Abstract
Background
Health care personnel and patients are at risk to acquire severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in health care settings, including in outpatient clinics and ancillary care areas.
Methods
Between May 1, 2020, and January 31, 2021, we identified clusters of 3 or more coronavirus disease 2019 (COVID-19) cases in which nosocomial transmission was suspected in a Veterans Affairs health care system. Asymptomatic employees and patients were tested for SARS-CoV-2 if they were identified as being at risk through contact tracing investigations; for 7 clusters, all personnel and/or patients in a shared work area were tested regardless of exposure history. Whole-genome sequencing was performed to determine the relatedness of SARS-CoV-2 samples from the clusters and from control employees and patients.
Results
Of 14 clusters investigated, 7 occurred in community-based outpatient clinics, 1 in the emergency department, 3 in ancillary care areas, and 3 on hospital medical/surgical wards that did not provide care for patients with known COVID-19 infection. Eighty-one of 82 (99%) symptomatic COVID-19 cases and 31 of 35 (89%) asymptomatic cases occurred in health care personnel. Sequencing analysis provided support for several transmission events between coworkers and in 2 cases supported transmission from health care personnel to patients. There were no documented transmissions from patients to personnel.
Conclusions
Clusters of COVID-19 with nosocomial transmission predominantly involved health care personnel and often occurred in outpatient clinics and ancillary care areas. There is a need for improved measures to prevent transmission of SARS-CoV-2 by health care personnel in inpatient and outpatient settings.
Keywords: health care personnel, outpatients, SARS-CoV-2, transmission
Patients and health care personnel are at risk to acquire severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in health care settings [1]. In hospitals, infection control measures including universal masking, use of appropriate personal protective equipment during patient care, and pre-admission and preprocedure screening are commonly used to minimize the risk for acquisition of SARS-CoV-2 [1–4]. Recent evidence suggests that these measures are effective in reducing, but not eliminating, the risk for SARS-CoV-2 transmission [4–12]. For example, transmission has been reported when COVID-19 cases are not recognized because admission screening results are negative or when personnel in the presymptomatic stage of COVID-19 provide patient care [7–8]. Infected personnel can also transmit SARS-CoV-2 to coworkers despite universal masking [4–12]. Exposures to infected coworkers may occur in areas such as breakrooms or in clinical areas where personnel work without adequate physical distancing [1,3–5,12].
Although nosocomial transmission of SARS-CoV-2 is often suspected, the actual source of acquisition is frequently unclear, particularly in the setting of widespread community transmission. In the VA Northeast Ohio Healthcare System, a majority of personnel with COVID-19 denied higher-risk exposures to SARS-CoV-2 at work or in the community but often worked in the same area as infected coworkers or patients with the potential for repeated brief interactions [4,12]. In addition to clusters of COVID-19 on hospital wards, our infection control program investigated multiple clusters of cases in outpatient clinics and ancillary care areas. Such areas could potentially present a relatively high risk for transmission because acutely infected patients are often evaluated in these areas, asymptomatic patients are not routinely screened before visits, and personnel often share workspaces. In the current study, we performed whole-genome sequencing to investigate several clusters of suspected nosocomial transmission of SARS-CoV-2 in outpatient and inpatient settings in the VA Northeast Ohio Healthcare System.
METHODS
Study Setting
The VA Northeast Ohio Healthcare System includes a 215-bed acute care hospital, an adjacent long-term care facility, and 13 community-based outpatient clinics. Of 5630 total system employees during the study, 793 (14.1%) worked in the community-based outpatient clinics, 4597 (81.7%) worked in the hospital, and 240 (4.3%) worked in the long-term care facility. During the study period, the hospital had a dedicated 22-bed COVID-19 ward, and 8 beds in the intensive care unit were dedicated to COVID-19 patients. All hospital admissions as well as patients undergoing selected surgical and medical procedures were screened for COVID-19 symptoms and tested for SARS-CoV-2 by nasopharyngeal swab reverse transcriptase polymerase chain reaction (RT-PCR).
