Abstract
Background
Although mRNA-based severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines report >90% efficacy, breakthrough infections occur. Little is known about their effectiveness against SARS-CoV-2 variants, including the highly prevalent B.1.427/B.1.429 variant.
Methods
In this quality improvement project, we collected demographic and clinical information from post-vaccine SARS-CoV-2 cases (PVSCs), defined as healthcare personnel (HCP) with positive SARS-CoV-2 nucleic acid amplification test after receiving ≥1 vaccine dose. Available specimens were tested for L452R, N501Y, and E484K mutations using reverse-transcription polymerase chain reaction. Mutation prevalence was compared among unvaccinated, early post-vaccinated (≤14 days after dose 1), partially vaccinated (positive test >14 days after dose 1 and <14 days after dose 2), and fully vaccinated (>14 days after dose 2) PVSCs.
Results
From December 2020 to April 2021, ≥23 090 HCP received ≥1 dose of an mRNA-based SARS-CoV-2 vaccine, and 660 HCP cases of SARS-CoV-2 occurred, of which 189 were PVSCs. Among the PVSCs, 114 (60.3%), 49 (25.9%), and 26 (13.8%) were early post-vaccination, partially vaccinated, and fully vaccinated, respectively. Of 261 available samples from vaccinated and unvaccinated HCP, 103 (39.5%), including 42 PVSCs (36.5%), had the L452R mutation presumptive of B.1.427/B.1.429. When adjusted for community prevalence of B.1.427/B.1.429, PVSCs did not have significantly elevated risk of B.1.427/B.1.429 compared with unvaccinated HCP.
Conclusions
Most PVSCs occurred prior to expected onset of full, vaccine-derived immunity. Presumptive B.1.427/B.1.429 was not more prevalent in post-vaccine cases than in unvaccinated SARS-CoV-2 HCP. Continued infection control measures, particularly <14 days post-vaccination, and continued variant surveillance in PVSCs are imperative to control future SARS-CoV-2 surges.
Keywords: post-vaccination SARS-CoV-2, breakthrough COVID-19, B.1.427/B.1.429, L452R, SARS-CoV-2 variant
In this retrospective report, 189 of 660 SARS-CoV-2 cases detected in healthcare personnel at an academic medical center occurred post-vaccination. Incidence of the L452R mutation, consistent with the B.1.427/B.1.429 variant of concern, did not vary by vaccination status.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected 120 million people worldwide, causing at least 2.6 million deaths since December 2019 [1]. In the United States, Pfizer and Moderna developed mRNA-based vaccines and obtained emergency use authorization in December 2020. Vaccine administration to elderly individuals, healthcare workers, and other first responders began shortly thereafter. In clinical trials and observational studies after vaccine rollout, both mRNA vaccines proved effective in preventing symptomatic and severe disease [2–4]. In clinical trials, the vaccines demonstrated 52%–95% efficacy against symptomatic disease 14 days after the first dose and 95% efficacy 7 days after the second dose [3, 4]. Early observational reports in vaccinated healthcare personnel (HCP) suggest 80% effectiveness >14 days after the first dose and 90% >14 days after the second dose [5–7]. Emerging evidence indicates that the vaccines may also prevent asymptomatic infection [8], a potential source of transmission.
Concerns about emergence of SARS-CoV-2 variants, including variants first reported in the United Kingdom (B.1.1.7), South Africa (B.1.351), Brazil (P.1), California (B.1.427/B.1.429), and India (B.1.617) [9, 10], have dampened hopes for a rapid end to the coronavirus disease 2019 (COVID-19) pandemic through vaccination. The B.1.427/B.1.429 variant, which carries a L452R substitution mutation in the spike protein [11], was first detected in the spring of 2020 and by January 2021 accounted for 35%–50% of all SARS-CoV-2 cases detected in California [9, 12]. Studies indicate reduced neutralizing activity of naturally and vaccine-acquired monoclonal antibodies against these variants, including B.1.427/B.1.429 [13–18], and possibly increased transmissibility [11, 19] and virulence. Humoral immunity, however, is not the only factor in determining protection against infection [20], and more real-world data are needed to determine the effectiveness of the available SARS-CoV-2 vaccines against SARS-CoV-2 and its variants.
Stanford Health Care (SHC) began vaccinating HCP against SARS-CoV-2 on 18 December 2020, during a surge in COVID-19 cases when the B.1.427/B.1.429 variant was rapidly spreading [9]. We performed a retrospective, quality improvement project of post-vaccine SARS-CoV-2 cases (PVSCs) to help inform infection control recommendations for vaccinated HCP. Our aims were to define and characterize post-vaccine SARS-CoV-2 infections in HCP, evaluate our HCP vaccination program, and determine the role of variants in causing PVSCs.
