), 1267–1271. 10.1016/j.jpsychires.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher L, Grunebaum MF, Sullivan GM, Burke AK, Cooper TB, Mann JJ, Oquendo MA, 2014. Association of testosterone levels and future suicide attempts in females with bipolar disorder. J. Affect. Disord 166, 98–102. 10.1016/j.jad.2014.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack S, 2014. Differentiating suicide ideators from attempters: violence--a research note. Suicide Life Threat. Behav 44(1), 46–57. 10.1111/sltb.12054. [DOI] [PubMed] [Google Scholar]
- Szczepankiewicz A, Narozna B, Rybakowski JK, Kliwicki S, Czerski P, Dmitrzak-Weglarz M, Skibinska M, Twarowska-Hauser J, Pawlak J, 2018. Genes involved in stress response influence lithium efficacy in bipolar patients. Bipolar Disorders. 10.1111/bdi.12639. [DOI] [PubMed] [Google Scholar]
- Taddei ML, Chiarugi P, Cirri P, Buricchi F, Fiaschi T, Giannoni E, Talini D, Cozzi G, Formigli L, Raugei G, Ramponi G, 2002. Beta-catenin interacts with low-molecular-weight protein tyrosine phosphatase leading to cadherin-mediated cell-cell adhesion increase. Cancer Research 62(22), 6489–6499. [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J, 2011. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. Journal of Neurochemistry 119(2), 275–282. 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JB, Xue C, Benice T, Xue L, Morris SW, Raber J, 2012. Anaplastic lymphoma kinase and leukocyte tyrosine kinase: functions and genetic interactions in learning, memory and adult neurogenesis. Pharmacol. Biochem. Behav 100(3), 566–574. 10.1016/j.pbb.2011.10.024. [DOI] [PubMed] [Google Scholar]
- Willour VL, Seifuddin F, Mahon PB, Jancic D, Pirooznia M, Steele J, Schweizer B, Goes FS, Mondimore FM, Mackinnon DF, Bipolar Genome Study C, Perlis RH, Lee PH, Huang J, Kelsoe JR, Shilling PD, Rietschel M, Nothen M, Cichon S, Gurling H, Purcell S, Smoller JW, Craddock N, DePaulo JR Jr., Schulze TG, McMahon FJ, Zandi PP, Potash JB, 2012. A genome-wide association study of attempted suicide. Molecular Psychiatry 17(4), 433–444. 10.1038/mp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X, 2011. Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics 89(1), 82–93. 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai CC, Goncalves VF, Tiwari AK, Gagliano SA, Hosang G, de Luca V, Shaikh SA, King N, Chen Q, Xu W, Strauss J, Breen G, Lewis CM, Farmer AE, McGuffin P, Knight J, Vincent JB, Kennedy JL, 2015. A genome-wide association study of suicide severity scores in bipolar disorder. J. Psychiatr. Res 65, 23–29. 10.1016/j.jpsychires.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang H, Pao LI, Zhou A, Brace AD, Halenbeck R, Hsu AW, Bray TL, Hestir K, Bosch E, Lee E, Wang G, Liu H, Wong BR, Kavanaugh WM, Williams LT, 2014. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proceedings of the National Academy of Sciences of the United States of America 111(44), 15741–15745. 10.1073/pnas.1412009111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga DG, Puts DA, Jordan CL, Breedlove SM, 2008. The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Hormones and Behavior 53(5), 613–626. 10.1016/j.yhbeh.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo C, Shin S, Keles S, 2015. atSNP: transcription factor binding affinity testing for regulatory SNP detection. Bioinformatics 31(20), 3353–3355. 10.1093/bioinformatics/btv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


