Abstract
Background
With reduced community mobility, household infections may become increasingly important in SARS-CoV-2 transmission dynamics.
Methods
We investigate the intra-household transmission of COVID-19 through the secondary-attack rate (SAR) and household reproduction number (Rh). We estimate these using (i) data from 29 prior studies (February–August 2020), (ii) epidemiologically linked confirmed cases from Singapore (January–April 2020) and (iii) widespread-testing data from Vo’ (February–March 2020). For (i), we use a Bayesian random-effects model that corrects for reverse transcription–polymerase chain reaction (RT–PCR) test sensitivity and asymptomatic cases. We investigate the robustness of Rh with respect to community transmission rates and mobility patterns.
Results
The corrected pooled estimates from prior studies for SAR and Rh are 24% (20–28%) and 0.34 (0.30–0.38), respectively. Without corrections, the pooled estimates are: SAR = 18% (14–21%) and Rh = 0.28 (0.25–0.32). The corrected estimates line up with direct estimates from contact-tracing data from Singapore [Rh = 0.32 (0.22–0.42)] and population testing data from Vo’ [SAR = 31% (28–34%) and Rh = 0.37 (0.34–0.40)]. The analysis of Singapore data further suggests that the value of Rh (0.22–0.42) is robust to community-spread dynamics; our estimate of Rh stays constant whereas the fraction of infections attributable to household transmission (Rh/Reff) is lowest during outbreaks (5–7%) and highest during lockdowns and periods of low community spread (25–30%).
Conclusions
The three data-source types yield broadly consistent estimates for SAR and Rh. Our study suggests that household infections are responsible for a large fraction of infections and so household transmission may be an effective target for intervention.
Keywords: Household transmission, secondary-attack rate, coronavirus
Key Messages
With reduced mobility, household infections are becoming increasingly important in the transmission of SARS-CoV-2.
We study two separate quantities that track household transmission: household secondary-attack rate and intra-household reproduction number.
We pool estimates for both quantities from household-infection studies and add (upwards) corrections for low test sensitivity and lack of asymptomatic testing.
These corrected estimates line up with central estimates from two disjointed data sources: blanket-testing data from Italy and contact-tracing data from Singapore.
Our analysis suggests that interventions targeted towards reducing household transmission (e.g. early isolation, encouraging household hygiene, etc.) may be effective at reducing the spread of the virus.
Background
Social distancing and lockdowns reduce community transmission of SARS-CoV-2 but do not directly address household transmission. When social restrictions are in peak use, household infections become increasingly important in percentage terms of virus transmission. Prior studies1–13 find that household members have a higher risk of infection compared with other contacts, with spouses being most likely and children being least likely to get infected. Existing meta-estimates of the secondary-attack rate (SAR) for COVID-19 fall in the 15–19% range.14 In comparison, estimates of SAR for the MERS-CoV and SARS-CoV-1 epidemics were 3–3.5× and 2–2.5× smaller, respectively.14 In addition, estimates of SAR for COVID-19 do not account for selective testing of (symptomatic) household members and test sensitivity, and can thus underestimate viral spread.
Methods
The dynamics of disease spread are described via the effective reproduction number R, which measures the average number of new infections caused by each infected person i. To quantify household transmission, we decompose R into two components: R = Rc + Rh. The community (respectively, intra-household) reproduction number Rc (respectively, Rh) is the average number of infections caused by an infected individual outside (respectively, inside) their household. The ratio Rh/R measures the fraction of transmission occurring within households. In addition to Rh, we consider household SAR: the probability that an infected person i infects a specific household member j. SAR measures the prevalence of infection among susceptible individuals, whereas Rh measures the growth of infection within households.
Estimation methodology
Estimating these metrics requires household-level data. Specifically, we need:
Test results (positive and negative) for all household members.
Properly attributed household-infection data, i.e. identifying primary cases and constructing household-transmission chains.
End-of-study outcomes—if there are still active cases at termination, the data may undercount the number of infections attributed to the case.
Assuming these are available, SAR can be estimated as the ratio of secondary household cases to susceptible household members, and Rh can be estimated as the ratio of secondary household infections to total household cases (unlike household SAR, Rh does not require negative case counts).
Challenges in estimation
In practice, estimating Rh and SAR is made difficult by (i) asymptomatic infections (asymptomatic cases may constitute 18–43% of all infections,15–19 yet studies predominantly test symptomatic individuals) and (ii) the low sensitivity of standard tests [reverse transcription–polymerase chain reaction (RT–PCR) tests have near-perfect specificity,20 but low and time-varying sensitivity—average sensitivity in the 10 days following symptom onset is estimated as 83% by21 and 70% by22]. Sensitivity also varies between different swab types21 and testing facilities.22
Many recent studies of household transmission do not adjust estimates of SAR for low test sensitivity and asymptomatic cases. To address this, we adjust all our estimates based on literature-inferred priors for sensitivity and asymptomatic rates.
Data sources and estimation procedure
We propose two approaches for estimating Rh and SAR while accounting for asymptomatic cases and test sensitivity: (i) adjusting and aggregating estimates from prior work and (ii) constructing (corrected) direct estimates from COVID case data. For the former, we use a random-effects model to pool estimates from previous studies, correcting for sensitivity and asymptomatic cases. For the latter, we estimate Rh and SAR using a blanket-testing data set from Vo’, Italy,15 and estimate Rh using a data set of epidemiologically linked cases based on scraping publicly available contact-tracing data from Singapore.23,24 As estimates from these distinct data sources (previous studies, blanket testing and contact tracing) rely on different assumptions, general agreement between them supports the robustness of our conclusions.
Estimates from previous studies
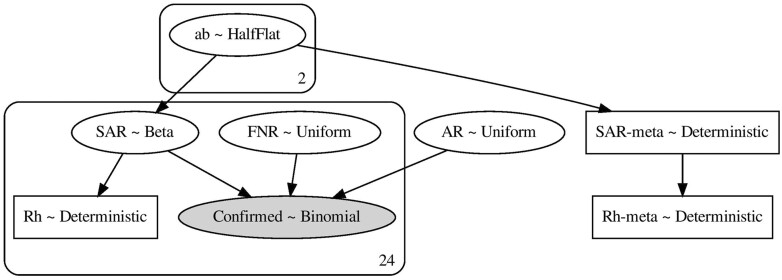
We found 29 household-transmission studies1–9,11–13,25–41 satisfying our selection criteria (see Supplementary Figure S1, available as Supplementary data at IJE online, for PRISMA42 diagram). Of these studies, nine tested all household contacts multiple times over the observation period. The remaining 21 performed single testing and require adjustments to correct for the undercounting of secondary cases due to RT–PCR false negatives. Out of them, eight tested only symptomatic contacts and thus require corrections that account for the selective testing. We use a Bayesian random-effects model with a per-study false-negative rate (FNR, equal to 1-sensitivity) and a global asymptomatic rate (AR): both are sampled from study-informed weak priors. We model the probability of the ith study with Ii index cases and Ni household contacts observing Pi positive tests as Binomial[n = Ni, p = SARi · (1 – FNRi) · (1 – AR)]. By fitting the random-effects model, we obtain a posterior distribution for SARi conditioned on the data. We compute corrected counts of secondary cases as the product of and Ni, where is a random sample from the posterior probability of SAR from the ith study. Using these corrected counts, we can estimate the corrected household reproduction number for the ith study, (Rh)i, as See Figure 1 for a graphical representation of the model (a textual description is given in the Supplementary Material, available as Supplementary data at IJE online).
