Abstract
The circulation of P.1 SARS-CoV-2 lineage becomes a challenge of pandemic control. Among the COVID-19 cases reported in Brazil, P.1 is estimated 54% more fatal than non-P.1 significantly. Considering the transmission advantage of P.1, we raise concerns regarding the rapid growth in critical patients.
Keywords: COVID-19, P.1 lineage, Brazil, case fatality, statistical modelling
Characterizing the role of mutation activities associated with the clinical severity of disease is of public health importance for informing healthcare and treatment strategies. In December 2020, the SARS-CoV-2 strains carrying several novel genetic mutations including K417T, E484K and N501Y were detected in Brazil, then classified as P.1 lineage, which is a descendant of previous B.1.1.28 lineage, and circulating or dominant in some Brazilian regions (e.g. Manaus) since 2021. The rapid spread of P.1 variants coincided with increased incidence of hospitalization,1 which suspects possible links to more severe clinical outcomes. In this work, we adopted a statistical framework to infer the association between COVID-19 case fatality risk (CFR) and P.1 SARS-CoV-2 lineage in Brazil.
Using the national-wide COVID-19 surveillance (Figure 1A and B) and SARS-CoV-2 sequencing data in Brazil (Supplementary Information S1), we reconstructed the variant-specified case fatality ratio on a real time basis by adopting the statistical framework in previous study2 (Supplementary Information S2). The risk ratio (ζ) between the fatality risks of P.1 lineage against non-P.1 lineages are defined as the ratio between variant-specified CFR. In other words, the CFR of P.1 is ζ-fold of the CFR of non-P.1. Thus, if ζ > 1, the P.1 lineage may be more fatal than the non-P.1 lineages and vice versa.
Figure 1.
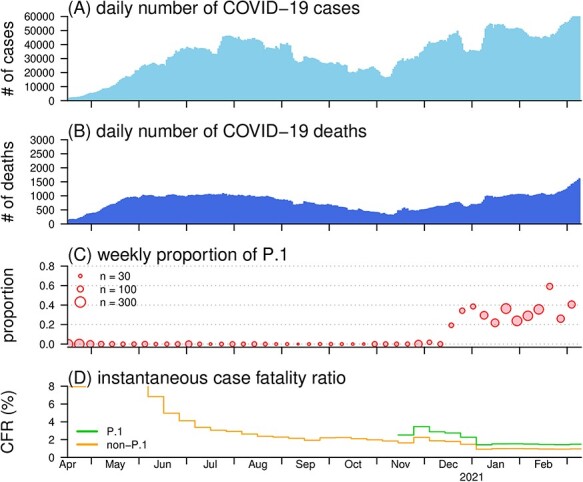
The daily number of COVID-19 cases (A) and deaths (B), proportion of the P.1 SARS-CoV-2 variants (C), and the reconstructed CFR (D) in Brazil. Panel (A) and (B) show the daily number of COVID-19 cases and deaths time series in Brazil, respectively. Panel (C) shows the observed proportion of P.1 variants among all samples of SARS-CoV-2 strains. In the legend, the size of the dot indicates the weekly sample size of the observed SARS-CoV-2 strains. Panel (D) shows the reconstructed CFR of P.1 (in green) and CFR of non-P.1 (in orange)
We observe that the proportion of P.1 variants increased rapidly since December 2020 and trended to dominance (>50%) after February 2021 in Brazil (Figure 1C). For the CFR reconstruction, we find that the CFR gradually decreased before October 2020 and stabilized at 1.3% [95% confidence interval (CI): 1.1, 1.5], see Figure 1D. We estimate that CFR of P.1 variants appear higher than that of non-P.1 variants by a factor of ζ = 1.54 (95%CI: 1.13, 2.02), which indicates an increased mortality risks among the COVID-19 cases. The increase in CFR associated with P.1 lineage appears in line with the evidence from recent literature.3 For validation, we repeat the estimating process of ζ with alternative setting and modelling schemes (Supplementary Information S3) and find that the ζ estimates are consistently and significantly larger than 1 in similar scales (data not shown). Some strengths and limitations, mainly due to lack of data or information, about this study are discussed in Supplementary Information S4, some of which is pointed out in.4
Besides the transmission advantage of P.1 variants found in existing literature,3 P.1 variants may also escape the neutralizing antibodies induced by vaccination or previous infection.5 A higher mortality risk will reaffirm the growing volumes of COVID-19 patients with critical status, which suggests increasing burdens to the local healthcare system and medical supply in Brazil. As such, we recommend to re-enforce the disease control measures to flatten the epidemic curve, e.g. nonpharmaceutical interventions and vaccination campaigns, and to optimize the planning and allocation of medical resources. Concerning the challenges in controlling the spatial spreading of COVID-19, we highlight the potential risks of P.1 variants when it invades other places having population with vulnerability.