Personnel providing care for patients with suspected or known COVID-19 wore gloves, a gown, a respirator, and face shield; medical procedure facemasks and eye protection were worn during care of other patients and during interactions with coworkers. Personnel were required to wear facemasks unless in a workspace behind closed doors. It was recommended that personnel eat meals alone at their desks and not sit together during break periods. All personnel were screened for COVID-19 symptoms on entry to the health care facility. Compliance of personnel with control measures including physical distancing and personal protective equipment use was monitored by infection control staff, and feedback was provided to individuals and supervisors. Patients were required to wear cloth facemasks when out of their room or when personnel entered. No visitors were allowed in the hospital, and family members were not allowed to attend outpatient clinic visits. Testing for COVID-19 was performed using commercial RT-PCR assays. For personnel with symptoms concerning for COVID-19 infection, including mild symptoms such as sore throat and nasal congestion, testing was available in the hospital and outpatient clinics and was recommended.
Patient Consent
The study protocol was approved by the Cleveland VA Medical Center’s Institutional Review Board with a waiver of informed consent.
Contact Tracing Investigations
The Infection Control Department conducted contact tracing in accordance with Centers for Disease Control and Prevention (CDC) recommendations [1,13]. Higher-risk exposures were defined as 15 minutes or more of continuous or cumulative contact within 6 feet without wearing both a facemask and eye protection occurring within 2 days before symptom onset through the time when the source individual met criteria for discontinuation of transmission-based precautions [13]. Contacts that included contact within 6 feet but for less than 15 minutes or while wearing both a facemask and eye protection were classified as lower-risk exposures. Personnel were questioned regarding contacts with coworkers both at work and in the community. Asymptomatic employees or patients were offered testing if they were identified as being at risk through contact tracing investigations. In clusters with large numbers of cases, surveillance nasopharyngeal swab testing was recommended for all personnel on a ward or in an outpatient clinic regardless of exposure history.
For the purposes of the study, we identified clusters of 3 or more COVID-19 cases between May 1, 2020, and January 31, 2021, in which nosocomial transmission was suspected. The study was initiated on May 1, 2020, when testing availability increased, allowing testing of contacts. Clusters in the long-term care facility were excluded. Clusters in which 3 or more nasopharyngeal swab specimens were available were included in the sequencing analysis. For comparison, we sequenced specimens from 10 employees with COVID-19 with no known exposures related to the clusters as well as 17 patients with COVID-19 after community exposures.
Sequencing
RNA was extracted from positive nasopharyngeal swab specimens with the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. Using AmpliSeq cDNA synthesis (Illumina, San Diego, CA, USA), isolated RNA samples were reverse-transcribed to make cDNA libraries for sequencing. Libraries for SARS-CoV-2 genomic sequencing were prepared using the AmpliSe Library PLUS kit (Illumina) with the SARS-CoV-2 community panel (Illumina). This panel consists of 237 SARS-CoV-2-specific primer pairs and 10 human gene expression control primer pairs. The resulting amplicons cover >99% of the viral genome, with amplicons ranging from 125 to 275 bp. The NextSeq Mid Output reagent kit, version 2 (Illumina, San Diego, CA, USA), was used for sequencing with a read length of 2 × 150 bp on an Illumina NextSeq550 (Illumina, San Diego, CA, USA).
Data Analysis
The raw sequencing data FASTQ file was uploaded to the BaseSpace sequence hub, and a consensus FASTA file was generated with SARS-CoV-2 reference sequence (NC_045512.2 SARS-CoV-2 Wuhan-Hu-1, complete genome) using the DRAGEN COVID lineage App (Illumina, San Diego, CA, USA) with default parameter. Stringent filtering criteria were used including only specimens with a minimum coverage of ≥95% and 100× median coverage depth in the final analysis. The clades were determined using Nextcladebeta (version 0.14.2; https://clades.nextstrain.org/).
The consensus FASTA files were downloaded and processed through Bionumerics 7.6 (Applied Maths, Austin, TX, USA) for cluster and single nucleotide polymorphism (SNP) analysis. The SARS-CoV-2 plugin tool was used to analyze the SARS-CoV-2 genomic sequences. Sequences were analyzed for SNP differences relative to the NCBI reference sequence for SARS-CoV-2 (NC_045512). Sequences with <2 SNP differences were considered related if they belonged to the same clade. Sequences with 3 to 4 SNP differences were deemed to be possibly related if contact tracing indicated a plausible transmission event and the sequences were of the same clade designation. Using the advanced clustering tools, a similarity matrix was calculated based on the similarity coefficient between the isolates. The results of the similarity matrix were then used as input data in the Complete linkage clustering algorithm to generate dendrograms and calculate SNP differences. Only samples that met our strict filtering criteria were used to generate the dendrograms.