METHODS
On 18 December 2020, SHC began vaccinating HCP with the Pfizer mRNA SARS-CoV-2 vaccine, initially to patient-facing frontline HCP in high-acuity settings, expanding to all patient-facing frontline HCP on 28 December, and finally to all health system employees on 8 January 2021. The Moderna mRNA vaccine became available on 22 January 2021. All HCP were required to self-monitor for COVID-related symptoms and attest to the absence of those symptoms every day when they entered the medical campus using a mandatory digital application. If any symptoms were reported, HCP underwent testing through Occupational Health. Asymptomatic HCP with high-risk occupational or community exposures (eg, household contacts, part of contact tracing effort after a work-related exposure) also were tested by Occupational Health. In addition, all HCP working on campus were strongly encouraged to participate in voluntary weekly asymptomatic testing using self-collected swabs that were processed using the Color platform [21]. HCP tested at non-SHC facilities were required to report any positive tests to Occupational Health.
Data Collection
PVSCs were defined as individuals identified by Occupational Health case notification and chart extraction with a positive SARS-CoV-2 nucleic acid amplification test (NAAT) after receiving at least 1 dose of a SARS-CoV-2 vaccine. Initial respiratory SARS-CoV-2 NAAT was conducted on a variety of platforms [22–24] including a previously described laboratory-developed reverse-transcription quantitative polymerase chain reaction (RT-qPCR) targeting the envelope gene (E gene) on the Rotor-Gene Q (Qiagen, Germantown, MD); a laboratory-developed RT-qPCR assay using a PerkinElmer kit targeting the ORF1ab and nucleocapsid gene ([N gene] PerkinElmer, San Jose, CA); Panther Fusion SARS-CoV-2 (Hologic, Marlborough, MA), a high-throughput RT-qPCR method targeting open reading frame 1ab (ORF1ab); Aptima SARS-CoV-2 (Panther System, Hologic), a transcription mediated amplification method targeting ORF1ab; GeneXpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA), a rapid RT-qPCR method targeting both E and N genes; cobas Liat SARS-CoV-2 & Influenza A/B (Roche, Indianapolis, IN), a point-of-care RT-PCR method targeting ORF1ab and N gene; and e-Plex SARS-CoV-2 (Genmark, Carlsbad, CA), a rapid RT-PCR method targeting the N gene. Screening tests performed on Color used a SARS-CoV-2 RT-LAMP Diagnostic Assay [21].
All HCP with positive SARS-CoV-2 tests were contacted within 24 hours of their result by an Occupational Health provider who documented demographic information as well as presence and date of onset of COVID-19 symptoms in a standardized note. For PVSCs, data collected also included dates of reported vaccine doses, household exposures to SARS-CoV-2, previous history of a COVID-19 diagnosis prior to vaccination, and immunocompromising conditions or medications.
Between 1 December 2020 and 28 February 2021, all available specimens that tested positive for SARS-CoV-2 by NAAT with an RT-qPCR cycle threshold (Ct) ≤30 or transcription-mediated amplification relative light units ≥1100 were subject to multiplex allele-specific genotyping RT-qPCR targeting 3 spike mutations associated with known variants of concern, including N501Y (B.1.1.7, B.1.351, P.1), E484K (B.1.351, P.1), and L452R (B.1.427/B.1.429) [25]. The screening threshold was changed to Ct ≤34 for specimens collected after 28 February.
Weekly variant prevalence from positive samples in the surrounding community was provided to us by the Stanford Clinical Virology Laboratory, which serves patients throughout the Bay Area and surrounding locales.
Available samples underwent SARS-CoV-2 whole-genome amplicon-based sequencing as previously described [25]. In brief, long-range PCR was used for target enrichment, followed by NEBNext library preparation (New England BioLabs, Ipswich, MA) and single-end 150-cycle sequencing on an Illumina MiSeq using MiSeq reagent kit V3 (Illumina, San Diego, CA). Whole-genome sequences with at least 90% genome coverage to a depth of at least 10 reads were accepted. Mutation calling required a depth of at least 12 reads with a minimum variant frequency of 20%. Genomes were assembled using National Center for Biotechnology Information NC_045512.2 as a reference, and pangolin was used for PANGO lineage assignment [26].
Data collection for this quality improvement project was approved by hospital privacy compliance and deemed to not be human subjects research by the Stanford University School of Medicine Panel on Human Subjects in Medical Research.
Exposures and Outcomes
The primary exposure was vaccination status at time of positive test, defined as unvaccinated, early post-vaccination (positive test ≤ 14 days after the first vaccine dose), partially vaccinated (positive test >14 days after the first vaccine dose to ≤ 14 days after the second vaccine dose), and fully vaccinated (positive test >14 days after the second vaccine dose).
The primary outcome was the presence of isolated L452R mutation consistent with presumptive B.1.427/B.1.429 [25]. If no L452R, N501Y, or E484K mutations were identified, the infection was designated as wild-type SARS-CoV-2.
Statistical Analyses
Statistical analyses were performed using SPSS Version 27. The χ2 test, Fisher exact test, independent samples T test, and a modified Poisson regression model (log-Poisson generalized linear model with robust standard errors) [27] were used to compare individuals with and without presumptive B.1.427/B.1.429 by vaccination status while adjusting for variant prevalence in the general population.
RESULTS
Of approximately 30 000 eligible HCP, at least 23 090 received ≥1 dose of the Pfizer (n = 20 559) or Moderna (n = 2170) vaccines, and at least 22 271 completed the 2-shot series between 18 December 2020 and 2 April 2021. In that same period, 51 638 SARS-CoV-2 NAATs were performed among 15 103 HCP within the SHC system; 15 759 of these tests were performed by Occupational Health and 35 879 through the Color screening platform, with 6029 individuals testing once, 2587 testing twice, and 6487 testing 3 or more times.