Figure 1.
A schematic of the Bayesian graphical model for computing the pooled estimates of SAR and Rh. Inference was performed via MCMC sampling using PyMC 3.4 with the built-in NUTS sampler. The sampler used eight chains with 2000 iterations each. The burn-in period is 2000. MCMC, Markov Chain Monte Carlo; NUTS, No U-Turn Sampler; PyMC, Python package for Bayesian statistical modelling.
Assumptions/limitations
The model assumes that all infections among household contacts are attributed to the index case. This assumption might inflate the SAR estimate (it treats tertiary transmissions as secondary) but is inevitable, since studies do not distinguish between secondary and tertiary cases. Another limitation is that most studies do not stratify based on household sizes and thus the model treats infection probability as independent of household size.
Direct estimate from contact-tracing data
We scraped a dashboard24 of Singapore’s contact-tracing data23 and extracted metadata for each positive case, including an undirected 6588-patient graph providing information about epidemiologically linked cases. Confirmation dates for cases ranged from 23 January to 19 April 2020.
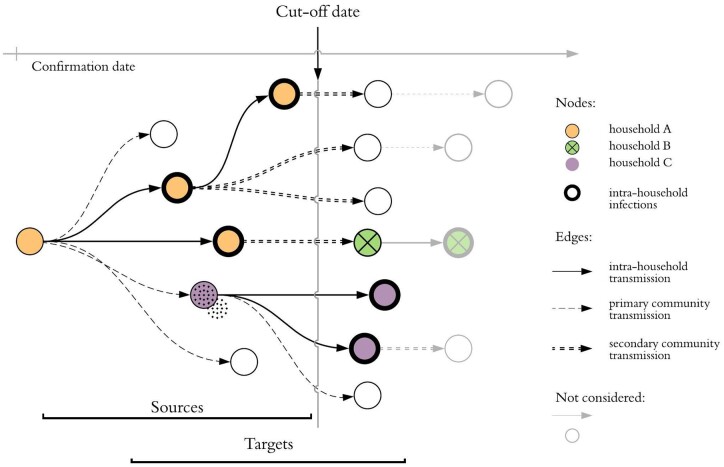
We consider two cases epidemiologically linked if there is a direct edge between them (Case i—Case j) or if they are connected via a cluster (Case i—Cluster A—Case j). We then label each case as a ‘source’ case, a ‘target’ case or both. A source case is any case with a confirmation date before a given cut-off date t. For each source, the corresponding target cases are the epidemiologically linked cases that have a confirmation date between 0 and 14 days after that of the source. Target cases with confirmation date ≤t (i.e. before the cut-off date) are thus labelled as both source and target. A schematic of the labelling procedure is shown in Figure 2.
Figure 2.
Schematic infection graph: nodes represent positive cases and their horizontal position indicates confirmation date. We consider the subset of the graph containing cases with confirmation date prior to the cut-off date: target nodes are infected by sources. R can be estimated as R = (number of targets)/(number of sources) and hence R = 13/7 for this cluster. There are three distinct households in the cluster and thick borders denote secondary household infections. The household reproductive number is Rh = 5/7. R, reproduction number; Rh, intra-household reproduction number.
We selected a cut-off date t of 27 March 2020 based on two criteria: (i) t should pre-date the large worker dormitory outbreaks in Singapore, which made accurate contact tracing difficult;43 and (ii) t should be more than 2 weeks prior to the last confirmation date in the data set, to avoid undercounting target cases due to end-of-study truncation bias. The resulting subset of the infection graph contains 710 cases: 417 sources and 599 targets (306 nodes are labelled as both source and target).
The average effective reproduction rate Reff can be estimated as the ratio of target cases to source cases. When there is a direct edge between two cases, the edge annotation reflects the relationship between them. Rh can be thus estimated as the ratio of the total number of household targets to the total number of source cases. Since there is no annotation for ‘household’, we use the annotation ‘family member’ as a proxy. This assumption may inflate or deflate Rh: we do not observe partners and roommates, but we do observe family members not residing in the same household. We obtain upper and lower reproduction number central estimates by varying how the untraced cases (singletons) are labelled.
Assumptions/limitations
Just as for the prior studies, the Singapore contact-tracing data cannot be used to distinguish between secondary and tertiary cases in a household, as infected household members are typically interconnected with an undirected edge in the graph. However, unlike the prior study data, the Singapore data set does not contain information about household sizes of confirmed cases, making the estimation of SAR impossible. Asymptomatic transmission is not a substantial issue for counting the number of household infections, as all household members of confirmed cases are typically tested. We do, however, correct for test sensitivity assuming an FNR of 20%.
Direct estimates from blanket-testing data. Most of the population in the town of Vo’, Italy, was tested both at the start of a lockdown and 14 days later.15 The two phases of testing covered 86% (2812 subjects) and 72% (2343 subjects) of the population in Vo’, respectively. After filtering for truncation bias (cf. Appendix), there are 53 primary cases, 23 secondary cases and 84 susceptible individuals.
The household SAR is again estimated as the ratio of secondary cases to total susceptible individuals, and Rh as the ratio of targets to sources (as was done for Singapore). We label the cases that are confirmed during the first round of testing (73 cases) as sources and the secondary cases attributed to one of the source cases (23 cases) as targets. The second testing date thus acts as a cut-off date for labelling cases as sources and targets.
We adjust our estimates for test sensitivity by using point estimates of sensitivity and AR derived from the data. Asymptomatic cases were 41% of the confirmed cases and, out of subjects residing in households with at least one confirmed case, four have experienced symptoms but have tested negative. This yields 5.7 [4 × (1 + 0.41)] expected false negatives, which corresponds to a test sensitivity of 78%.