RESULTS
Contact Tracing Investigations
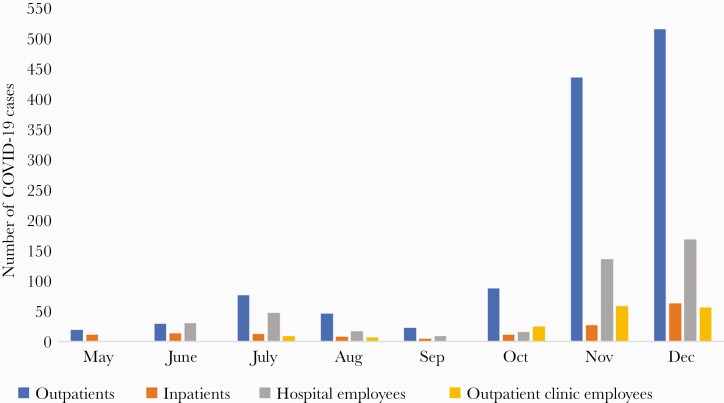
During the 8-month study period, 1388 patients and 584 employees were diagnosed with COVID-19 (Figure 1). The employees included 402 (68.8%) hospital employees and 182 (31.2%) employees based in outpatient clinics. The patients with COVID-19 included 1236 (89.0%) outpatients and 152 (11.0%) hospitalized patients. The peak in cases in November–December 2020 coincided with the peak in COVID-19 cases occurring in the community in Northeast Ohio. Only 4 of the 152 (2.6%) hospitalized patients with COVID-19 were suspected to have a health care–associated infection based on history of exposure to personnel with COVID-19 and duration of hospitalization >5 days.
Figure 1.
Number of COVID-19 cases diagnosed in outpatients, inpatients, hospital employees, and employees based in community outpatient clinics. Abbreviation: COVID-19, coronavirus disease 2019.
During the study, the Infection Control Department investigated 14 clusters of COVID-19 infections where nosocomial transmission was suspected. Table 1 shows the location of the clusters and the numbers of personnel and patients involved including the initial cases and the number of asymptomatic individuals screened. In each of the clusters, the initial cases were employees diagnosed with COVID-19 after known or suspected community exposures, followed by suspected transmission to coworkers. Of the clusters investigated, 3 were in inpatient wards, 7 were in community-based outpatient clinics, 1 was in the emergency department, and 3 were in ancillary care areas including radiology, sleep lab, and vascular lab. In multiple areas, it was noted that computer workstations were separated by <6 feet, and efforts were made to increase spacing between employees. No clusters were linked to contacts during shared meals or to exposures in break rooms. No clusters of infections occurred among personnel working on the COVID-19 ward or intensive care unit.
Table 1.
