SUMMARY
Chitin, a major component of fungal cell walls, has been associated with allergic disorders such as asthma. However, it is unclear how mammals recognize chitin and the principal receptor(s) on epithelial cells that sense chitin remain to be determined. In this study, we show that LYSMD3 is expressed on the surface of human airway epithelial cells and demonstrate that LYSMD3 is able to bind chitin, as well as β-glucan, on the cell walls of fungi. Knockdown or knockout of LYSMD3 also sharply blunts the production of inflammatory cytokines by epithelial cells in response to chitin and fungal spores. Competitive inhibition of the LYSMD3 ectodomain by soluble LYSMD3 protein, multiple ligands, or antibody against LYSMD3 also blocks chitin signaling. Our study reveals LYSMD3 as a mammalian pattern recognition receptor (PRR) for chitin and establishes its role in epithelial cell inflammatory responses to chitin and fungi.
Graphical abstract
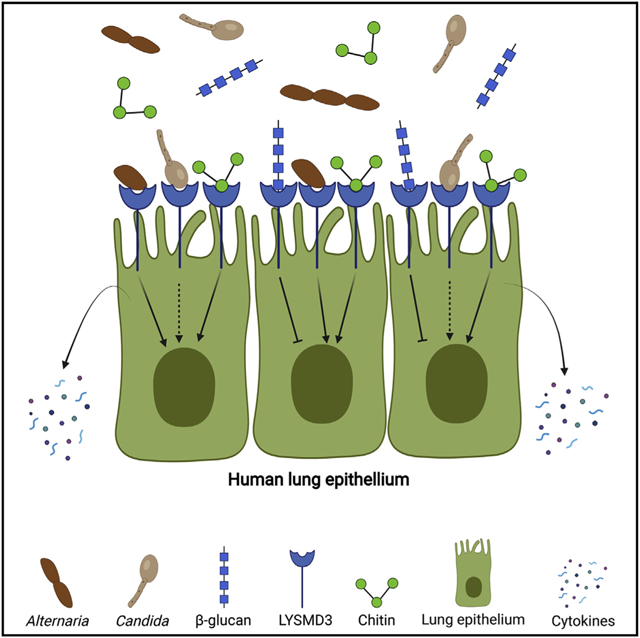
In brief
Mammalian chitin recognition is poorly understood but significant because chitin is ubiquitous and linked to disease. Here, He et al. reveal that LYSMD3 on human lung epithelial cells binds to chitin and fungal spores and mediates production of cytokines. They find that LYSMD3 is also able to bind β-glucan.
INTRODUCTION
Allergic responses have been linked with environmental triggers thought to result from airway inflammatory responses to ubiquitous pathogen-associated molecules (Umetsu et al., 2002). Human fungal pathogens, including the allergenic species Alternaria alternata and Aspergillus fumigatus, have protective cell walls containing structural polymers of two types depending on their solubility in alkaline solutions (Fontaine et al., 2000). While soluble components such as galactomannan play important roles in cell wall integrity and host immune responses, alkali-insoluble complexes of chitin and β-glucan are more abundant (Cantu et al., 2009; Latgé et al., 1994). Considerable work has demonstrated that fungal β-glucan is recognized by Dectin-1, among other receptors, and elicits strong host immune responses (Brown et al., 2007; Rosas et al., 2008; Taylor et al., 2007). In contrast, there are conflicting data on how chitin is sensed and the mechanisms that underpin ensuing inflammatory processes.
Chitin is the second most abundant polysaccharide in nature after cellulose. It is present in the cell walls or exoskeletons of organisms such as fungi, crustaceans, parasites, and insects (Lee, 2009). Chitin exposure has been linked to allergic disease, prompting study of chitin as an elicitor of Th2 immunity (Cartier et al., 2004; Van Dyken et al., 2011). Chitin particles induce accumulation of IL-4-producing innate immune cells in mouse models. Such accumulation is abolished when mice are challenged with chitin after they are pretreated with recombinant acidic mammalian chitinase (AMCase) or if they overexpress AMCase (Reese et al., 2007; Semenuk et al., 2001). These results suggest that innate immune responses induced by chitin support the development of Th2-mediated allergic Inflammation.
Airway epithelial cells represent the first line of defense against inflammatory stimuli and antigens. Activation of epithelial cells is a characteristic of asthma and allergic rhinitis and is associated with allergic sensitization (Wang et al., 2008). Cytokines produced by respiratory epithelial cells, including interleukin (IL)-25, IL-33, and thymic stromal lymphopoietin (TSLP), play a role in regulating type 2 immunity (Uchida et al., 2017). Direct effects of chitin on airway epithelial cells are believed to contribute to allergic airway diseases such as asthma (Khosravi and Erle, 2016). Inhaled chitin stimulates epithelial cell release of Th2 cytokines IL-25, IL-33, and TLSP, which induce secretion of IL-5 and IL-13 in innate lymphoid type 2 cells (Van Dyken et al., 2014). However, IL-17A-producing γδ T cells are also associated with prolonged influx of neutrophils at the site of chitin challenge. Thus, chitin elicits complex and potentially pathological pathways during the early inflammatory response, underscoring the need for elucidation of the mechanism of chitin recognition. While fragments of chitin elicit innate and prolonged Th2-driven inflammation in humans and animal models, it is unclear how host cells recognize chitin fragments and propagate the signaling events required for immune cell recruitment. In particular, the pattern recognition receptors (PRRs) of epithelial cells that mediate these responses are poorly understood.
The lysin motif (LysM) is a carbohydrate binding module conserved across all kingdoms of life. In prokaryotic and eukaryotic proteins, LysM domains bind to chitin and peptidoglycan (Iizasa et al., 2010; Visweswaran et al., 2012; Willmann et al., 2011; Wong et al., 2014). LysM domains are found in various extracellular proteins and receptors. In plants, LysM domain-containing cell-surface receptors such as OsCEBiP, AtCERK1, AtLYK4, and AtLYK5 promote chitin recognition and trigger innate immune responses against chitin-containing pathogens such as fungi (Cao et al., 2014; Wan et al., 2008). LYSMD3 is a human transmembrane protein containing an evolutionarily conserved extracellular LysM domain (Yokoyama et al., 2018). Here, we investigated whether LYSMD3 on human lung epithelial cells mediates recognition of chitin and promotes early inflammatory responses.
RESULTS
LYSMD3 on human airway epithelium
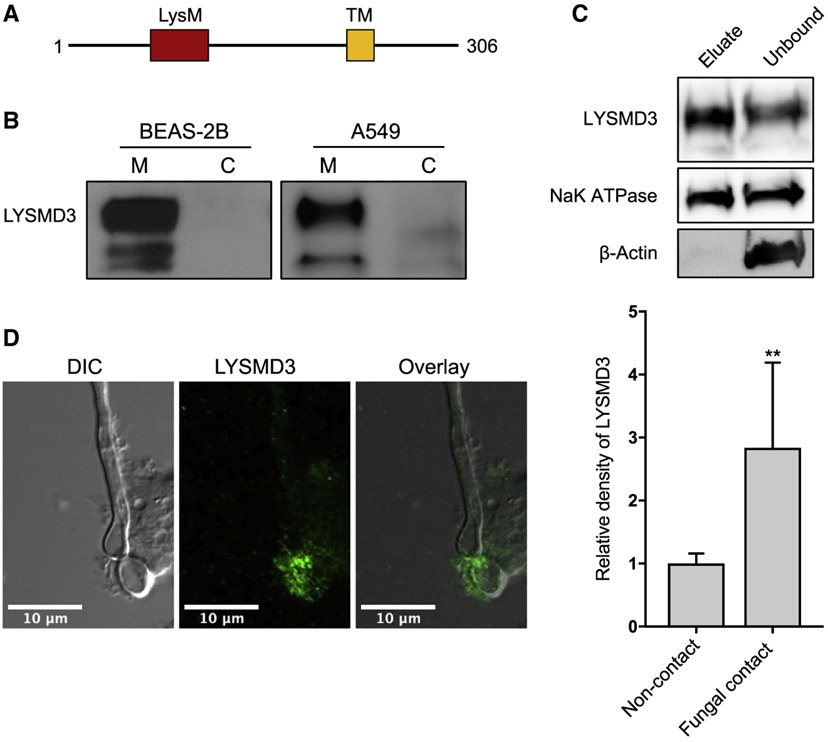
We first defined the subcellular localization of LYSMD3 in epithelial cells. In silico analysis predicts that LYSMD3 contains a transmembrane domain (Figure 1A). LYSMD3 has been detected in the Golgi of HeLa cells and mouse embryonic fibroblast cells (Yokoyama et al., 2018), and in the plasma membrane of HeLa cells (Serebrenik et al., 2018). By using biochemical fractionation, we observed LYSMD3 expression in the membrane compartment (plasma membrane and internal membranes) of human bronchial epithelial cells (BEAS-2B) and alveolar type II epithelial cells (A549) (Figure 1B). LYSMD3 was not detectable in the cytosolic fraction (Figure 1B). We next biotinylated cell-surface proteins and demonstrated by western blot that LYSMD3 is expressed on the surface of unstimulated BEAS-2B lung epithelial cells (Figure 1C). Our results are consistent with the findings of Yokoyama and colleagues that LYSMD3 is associated with the membrane compartment and not the nucleus or cytoplasm (Yokoyama et al., 2018). We now extend those findings and reveal that LYSMD3 is displayed on the surface of human lung epithelium.