Results
Estimates from previous studies
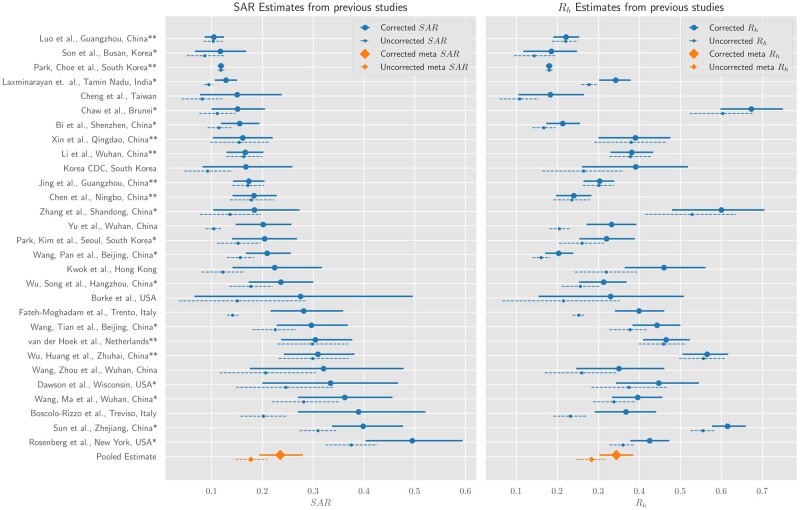
Fitting a random-effects model to the data from the 29 studies yields a pooled corrected SAR estimate of 24% (95% confidence interval: 20–28%) and an Rh estimate of 0.34 (0.3–0.38). Without corrections for asymptomatic cases and test sensitivity, the pooled estimates for SAR and Rh are 18% (14–21%) and 0.28 (0.25–0.32). Study-level SAR and Rh are shown in Figure 3 and Table 1, and precise counts and estimates in Table 2. We find substantial heterogeneity in the SAR and Rh estimates across studies. Moreover, the relative orderings of studies by SAR and Rh are considerably different. To gain further insight, we stratify the studies based on location (see Supplementary Figures S2 and S3, available as Supplementary data at IJE online), average household size (see Supplementary Figure S4, available as Supplementary data at IJE online) and stringency of the mitigation strategy (see Supplementary Figure S5, available as Supplementary data at IJE online). Both metrics are smaller in East Asian countries [22 studies, SAR: 21% (17–25%), Rh: 0.30 (0.26–0.34)] compared with other countries [7 studies: SAR: 33% (20–46%), Rh: 0.48 (0.38–0.57)] (see Table 1). More stringent preventative measures correspond to decreases in both metrics: for 13 studies that placed contacts in quarantine, SAR and Rh were 21% (16–28%) and 0.30 (0.24–0.36) compared with 26% (20–32%) and 0.43 (0.37–0.49) for 16 studies that did not (see Table 1). A subgroup analysis based on average household size partially explains why the relative ordering of studies varies when ranking by SAR and by Rh: the 10 studies with small average household sizes (<2.9 members) had higher SAR: 29% (19–40%), but lower Rh: 0.34 (0.25–0.41) than the 9 studies with large average household size (>4) where SAR: 22% (15–30%) and Rh: 0.48 (0.40–0.57) (see Table 1). Other potential causes contributing to heterogeneity include both intrinsic demographic variation (e.g. age distributions) and extrinsic factors (e.g. ventilation and hygiene). Graphical illustrations of all subgroup analyses are given in the Supplementary data at IJE online.
Figure 3.
Adjusted literature estimates for SAR and Rh. Dashed lines show estimates from the original studies. Solid lines show 95% credible intervals from a Bayesian hierarchical model, which adjusts estimates for false negatives and asymptomatics where appropriate. The meta-estimates refer to the pooled estimate for SAR and Rh. In the left plot, the meta-estimate (orange) are the model’s credible intervals for the pooled SAR. If a study has a single asterisk, this means it was unnecessary to adjust for asymptomatics (only false negatives). The double asterisk means no adjustment was necessary. Rh, intra-household reproduction number; SAR, household secondary-attack rate.
Table 1.
Literature estimates: this table contains study-level data for the number of index cases, secondary cases and total household contacts
| Name | Index cases | Secondary infections | Household contacts | Average household | SAR (corrected) | SAR (uncorrected) | Rh (corrected) | Rh (uncorrected) | Correction | Contacts quarantined |
|---|---|---|---|---|---|---|---|---|---|---|
| Bi et al., Shenzhen, China* | 391 | 77 | 686 | 2.75 | 16% (12–20%) | 11% (9–14%) | 0.21 (0.17––0.26) | 0.17 (0.14–0.20) | FNR | False |
| Boscolo-Rizzo et al., Treviso, Italy | 179 | 54 | 269 | 2.5 | 36% (25–48%) | 20% (15–24%) | 0.35 (0.28–0.42) | 0.23 (0.19–0.27) | AR/FNR | True |
| Burke et al., USA | 10 | 2 | 19 | 2.9 | 22% (7–40%) | 14% (4–25%) | 0.29 (0.14–0.44) | 0.21 (0.09–0.33) | AR/FNR | True |
| Chaw et al., Brunei* | 19 | 28 | 264 | 14.89 | 15% (10–21%) | 11% (8–15%) | 0.67 (0.59–0.75) | 0.60 (0.52–0.68) | FNR | True |
| Chen et al., Ningbo, China** | 157 | 49 | 272 | 2.73 | 18% (14–23%) | 18% (14–22%) | 0.24 (0.20–0.28) | 0.24 (0.19–0.28) | – | False |
| Cheng et al., Taiwan | 100 | 10 | 151 | 2.51 | 15% (8–23%) | 8% (4–12%) | 0.18 (0.10–0.26) | 0.11 (0.06–0.15) | AR/FNR | True |
| Dawson et al., Wisconsin, USA* | 26 | 16 | 64 | 3.46 | 31% (19–43%) | 23% (15–32%) | 0.43 (0.33–0.52) | 0.36 (0.27–0.45) | FNR | False |
| Fateh-Moghadam et al., Trento, Italy | 1489 | 500 | 3546 | 3.38 | 27% (21–34%) | 14% (13–15%) | 0.39 (0.33–0.45) | 0.25 (0.24–0.27) | AR/FNR | True |
| Jing et al., Guangzhou, China** | 215 | 93 | 542 | 3.52 | 17% (14–20%) | 17% (14–20%) | 0.30 (0.27–0.34) | 0.30 (0.26–0.34) | – | False |
| Korea CDC, South Korea | 30 | 9 | 119 | 4.96 | 17% (8–26%) | 9% (5–14%) | 0.39 (0.27–0.53) | 0.26 (0.16–0.36) | AR/FNR | False |