COVID-19 Clusters in a Health Care System With Suspected Nosocomial Transmission Based on Contact Tracing
| Cluster | Setting: Initial Cases and Contacts | Dates Initial to Final Case | No. Symptomatic COVID-19 Cases | No. Asymptomatic Cases/No. Screened (%) |
|---|---|---|---|---|
| A | Medical ward: 2 nurses with multiple coworker and patient contacts | 6/19/20–7/1/20 | 13 employees 1 patient |
1/36 (2.8) employees 1/31 (3.2) patients |
| B | Outpatient clinic: optometrist with multiple patient contacts | 10/26/20–10/28/20 | 1 employee 0 patients |
2/10 (20) patients |
| C | Medical ward: nursing assistant with multiple coworker contacts | 11/2/20–11/5/20 | 4 employees 0 patients |
2/87 (2.3) employees |
| D | Outpatient clinic: 4 nurses in shared workspace | 10/21/20–10/28/20 | 4 employees 0 patients |
N/A |
| E | Outpatient clinic: 6 employees working in different areas infected | 10/29/20–11/10/20 | 6 employees 0 patients |
9/114 (7.9) employees |
| F | Outpatient clinic: physical therapist with patient and coworker contacts | 11/3/20–12/1/20 | 13 employees 0 patients |
3/85 (3.5) employees 1/15 (6.7) patients |
| G | Spinal cord injury unit: nurse with coworker and patient contacts | 10/15/20–11/10/20 | 2 employees 0 patients |
5/85 (5.9) employees 0/26 (0) patients |
| H | Emergency department: 6 coworkers in shared workspace infected | 7/8/20–7/25/20 | 6 employees 0 patients |
3/96 (3.1) employees |
| I | Outpatient clinic: nurse with coworker contacts | 11/5/20–11/30/20 | 9 employees 0 patients |
6/109 (3.7) employees |
| J | Sleep lab: nursing assistant with coworker exposures | 11/6/20–11/10/20 | 3 employees 0 patients |
0/22 (0) employees |
| K | Outpatient clinic: 2 medical technologists with coworker contacts | 12/24/20–1/5/20 | 5 employees 0 patients |
3/26 (11.5) employees |
| L | Radiology: 4 staff cases with coworker contacts | 11/27/20–12/1/20 | 4 employees 0 patients |
0/16 (0) employees |
| M | Vascular lab: lab technician with coworker contacts | 12/10/20–12/31/20 | 7 employees 0 patients |
0/12 (0) employees |
| N | Outpatient clinic: 3 nurses in shared workspace with coworker contacts | 12/9/20–12/29/20 | 4 employees 0 patients |
1/32 (3.1) employees |
Abbreviation: COVID-19, coronavirus disease 2019.
Of 82 total symptomatic COVID-19 cases included in the 14 clusters, 81 (99%) occurred in personnel working with infected coworkers and 1 (1%) occurred in a patient receiving care from infected personnel (cluster A). In 6 of the clusters (C, E, F, G, H, I), screening of all personnel and/or patients was performed regardless of documented exposure to a case. Of 802 total asymptomatic individuals screened, 35 (4.4%) tested positive, including 31 of 720 (4.3%) employees and 4 of 82 (4.9%) patients. Asymptomatic individuals were more likely to test positive if they had a higher-risk exposure than if they worked in the same area but did not report a higher-risk exposure (18 of 201, 6.0%, vs 19 of 601, 3.8%; P = 0.048). Of the 112 total employees testing positive in the cluster investigations, 70 (62.5%) were nurses, 38 (33.9%) were ancillary staff, and 4 (3.6%) were physicians.
Sequencing Analysis
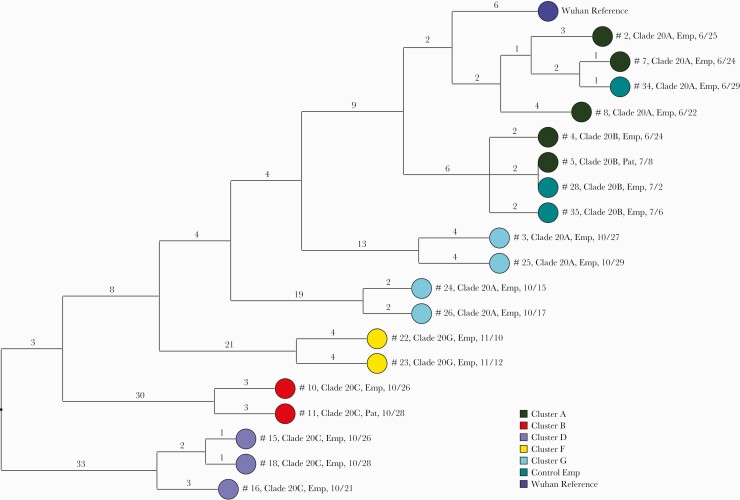
Of 90 samples submitted for sequencing, 53 (59%) had minimum coverage of 95% and 100× median coverage depth and were included in the analysis. Of the 14 clusters, 8 had >2 sequencing results available for analysis; 1 cluster was reported previously [10], and therefore 7 were included in this investigation. In 5 of the 7 clusters, the sequencing analysis provided evidence of SARS-CoV-2 transmission. Figure 2 provides a dendrogram displaying the SNP differences between the viral sequences that were related or possibly related in the 5 clusters with evidence of transmission; 3 control hospital employee sequences that were related to the cluster A sequences are also shown.