Figure 1. LYSMD3 is located in the plasma membrane of human airway epithelial cells and in sites of fungal-epithelial interactions.
(A) Schematic illustration of the predicted domains of human LYSMD3.
(B) Immunoblot analysis of membrane (M) and cytosolic (C) fractions from BEAS-2B and A549 cells probed with anti-LYSMD3.
(C) Immunoblot analysis of biotinylated cell-surface proteins (eluate, 25 μL) and unbound lysate (25 μL) from BEAS-2B cells
(D)Representative images of BEAS-2B cells infected with live C. albicans SC5314 and stained for LYSMD3 (green). Scale bars, 10 μm. Accompanying histogram (right) depicts enrichment in LYSMD3 at sites of contact with the fungus, compared to sites not in contact with the fungus. Results were calculated from seven pairs of fungal contact regions and non-contact regions. IgG control staining during C. albicans infection of lung epithelial cells revealed negligible staining (data not shown). **p < 0.01 (Student’s t test). Data are from one representative experimental of three performed with similar results (mean and SD of seven samples per group).
Chitin is exposed at the bud sites of live yeast (Gantner et al., 2005) and influences host recognition of Candida albicans in vitro and in vivo (Marakalala et al., 2013; Mora-Montes et al., 2011). We used live C. albicans to investigate localization of endogenous LYSMD3 with this fungus upon binding to epithelial cells. By confocal imaging, we found that LYSMD3 accumulated chiefly at sites in contact with C. albicans in the areas of fungal budding (Figure 1D). The density of LYSMD3 signal was enriched at areas in contact with C. albicans compared to areas not in contact. Thus, LYSMD3 may bind chitin since the receptor accumulates in areas of putative chitin enrichment.
LYSMD3 binding of chitin
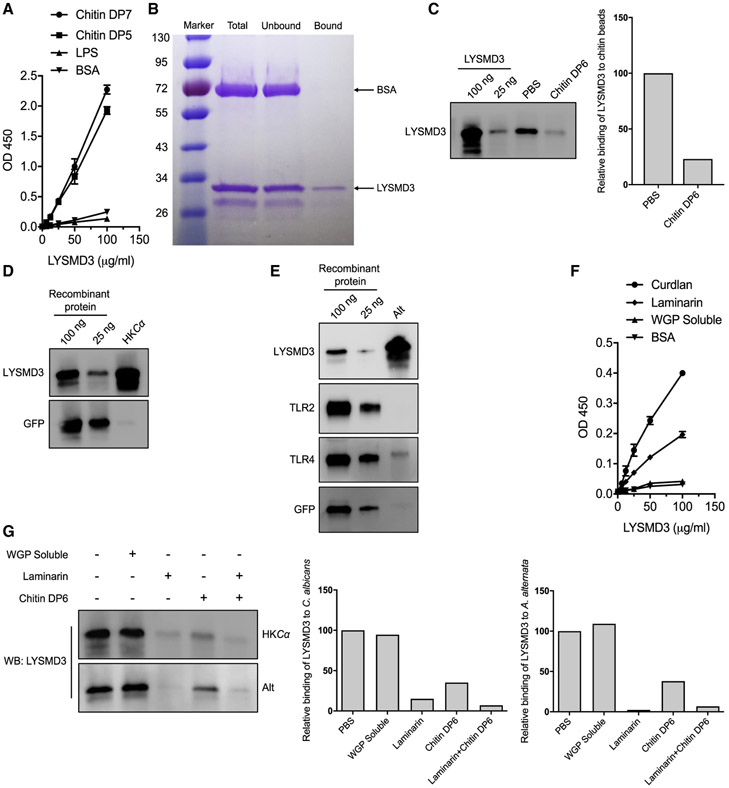
Since some lysin motif (LysM)-containing receptors in plants bind chitin oligosaccharides (Liu et al., 2012; Miya et al., 2007), we first investigated whether human LYSMD3 could bind soluble chitin oligosaccharides. By using an ELISA assay for protein binding to immobilized chitin, we found that the purified ectodomain of human LYSMD3 is able to directly bind chitin oligosaccharides with different degrees of polymerization of 5–7 (DP5–7) but does not bind bovine serum albumin (BSA) or LPS (Figure 2A; data not shown). Consistent with our ELISA results, pull-down assays demonstrated that the LYSMD3 ectodomain co-immunoprecipitated with chitin beads (50–70 μm) (Figure 2B), suggesting LYSMD3 can also interact with a large, insoluble particulate of chitin. Finally, chitin oligomers inhibited binding of LYSMD3 to these chitin beads (Figure 2C), indicating that the interaction of LYSMD3 with chitin beads is specific. Notably, we found that the LYSMD3 ectodomain did not bind E. coli peptidoglycan (PGN-ECndss) (Figure S1A), although some LysM domains are reportedly able to bind peptidoglycan (Visweswaran et al., 2012; Willmann et al., 2011).
Figure 2. LYSMD3 is a chitin and β-glucan binding protein.
(A) Direct binding of recombinant (r) LYSMD3 to immobilized carbohydrates or bovine serum albumin (BSA), determined by ELISA. The plate was coated with chitin DP5, DP7, LPS-EB, or BSA. An increasing amount ectodomain of LYSMD3 was added, and the amount bound was detected with the LYSMD3 antibody. See also Figure S1A.
(B) Direct binding of rLYSMD3 to chitin beads. A mixture of purified ectodomain of LYSMD3 and BSA was incubated with chitin magnetic beads. LYSMD3 bound chitin, while BSA remained in the soluble fraction (Unbound). Samples were assayed by SDS-PAGE and Coomassie blue stain.
(C) rLYSMD3 (150 ng) was incubated with chitin beads in the presence of chitin DP6 (3 mM). Bound LYSMD3 was detected by western using anti-LYSMD3. rLYSMD3 was run in the first two lanes. Western bands were quantified by densitometry using VisionWorks 9. Binding was normalized to the value without carbohydrate as 100%.
(D) rLYSMD3 and GFP were incubated (2.5 μg per sample) with suspensions of 1 × 107 heat-killed C. albicans SC5314 (HKCα). Proteins bound to fungal cell wall were detected by immunoblotting with anti-LYSMD3 or anti-GFP antibody. rLYSMD3 and GFP were run in the first two lanes.
(E) rLYSMD3, TLR2, TLR4 and GFP were incubated (2.5 μg per sample) with suspensions of 5 × 106 live Alternaria alternata (Alt) spores. Proteins bound to fungal cell wall were detected by immunoblot with anti-LYSMD3, anti-TLR2, anti-TLR4, or anti-His tag (GFP) antibody. rLYSMD3, TLR2, TLR4, and GFP were run in the first two lanes.
(F) Binding of rLYSMD3 to immobilized β-glucans or BSA by ELISA. See also Figures S1B-S1D.
(G) 100 ng of rLYSMD3 ectodomain was incubated with heat-killed C. albicans SC5314 (HKCα) or A. alternata spores in the presence of soluble WGP, laminarin, or chitin DP6 (2 mg/mL). Bound LYSMD3 was detected by western and quantified by densitometry. Binding was normalized to the value without carbohydrate as 100%.
Data are from one representative of three independent experiments (mean and SD of three samples per group in A and F).
LYSMD3 binding to fungal cells
Since fungi harbor chitin in their cell wall, we investigated whether LYSMD3 recognizes fungal cells. We found that the LYSMD3 ectodomain bound heat-killed C. albicans yeasts (Figure 2D), which have increased surface-exposed chitin (Mora-Montes et al., 2011). We also found that the LYSMD3 ectodomain bound live Alternaria spores (Figure 2E), whereas TLR2 did not bind the spores and TLR4 bound them only weakly. Given that chitin only constitutes a small portion (~3%–4%) of the fungal cell wall and is mainly present in the inner layer of live cells (Gow et al., 2017), we tested whether LYSMD3 also binds other fungal cell wall components. Eukaryotic LysM domains reportedly bind mainly chitin and peptidoglycan (Iizasa et al., 2010; Willmann et al., 2011); however, we found no evidence for LYSMD3 binding of peptidoglycan (Figure S1A).