Between 18 December 2020 and 2 April 2021, Occupational Health identified 660 HCP with SARS-CoV-2 (Table 1), 189 of whom were PVSCs (<1% of HCP who received at least 1 dose of vaccine). Of these 189 PVSCs, 173 (91.5%) received at least 1 dose of the Pfizer and 15 (7.9%) received the Moderna vaccine; 1 PVSC vaccinated outside of the SHC system had no documentation of vaccine brand (Table 2).
Table 1.
Severe Acute Respiratory Syndrome Coronavirus 2 Cases Among Healthcare Personnel, 18 December 2020 to 2 April 2021
| Variable | Total, n = 660 | Unvaccinated, n = 471 | Early Post-Vaccination, n = 114 | Partially Vaccinated, n = 49 | Fully Vaccinated, n = 26 |
|---|---|---|---|---|---|
| Age, mean (SD), years | 37.5 (10.6) | 36.1 (10.0) | 39.8 (10.8) | 44.0 (12.6) | 39.1 (9.5) |
| Sex | |||||
| Female | 461 (69.8%) | 336 (71.3%) | 75 (65.8%) | 32 (65.3%) | 18 (69.2%) |
| Male | 199 (30.2%) | 135 (28.7%) | 39 (34.2%) | 17 (34.7%) | 8 (30.8%) |
| Mutation identified by reverse-transcription quantitative polymerase chain reactiona | |||||
| E484K | 3 (1.1%) | 3 (2.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| L452R | 103 (39.5%) | 61 (41.8%) | 22 (29.7%) | 10 (43.5%) | 10 (55.6%) |
| N501Y | 16 (6.1%) | 13 (8.9%) | 0 (0.0%) | 2 (8.7%) | 1 (5.6%) |
| No mutation | 139 (53.3%) | 69 (47.3%) | 52 (70.3%) | 11 (47.8%) | 7 (38.9%) |
| PANGO lineageb | |||||
| B.1.1.7 | 10 (7.5%) | 8 (17.0%) | 0 (0.0%) | 2 (10.5%) | 1 (11.1%) |
| B.1.427 | 26 (19.4%) | 5 (10.6%) | 13 (22.0%) | 5 (26.3%) | 3 (33.3%) |
| B.1.429 | 9 (6.7%) | 2 (4.3%) | 4 (6.8%) | 1 (5.3%) | 2 (22.2%) |
| B.1.617 | 1 (0.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (11.1%) |
| Not variant of concern/interest | 88 (65.7%) | 32 (68.0%) | 42 (71.2%) | 11 (57.9%) | 2 (22.2%) |
| Cycle threshold value,c mean (SD) | 23.5 (7.6) | 23.0 (7.4) | 22.6 (7.0) | 27.7 (8.7) | 28.5 (7.4) |
Abbreviation: SD, standard deviation.
aMutation analysis was performed on n = 261 healthcare personnel (HCP). Mutation analysis was not available for 149 (22.6%) who were tested outside the Stanford Health Care laboratory, 43 (6.5%) did not meet cycle threshold (Ct) or relative light unit (RLU) value criteria, 114 (17.3%) with insufficient quantity, 6 (0.9%) that failed amplification, and 87 (13.2%) that were discarded or otherwise unavailable.
bPANGO lineage as determined by next-generation sequencing (NGS) was available for n = 134 samples; NGS was unavailable for n = 148 HCP who were tested by Color or outside of Stanford Health Care laboratory, n = 43 with high Ct values, n = 13 samples with low transcription-mediated amplification RLU, n = 6 samples with polymerase chain reaction amplification failure, n = 114 with insufficient quantity, and n = 215 that were otherwise unavailable.
cCt value available for n = 283 samples.
Table 2.