Figure 2.
Dendrogram displaying the SNP differences between SARS-CoV-2 viral sequences that were related (<2 SNP differences) or possibly related (3–5 SNP differences) in the 5 transmission clusters and in 3 employee controls with COVID-19 infection. Clades were determined based on the Nextstrain classification system. The Wuhan-Hu-1 reference genome is shown for comparison. Dates indicate the date of specimen collection. Abbreviations: COVID-19, coronavirus disease 2019; Emp, employee; Pat, patient; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SNP, single nucleotide polymorphism.
Cluster A involved 2 medical wards with shared personnel. In cluster A, there were 2 distinct transmission clusters based on the sequencing results. In the first cluster, an employee (#4), a patient (#5) cared for by the employee and by other infected staff members, and 2 employee controls (#28 and #35) with symptomatic COVID-19 were infected by a related 20.B clade virus. In the second cluster, an employee (#7) and employee control (#34) with symptomatic COVID-19 were infected by a related virus, and 2 additional employees (#2 and #8) were infected by possibly related viruses (3–4 SNP differences). Based on chart review and interviews, there were no direct contacts between the employee controls and the cluster A employees with related viruses. Two cluster A sequences were unrelated to the other sequences in cluster A.
In cluster B, an optometrist (#10) wearing a facemask evaluated multiple patients 1–2 days before onset of COVID-19 symptoms, and 1 patient (#11) was subsequently infected with a possibly related clade 20.C virus (3 SNP difference). In cluster D, 2 nurses working in proximity in an outpatient clinic had related SARS-CoV-2 viruses (#15 and #18), and a third nurse (#16) had a possibly related virus with 3 SNP differences; a fourth nurse was infected with an unrelated virus. In cluster F, a physical therapist implicated as a possible source of transmission had a SARS-CoV-2 virus that was distinct from 2 subsequently infected coworkers, but the coworkers (#22 and #23) had possibly related viruses (4 SNP differences). In cluster G, a nurse index case (#24) on a spinal cord injury unit was infected with a virus related to a second employee (#26) with a high-risk exposure; 2 employees (#3 and #25) later diagnosed on the same unit were infected with possibly related (4 SNP difference) viruses that were distinct from the index case virus. For 2 of the suspected clusters (cluster C and cluster E), there was no evidence of transmission based on sequence analysis.
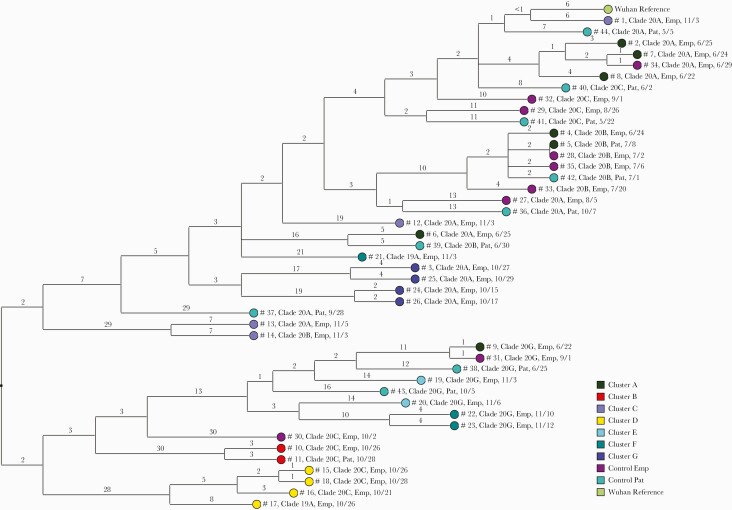
Figure 3 provides a dendrogram displaying the SNP differences between all the viral sequences, including 17 patient control samples with COVID-19 after community exposures and 10 employee controls. One of the patient control samples (#42) was related to 2 cluster A samples (#4 and #5) and 2 of the employee controls (#28 and #35). None of the other patient control samples were related to the other samples from the clusters or from the employee controls.