In addition to chitin, the plant LysM receptor AtCERK1 recognizes non-branched β-1,3-glucan oligosaccharides (Mélida et al., 2018). Because heat-killing of Candida exposes both chitin and β-glucan, we assayed binding of the LYSMD3 ectodomain to β-glucan. We found that the LYSMD3 ectodomain bound curdlan, a particulate linear β-(1,3)-glucan without branches, and laminarin, a linear soluble β-(1-3)-glucan with β-(1-6)-linkages (Figure 2F). Notably, LYSMD3 showed no affinity for soluble whole-glucan particles (WGP) (Figure 2F). We validated these findings with a second, commercial source of purified LYSMD3 ectodomain (Figures S1B-S1D).
In competition assays, we analyzed the ability of different soluble chitin or β-glucan preparations to compete with the binding of LYSMD3 to fungal cells. As expected, chitin oligosaccharides and laminarin, alone or together, competed the binding of LYSMD3 to killed Candida yeast and Alternaria spores (Figure 2G). In contrast, soluble WGP again had no effect. These data buttressed the idea that LYSMD3 binds chitin and β-glucan and suggested that the observed binding of LYSMD3 to fungal cells is likely mediated through recognition of chitin or β-glucan, or both. Taken together, our findings indicate that chitin and β-glucan are ligands for LYSMD3.
Role of LYSMD3 in the innate response of human lung epithelial cells to chitin
In plants, recognition by LysM receptors of chitin oligosaccharides of at least seven N-acetyl glucosamine repeats triggers strong immune responses (Hamel and Beaudoin, 2010). However, little information is available on the chitin structure recognized by mammals. Chitin oligosaccharides reportedly induce size-dependent immune stimulation of human and mouse immune cells and of mice (Fuchs et al., 2018), but the immune effect of oligomeric chitin fragments on epithelial cells has not been described. The findings that plant LysM receptors recognize chitin oligomers, and that AMCase is secreted in airways (Van Dyken et al., 2017), prompted us to evaluate the immune effects of chitin oligosaccharides on human airway epithelial cells. We tested chitin oligomers with defined lengths of DP 2–7 and found that chitin hexamers (DP6) or heptamers (DP7) elicited IL-6 and IL-8 production in BEAS-2B cells, whereas smaller oligomers (DP <6) elicited little or no activity (Figure 3A). These findings are in line with the size-dependent chitin sensing and downstream activation of defense mechanisms via conserved LysM receptors in plants (Hamel and Beaudoin, 2010).
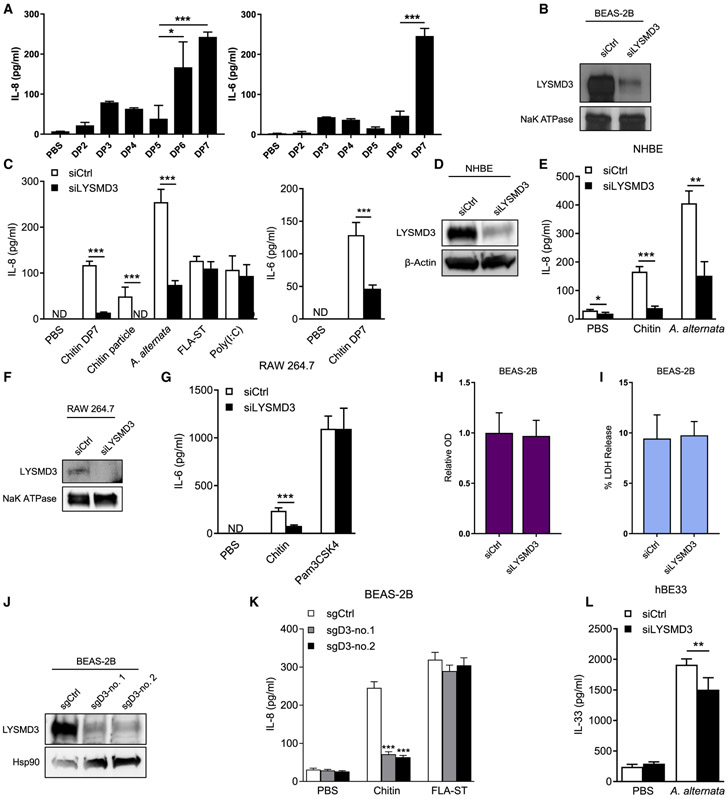
Figure 3. LYSMD3 mediates the expression of pro-inflammatory products upon stimulation with chitin or chitin containing fungi.
(A) ELISA of cytokines in supernatants of BEAS-2B cells stimulated for 24 h with 500 μM chitin oligomers.
(B) Immunoblot of LYSMD3 in membrane protein fraction of BEAS-2B cells 48 h after transfection with control siRNA or a siRNA pool specific for LYSMD3. NaK ATPase is loading control. See also Figure S2C.
(C) ELISA of cytokines in supernatants of BEAS-2B cells transfected with siRNAs as in (B), then stimulated for 24 h with chitin DP7 (500 μg/mL), chitin particles (500 μg/mL), Alternaria spores (2 × 105/mL), FLA-ST (300 ng/mL), or Poly(I:C) (3 μg/mL). See also Figures S2D and S2E.
(D) Immunoblot of LYSMD3 in cell lysates of NHBE cells 48 h after transfection with control siRNA or siRNA specific for LYSMD3. β-Actin is loading control.
(E) ELISA of IL-8 in supernatants of NHBE cells transfected with siRNA as in (D) and then stimulated for 24 h with chitin DP7 (500 μg/mL) or A. alternata spores (2 × 105/mL).
(F) Immunoblot of LYSMD3 in membrane protein fraction of RAW 264.7 cells 48 h after transfection with control siRNA or siRNA specific for LYSMD3.
(G) ELISA of IL-6 in supernatants of RAW 264.7 cells transfected with siRNA as in (F) and then stimulated for 24 h with chitin DP7 (100 μg/mL) or Pam3CSK4 (100 ng/mL).
(H) MTT assay 48 h after transient transfection of siRNA asin(B)in BEAS-2B cells. Cell growth rate is expressed as absorbance at 570 and was normalized to the value with control siRNA as 1.
(I) LDH cytotoxicity assay 48 h after transient transfection of siRNAs as in (B) in BEAS-2B cells.
(J) Expression of LYSMD3 protein in BEAS-2B cell with CRISPR-Cas9 knockout of LYSMD3 (sgD3). Hsp90 is the loading control.
(K) ELISA of IL-8 in supernatants of BEAS-2B cell knockout of LYSMD3 as in (J), unstimulated or stimulated for 24 h with chitin DP7 (500 μg/mL) or FLA-ST (300 ng/mL). See also Figure S2F.
(L) ELISA of IL-33 in supernatants of hBE33 cells transfected with siRNA as in (D) and then stimulated for 1 h with Alternaria extract (200 μg/mL).
*p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s t test). Data are representative of three experiments (mean and SD of three samples per group in A, C, and E, or four samples per group in G, K, and I, or eight samples per group in H and L).
We next investigated the effect of LYSMD3 knockdown on the expression of cytokines in lung epithelial cells when stimulated with chitin or chitin-containing fungi. Knockdown of LYSMD3 in BEAS-2B cells using small interfering RNAs (siRNAs) (Figure 3B) nearly abolished production of IL-6 and IL-8 in response to chitin oligomers (DP7) or chitin particles (Figure 3C). Knockdown of LYSMD3 also reduced IL-8 production induced by Alternaria spores (Figure 3C). Similar results were obtained using primary human bronchial epithelial (NHBE) cells (Figures 3D and 3E) and human alveolar type II epithelial cells (A549) (Figures S2A and S2B). To solidify our results, we used another non-overlapping siRNA to silence LYSMD3 (Figure S2C); this LYSMD3 knockdown also decreased chitin-induced IL-8 production in BEAS-2B cells (Figure S2D).
To determine the specificity of LYSMD3 signaling, we investigated whether knockdown of LYSMD3 affected cytokine production induced by other PRR ligands. Knockdown of LYSMD3 had no effect on cytokine production induced by the TLR3 ligand Poly (I:C) or TLR5 agonist FLA-ST (Figures 3C and S2E). In mouse macrophages, silencing of LYSMD3 blunted production of IL-6 after stimulation with chitin but had no effect on IL-6 production in response to Pam3CSK4 (Figures 3F and 3G). To exclude that knockdown of LYSMD3 reduced viability or fitness and thus response to certain innate immune stimuli, we performed MTT and LDH cytotoxicity assays. LYSMD3 knockdown had no effect on bronchial epithelial cell viability or proliferation (Figures 3H and 3I), indicating LYSMD3 is not essential for bronchial epithelial cell viability and membrane function integrity.