| Kwok et al., Hong Kong | 53 | 24 | 206 | 4.88 | 23% (14–32%) | 12% (8–16%) | 0.47 (0.36–0.56) | 0.32 (0.25–0.40) | AR/FNR | True |
| Laxminarayan et. al., Tamin Nadu, India* | 997 | 380 | 4066 | 5.07 | 13% (11–15%) | 9% (8–10%) | 0.34 (0.30–0.38) | 0.28 (0.26–0.29) | FNR | False |
| Li et al., Wuhan, China** | 105 | 64 | 392 | 4.73 | 17% (13–20%) | 16% (13–20%) | 0.38 (0.33–0.43) | 0.38 (0.33–0.43) | – | True |
| Luo et al., Guangzhou, China** | 347 | 96 | 946 | 3.72 | 10% (9–12%) | 10% (8–12%) | 0.22 (0.19–0.25) | 0.22 (0.19–0.25) | – | True |
| Park, Choe et al., South Korea** | 5706 | 1250 | 10 592 | 2.85 | 12% (11–12%) | 12% (11–12%) | 0.18 (0.17–0.19) | 0.18 (0.17–0.19) | – | True |
| Park, Kim et al., Seoul, South Korea* | 97 | 34 | 225 | 3.31 | 21% (14–27%) | 15% (11–20%) | 0.32 (0.25–0.39) | 0.26 (0.21–0.32) | FNR | True |
| Rosenberg et al., New York, USA* | 229 | 131 | 343 | 2.49 | 47% (39–56%) | 37% (32–42%) | 0.41 (0.37–0.46) | 0.36 (0.33–0.39) | FNR | False |
| Son et al., Busan, Korea* | 108 | 16 | 212 | 2.96 | 12% (7–17%) | 8% (5–12%) | 0.18 (0.12–0.25) | 0.14 (0.09–0.20) | FNR | False |
| Sun et al., Zhejiang, China* | 148 | 189 | 598 | 5.04 | 41% (34–48%) | 31% (28–35%) | 0.62 (0.58–0.66) | 0.56 (0.53–0.58) | FNR | False |
| Wang, Ma et al., Wuhan, China* | 85 | 47 | 155 | 2.82 | 38% (28–48%) | 29% (22–36%) | 0.41 (0.34–0.47) | 0.34 (0.29–0.40) | FNR | True |
| Wang, Pan et al., Beijing, China* | 585 | 111 | 714 | 2.22 | 21% (17–26%) | 16% (13–18%) | 0.20 (0.17–0.24) | 0.16 (0.14–0.18) | FNR | True |
| Wang, Tian et al., Beijing, China* | 124 | 77 | 335 | 3.7 | 30% (23–38%) | 23% (18–27%) | 0.45 (0.39–0.51) | 0.38 (0.33–0.42) | FNR | False |
| Wang, Zhou et al., Wuhan, China | 25 | 10 | 43 | 2.72 | 35% (18–52%) | 21% (12–31%) | 0.37 (0.25–0.48) | 0.27 (0.17–0.36) | AR/FNR | False |
| Wu, Huang et al., Zhuhai, China** | 35 | 48 | 148 | 5.22 | 32% (24–38%) | 30% (24–38%) | 0.57 (0.51–0.62) | 0.56 (0.50–0.61) | – | False |
| Wu, Song et al., Hangzhou, China* | 144 | 50 | 280 | 2.94 | 24% (17–31%) | 18% (14–22%) | 0.32 (0.26–0.38) | 0.26 (0.21–0.30) | FNR | False |
| Xin et al., Qingdao, China** | 31 | 19 | 125 | 5.03 | 16% (10–23%) | 16% (10–21%) | 0.39 (0.30–0.48) | 0.38 (0.29–0.47) | – | False |
| Yu et al., Wuhan, China | 560 | 143 | 1396 | 3.49 | 20% (14–26%) | 10% (9–12%) | 0.33 (0.27–0.39) | 0.20 (0.18–0.23) | AR/FNR | True |
| Zhang et al., Shandong, China* | 11 | 12 | 93 | 9.45 | 19% (10–28%) | 14% (7–20%) | 0.60 (0.48–0.71) | 0.53 (0.42–0.64) | FNR | False |
| van der Hoek et al., Netherlands** | 54 | 47 | 155 | 3.87 | 30% (23–37%) | 29% (22–35%) | 0.46 (0.40–0.52) | 0.45 (0.39–0.50) | – | False |
| Global meta-estimate | 12060 | 3586 | 26 956 | 3.23 | 24% (20–28%) | 18% (14–21%) | 0.34 (0.30–0.38) | 0.28 (0.25–0.32) | ||
| China | 2963 | 1085 | 6725 | 3.27 | 24% (19–30%) | 19% (15–24%) | 0.35 (0.30–0.41) | 0.30 (0.26–0.35) | ||
| Not China | 9097 | 2501 | 20 231 | 3.22 | 24% (17–32%) | 17% (12–22%) | 0.35 (0.28–0.42) | 0.27 (0.21–0.33) | ||
| East Asia | 9076 | 2456 | 18 494 | 3.04 | 21% (17–26%) | 16% (13–20%) | 0.30 (0.26–0.34) | 0.25 (0.21–0.29) | ||
| Not East Asia | 2984 | 1130 | 8462 | 3.83 | 33% (21–46%) | 23% (14–34%) | 0.48 (0.39–0.57) | 0.39 (0.29–0.50) | ||
| Small household | 7467 | 1741 | 13 244 | 2.77 | 29% (19–40%) | 20% (13–27%) | 0.34 (0.25–0.41) | 0.26 (0.20–0.33) | ||
| Medium households | 3164 | 1072 | 7701 | 3.43 | 23% (16–30%) | 18% (12–23%) | 0.35 (0.29–0.42) | 0.30 (0.24–0.36) | ||
| Large households | 1429 | 773 | 6011 | 5.21 | 22% (15–30%) | 18% (12–25%) | 0.48 (0.40–0.57) | 0.42 (0.33–0.52) | ||
| Contacts quarantined | 9335 | 2363 | 18 875 | 3.02 | 21% (16–28%) | 14% (11–18%) | 0.30 (0.24–0.36) | 0.23 (0.19–0.27) | ||
| Contacts not quarantined | 2725 | 1223 | 8081 | 3.97 | 26% (20–32%) | 21% (15–26%) | 0.43 (0.37–0.49) | 0.38 (0.32–0.44) |
It includes estimates and 95% confidence intervals of study-level SAR and Rh (with and without AR/FNR adjustments). The weight columns contain the contribution of each study towards the meta-estimate. A high SAR value does not always imply a high Rh value, or vice versa. SAR measures the prevalence of infection among susceptible individuals, whereas Rh measures the growth of infection within households. The relationship between the two measures is described by the equation: SAR = Rh × (#total infected)/(#susceptible). Assuming a fixed ratio of primary to secondary infections, SAR is inversely proportional to the relative number of susceptible individuals. Studies that have larger numbers of susceptible members (larger average household size) tend to have smaller SAR values. Conversely, studies that have smaller household sizes tend to have larger SAR values.