Figure 3.
Dendrogram displaying the single nucleotide polymorphism differences between SARS-CoV-2 viral sequences of individuals implicated in suspected transmission clusters based on contact tracing and in 17 patients with COVID-19 after community exposures and 10 employee controls. Clades were determined based on the Nextstrain classification system. The Wuhan-Hu-1 reference genome is shown for comparison. Abbreviations: COVID-19, coronavirus disease 2019; Emp, employee; Pat, patient; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SNP, single nucleotide polymorphism.
DISCUSSION
During the study, the infection control program in our health care system investigated multiple clusters of COVID-19 with suspected transmission based on contact tracing. Nearly all the symptomatic and asymptomatic cases in the clusters occurred in health care personnel (81 of 82 symptomatic cases and 31 of 35 asymptomatic cases). Contact tracing investigations suggested that initial cases in employees were acquired in the community, with subsequent transmission to coworkers. Sequencing analysis provided support for several transmission events between coworkers and in 2 cases supported transmission from health care personnel to patients. However, sequencing also demonstrated that some individuals linked to the clusters based on contact tracing were infected with unrelated viruses. There were no documented transmissions from patients to personnel. Our findings are consistent with recent evidence that health care personnel are more likely to acquire SARS-CoV-2 from infected coworkers than from infected patients in settings with good infection control measures in place and that nosocomial acquisition by patients is uncommon [4,10,14–15].
One notable finding from our study was that only 3 of the 14 clusters with suspected transmission occurred on hospital wards. Seven of the clusters occurred in community-based outpatient clinics, 1 in the emergency department, and 3 in ancillary care areas. Based on contact tracing investigations, the outpatient clinics and ancillary care areas were considered relatively high risk for transmission among coworkers because personnel often shared work areas and break areas and had computer stations separated by <6 feet. Similar concerns regarding inadequate physical distancing of work and break areas were reported in a recent observational study on a general medical ward [16]. In response to the clusters, the infection control program made efforts to reinforce compliance with masking and eye protection and to increase spacing in work and break areas. For example, in areas where computer workstations were separated by <6 feet, new workstations were created to provide better spacing between employees.
The outpatient clinic and ancillary care settings could also present a relatively high risk for acquisition of SARS-CoV-2 from patients. In these areas, acutely infected patients with relatively high viral burden are often seen by providers and asymptomatic outpatients are not routinely screened for SARS-CoV-2. However, the contact tracing investigations and the sequencing analysis suggested that transmission from patients was uncommon.
Based on the sequencing analysis, 3 control employees with symptomatic COVID-19 were infected with viruses related to cluster A employees in the absence of known exposures. It is possible that the SARS-CoV-2 variant associated with cluster A was widely circulating with community acquisition by multiple personnel rather than nosocomial transmission. Alternatively, there may have been interactions between the control and cluster A employees that were not recollected. Because employees were not routinely screened for SARS-CoV-2, it is also plausible that employees with asymptomatic shedding of the viruses may have served as intermediate sources of transmission linking the control and cluster A employees. Previous studies with other pathogens have demonstrated that many transmissions in hospitals that are identified using highly discriminatory typing methods occur in the absence of shared ward exposure [17–18]. For example, Eyre et al. [17] reported that 9% of Clostridioides difficile transmissions based on whole-genome sequencing occurred in patients who shared time in the hospital but were never on the same ward.
The estimated mutation rate of SARS-CoV-2 is 2.5 nucleotides per month [11]. Based on this mutation rate, genetic relatedness has typically been defined as 0 to 1 or 0 to 2 SNP differences in cases with plausible epidemiological links [5,8–11]. In the current analysis, we identified several instances where there were plausible epidemiological links between cases with 3 to 4 SNP differences. We deemed these cases to be possibly related. However, further studies will be needed to clarify whether a cutoff of 2 SNP differences is required to define transmission events.
Our study has several limitations. We did not sequence all viruses from the clusters because some samples were not available or did not meet the stringent requirements for quality of sequencing. In addition, we only sequenced a small sample of control employee and patient samples. Thus, we cannot be certain that some of the transmission events did not represent concurrent acquisition of related viruses widely circulating in the community. Finally, it is possible that we underestimated the sequence relatedness because we used strict filtering criteria and both the Nextclade and Bionumerics 7.6 phylogenetic tree methodologies for concurrence.