Because gene silencing may have off-target effects, we also eliminated LYSMD3 with CRISPR. We used two single-guide RNAs (sgRNAs) to knock out LYSMD3 in BEAS-2B cells (Figure 3J). Consistent with siRNA results, both LYSMD3 sgRNAs decreased IL-8 production in response to chitin but had no effect on IL-8 secretion in response to Flagellin; a non-targeting sgRNA control had no effect on response to chitin (Figure 3K). The knockout of LYSMD3 also had no effect on IL-8 production induced by peptidoglycan from either gram-positive Bacillus subtilis (PGN-BS) or gram-negative E. coli K12 (PGN-EK) (Figure S2F). Thus, LYSMD3 in human lung epithelial cells promotes innate immune responses to chitin but not peptidoglycan.
Interleukin (IL)-33, stored in the nuclei of airway epithelial cells, is released quickly into the airway lumen when the cells are exposed to Alternaria antigen extract, but not to other common aeroallergens (Bartemes and Kita, 2018). The mechanisms that mediate IL-33 release are a subject of intense research. We found that knockdown of LYSMD3 reduced Alternaria extract-induced IL-33 release by IL-33-producing human bronchial epithelial cells (HBE33) (Figure 3L). Thus, LYSMD3 may have pathological relevance in the context of fungal innate immunity and allergic inflammation.
Antagonism of chitin-induced signaling via LYSMD3
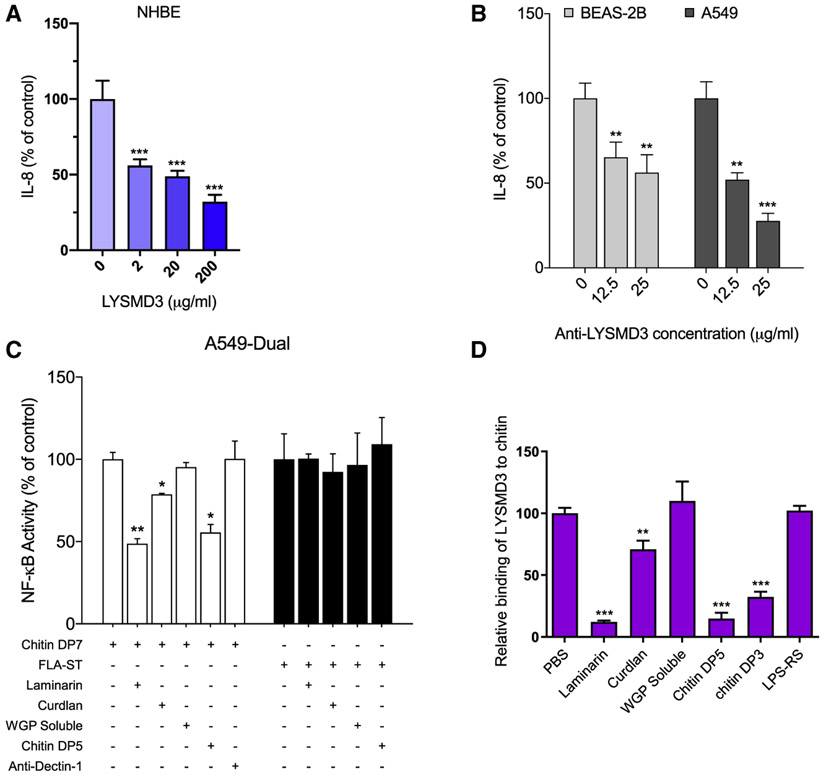
We sought to identify antagonists that interfere with LYSMD3 sensing, signaling, and innate response to chitin. First, we found that addition of the soluble LYSMD3 ectodomain to primary human bronchial epithelial cells retarded IL-8 production induced by chitin, whereas the ectodomain had no effect on secretion of IL-8 induced by flagellin (FLA-ST) (Figures 4A and S3). Treatment with a polyclonal antibody against the LYSMD3 ectodomain also inhibited IL-8 production induced by chitin in both BEAS-2B and A549 cells. This antibody acted in a concentration-dependent manner whereas control polyclonal antibody had no effect (Figure 4B).
Figure 4. Antagonism of LYSMD3 ectodomain function impairs epithelial cell inflammatory responses induced by chitin.
(A) ELISA of IL-8 in supernatants of NHBE cells stimulated for 24 h with chitin DP7 (500 μg/mL) in the presence of soluble LYSMD3 ectodomain. IL-8 concentration in cell-free supernatant in the absence of the LYSMD3 ectodomain was normalized to 100%. See also Figure S3.
(B) ELISA of IL-8 in supernatants of BEAS-2B or A549 cells stimulated for 24 h with chitin DP7 (500 μg/mL) after pre-incubation with LYSMD3 antibody (low endotoxin, azide free) or isotype control. IL-8 concentrations in supernatants were normalized to 100% as in (A).
(C) A549-Dual cells were stimulated with chitin DP7 (500 μg/mL) or FLA-ST (300 ng/mL) for 24 h after preincubation with laminarin (1 mg/mL), curdlan (1 mg/mL), soluble WGP (1 mg/mL), chitin DP5 (200 μM), Dectin-1 blocking antibody (3 μg/mL), or corresponding isotype control. NF-μB activation was assayed by optical density (OD) at 655 nm using QUANTI-Blue. NF-κB activities were normalized to 100% in the absence of inhibitors as in (A).
(D) Relative binding of rLYSMD3 (50 μg/mL) to immobilized chitin DP7 in the presence of laminarin (100 μg/mL), curdlan (500 μg/mL), soluble WGP (500 μg/mL), chitin DP5 (200 μM), chitin DP3 (200 μM), or LPS-RS (500 μg/mL), as determined by ELISA. Binding was normalized 100% in the absence of carbohydrates as in (A).
*p < 0.05, **p < 0.01, and ***p < 0.001 (Student’s t test). Data are from one representative experiment of three performed (mean and SD of four samples per group in A or three samples per group in B, C, and D).
It is unclear whether LYSMD3 on epithelial cells plays a role in responding immunologically to β-glucan. We found above that LYSMD3 bound to β-glucan in addition to chitin (Figures 2F and 2G). Others have reported that airway epithelial cells are unresponsive or respond poorly to β-glucan, which is consistent with low or no expression of Dectin-1 (Heyl et al., 2014; Lee et al., 2009; Mayer et al., 2007). We tested activation of nuclear factor κB (NF-κB) in A549-Dual reporter cells and found that it was not induced by even large amounts (1 mg/mL) of various forms of pure β-glucan including curdlan, laminarin, and whole-glucan particles (soluble WGPd) or by chitin DP5 at 200 μM (data not shown). However, preincubation of A549 cells with curdlan, laminarin, or chitin DP5 significantly reduced the level of NF-κB activation induced by a chitin stimulus (Figure 4C). Curdlan, laminarin, and chitin DP5 also were able to bind the ectodomain of LYSMD3 and block chitin binding to LYSMD3 (Figure 4D), whereas soluble WGP, which has no affinity for LYSMD3, had no effect. Preincubation of A549 cells with curdlan, laminarin, or chitin DP5 likewise did not affect the induction of NF-κB activation by Flagellin (Figure 4C), suggesting that the blocking effect of these products was not due to steric masking of specific cell-surface receptors. Additionally, the inhibitory effects of curdlan, laminarin, or chitin DP5 were not likely due to interaction with Dectin-1 because neither Dectin-1 blocking antibody nor antagonism of the receptor with soluble WGP had any effect on NF-κB activation in response to chitin (Figure 4C). Moreover, Dectin-1 does not bind chitin as shown here (Figure S1C) and in multiple other studies (Gantner et al., 2005; Gour et al., 2018; Mora-Montes et al., 2011). Thus, Dectin-1 is not involved in recognition of chitin by A549 cells and the inhibition of chitin-induced NF-κB activation by β-glucan is independent of its blockade of Dectin-1. Taken together, our findings suggest that the inhibition of chitin signaling by curdlan, laminarin, and chitin DP5 is due to their direct binding to LYSMD3 and blockade of chitin binding to this receptor.