Characteristics of Post-Vaccine Severe Acute Respiratory Syndrome Coronavirus 2 Cases Among Healthcare Personnel
| Variable | Total, N = 189 | No Mutation Identified (Wild-Type), N = 70a (62.5%) | L452R (Presumptive B.1.427/B.1.429), N = 42a (37.5%) | P Value |
|---|---|---|---|---|
| Age, years | ||||
| Mean (SD) | 40.8 (11.2) | 42.5 (11.6) | 40.8 (10.1) | .424 |
| Median (IQR) | 38 (32–48) | |||
| Sex | ||||
| Male | 64 (33.9%) | 18 (25.7%) | 13 (31.0%) | .549 |
| Female | 125 (66.1%) | 52 (74.3%) | 29 (69.0%) | |
| Professional roleb | ||||
| Patient facing | 129 (68.3%) | 45 (66.2%) | 31 (73.8%) | .400 |
| Non-patient facing | 55 (29.1%) | 23 (33.8%) | 11 (26.2%) | |
| Unknown | 5 (2.6%) | |||
| Immunocompromised | ||||
| Yes | 7 (3.7%) | 3 (4.5%) | 2 (5.4%) | .846 |
| No | 166 (87.8%) | 63 (95.5%) | 35 (94.6%) | |
| Unknown | 16 (8.5%) | |||
| Tested positive after second vaccine dose | ||||
| Yes | 39 (20.6%) | 10 (14.3%) | 14 (33.3%) | .017 |
| No | 150 (79.4%) | 60 (85.7%) | 28 (66.7%) | |
| Days from dose 1 to symptom onset (n = 151)c | ||||
| Mean (SD) | 16.8 (23.3) | 11.6 (17.4)c1 | 24.2 (29.0)c2 | .008 |
| Median (IQR) | 8 (3–18) | |||
| Days from dose 1 to positive NAAT | ||||
| Mean (SD) | 19.4 (22.0) | 14.7 (17.7) | 25.8 (28.0) | .011 |
| Median (IQR) | 11 (7–22) | |||
| Days from dose 2 to symptom onset (n = 31)d | ||||
| Mean (SD) | 30.0 (27.8) | 25.0 (22.0)d1 | 36.6 (32.0)d2 | .350 |
| Median (IQR) | 24 (4-60) | |||
| Days from dose 2 to positive NAAT (n = 39)e | ||||
| Mean (SD) | 31.3 (25.5) | 29.6 (22.2)e1 | 35.5 (28.2)e2 | .573 |
| Median (IQR) | 24 (7-55) | |||
| Vaccination status at time of positive NAAT | ||||
| Early post-vaccination | 114 (60.3%) | 52 (74.3%) | 22 (52.4%) | .020f |
| Partially vaccinated | 49 (25.9%) | 11 (15.7%) | 10 (23.8%) | |
| Fully vaccinated | 26 (13.8%) | 7 (10.0%) | 10 (23.8%) | |
| Vaccine brand | ||||
| Pfizer | 173 (91.5%) | 66 (94.3%) | 39 (92.9%) | .762 |
| Moderna | 15 (7.9%) | 4 (5.7%) | 3 (7.1%) | |
| Unknown | 1 (0.5%) | |||
| Cycle threshold value (n = 101)g | ||||
| Mean (SD) | 24.9 (8.0) | 21.1 (4.5)g1 | 19.3 (4.4)g2 | .133 |
| Median (IQR) | 22 (17–31) | |||
| Experienced SARS-CoV-2 symptoms | ||||
| Yes | 157 (83.1%) | 63 (92.6%) | 38 (95.0%) | .631 |
| No | 26 (13.8%) | 5 (7.4%) | 2 (5.0%) | |
| Unknown | 6 (3.2%) | |||
| Household contact | ||||
| Yes | 79 (41.8%) | 32 (50.0%) | 21 (55.3%) | .607 |
| No | 94 (49.7%) | 32 (50.0%) | 17 (44.7%) | |
| Unknown | 16 (8.5%) | |||
| Previously positive for SARS-CoV-2 | ||||
| Yes | 4 (2.1%) | 2 (3.1%) | 0 | .533 |
| No | 167 (88.4%) | 63 (96.9%) | 37 (100%) | |
| Unknown | 18 (9.5%) |
Abbreviations: IQR, interquartile range; NAAT, nucleic acid amplification test; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation.
aMutation data available for n = 115 post-vaccine SARS-CoV-2 cases (PVSCs); 70 with no mutation, 42 with isolated L452R mutation presumptive of B.1.427/B.1.429 variant, and 3 with isolated N501Y mutation. Only samples with L452R and no other mutations are compared here due to few N501Y mutations. Mutation data not available for n = 32 with cycle threshold (Ct) values not meeting criteria, n = 27 with SARS-CoV-2 test done outside Stanford Health Care system, n = 15 with samples otherwise not available for mutation testing.
bPatient-facing roles = physician/physician assistant/nurse practitioner (n = 22, 11.7%), nursing (n = 42, 22.2%), medical assistant (n = 17, 9.0%), respiratory therapy/physical therapy/occupational therapy (n = 5, 2.6%), other (n = 43, 22.8%). Non-patient facing roles = housekeeping/food services (n = 22, 11.7%); other (33, 17.4%).
cn = 151 includes symptomatic PVSCs with known date of symptom onset; c1n = 62; c2n = 37.
dn = 31 includes symptomatic PVSCs who tested positive after dose 2 and had known date of symptom onset; d1n = 9; d2n = 11.
en = 39 includes all PVSCs who tested positive after dose 2; e1n = 10; e2n = 14.
f P value reflects extended Mantel-Haenszel χ2 for linear trend, univariate analysis unadjusted for rising prevalence of B.1.427/B.1.429 from December 2020 to March 2021.
gCt values not available for individuals tested outside the Stanford Health Care system or by transcription-mediated amplification; only samples with Ct<30 (before 1 March 2021) or Ct<34 (after 1 March 2021) included in variant analysis; g1n = 45; g2n = 19.
The median age of PVSCs was 38 years (interquartile range [IQR], 32–48) and 125 (66.1%) were female; 42 (22.2%) were nurses (Table 2). Few individuals had immunocompromising conditions (n = 7, 3.7%). Seventy-six (41.8%) reported a household contact with SARS-CoV-2 infection at the time of their positive NAAT.