Table 2.
Estimates for effective reproductive number Reff and household reproductive number Rh, computed for different values of cut-off date: for each cut-off date, we count the number of cases assigned as sources and targets
| Cut-off date | Sources | Targets | Household targets | Untraced | Reff | R h | Ratio |
|---|---|---|---|---|---|---|---|
| Jan-26 | 4 | 16 | 3 | 0 | 4.00 [4.00–4.00] | 0.94 (0.64–1.23) | 0.23 |
| Jan-27 | 7 | 16 | 3 | 0 | 2.29 [2.29–2.29] | 0.54 (0.34–0.73) | 0.23 |
| Jan-28 | 8 | 22 | 4 | 0 | 2.75 [2.75–2.75] | 0.62 (0.43–0.82) | 0.23 |
| Jan-29 | 12 | 22 | 4 | 0 | 1.83 [1.83–1.83] | 0.42 (0.26–0.57) | 0.23 |
| Jan-30 | 16 | 27 | 4 | 0 | 1.69 [1.69–1.69] | 0.31 (0.18–0.45) | 0.19 |
| Jan-31 | 17 | 27 | 4 | 0 | 1.59 [1.59–1.59] | 0.29 (0.17–0.42) | 0.19 |
| Feb-1 | 18 | 27 | 4 | 0 | 1.50 [1.50–1.50] | 0.28 (0.16–0.40) | 0.19 |
| Feb-2 | 18 | 27 | 4 | 0 | 1.50 [1.50–1.50] | 0.28 (0.16–0.40) | 0.19 |
| Feb-3 | 22 | 35 | 7 | 0 | 1.59 [1.59–1.59] | 0.40 (0.26–0.54) | 0.25 |
| Feb-4 | 26 | 35 | 7 | 0 | 1.35 [1.35–1.35] | 0.34 (0.21–0.46) | 0.25 |
| Feb-5 | 28 | 35 | 7 | 1 | 1.25 [1.21–1.29] | 0.31 (0.17–0.46) | 0.25 |
| Feb-6 | 31 | 37 | 7 | 2 | 1.19 [1.12–1.26] | 0.28 (0.15–0.42) | 0.24 |
| Feb-7 | 33 | 37 | 7 | 4 | 1.12 [1.00–1.24] | 0.27 (0.14–0.39) | 0.24 |
| Feb-8 | 38 | 42 | 8 | 5 | 1.11 [0.98–1.24] | 0.26 (0.15–0.38) | 0.24 |
| Feb-9 | 39 | 42 | 8 | 5 | 1.08 [0.95–1.21] | 0.26 (0.14–0.37) | 0.24 |
| Feb-10 | 41 | 42 | 8 | 6 | 1.02 [0.89–1.17] | 0.24 (0.13–0.35) | 0.24 |
| Feb-11 | 43 | 64 | 11 | 6 | 1.49 [1.31–1.63] | 0.32 (0.21–0.43) | 0.21 |
| Feb-12 | 47 | 67 | 13 | 6 | 1.43 [1.26–1.55] | 0.35 (0.24–0.45) | 0.24 |
| Feb-13 | 56 | 68 | 13 | 6 | 1.21 [1.10–1.32] | 0.29 (0.19–0.39) | 0.24 |
| Feb-14 | 61 | 71 | 14 | 7 | 1.16 [1.04–1.28] | 0.29 (0.18–0.39) | 0.25 |
| Feb-15 | 67 | 72 | 15 | 7 | 1.07 [0.97–1.18] | 0.28 (0.18–0.38) | 0.26 |
| Feb-16 | 69 | 73 | 15 | 7 | 1.06 [0.96–1.16] | 0.27 (0.17–0.37) | 0.26 |
| Feb-17 | 72 | 73 | 15 | 7 | 1.01 [0.92–1.11] | 0.26 (0.17–0.36) | 0.26 |
| Feb-18 | 75 | 74 | 15 | 7 | 0.99 [0.90–1.08] | 0.25 (0.16–0.34) | 0.25 |
| Feb-19 | 77 | 74 | 15 | 7 | 0.96 [0.88–1.05] | 0.24 (0.15–0.34) | 0.25 |
| Feb-20 | 78 | 74 | 15 | 8 | 0.95 [0.86–1.05] | 0.24 (0.15–0.33) | 0.25 |
| Feb-21 | 80 | 74 | 15 | 8 | 0.93 [0.84–1.02] | 0.23 (0.14–0.32) | 0.25 |
| Feb-22 | 81 | 74 | 15 | 9 | 0.91 [0.82–1.02] | 0.23 (0.14–0.32) | 0.25 |
| Feb-23 | 82 | 74 | 15 | 9 | 0.90 [0.81–1.01] | 0.23 (0.14–0.32) | 0.25 |
| Feb-24 | 82 | 74 | 15 | 9 | 0.90 [0.81–1.01] | 0.23 (0.14–0.32) | 0.25 |
| Feb-25 | 82 | 74 | 15 | 9 | 0.90 [0.81–1.01] | 0.23 (0.14–0.32) | 0.25 |
| Feb-26 | 84 | 130 | 24 | 10 | 1.55 [1.38–1.67] | 0.36 (0.27–0.45) | 0.23 |
| Feb-27 | 87 | 131 | 24 | 10 | 1.51 [1.35–1.62] | 0.34 (0.26–0.43) | 0.23 |
| Feb-28 | 90 | 131 | 24 | 10 | 1.46 [1.31–1.57] | 0.33 (0.25–0.42) | 0.23 |
| Feb-29 | 92 | 132 | 24 | 10 | 1.43 [1.29–1.54] | 0.33 (0.24–0.41) | 0.23 |
| Mar-1 | 97 | 133 | 24 | 10 | 1.37 [1.24–1.47] | 0.31 (0.23–0.39) | 0.23 |
| Mar-2 | 100 | 134 | 24 | 10 | 1.34 [1.22–1.44] | 0.30 (0.22–0.38) | 0.22 |
| Mar-3 | 100 | 134 | 24 | 11 | 1.34 [1.21–1.45] | 0.30 (0.22–0.38) | 0.22 |
| Mar-4 | 102 | 134 | 24 | 12 | 1.31 [1.18–1.43] | 0.29 (0.22–0.37) | 0.22 |
| Mar-5 | 110 | 134 | 24 | 14 | 1.22 [1.08–1.35] | 0.27 (0.20–0.35) | 0.22 |
| Mar-6 | 121 | 136 | 25 | 17 | 1.12 [0.99–1.26] | 0.26 (0.19–0.33) | 0.23 |
| Mar-7 | 126 | 148 | 26 | 18 | 1.17 [1.03–1.32] | 0.26 (0.18–0.33) | 0.22 |
| Mar-8 | 133 | 149 | 27 | 20 | 1.12 [0.97–1.27] | 0.25 (0.18–0.32) | 0.23 |
| Mar-9 | 142 | 149 | 27 | 21 | 1.05 [0.91–1.20] | 0.24 (0.17–0.31) | 0.23 |