DISCUSSION
Chitin, chitinase, and chitinase-like proteins have been associated with a number of human pathologies. The mechanisms that underpin these pathologies are unclear (Ziatabar et al., 2018). Large chitin particles reportedly induce chitinase expression in epithelial (Lalaker et al., 2009) and immune cells (Fuchs et al., 2018) and in vivo (Kim et al., 2015). Interaction between inhaled chitin-containing fungi and airway epithelium is an early event that can shape the immunological outcome (Roy and Klein, 2013), but the mode of recognition of chitin particles or chitin in fungal cell walls by lung epithelial cells remains obscure. Herein, we report that LYSMD3 on human lung epithelial cells mediates chitin recognition. Our elucidation and characterization of LYSMD3 as a human epithelial cell receptor for chitin provides insight into mammalian innate immune responses to chitin, chitin-containing organisms, and allergic inflammation.
We demonstrate that LYSMD3 binds chitin and also β-glucan from fungi. These moieties are PAMPs covalently cross-linked to one another in the fungal cell wall. A. fumigatus chitin linked to β-glucan induce enhanced immune responses compared with single cell wall polysaccharides (Dubey et al., 2014). Chitin-β-glucan particles also induce recruitment of eosinophils and neutrophils, chitinase activity, and production of tumor necrosis factor (TNF)-α and TSLP in mouse lungs. Composite PAMPs may induce synergy between individual receptors. Alternatively, a single receptor may recognize composite PAMPs better than individual PAMPs. This receptor is likely not Dectin-1 since mice lacking the receptor display increased eosinophil recruitment in response to inhaled Aspergillus conidia (Amarsaikhan and Templeton, 2015). We are not aware of receptors other than LYSMD3 that can interact with both chitin and β-glucan. LYSMD3 might recognize composite fungal chitin-β-glucan more efficiently than either chitin or β-glucan alone and this function warrants further study.
Dectin-1 has been implicated in mediating immune responses to chitin, but studies have yielded conflicting results. Although treatment of mouse peritoneal macrophages with large amounts of the Dectin-1 blocker laminarin (1 mg/mL) blunted TNF-α and IL-10 production in response to chitin (Da Silva et al., 2008), there is no direct evidence that Dectin-1 binds chitin or mediates a response to chitin. For example, Dectin-1 reconstitution in HEK293T does not result in NF-κB activation in response to chitin and macrophages from Dectin-1-deficient mice produce TNF-α in response to chitin as efficiently as macrophages from wild-type mice (Fuchs et al., 2018). In another study (Wagener et al., 2014) that conflicts with the findings of Da Silva et al. (2008), the use of laminarin in low amounts (100 μg/mL) sufficient to block Dectin-1 had no effect on chitin-induced IL-10 production in mouse macrophages.
The conflicting findings from the studies above suggest that blockade of another, non-Dectin-1 receptor with a relative low affinity for laminarin might be responsible for the abolished chitin responsiveness that was observed in immune cells. We have shown here that laminarin directly binds LYSMD3 and is able to block its activity. Furthermore, knockdown of LYSMD3 specifically inhibited chitin-induced IL-6 production in mouse macrophages. Similarly, in human bronchial epithelial cells, curdlan and laminarin treatment reduced IL-8 release induced by the allergenic fungus Fusarium proliferatum by 20% and 53%, respectively, while Dectin-1 blocking antibody showed less inhibitory effect than β-glucan (Yeh et al., 2017). These results suggest that other, non-Dectin-1 receptors blocked by curdlan and laminarin may promote recognition of F. proliferatum by airway epithelial cells. Collectively, our results provide an explanation for previous conflicting reports about the role of Dectin-1 in chitin signaling and suggest instead that laminarin blockade of LYSMD3 (rather than Dectin-1) accounts for the reduced innate immune response to chitin or chitin-containing fungi in immune cells and epithelial cells.
Further studies will be required to determine the functional role of LYSMD3 in vivo. A recent study of mice deficient in LYSMD3 failed to detect a phenotype in the response to a wide variety of pathogens (Yokoyama et al., 2018). The study explored 10 different bacteria, two viruses, LPS, and two fungi, including Cryptococcus neoformans and A. fumigatus. That study yielded results consistent with our finding that LYSMD3 does not bind to peptidoglycan. Studies that were designed to address chitin were limited to analysis of mouse weight loss, number of fungi in lung, and total cell counts after pulmonary infection with the two fungi. However, the function of chitin in the pathogenicity of the these fungal species or the host response to them remains elusive (Chai et al., 2011). C. neoformans has a polysaccharide capsule that surrounds its cell wall (Zaragoza et al., 2009), and cell wall chitin of C. neoformans is mainly deacetylated to chitosan (Hole et al., 2020). Resting A. fumigatus conidia are covered by a hydrophobic protein layer that masks carbohydrate (Bain et al., 2015). The negative data by Yokoyama et al. emphasize the importance of choosing the right model in assaying the role of LYSMD3 in response to chitin in vivo and underscore the potential value of models that involve microbial chitin induced allergic inflammation.
The study by Yokoyama et al. (2018) also reminds us that differences between murine and human LYSMD3 may influence interpretation of in vivo relevance.Mice and humans may respond differently to chitin. Chitin was found to have size-dependent effects on mouse cells (Da Silva et al., 2008) and in mice (Kim et al., 2015), but we did not observe this difference in human cells. We found that chitin oligosaccharides and chitin particles were both recognized by LYSMD3 and induced similar immune responses in human epithelial cells. Also, in a previous report, chitin stimulation of human PBMC led to secretion of the anti-inflammatory cytokine IL-10 and the pro-inflammatory cytokines IL-6 and TNF-α, whereas in mouse macrophages only IL-10 was induced by chitin (Wagener et al., 2014). In addition, Dectin-1, mannose receptor, NOD2 and TLR9 have all been described as receptors for chitin in mouse cells, whereas none of these receptors have been implicated in chitin recognition in human cells. Finally, mice lack LYSMD3 isoforms 2 and 3, which may be required for its function in humans. Thus, the biological role of mouse LYSMD3 remains unclear and may differ from that of human LYSMD3. A definitive study of human LYSMD3 interactions with chitin in vivo may require that the human receptor be expressed in the mouse.
STAR☆METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Xin He (xinhe@vt.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary cells and cell culture
BEAS-2B, A549 and RAW 264.7 cells were obtained from ATCC. NHBE cells were obtained from Lonza. A549-Dual and HEK-Blue hNOD2 cells were purchased from InvivoGen. hBE33 cells containing the human IL-33 gene were developed as previously described (Uchida et al., 2017). A549, RAW 264.7 and HEK-Blue hNOD2 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS). BEAS-2B, NHBE and hBE33 cells were maintained in bronchial epithelial growth media (BEGM) supplemented with defined growth factors and retinoic acid (Lonza). All cells were propagated at 37°C and 5% CO2 in a humidified incubator.
Fungal strain and growth conditions
C. albicans strain SC5314 (ATCC MYA-2876) was grown in YPD medium at 30°C with 300 rpm agitation for 18 h. The cells were collected by centrifugation and washed twice in phosphate-buffered saline (PBS). Heat-killed C. albicans cells were prepared by incubation at 65°C for 1 h to expose chitin on the cell surface. Alternaria alternata (ATCC 66981) was cultured on potato dextrose agar (0.4% potato starch, 2% dextrose, 1.5% agar) and incubated at 25°C in the absence of light. Spores were collected using gentle agitation in PBS and counted on a hemacytometer.
METHOD DETAILS
Expression and purification of recombinant LYSMD3
The ectodomain of human recombinant LYSMD3 was synthesized and codon-optimized for expression in E. coli (GenScript) and subcloned into His-Trx fusion vector pET32a. The plasmid was transformed into E. coli BL21(DE3) cells. Cells were grown at 37°C to mid log phase (OD600 = 1.2), induced with IPTG and harvested by centrifugation 4 h after induction. Cell pellets were resuspended in lysis buffer and lysed by sonication. The protein was purified using Ni-NTA column chromatography. The His-Trx tag of rLYSMD3 was removed by TEV protease cleavage. Endotoxin was removed from rLYSMD3 by size exclusion chromatography. The endotoxin level is < 0.01 EU/μg of the protein as determined by the LAL assay. Protein purity was analyzed by SDS-PAGE (Figure S4A). Protein identity was confirmed by western blot and mass spec. Expression and purification was performed by GenScript. The absence of peptidoglycan contaminants in rLYSMD3 was confirmed with HEK-Blue hNOD2 cells (Figure S4B). A separate commercial source of purified rLYMD3 (6xHis-Met 1-Gln 142) (Proteintech) was used in some assays to validate results (Figures S1B-S1D).