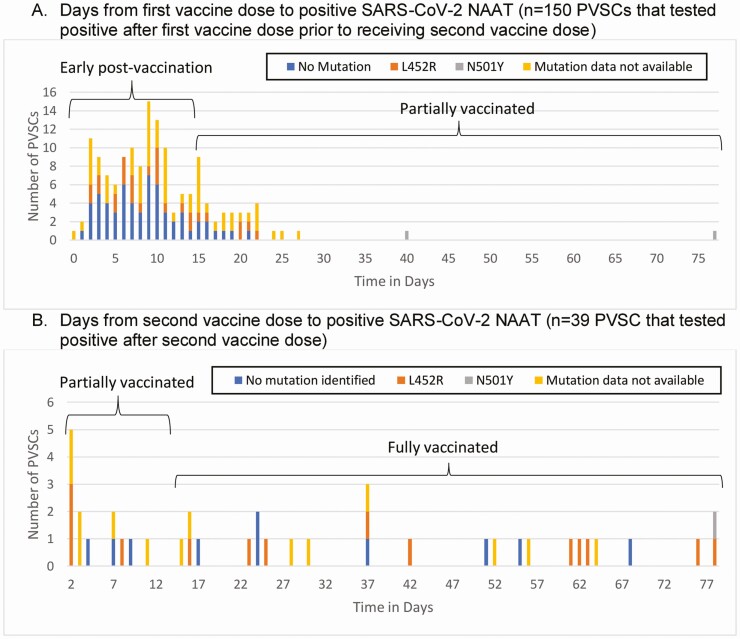
The majority (n = 114, 60.3%) of PVSCs occurred early post-vaccination (Figure 1A); 49 (25.9%) occurred while partially vaccinated, and 26 (13.8%) occurred when fully vaccinated (Figure 1B). Of the 183 PVSCs, 157 (85.8%) experienced symptoms and 151 reported the date of symptom onset; of these, 104 (68.9%) developed symptoms in the early post-vaccination period within 14 days of the first dose.
Figure 1.
Time from vaccination to coronavirus disease 2019 symptom onset and positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid amplification test (NAAT). A, Days from first vaccine dose to positive SARS-CoV-2 NAAT (n = 150 PVSCs who tested positive after first vaccine dose prior to receiving second vaccine dose). B, Days from second vaccine dose to positive SARS-CoV-2 NAAT (n = 39 PVSCs who tested positive after second vaccine dose). Abbreviation: PVSC, post-vaccine SARS-CoV-2 cases.
The majority of PVSCs (n = 140, 74.1%) received their first vaccine in December 2020 with the greatest number vaccinated on 22–23 December 2020 (n = 46, 24.3%), corresponding with the highest daily administration of vaccines (1940 doses given on 22 December 2020 and 1934 given on 23 December 2020). Of the PVSCs vaccinated on these dates, 21 (45.7%) tested positive >14 days after their vaccination. The majority of positive tests in PVSCs occurred in December 2020 and early January 2021, when most PVSCs were ≤ 14 days from their first vaccine dose, coincident with the winter surge in COVID-19 in northern California. The B.1.427/B.1.429 variant rose in prevalence from representing 24.8% of SARS-CoV-2 cases detected by Stanford laboratory in December 2020 to 62.5% of cases by March 2021 [25].
Symptoms in the 157 symptomatic PVSCs began a median of 8 days (IQR, 3–18) after the first vaccination dose. Eighteen PVSCs developed symptoms of COVID-19 prior to or on the day of their first vaccine dose. Two PVSCs, both of whom tested positive ≤ 14 days after their first vaccine dose, were hospitalized with wild-type virus; there were no deaths. Of the 26 patients with no symptoms, 7 PVSCs (26.9%) had at least 1 repeat negative NAAT within 48 hours of their positive test. PVSCs who reported symptoms of COVID-19 had lower mean Ct values (24.2 vs 30.9, P = .011) than those without symptoms but did not differ in terms of age (40.7 vs 40.3 years, P = .879) or time from first vaccine dose to positive test (19.3 vs 21.8 days, P = .572).
Four PVSCs (2.2%) had previous documentation of SARS-CoV-2 infection between 22 and 98 days before their vaccinations; 2 of these reported new onset of symptoms at the time of their positive post-vaccination test. Two of the 4 with prior infections had specimens available for mutation screening, and neither had evidence of a variant of concern.
Of the 7 immunocompromised PVSCs, 3 were positive for SARS-CoV-2 early post-vaccination, 2 were partially vaccinated, and 2 were fully vaccinated; 6 were symptomatic and none required hospitalization.
Partially and fully vaccinated PVSCs had higher mean Ct values than unvaccinated or early post-vaccination individuals (27.9 vs 22.9, P < .001, of n = 283 individuals with known Ct value) and were older (mean age, 42.3 vs 36.8 years, P < .001; Table 3).
Table 3.