| Mar-10 | 148 | 155 | 29 | 22 | 1.05 [0.91–1.20] | 0.24 (0.18–0.31) | 0.23 |
| Mar-11 | 155 | 158 | 29 | 24 | 1.02 [0.88–1.17] | 0.23 (0.17–0.30) | 0.23 |
| Mar-12 | 163 | 167 | 31 | 26 | 1.02 [0.88–1.18] | 0.24 (0.18–0.30) | 0.23 |
| Mar-13 | 174 | 178 | 32 | 30 | 1.02 [0.87–1.20] | 0.23 (0.17–0.29) | 0.22 |
| Mar-14 | 181 | 183 | 34 | 38 | 1.01 [0.84–1.22] | 0.23 (0.18–0.29) | 0.23 |
| Mar-15 | 190 | 199 | 43 | 42 | 1.05 [0.86–1.27] | 0.28 (0.22–0.35) | 0.27 |
| Mar-16 | 201 | 203 | 44 | 46 | 1.01 [0.82–1.24] | 0.27 (0.21–0.33) | 0.27 |
| Mar-17 | 220 | 216 | 51 | 67 | 0.98 [0.75–1.29] | 0.29 (0.23–0.35) | 0.3 |
| Mar-18 | 237 | 222 | 53 | 87 | 0.94 [0.69–1.30] | 0.28 (0.23–0.33) | 0.3 |
| Mar-19 | 253 | 228 | 57 | 110 | 0.90 [0.63–1.34] | 0.28 (0.22–0.34) | 0.31 |
| Mar-20 | 269 | 242 | 62 | 140 | 0.90 [0.59–1.42] | 0.29 (0.23–0.34) | 0.32 |
| Mar-21 | 282 | 256 | 64 | 168 | 0.91 [0.57–1.50] | 0.28 (0.23–0.34) | 0.31 |
| Mar-22 | 294 | 265 | 67 | 209 | 0.90 [0.53–1.61] | 0.28 (0.23–0.34) | 0.32 |
| Mar-23 | 306 | 300 | 79 | 224 | 0.98 [0.57–1.71] | 0.32 (0.26–0.38) | 0.33 |
| Mar-24 | 338 | 337 | 86 | 265 | 1.00 [0.56–1.78] | 0.32 (0.23–0.41) | 0.32 |
| Mar-25 | 368 | 572 | 94 | 292 | 1.55 [0.87–2.35] | 0.32 (0.23–0.41) | 0.21 |
| Mar-26 | 395 | 585 | 102 | 319 | 1.48 [0.82–2.29] | 0.32 (0.22–0.42) | 0.22 |
| Mar-27 | 417 | 599 | 108 | 366 | 1.44 [0.77–2.31] | 0.32 (0.22–0.42) | 0.23 |
| Mar-28 | 444 | 2105 | 109 | 387 | 4.74 [2.53–5.61] | 0.31 (0.21–0.40) | 0.06 |
| Mar-29 | 465 | 2120 | 117 | 402 | 4.56 [2.45–5.42] | 0.31 (0.22–0.40) | 0.07 |
| Mar-30 | 489 | 2333 | 127 | 419 | 4.77 [2.57–5.63] | 0.32 (0.24–0.41) | 0.07 |
| Mar-31 | 520 | 2566 | 134 | 446 | 4.93 [2.66–5.79] | 0.32 (0.24–0.40) | 0.07 |
| Apr-1 | 565 | 2658 | 137 | 461 | 4.70 [2.59–5.52] | 0.30 (0.22–0.38) | 0.06 |
| Apr-2 | 608 | 3228 | 142 | 473 | 5.31 [2.99–6.09] | 0.29 (0.22–0.37) | 0.05 |
| Apr-3 | 669 | 3274 | 147 | 498 | 4.89 [2.81–5.64] | 0.27 (0.20–0.34) | 0.06 |
| Apr-4 | 688 | 3276 | 147 | 501 | 4.76 [2.76–5.49] | 0.27 (0.20–0.34) | 0.06 |
| Apr-5 | 783 | 3293 | 157 | 526 | 4.21 [2.52–4.88] | 0.25 (0.19–0.31) | 0.06 |
| Apr-6 | 839 | 3298 | 159 | 537 | 3.93 [2.40–4.57] | 0.24 (0.18–0.29) | 0.06 |
| Apr-7 | 975 | 3835 | 163 | 595 | 3.93 [2.44–4.54] | 0.21 (0.16–0.26) | 0.05 |
Among the targets, we identify cases that are linked to infected household members. The ‘Untraced’ column contains the number of cases in the data with a confirmation date prior to the cut-off date that are not linked to any other cases. In the case of Reff, the square brackets are not confidence intervals, but rather upper and lower bounds on Reff, depending on how the untraced cases are labelled (labelling as sources yields a lower bound and labelling as targets yields an upper bound on the central estimate for Reff).
Estimates from Singapore contact-tracing data
Our estimate for the reproduction number R (see ‘Methods’) is (number of target nodes)/(number of source nodes) = 599/417 = 1.44 (0.77–2.31). There are 108 household infections in the graph, yielding an uncorrected estimate of Rh and binomial confidence intervals of 108/417 = 0.26 (0.23–0.29). After correcting for test sensitivities, the resulting corrected estimate for Rh is 0.36 (0.24–0.48).
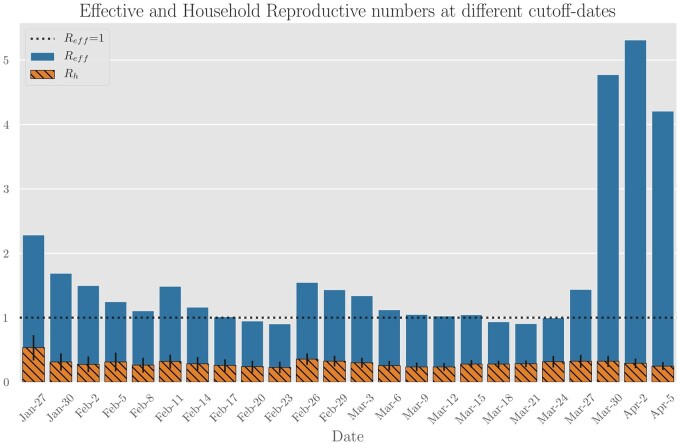
We repeated the calculations for various values of cut-off date t to ensure that our estimates are robust to the choice of t. Figure 4 contains estimates for cut-off dates other than 27 March; estimates for Reff range between 0.90 and 4.93, and estimates of corrected Rh ranges from 0.19 to 0.34. The fraction of cases attributable to household infection (Rh/R) ranges from 0.20 to 0.30.