Quality control of chitin
The purity of the chitin oligosaccharides was > 90% as stated by the manufacturers (Elicityl and IsoSep). The degree of acetylation of the chitin oligosaccharides was > 90% as demonstrated by Fuchs et al.l (Fuchs et al., 2018). Chitin from crab shells was placed in endotoxin-free sterile PBS and sonicated with a Branson Ultrasonics Sonicator at 25% output power for 5 min. Sonication was repeated two more times for a total of three 5-minute intervals. Afterward, the suspensions were passed through a 10 μm cell strainer. All chitin preparations were demonstrated to be endotoxin-free.
siRNA transfection
BEAS-2B and A549 were transfected with siRNA duplexes through the use of Lipofectamine RNAiMAX transfection reagent. Transfecting siRNA into NHBE, hBE33 or RAW 264.7 cells was performed using the HiPerFect transfection reagent. X-tremeGENE HP DNA transfection reagent was used for plasmid transfection.
CRISPR-Cas9-mediated LYSMD3 gene knockout
BEAS-2B cells were infected with non-targeting control or LYSMD3-sgRNAs Edit-R All-in-one Lentiviral particles to establish stable LYSMD3 KO cells. Cells were selected with 1 μg/ml puromycin for 4 days.
Chitin binding assay
Chitin-binding assay was performed according to a previously described method (Cadoret et al., 2014) with modifications. Briefly, 100 μl of chitin magnetic beads in chitin-binding buffer (NaCl 500 mM, Tris-HCI 20 mM, EDTA 1 mM, Tween-20 0.1%, pH 8) was incubated with a 200 μl solution containing recombinant LYSMD3 ectodomain and BSA proteins at 200 μg/ml each in chitin-binding buffer. The sample was incubated at 4°C with agitation for 1 hour. Separation of the supernatant (unbound fraction) from the beads (bound fraction) was performed by applying a magnet. After collection of the supernatant, the beads were washed three times with 200 μl of chitin-binding buffer. 15 μl of samples from the total, unbound and bound fractions were analyzed using SDS-PAGE and Coomassie blue staining.
LYSMD3-binding ELISA
Nunc MaxiSorp 96-well ELISA plates (ThermoFisher Scientific) were coated with 1 μg chitin oligosaccharides, 20 μg LPS-EB, 4 μg curdlan, 4 μg laminarin or 4 μg soluble WGP in carbonate buffer, pH 9.5 or 5 μg BSA in PBS overnight at 4°C. Wells were blocked with StartingBlock T20 (TBS) Blocking Buffer (ThermoFisher Scientific) and then incubated with human recombinant LYSMD3 ectodomain at increasing concentration for 1 h at room temperature. After washing, recombinant LYSMD3 was detected by polyclonal LYSMD3 antibody, 1/3000 dilution, followed by mouse anti-rabbit IgG monoclonal antibody conjugated to HRP, 1/20000 dilution, and addition of tetramethylbenzidine (TMB). Stop solution was added and the optical density was measured at 450 nm.
Alternatively, chitin or WGP-coated wells were incubated with commercial, recombinant human LYSMD3 (6xHis-Met 1-Gln 142), Dectin-1 or BSA at 4 μg/ml, followed by rinsing and addition of anti-His-HRP monoclonal antibody and processed as described above.
Fungal binding assay
The assay was adapted from a previously described method (Vera et al., 2009). In order to expose cell wall PAMPs, fungal spores were suspended in complete RPMI 1640 medium and incubated for 2 h at 30°C to induce germination. Recombinant proteins (2.5 μg per assay) were incubated for 1 h at 4°C under rotation with a suspension of heat-killed C. albicans (1 × 107) or live Alternaria alternata spores (5 × 106) in binding buffer (TBS, 1% BSA, 5 mM CaCl2) to a final volume of 0.5 ml. Following incubation, cells were collected using Spin-X centrifuge tube filter (Corning) by centrifugation and washed thoroughly with TBS plus 5 mM CaCl2 to remove nonspecifically bound proteins. Cells were then resuspended in 60 μL Laemmli’s sample buffer and denatured by heating at 100°C for 15 min. Next, 30 μL of the lysate and pure recombinant proteins (25 or 100 ng) were separated by 10% SDS-PAGE followed by western blot analysis.
Confocal microscopy
BEAS-2B cells were infected with live C. albicans for 2.5 h. Cells were then fixed in 3% paraformaldehyde, blocked in BlockAid Blocking Solution (Thermo Fisher Scientific) with Human BD Fc Block (BD Biosciences) and incubated with antibodies against LYSMD3 (Proteintech) followed by an AlexaFluor 488-labeled goat anti-rabbit antibody. The samples were then taken for microscopic observation. Technical controls including omission of all immunostaining and omission of the primary antibody were performed. Mean Fluorescence Intensity (MFI) with these controls were not statistically distinguishable from irrelevant primary IgG control (data not shown). Images were acquired with an Olympus FV1000 confocal microscope with PlanApo 60X oil (NA = 1.4) super-corrected objective lens. Pixels were sampled at a spatial rate sufficient to meet Nyquist criteria (103 nm linear pixel size in the sample xy plane). Excitation was via 473 nm diode laser line (nominal 20mW) at 1% power with PMT voltage set to minimize saturation and photobleaching. Pinhole aperture size was set at 1 Airy Unit. Datasets were acquired as z stacks through the focal depth of the specimen (11 μm total depth with 1 μm plane spacing). DIC images were also acquired simultaneously.
Contact site data analysis
All image analysis was conducted in ImageJ (Fiji ImageJ ver. 2.1.0/1.53.h). Contact sites between live C. albicans yeast and/or hyphae and BEAS-2B cells were defined based on the DIC images to avoid biasing Region of Interest (ROI) selection with respect to staining patterns. ROIs were defined as polygons drawn around the fungal/cell contact in ROI Manager. Due to the fact that contiguous contact site membrane can exist on multiple focal planes, we conducted all measurements of Mean Fluorescence Intensity (MFI) of immune-stained target signal from the ROI applied to a maximum intensity z-projection image. For non-contact site membrane comparisons, we selected ROIs in regions distal from the contact site ROIs but on the same cell.
ELISA
BEAS-2B, NHBE, A549 or RAW 264.7 cells were stimulated for 24 h with chitin, Alternaria spores, Pam3CSK4, FLA-ST, Poly(I:C) or PGN. hBE33s were stimulated for 1 h with Alternaria extract. Supernatants were harvested and the amount of IL-6, IL-8, IL-33 or RANTES secreted was measured with ELISA kits according to the manufacturer’s recommendations (R&D Systems or BioLegend).
Protein preparation
Total cellular protein was isolated with RIPA Buffer. Fractions of membrane-and-cytoplasmic proteins from cultured cells were obtained by using Mem-PER plus membrane protein extraction kit (Thermo Fisher Scientific). Biotinylation of the cell surface proteins of BEAS-2B cells was performed with the Cell Surface Protein Isolation Kit (BioVision) according to the manufacturer’s protocol. Briefly, cells were grown in a T75 flask until they reached confluency. The cells were then washed with PBS and incubated with Sulfo-NHS-SS-biotin for 30 min at 4°C. A quenching solution was added, and cells were lysed with lysis buffer (500 μL) containing the Halt protease inhibitor cocktail (Thermo Fisher Scientific). The biotinylated cell surface proteins were isolated with streptavidin beads, eluted by the elution buffer (100 μl) with DTT, and analyzed by western blotting.
MTT assay
MTT assays were carried out using MTT Cell Proliferation Assay Kit (Cayman Chemical) following the manufacturer’s instructions.