Age and Cycle Threshold Value by Vaccination Status
| Variable | Unvaccinated + Early Post-Vaccination | Partially + Fully Vaccinated | P Value |
|---|---|---|---|
| Age, mean (SD), years | 36.8 (10.2) | 42.3 (11.8) | <.001 |
| Cycle threshold value (n = 98), mean (SD) | 22.9 (7.3) | 27.9 (8.2) | <.001 |
T tests comparing age and cycle threshold value by vaccination status. Early post-vaccination = positive severe acute respiratory syndrome coronavirus 2 nucleic acid amplification test <14 days from vaccine dose 1, partially vaccinated = >14 days from vaccine dose 1 and <14 days from vaccine dose 2, fully vaccinated = >14 days from vaccine dose 2.
Abbreviation: SD, standard deviation.
Tests for novel variant-associated mutations were performed on samples from 261 infected HCP including 115 PVSCs (Table 1). Sixteen HCP (6.1%) were found to have the N501Y mutation, including 2 partially and 1 fully vaccinated PVSCs. Three E484K mutations were found in unvaccinated individuals. Samples with isolated L452R mutations, presumed to be B.1.427/B.1.429 lineage, were found in 103 (39.5%) HCP samples including 42 (36.5%) PVSCs.
In unadjusted analysis, PVSCs infected with L452R-containing viruses were more likely than those with no identified mutations to have tested positive after the second vaccine dose or to be partially or fully vaccinated at the time of a positive test (Table 2). This variant was not associated with age, gender, job role, brand of vaccine received, immunocompromised status, or symptomatic infection. In multivariate analysis, when controlling for community prevalence of L452R mutation the week prior to a positive test (when exposure likely occurred), vaccination status at the time of positive test was not significantly associated with presumptive B.1.427/B.1.429 (Table 4).
Table 4.
Risk Ratios for Infection With Presumptive B.1.427/B.1.429 Among Healthcare Personnel by Vaccination Status and Adjusted for Community Prevalence of L452R Mutation at Time of Infection
| Vaccination Status at Time of Positive Test | n | Presumptive B.1.427/B.1.429, n (%) | Concomitant Community Prevalence of L452R Median (Quartiles) | Unadjusted RR (95% CI) | Adjusted RR (95% CI) |
|---|---|---|---|---|---|
| Unvaccinated | 130 | 61 (46.9) | 41.4% (32.8%, 47.4%) | Ref. | Ref. |
| Early post-vaccination | 74 | 22 (29.7) | 36.2% (32.8%, 41.4%) | 0.63 (.43–.94) | 0.70 (.47–1.05) |
| Partially vaccinated | 21 | 10 (47.6) | 41.4% (32.8%, 45.7%) | 1.02 (.63–1.65) | 1.05 (.65–1.70) |
| Fully vaccinated | 17 | 10 (58.8) | 51.8% (50.7%, 57.7%) | 1.25 (.81–1.94) | 1.05 (.65–1.68) |
Modified Poisson regression model (log-Poisson generalized linear model with robust standard errors) comparing individuals with and without presumptive B.1.427/B.1.429 by vaccination status at time of positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid amplification test (NAAT) and adjusted for percent of total positive SARS-CoV-2 tests with isolated L452R mutation presumptive of B.1.427/B.1.429 within Stanford Health Care during week of infection. Early post-vaccination = positive SARS-CoV-2 NAAT <14 days from vaccine dose 1, partially vaccinated = positive SARS-CoV-2 NAAT >14 days from vaccine dose 1 and <14 days from vaccine dose 2, fully vaccinated = positive SARS-CoV-2 NAAT >14 days from vaccine dose 2.
Abbreviations: CI, confidence interval; RR, risk ratio.
Next-generation sequencing was available for 134 infected HCP, including 87 of the 189 PVSCs (Table 1). Of 31 individuals with L452R mutation and available sequencing results, 30 (96.8%) were confirmed to be the B.1.427/B.1.429 variant and 1 was the B.1.617 variant (Table 5) [28].
Table 5.
PANGO Lineage Identified by Next-Generation Sequencing Compared With Mutations Identified by Reverse-Transcription Polymerase Chain Reaction Among Severe Acute Respiratory Syndrome Coronavirus 2–Infected Healthcare Personnel
| Mutation Identified by Reverse-Transcription Polymerase Chain Reaction | ||||||
|---|---|---|---|---|---|---|
| L452R | N501Y | E484K | No Mutation | Mutation Data Not Available | Total | |
| PANGO Lineage by Next-Generation Sequencing | ||||||
| B.1.1.7 | 0 | 11 | 0 | 0 | 0 | 11 |
| B.1.427 | 22 | 0 | 0 | 0 | 4 | 26 |
| B.1.429 | 8 | 0 | 0 | 0 | 1 | 9 |
| B.1.617 | 1a | 0 | 0a | 0 | 0 | 1 |
| Not variant of concern/interest | 0 | 0 | 1 | 57 | 29 | 87 |
| Total | 31 | 11 | 1 | 57 | 34 | 134 |
Next-generation sequencing (NGS) available for n = 134 healthcare personnel (HCP); NGS was unavailable for n = 148 HCP who were tested by Color or outside of Stanford Health Care laboratory, n = 43 with high cycle threshold values, n = 13 samples with low transcription-mediated amplification relative light unit, n = 6 samples with polymerase chain reaction amplification failure, n = 114 with insufficient quantity, n = 215 that were otherwise unavailable.
aThis specimen had a blunted E484K amplification curve that was revealed to represent an E484Q mutation by sequencing [28].