Supplementary Material
Contributor Information
Shi Zhao, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China; CUHK Shenzhen Research Institute, Shenzhen, China.
Jingzhi Lou, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China.
Lirong Cao, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China.
Hong Zheng, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China.
Zigui Chen, Department of Microbiology, Chinese University of Hong Kong, Hong Kong, China.
Renee W Y Chan, Department of Paediatrics, Chinese University of Hong Kong, Hong Kong, China; Hong Kong Hub of Pediatric Excellence, Chinese University of Hong Kong, Hong Kong, China; CUHK-UMCU Joint Research Laboratory of Respiratory Virus and Immunobiology, Chinese University of Hong Kong, Hong Kong, China; Li Ka Shing Institute of Health Sciences, Faculty of Medicine, Chinese University of Hong Kong, Hong Kong, China.
Benny C Y Zee, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China; CUHK Shenzhen Research Institute, Shenzhen, China.
Paul K S Chan, Department of Microbiology, Chinese University of Hong Kong, Hong Kong, China.
Marc K C Chong, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China; CUHK Shenzhen Research Institute, Shenzhen, China.
Maggie H Wang, JC School of Public Health and Primary Care, Chinese University of Hong Kong, Hong Kong, China; CUHK Shenzhen Research Institute, Shenzhen, China.
Authors’ contributions
S.Z. conceived the study, carried out the analysis and drafted the first manuscript. S.Z. and J.L. collected and processed the data. S.Z., L.C., M.K.C.C. and M.H.W. discussed the results. All authors critically read and revised the manuscript and gave final approval for publication.
Acknowledgements
This study is conducted using the resources of Alibaba Cloud Intelligence High Performance Cluster computing facilities, which is made free for COVID-19 research. The SARS-CoV-2 genetic sequences were retrieved from the global initiative on sharing all influenza data (GISAID) at http://platform.gisaid.org/ (accessed on 31 March 2021). The complete acknowledgment table could be found in online Supplementary Acknowledgment Tables. We thank the contributions of the submitting and the originating laboratories.
Funding
Chinese University of Hong Kong (CUHK) grant [PIEF/Ph2/COVID/06, 4054456]; Health and Medical Research Fund (HMRF) Commissioned Research [COVID190103, INF-CUHK-1] of Hong Kong, China; National Natural Science Foundation of China (NSFC) [31871340, 71974165].
Conflict of interests: M.H.W. is a shareholder of Beth Bioinformatics Co., Ltd. B.C.Y.Z. is a shareholder of Beth Bioinformatics Co., Ltd and Health View Bioanalytics Ltd. Other authors declared no competing interests.
Ethics approval and consent to participate
The COVID-19 number of cases and sequencing data are collected via public domains and thus neither ethical approval nor individual consent is applicable.
References
- 1. Martins AF, Zavascki AP, Wink PL et al. Detection of SARS-COV-2 lineage p. 1 in patients from a region with exponentially increasing hospitalisation rate, February 2021, Rio Grande do Sul, Southern Brazil. Eurosurveillance 2021; 26:2100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao S, Lou J, Chong MKC et al. Inferring the association between the risk of COVID-19 case fatality and N501Y substitution in SARS-COV-2. Viruses 2021; 13:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Faria NR, Mellan TA, Whittaker C et al. Genomics and epidemiology of the p. 1 SARS-COV-2 lineage in Manaus, Brazil. Science 2021; 372:815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ong SWX, Young BE, Lye DC. Lack of detail in population-level data impedes analysis of SARS-COV-2 variants of concern and clinical outcomes. Lancet Infect Dis 10.1016/S1473-3099(21)00201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffmann M, Arora P, Groß R et al. SARS-COV-2 variants b. 1.351 and p. 1 escape from neutralizing antibodies. Cell 2021; 184:2384–2393.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