Figure 4.
Aggregate estimates of effective and household reproduction numbers in Singapore based on contact-tracing data while varying the cut-off date (i.e. the date after which source infections are ignored). Household transmission appears to stay constant, with the intra-household reproduction number in the 0.2–0.3 range. The ratio of infections attributable to households decreases sharply at the end of March due to large outbreaks in migrant-worker dormitories. Even though their infections are not annotated as households, this suggests that cohabitation and proximity play a large role in transmission dynamics.
Estimates from Vo’s blanket-testing data
Our estimation procedure yields an uncorrected estimate for SAR of 27% (24–30%) and Rh of 0.32 (0.28–0.36). With the FNR and AR correction described in ‘Methods’, the corrected estimates are SAR = 31% (28–34%) and Rh = 0.37 (0.34–0.40).
A subgroup analysis suggests that testing did not decrease the household transmission due to early isolation. The SAR associated with 10 households where the first household member had symptoms a week prior to the test date is essentially unchanged at 26%. Our analysis may overestimate the SAR in Vo’ due to violations of single index case assumptions and the presence of additional household infections attributable to community spread. However, due to smaller-than-typical household sizes of 2.1, we expect that such violations are infrequent. We expect Vo’s SAR and Rh estimates to be larger than those of other locations due to an older population, lack of risk awareness and insufficient protective measures in early February.
The contribution of household transmission to R
Central estimates for Rh based on literature estimates, contact-tracing data and blanket-testing data varied from 0.36–0.39. We now estimate how much household transmission contributes to overall transmission levels, i.e. the ratio of the effective household reproductive number Rh to the total effective reproductive number R.
For Singapore, we estimate that Rh is 19–34% of R, meaning that household infections account for 19–34% of total disease transmission.
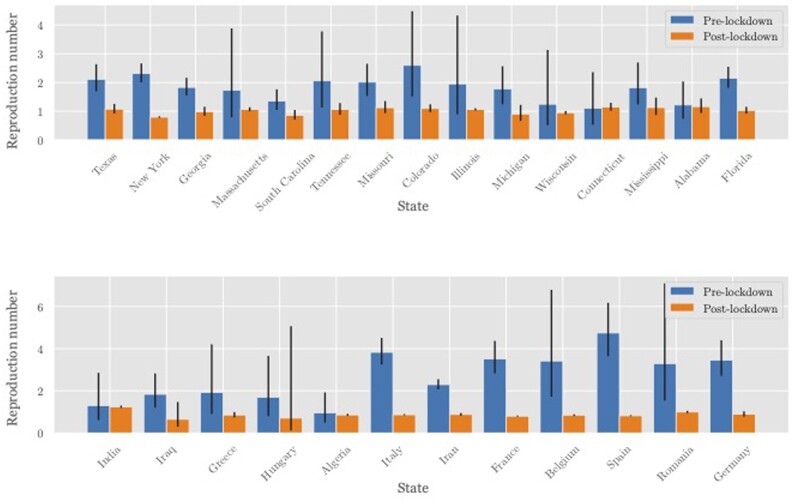
For other geographic regions, we model Rh = 0.3 pre lockdown (based on the previous section) and Rh = 0.3 · M post lockdown, where M is the time spent at home relative to pre lockdown (e.g. M = 1.11 for the USA44). We estimate the pre- and post-lockdown R from death-count data across regions where enough data were available for both time periods (Figure 5).
Figure 5.
Estimated values of the reproduction number R pre and post lockdown in a subset of US states (top) and other countries (bottom). The growth rate was estimated from daily death statistics to avoid testing bias. This was translated into a reproduction number R via the generation time distribution;3 95% confidence intervals are shown. R, reproduction number; US, United States of America.
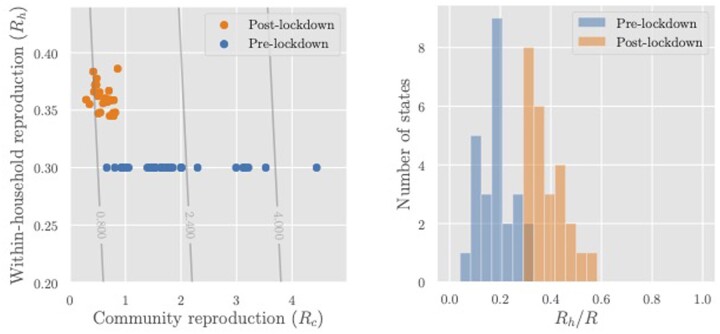
Figure 6 (left) plots the community reproduction number (R – Rh) against Rh for each region both pre and post lockdown. Figure 6 (right) shows a histogram of the estimated contribution of household transmission to the total reproduction number (Rh/R) pre and post lockdown. The share of R attributed to household transmission increased to 25–50% post lockdown, indicating that there may be meaningful benefits (in terms of overall transmission) from interventions that reduce Rh.
Figure 6.
Left: Reproduction numbers for community transmission (Rc) and intra-household transmission (Rh) for the regions whose R values are shown in Figure 4. The overlaid contour plot shows level sets of the overall reproduction number R = Rh + Rc. Right: Estimated share of transmission attributable to household infections (Rh/R). In both graphs, Rh = 0.3 pre lockdown is assumed. Post-lockdown Rh of an area is calculated by multiplying the pre-lockdown value with a mobility factor M obtained from Google’s estimates of the average time spent in residential areas. Rc, community reproduction number; Rh, intra-household reproduction number; R, reproduction number; M, mobility factor (increase in mobility post lockdown).
Discussion
Should non-pharmaceutical interventions (NPIs) target household transmission?
In order for households to be a fruitful target for policy interventions, household transmissions should (i) play a role in disease spread, (ii) be amenable to intervention (i.e. preventable in practice) and (iii) have potential for downstream community transmissions. Our estimates in the previous sections suggest that, for SARS-CoV-2, condition (i) is satisfied. We now turn our attention to the other two conditions.
(ii) Is household transmission inevitable? The data suggest that household transmission is not inevitable in the strictest sense—SAR is lower than 100%, even after adjustments.
There is also evidence that SAR can be reduced by behavioural interventions. Wang et al. al.35 found that the SAR was lower in households where people wore masks at home, cleaned regularly with disinfectant and avoided close contact with the primary cases. Li et al.32 found that the SAR was 0% for households where the primary case was isolated on symptom onset compared with 16.9% (uncorrected) without isolation. Our subgroup analysis of quarantine status also suggests that isolation of cases and quarantine can be effective for reducing household transmission.