DISCUSSION
In this cohort of HCP at an academic medical center in Northern California, similar to other reports elsewhere [5, 6], we found that SARS-CoV-2 infection post-vaccination is uncommon despite widespread community disease. We identified only 189 such cases out of >23 000 vaccinated HCP [29]. Not surprisingly, most of these cases occurred in the first 2 weeks after vaccination, before immunity is expected to develop. Whether these individuals were exposed before or after their vaccine dose cannot be known, although the dates of symptom onset combined with the typical incubation period of 5–9 days [30] suggest that some early cases were acquired prior to the first vaccine dose. In contrast, 26 PVSCs occurred >14 days after the second dose, when full immunity is expected. Although >85% of PVSC infections were symptomatic, only 2 PVSCs required hospitalization; both developed symptoms and tested positive within 14 days of the first vaccine dose. These findings are consistent with other real-world reports of excellent vaccine effectiveness >14 days after the first and second doses [5, 6, 31], particularly in preventing severe disease, and highlight the need to observe strict precautions until full immunity is achieved >14 days after the second vaccine dose.
Isolated L452R mutation presumptive of infection with B.1.427/B.1.429 variant was associated with partially and fully vaccinated status on univariate analysis of PVSCs. This finding appears to be attributable to the timing of most vaccinations early in December and the subsequent rising prevalence of B.1.427/B.1.429 from December 2020 to March 2021, rather than to reduced vaccine effectiveness against B.1.427/B.1.429. On multivariate analysis controlling for overall community prevalence of the variant, infection with presumptive B.1.427/B.1.429 was not significantly more common in PVSCs compared with unvaccinated HCP. Although this finding is reassuring, case numbers were small and further vigilance is warranted. Reduced in vitro neutralizing activity of naturally and vaccine-acquired antibodies against the B.1.427/B.1.429 variant compared with wild-type S protein has been reported [11, 18]. As more people become fully vaccinated, careful surveillance will be needed to determine if variants such as B.1.427/B.1.429 can escape vaccine-derived protection in vivo, and continued caution is needed, especially as states begin to relax restrictions intended to curb transmission.
Higher Ct values, which correspond with lower viral load, were identified in PVSCs positive >14 days after the first vaccine dose compared with unvaccinated and early post-vaccination cases, suggesting that lower levels of viral replication may afford some protection and reduction in transmissibility in people who contract SARS-CoV-2 post-vaccination. This finding is consistent with reports from a mass vaccination campaign in Israel [32]. Lower Ct values were seen in PVSCs who reported symptoms consistent with COVID-19 compared with the few who remained asymptomatic.
The peak in post-vaccine positive SARS-CoV-2 cases in late December 2020 to early January 2021 corresponds with a rise in county-wide cases and HCP cases at that time. We did not find any evidence to support transmission clusters or superspreading events contributing to post-vaccination SARS-CoV-2 infections within the healthcare system. Although several PVSCs had been vaccinated on the same 2 days in December, nearly half of these infections occurred more than 2 weeks after vaccination, suggesting that exposure occurred sometime after the date of vaccination. Further, 41.8% reported household contacts with known SARS-CoV-2 infection at the time of their positive test, suggesting community transmission was driving these cases rather than workplace exposure; the multiple cases in late December and early January may have been related to holiday celebrations. This highlights the importance of maintaining social distancing and diligent mask-wearing at work in the setting of increased prevalence of variants. Describing the phenomenon of PVSCs in this quality improvement project has allowed us to more effectively promote these measures within our system at a time when many are tempted to loosen precautions due to “pandemic fatigue” and a sense of security after being vaccinated.
We do note several limitations in this report. We did not obtain detailed demographic and clinical information from the comparison group of unvaccinated SARS-CoV-2–infected HCP and are unable to comment on differences in clinical course between vaccinated and unvaccinated SARS-CoV-2–positive HCP. Moreover, we cannot state with certainty that the 26 asymptomatic PVSCs remained asymptomatic since this information was only obtained on 2 occasions: within 24 hours of a positive test and at the end of the quarantine period. If symptoms developed in the interim period, they would not have been captured. We cannot estimate vaccine efficacy since some HCP may not have reported infections detected outside the SHC system. Also, since asymptomatic screening was not required, asymptomatic and mildly symptomatic cases may have been missed. Finally, although presence of L452R mutation in the absence of N501Y and E484K has been shown to be highly correlative with B.1.427/B.1.429 by sequencing during this time period [25], these mutations are occasionally detected sporadically in nonvariants of concern/interest lineages, and it is not possible to know the variant definitively in unsequenced samples. The identification of the B.1.617 variant in 1 sample with an L452R mutation is illustrative of the need for ongoing surveillance with sequencing in post-vaccine cases. Nevertheless, we believe our findings are an important contribution to the understanding of PVSCs in the context of rising prevalence of new SARS-CoV-2 variants.