LDH assay
Lactate dehydrogenase (LDH) assays were performed with Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Number of biological replicates is stated in each legend. Data are expressed as mean (SD). Pairwise comparisons between two groups were analyzed by unpaired Student’s t test. p values are indicated on graphs as appropriate: *p < 0.05, **p < 0.01 and ***p < 0.001. Detailed information can be found in the respective figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-LYSMD3 | Proteintech | Cat# 24313-1-AP |
| Anti-LYSMD3, Azide Free | Proteintech | N/A |
| Anti-HSP90 | Proteintech | Cat# 13171-1-AP; RRID: AB_2120924 |
| Anti-sodium potassium ATPase (HRP) | Abcam | Cat# ab185065 |
| Anti-β-Actin | Santa Cruz Biotechnology | Cat# sc-47778; RRID: AB_626632 |
| Mouse IgG2B Isotype Control antibody | R&D | Cat# MAB004; RRID: AB_357346 |
| Anti-Human TLR2 | R&D | Cat# AF2616; RRID: AB_416645 |
| Anti-Human TLR4 | R&D | Cat# MAB1478; RRID: AB_2240713 |
| Anti-Human Dectin-1 | R&D | Cat# MAB1859; RRID: AB_2081791 |
| Anti-GFP | Abcam | Cat# ab13970; RRID: AB_300798 |
| Rabbit IgG Isotype Control antibody | Thermo Fisher Scientific | Cat# 31235; RRID: AB_243593 |
| Anti-6x-His Tag (HRP) | Thermo Fisher Scientific | Cat# MA1-21315-HRP; RRID: AB_2536989 |
| IgA2 Isotype Control | InvivoGen | Cat# maba2-ctrl; RRID: AB_11124905 |
| Anti-hTLR2-IgA | InvivoGen | Cat# maba2-htlr2; RRID: AB_11142484 |
| Mouse Anti-Rabbit IgG (HRP) | GenScript | Cat# A01827 |
| Chemicals, peptides, and recombinant proteins | ||
| Chitin oligosaccharides DP2 | Elicityl | GLU432-90%; CAS: 35061-50-8 |
| Chitin oligosaccharides DP3 | Elicityl | GLU433-90%; CAS: 41708-93-4 |
| Chitin oligosaccharides DP4 | Elicityl | GLU434-90%; CAS: 2706-65-2 |
| Chitin oligosaccharides DP5 | Elicityl | GLU435-90%; CAS: 36467-68-2 |
| Chitin oligosaccharides DP6 | Elicityl | GLU436-90%; CAS: 6734-92-5 |
| Chitin oligosaccharides DP7 | Elicityl | GLU437-90%; CAS: 38854-46-5 |
| Chitoheptaose | IsoSep | 57/11; CAS: 38854-46-5 |
| Chitin from shrimp shells | Sigma-Aldrich | C9752; CAS: 1398-61-4 |
| Chitin Magnetic Beads | NEB | E8036S |
| LPS-EB Ultrapure | InvivoGen | tlrl-3pelps |
| LPS-RS Ultrapure | InvivoGen | tlrl-prslps |
| Pam3CSK4 | InvivoGen | tlrl-pms; CAS: 112208-00-1 |
| FLA-ST Ultrapure | InvivoGen | tlrl-epstfla |
| Poly(I:C) HMW | InvivoGen | tlrl-pic; CAS: 31852-29-6 |
| Laminarin | InvivoGen | tlrl-lam; CAS: 9008-22-4 |
| WGP Soluble | InvivoGen | tlrl-wgps |
| Curdlan | InvivoGen | tlrl-curd; CAS: 54724-00-4 |
| PGN-ECndss | InvivoGen | tlrl-ksspgn |
| PGN-EK | InvivoGen | tlrl-pgnek |
| PGN-BS | InvivoGen | tlrl-pgnb3 |
| QUANTI-Blue | InvivoGen | Cat# rep-qbs |
| Extracts of Alternaria | Greer Laboratories | Cat# XPMID3A2.5; Lot# 312142 |
| Recombinant Human LYSMD3-His | Proteintech | Cat# Ag19496 |
| Recombinant Human TLR2, CF | R&D | Cat# 2616-TR |
| Recombinant Human TLR4, CF | R&D | Cat# 1478-TR |
| Recombinant Human Dectin-1, CF | R&D | Cat# 1859-DC |
| Recombinant Aequorea victoria GFP-His | Sino Biological | Cat# 13105-S07E |
| Lipofectamine RNAiMAX Transfection Reagent |
Thermo Fisher Scientific | Cat# 13778 |
| HiPerFect Transfection Reagent | QIAGEN | Cat# 301704 |
| Puromycin | InvivoGen | Cat# ant-pr |
| Critical commercial assays | ||
| ELISA MAX Standard Set Human IL-6 | BioLegend | Cat# 430501 |
| ELISA MAX Standard Set Mouse IL-6 | BioLegend | Cat# 431301 |
| Human IL-8 DuoSet ELISA | R&D | Cat# DY208 |
| ELISA MAX Standard Set Human IL-8 | BioLegend | Cat# 431501 |
| Human IL-33 Quantikine ELISA Kit | R&D | Cat# D3300B |
| ELISA MAX Standard Set Mouse MCP-1 | BioLegend | Cat# 432701 |
| ELISA MAX Deluxe Set Human CCL5 | BioLegend | Cat# 440804 |
| Mem-PER Plus Membrane Protein Extraction Kit | Thermo Fisher Scientific | Cat# 89842 |
| Cell Surface Protein Isolation Kit | BioVision | Cat# K295 |
| MTT Cell Proliferation Assay Kit | Cayman Chemical | Cat# 10009365 |
| Pierce LDH Cytotoxicity Assay Kit | Thermo Fisher Scientific | Cat# 88953 |
| Experimental models: Cell lines | ||
| BEAS-2B | ATCC | CRL-9609 |
| A549 | ATCC | CCL-185 |
| RAW 264.7 | ATCC | TIB-71 |
| NHBE | Lonza | CC-2540 |
| A549-Dual | InvivoGen | a549d-nfis |
| HEK-Blue hNOD2 | InvivoGen | hkb-hnod2 |
| hBE33 cells | Uchida et al., 2017 | N/A |
| Experimental models: Organisms/strains | ||
| C. albicans | ATCC | ATCC MYA-2876 |
| A. alternata | ATCC | ATCC 66981 |
| Oligonucleotides | ||
| Control siRNA-A | Santa Cruz Biotechnology | Cat# sc-37007 |
| LYSMD3 siRNA (h) | Santa Cruz Biotechnology | Cat# sc-91992 |
| LYSMD3 siRNA (m) | Santa Cruz Biotechnology | Cat# sc-149191 |
| ON-TARGETplus Non- targeting siRNA #1 | Dharmacon | Cat# D-001810-01 |
| ON-TARGETplus Human LYSMD3 siRNA-Individual | Dharmacon | Cat# J-024521-09 |
| Recombinant DNA | ||
| Edit-R All-in-one Lentiviral sgRNA hEF1a Non-targeting Control #1 | Dharmacon | VSGC11964 |
| Edit-R Human LYSMD3 hEF1a All-in-one Lentiviral sgRNA-no. 1 |
Dharmacon | VSGH11936-247618492 |
| hEF1a All-in-one Lentiviral sgRNA-no. 2 | Dharmacon | VSGH11936-247547638 |
| Software and algorithms | ||
| Prism Version 8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| VisionWorks 9 | Analytik Jena | N/A |
| ImageJ | National Institutes of Health | https://imagej.net/ |
Highlights.