In conclusion, we found that clusters of COVID-19 with suspected transmission predominantly involved health care personnel and often occurred in outpatient clinics. Sequencing results provided evidence supporting multiple transmission events between coworkers and in 2 cases from health care personnel to patients. The findings contributed to development of improved infection control measures to limit nosocomial transmission of SARS-CoV-2, including efforts to increase spacing between coworkers.
Acknowledgments
Financial support. This work was supported by a Merit Review grant (CX001848) from the Department of Veterans Affairs to C.J.D.
Potential conflicts of interest. C.J.D. has received research funding from Clorox, PDI, and Pfizer. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Centers for Disease Control and Prevention. Interim infection prevention and control recommendations for healthcare personnel during the coronavirus disease 2019 (COVID-19) pandemic. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control-recommendations.html. Accessed 22 April 2021.
- 2.Seidelman JL, Lewis SS, Advani SD, et al. Universal masking is an effective strategy to flatten the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) healthcare worker epidemiologic curve. Infect Control Hosp Epidemiol 2020; 41:1466–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richterman A, Meyerowitz EA, Cevik M. Hospital-acquired SARS-CoV-2 infection: lessons for public health. JAMA 2020; 324:2155–6. [DOI] [PubMed] [Google Scholar]
- 4.Zabarsky TF, Bhullar D, Silva SY. What are the sources of exposure in healthcare personnel with coronavirus disease 2019 infection? Am J Infect Control 2021;. 49:392–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paltansing S, Sikkema RS, Man SJ, et al. Transmission of SARS-CoV-2 among healthcare workers and patients in a teaching hospital in the Netherlands confirmed by whole genome sequencing. J Hosp Infect 2021; 110:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sikkema RS, Pas SD, Nieuwenhuijse DF, et al. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis 2020; 20:1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klompas M, Baker MA, Rhee C, et al. A SARS-CoV-2 cluster in an acute care hospital. Ann Intern Med 2021; 174:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klompas M, Baker MA, Griesbach D, et al. Transmission of SARS-CoV-2 from asymptomatic and presymptomatic individuals in healthcare settings despite medical masks and eye protection. Clin Infect Dis 2021; ciab218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maltezou HC, Dedoukou X, Tseroni M, et al. SARS-CoV-2 infection in healthcare personnel with high-risk occupational exposure: evaluation of 7-day exclusion from work policy. Clin Infect Dis 2020; 71:3182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun KM, Moreno GK, Buys A, et al. Viral sequencing reveals US healthcare personnel rarely become infected with SARS-CoV-2 through patient contact. Clin Infect Dis 2021; ciab281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meredith LW, Hamilton WL, Warne B, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis 2020; 20:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan ER, Jones LD, Redmond SN, et al. Use of whole genome sequencing to investigate a cluster of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in emergency department personnel. Infect Control Hosp Epidemiol 2021; 1–3. doi: 10.1017/ice.2021.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Interim U.S. guidance for risk assessment and work restrictions for healthcare personnel with potential exposure to SARS-CoV-2. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-risk-assesment-hcp.html. Accessed 27 April 2021.
- 14.Baker MA, Fiumara K, Rhee C, et al. Low risk of COVID-19 among patients exposed to infected healthcare workers. Clin Infect Dis 2020; ciaa1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee C, Baker M, Vaidya V, et al. ; CDC Prevention Epicenters Program. Incidence of nosocomial COVID-19 in patients hospitalized at a large US academic medical center. JAMA Netw Open 2020; 3:e2020498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller SC, Pau S, Salinas AB, et al. Barriers to physical distancing among healthcare workers on an academic hospital unit during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol 2021; 1–7. doi: 10.1017/ice.2021.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre DW, Cule ML, Wilson DJ, et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 2013; 369:1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donskey CJ, Sunkesula VCK, Stone ND, et al. Transmission of Clostridium difficile from asymptomatically colonized or infected long-term care facility residents. Infect Control Hosp Epidemiol 2018; 39:909–16. [DOI] [PubMed] [Google Scholar]