CONCLUSIONS
Post-vaccination SARS-CoV-2 infection occurred in <1% of vaccinated HCP, with the great majority occurring prior to full, vaccine-derived immunity. Presumptive B.1.427/B.1.429 did not represent a significantly higher proportion of cases in vaccinated vs unvaccinated SARS-CoV-2–infected HCP, but the number of fully vaccinated PVSCs was small. Continued infection control measures in the workplace and in the community, including social distancing and wearing a mask, particularly in the early days post-vaccination, as well as continued variant surveillance in PVSCs is imperative in order to anticipate and control future surges of infection.
Notes
Acknowledgments. We thank Karen Bains and all of Stanford Health Care’s Occupational Health team, as well as all Stanford Health Care personnel for their tireless efforts throughout the pandemic.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Baltimore, MD: Johns Hopkins University; 2021. Available at: https://coronavirus.jhu.edu/map.html. Accessed 10 April 2021. [Google Scholar]
- 2. Baden LR, El Sahly HM, Essink B, et al. ; COVE Study Group . Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 2021; 384:403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Polack FP, Thomas SJ, Kitchin N, et al. ; C4591001 Clinical Trial Group . Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med 2021; 384:1412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keehner J, Horton LE, Pfeffer MA, et al. SARS-CoV-2 infection after vaccination in health care workers in California. N Engl J Med 2021; 384:1774–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daniel W, Nivet M, Warner J, Podolsky DK. Early evidence of the effect of SARS-CoV-2 vaccine at one medical center. N Engl J Med 2021; 384:1962–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thompson MG, Burgess JL, Naleway AL, et al. Interim estimates of vaccine effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines in preventing SARS-CoV-2 infection among health care personnel, first responders, and other essential and frontline workers—Eight U.S. Locations, December 2020–March 2021. MMWR Morb Mortal Wkly Rep 2021; 70:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tande AJ, Pollock BD, Shah ND, et al. Impact of the COVID-19 vaccine on asymptomatic infection among patients undergoing pre-procedural COVID-19 molecular screening. Clin Infect Dis 2021; ciab229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA 2021; 325:1324–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yadav PD, Sapkal GN, Abraham P, et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin Infect Dis 2021; ciab411. [DOI] [PubMed] [Google Scholar]
- 11. Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv 2021;2021.03.07.21252647. [Google Scholar]
- 12. Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 2017; 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collier DA, De Marco A, Ferreira I, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021; 593:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021; 27:622–5. [DOI] [PubMed] [Google Scholar]
- 15. Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 2021; 29:747–51.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starr TN, Greaney AJ, Dingens AS, Bloom JD. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. bioRxiv 2021; 2021.02.17.431683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020; 182:1284–1294.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McCallum M, Bassi J, Marco A, et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv 2021; 2021.03.31.437925. [Google Scholar]
- 19. Peng J, Liu J, Mann SA, et al. Estimation of secondary household attack rates for emergent spike L452R SARS-CoV-2 variants detected by genomic surveillance at a community-based testing site in San Francisco. Clin Infect Dis 2021; ciab283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarke A, Sidney J, Methot N, et al. Negligible impact of SARS-CoV-2 variants on CD4 (+) and CD8 (+) T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv 2021; 2021.02.27.433180. [Google Scholar]
- 21. SARS-CoV-2 LAMP Diagnostic Assay. Burlingame, CA: Color; [updated May 21, 2020. Version 1.2. Available at: https://www.color.com/wp-content/uploads/2020/05/Color-LAMP-Diagnostic-Assay_v1-2_Updated-052120.pdf. Accessed 10 April 2021.
- 22. Bulterys PL, Garamani N, Stevens B, et al. Comparison of a laboratory-developed test targeting the envelope gene with three nucleic acid amplification tests for detection of SARS-CoV-2. J Clin Virol 2020; 129:104427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25: 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 2020; 323:1967–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Miller JA, Verghese M, et al. Multiplex SARS-CoV-2 genotyping RT-PCR for population-level variant screening and epidemiologic surveillance. J Clin Microbiol 2021;JCM0085921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou G. A modified Poisson regression approach to prospective studies with binary data. Am J Epidemiol 2004; 159:702–6. [DOI] [PubMed] [Google Scholar]
- 28. Verghese M, Jiang B, Iwai N, et al. Identification of a SARS-CoV-2 variant with L452R and E484Q neutralization resistance mutations. J Clin Microbiol 2021; 59:e0074121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Novel Coronavirus (COVID-19). San Jose, CA: Santa Clara County Public Health; 2021 [updated 30 March 2021]. Available at: https://www.sccgov.org/sites/covid19/Pages/home.aspx. Accessed 30 March 2021. [Google Scholar]
- 30. Alene M, Yismaw L, Assemie MA, Ketema DB, Gietaneh W, Birhan TY. Serial interval and incubation period of COVID-19: a systematic review and meta-analysis. BMC Infect Dis 2021; 21:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amit S, Beni SA, Biber A, Grinberg A, Leshem E, Regev-Yochay G. Postvaccination COVID-19 among healthcare workers, Israel. Emerg Infect Dis 2021; 27:1220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petter E, Mor O, Zuckerman N, et al. Initial real world evidence for lower viral load of individuals who have been vaccinated by BNT162b2. medRxiv 2021:2021.02.08.21251329. [Google Scholar]