Human airway epithelial cells display the receptor LYSMD3 on their surface
LYSMD3 contains a LysM (chitin-binding) domain and is able to bind chitin and fungi
Chitin and fungi fuel cytokine release by binding LYSMD3 on airway epithelial cells
LYSMD3 also binds β-glucan and therefore may also sense β-glucan
ACKNOWLEDGMENTS
This work was supported by NIH R21AI115986 to C.B.L. and L.L. and by NIH R01AI130411 to B.S.K. The Graphical abstract was created with BioRender.com.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109392.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amarsaikhan N, and Templeton SP (2015). Co-recognition of β-glucan and chitin and programming of adaptive immunity to Aspergillus fumigatus. Front. Microbiol 6, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Gow NAR, and Erwig L-P (2015). Novel insights into host-fungal pathogen interactions derived from live-cell imaging. Semin. Immunopathol 37, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartemes KR, and Kita H (2018). Innate and adaptive immune responses to fungi in the airway. J. Allergy Clin. Immunol 142, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, O’Callaghan CA, Marshall AS, Gilbert RJ, Siebold C, Gordon S, Brown GD, and Jones EY (2007). Structure of the fungal beta-glucan-binding immune receptor dectin-1: implications for function. Protein Sci. 16, 1042–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret F, Ball G, Douzi B, and Voulhoux R (2014). Txc, a new type II secretion system of Pseudomonas aeruginosa strain PA7, is regulated by the TtsS/TtsR two-component system and directs specific secretion of the CbpE chitin-binding protein. J. Bacteriol 196, 2376–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu D, Greve LC, Labavitch JM, and Powell AL (2009). Characterization of the cell wall of the ubiquitous plant pathogen Botrytis cinerea. Mycol. Res 113, 1396–1403. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, and Stacey G (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier A, Lehrer SB, Horth-Susin L, Swanson M, Neis B, Howse D, and Jong M (2004). Prevalence of crab asthma in crab plant workers in Newfoundland and Labrador. Int. J. Circumpolar Health 63 (Suppl 2), 333–336. [DOI] [PubMed] [Google Scholar]
- Chai LYA, Vonk AG, Kullberg BJ, Verweij PE, Verschueren I, van der Meer JWM, Joosten LAB, Latgé J-P, and Netea MG (2011). Aspergillus fumigatus cell wall components differentially modulate host TLR2 and TLR4 responses. Microbes Infect. 13, 151–159. [DOI] [PubMed] [Google Scholar]
- Da Silva CA, Hartl D, Liu W, Lee CG, and Elias JA (2008). TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. J. Immunol 181, 4279–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey LK, Moeller JB, Schlosser A, Sorensen GL, and Holmskov U (2014). Induction of innate immunity by Aspergillus fumigatus cell wall polysaccharides is enhanced by the composite presentation of chitin and beta-glucan. Immunobiology 219, 179–188. [DOI] [PubMed] [Google Scholar]
- Fontaine T, Simenel C, Dubreucq G, Adam O, Delepierre M, Lemoine J, Vorgias CE, Diaquin M, and Latgé JP (2000). Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem 275, 27594–27607. [DOI] [PubMed] [Google Scholar]
- Fuchs K, Cardona Gloria Y, Wolz OO, Herster F, Sharma L, Dillen CA, Täumer C, Dickhöfer S, Bittner Z, Dang TM, et al. (2018). The fungal ligand chitin directly binds TLR2 and triggers inflammation dependent on oligomer size. EMBO Rep. 19, e46065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner BN, Simmons RM, and Underhill DM (2005). Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 24, 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gour N, Lajoie S, Smole U, White M, Hu D, Goddard P, Huntsman S, Eng C, Mak A, Oh S, et al. (2018). Dysregulated invertebrate tropomyosin-dectin-1 interaction confers susceptibility to allergic diseases. Sci. Immunol 3, eaam9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR, Latge JP, and Munro CA (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L-P, and Beaudoin N (2010). Chitooligosaccharide sensing and downstream signaling: contrasted outcomes in pathogenic and beneficial plant-microbe interactions. Planta 232, 787–806. [DOI] [PubMed] [Google Scholar]
- Heyl KA, Klassert TE, Heinrich A, Müller MM, Klaile E, Dienemann H, Grünewald C, Bals R, Singer BB, and Slevogt H (2014). Dectin-1 is expressed in human lung and mediates the proinflammatory immune response to nontypeable Haemophilus influenzae. MBio 5, e01492–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole CR, Lam WC, Upadhya R, and Lodge JK (2020). Cryptococcus neoformans chitin synthase 3 plays a critical role in dampening host inflammatory responses. MBio 11, e03373–e03319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa E, Mitsutomi M, and Nagano Y (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem 285, 2996–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi AR, and Erle DJ (2016). Chitin-induced airway epithelial cell innate immune responses are inhibited by carvacrol/thymol. PLoS ONE 11, e0159459–e0159459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LK, Morita R, Kobayashi Y, Eisenbarth SC, Lee CG, Elias J, Eynon EE, and Flavell RA (2015). AMCase is a crucial regulator of type 2 immune responses to inhaled house dust mites. Proc. Natl. Acad. Sci. USA 112, E2891–E2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaker A, Nkrumah L, Lee WK, Ramanathan M, and Lane AP (2009). Chitin stimulates expression of acidic mammalian chitinase and eotaxin-3 by human sinonasal epithelial cells in vitro. Am. J. Rhinol. Allergy 23, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé JP, Kobayashi H, Debeaupuis JP, Diaquin M, Sarfati J, Wieruszeski JM, Parra E, Bouchara JP, and Fournet B (1994). Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun 62, 5424–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG (2009). Chitin, chitinases and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med. J 50, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Yuk JM, Shin DM, and Jo EK (2009). Dectin-1 is inducible and plays an essential role for mycobacteria-induced innate immune responses in airway epithelial cells. J. Clin. Immunol 29, 795–805. [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Marakalala MJ, Vautier S, Potrykus J, Walker LA, Shepardson KM, Hopke A, Mora-Montes HM, Kerrigan A, Netea MG, Murray GI, et al. (2013). Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 9, e1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AK, Muehmer M, Mages J, Gueinzius K, Hess C, Heeg K, Bals R, Lang R, and Dalpke AH (2007). Differential recognition of TLR-dependent microbial ligands in human bronchial epithelial cells. J. Immunol 178, 3134–3142. [DOI] [PubMed] [Google Scholar]
- Mélida H, Sopeña-Torres S, Bacete L, Garrido-Arandia M, Jordá L, López G, Muñoz-Barrios A, Pacios LF, and Molina A (2018). Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 93, 34–49. [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, and Shibuya N (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes HM, Netea MG, Ferwerda G, Lenardon MD, Brown GD, Mistry AR, Kullberg BJ, O’Callaghan CA, Sheth CC, Odds FC, et al. (2011). Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun 79, 1961–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Liang HE, Tager AM, Luster AD, Van Rooijen N, Voehringer D, and Locksley RM (2007). Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas M, Liddiard K, Kimberg M, Faro-Trindade I, McDonald JU, Williams DL, Brown GD, and Taylor PR (2008). The induction of inflammation by dectin-1 in vivo is dependent on myeloid cell programming and the progression of phagocytosis. J. Immunol 181, 3549–3557. [DOI] [PubMed] [Google Scholar]
- Roy RM, and Klein BS (2013). Fungal glycan interactions with epithelial cells in allergic airway disease. Curr. Opin. Microbiol 16, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenuk T, Krist P, Pavlícek J, Bezouska K, Kuzma M, Novák P, and Kren V (2001). Synthesis of chitooligomer-based glycoconjugates and their binding to the rat natural killer cell activation receptor NKR-P1. Glycoconj. J 18, 817–826. [DOI] [PubMed] [Google Scholar]
- Serebrenik YV, Hellerschmied D, Toure M, López-Giráldez F, Brookner D, and Crews CM (2018). Targeted protein unfolding uncovers a Golgi-specific transcriptional stress response. Mol. Biol. Cell 29, 1284–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, and Brown GD (2007). Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol 8, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M, Anderson EL, Squillace DL, Patil N, Maniak PJ, Iijima K, Kita H, and O’Grady SM (2017). Oxidative stress serves as a key checkpoint for IL-33 release by airway epithelium. Allergy 72, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umetsu DT, McIntire JJ, Akbari O, Macaubas C, and DeKruyff RH (2002). Asthma: an epidemic of dysregulated immunity. Nat. Immunol 3, 715–720. [DOI] [PubMed] [Google Scholar]
- Van Dyken SJ, Garcia D, Porter P, Huang X, Quinlan PJ, Blanc PD, Corry DB, and Locksley RM (2011). Fungal chitin from asthma-associated home environments induces eosinophilic lung infiltration. J. Immunol 187, 2261–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF, McKenzie AN, Krummel MF, Liang HE, and Locksley RM (2014). Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity 40, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Liang HE, Naikawadi RP, Woodruff PG, Wolters PJ, Erle DJ, and Locksley RM (2017). Spontaneous chitin accumulation in airways and age-related fibrotic lung disease. Cell 169, 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera J, Fenutría R, Cañadas O, Figueras M, Mota R, Sarrias M-R, Williams DL, Casals C, Yelamos J, and Lozano F (2009). The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc. Natl. Acad. Sci. USA 106, 1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GRR, Dijkstra BW, and Kok J (2012). A genetically engineered protein domain binding to bacterial murein, archaeal pseudomurein, and fungal chitin cell wall material. Appl. Microbiol. Biotechnol 96, 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener J, Malireddi RKS, Lenardon MD, Köberle M, Vautier S, MacCallum DM, Biedermann T, Schaller M, Netea MG, Kanneganti T-D, et al. (2014). Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 10, e1004050–e1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, and Stacey G (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bai C, Li K, Adler KB, and Wang X (2008). Role of airway epithelial cells in development of asthma and allergic rhinitis. Respir. Med 102, 949–955. [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman M-A, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono J-J, Cullimore JV, et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 108, 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JEMM, Alsarraf HMAB, Kaspersen JD, Pedersen JS, Stougaard J, Thirup S, and Blaise M (2014). Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J. 281, 1196–1208. [DOI] [PubMed] [Google Scholar]
- Yeh CC, Horng HC, Chou H, Tai HY, Shen HD, Hsieh SL, and Wang PH (2017). Dectin-1-mediated pathway contributes to Fusarium proliferatum-induced CXCL-8 release from human respiratory epithelial cells. Int. J. Mol. Sci 18, 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama CC, Baldridge MT, Leung DW, Zhao G, Desai C, Liu TC, Diaz-Ochoa VE, Huynh JP, Kimmey JM, Sennott EL, et al. (2018). LysMD3 is a type II membrane protein without an in vivo role in the response to a range of pathogens. J. Biol. Chem 293, 6022–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, and Casadevall A (2009). The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol 68, 133–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziatabar S, Zepf J, Rich S, Danielson BT, Bollyky PI, and Stern R (2018). Chitin, chitinases, and chitin lectins: Emerging roles in human pathophysiology. Pathophysiology 25, 253–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.