(iii) Is household transmission contained? Interventions targeting household transmission would also have little effect if secondary cases resulted in no downstream community infections. Since we cannot reliably attribute community infections to primary vs secondary household cases in our data, we instead discuss the key factors distinguishing secondary cases from primary cases in terms of downstream effects:
Demographic variability, e.g. household infections may skew towards children, whose transmission dynamics differ from those of adults.45,46
Community exposure, e.g. household infections are more likely to be from high- to low-exposure individual(s), who may have a lesser effect on community transmission. Contact patterns47 and the large number of essential workers48 suggest that this will dampen but not nullify the effect of household transmission.
Early isolation : household members may notice the source’s symptoms and self-isolate early, preventing downstream community transmission. However, this is in fact a household NPI, and thus its success actually supports rather than detracts from the effectiveness of household interventions.
Overall, our results suggest that households may indeed be a worthwhile intervention point and motivate further study into quantifying containment.
Implications for modelling
Our estimates can also inform simulated models of SARS-CoV-2 dynamics (e.g.46,49–51). These models often inform policy17,52,53 but common estimates of average transmission risk are often in the 0.5–0.8 range.54 Our estimates of household SAR suggest that these estimates are likely implausible, as households present, on average, one of the highest risks of infection among contacts (high duration indoors); household SAR is thus a likely upper bound on the average transmission risk.
Limitations and conclusions
Being an observational study, the primary limitation of our work lays in the gathered data and, more specifically, the assumptions that we must put on the data to facilitate valid estimation of Rh and SAR. We have outlined these assumptions and their associated drawbacks explicitly throughout Sections 2 and 3. Conversely, an advantage of our work is that we obtain three separate—yet broadly agreeing—estimates based on rather orthogonal modelling assumptions.
Another limitation stems from heterogeneity in the global response to the COVID-19 pandemic; each data source that we consider is inevitably influenced by health policy at the time and place where the data were collected. However, we are able to partially explain this heterogeneity in terms of geography and household policy. Also, the stability of our estimates over time (cf. Figure 4 and related discussion) and the data source lends them some credence.
Our work has presented a set of data sources and corresponding methods for estimating household transmission of SARS-CoV-2. Specifically, we estimate the intra-household reproduction number (Rh) and the household SAR using contact-tracing data from Singapore, widespread-testing data from Vo’ and aggregated data from prior studies, applying the necessary corrections for test sensitivity and asymptomatic cases. Our estimates suggest that household transmission constitutes a stable and significant component of overall transmission and is also not inevitable, making it a promising target for further research and intervention design. Relatedly, our results encourage further study into understanding and explaining the observed heterogeneity in household transmission.
Supplementary data
Supplementary data are available at IJE online.
Ethics approval
Not applicable (no human subjects used), as we performed the study from publicly available observational data.
Funding
None.
Data availability
Outside of what was directly downloaded from cited works, the data for this work (namely formatted contact-tracing data from Singapore) and scripts for reproducing the results are available at https://github.com/Andrewilyas/covid-household-transmission. The data were scraped and processed from the following web dashboard: https://www.againstcovid19.com/singapore/dashboard.
Supplementary Material
Acknowledgements
The authors would like to thank Steve Yadlowsky and Mathew Kiang for comments on a draft of this manuscript.
Conflict of interest
None declared.
References
- 1. Wang X, Pan Y, Zhang D. et al. Basic epidemiological parameter values from data of real-world in mega-cities: the characteristics of COVID-19 in Beijing, China. BMC Infect Dis 2020;20:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y, Wang AH, Yi B. et al. Epidemiological characteristics of infection in COVID-19 close contacts in Ningbo city. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:667–71. [DOI] [PubMed] [Google Scholar]
- 3. Fateh-Moghadam P, Battisti L, Molinaro S. et al. Contact tracing during Phase I of the COVID-19 pandemic in the Province of Trento, Italy: key findings and recommendations. BioRxiv, 29 July 2020; doi:2020:2020.07.16.20127357, preprint: not peer-reviewed. [Google Scholar]
- 4. Bi Q, Wu Y, Mei S. et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis 2020;20:911–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo L, Liu D, Liao X. et al. Contact settings and risk for transmission in 3410 close contacts of patients with COVID-19 in Guangzhou, China : a prospective cohort study. Ann Intern Med 2020;173:879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang JZ, Zhou P, Han DB. et al. Investigation on a cluster epidemic of COVID-19 in a supermarket in Liaocheng, Shandong province. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41:2024–28. [DOI] [PubMed] [Google Scholar]
- 7. Chaw L, Koh WC, Jamaludin SA, Naing L, Alikhan MF, Wong J.. Analysis of SARS-CoV-2 transmission in different settings, among cases and close contacts from the Tablighi cluster in Brunei Darussalam. Emerg Infect Dis 2020;26(11):2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng H-Y, Jian S-W, Liu D-P, Ng T-C, Huang W-T, Lin H-H, Taiwan COVID-19 Outbreak Investigation Team. Contact tracing assessment of COVID-19 transmission dynamics in Taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med 2020;180:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Korea CDC. Coronavirus disease-19: summary of 2,370 contact investigations of the first 30 cases in the Republic of Korea. Osong Public Health Res Perspect 2020;11:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Böhmer MM, Buchholz U, Corman VM. et al. Investigation of a COVID-19 outbreak in Germany resulting from a single travel-associated primary case: a case series. Lancet Infect Dis 2020;20:920–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burke RMMidgley CMDratch Aet al. . Active Monitoring of Persons Exposed to Patients with Confirmed COVID-19- United States, January-February 2020. MMWR Morb Mortal Wkly Rep 2020;69:245–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Y, Song S, Kao Q, Kong Q, Sun Z, Wang B.. Risk of SARS-CoV-2 infection among contacts of individuals with COVID-19 in Hangzhou. Public Health 2020;185:57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park YJ, Choe YJ, Park O. et al. ; COVID-19 National Emergency Response Center, Epidemiology and Case Management Team. Contact tracing during Coronavirus disease outbreak, South Korea, 2020. Emerg Infect Dis 2020;26:2465–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madewell ZJ, Yang Y, Longini IM Jr, Halloran ME, Dean NE.. Household transmission of SARS-CoV-2: a systematic review and Meta-analysis. JAMA Netw Open 2020;3:e2031756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lavezzo E, Franchin E, Ciavarella C. et al. ; Imperial College COVID-19 Response Team. Suppression of COVID-19 outbreak in the municipality of Vo and Italy. Nature 2020;584:425–29.32604404 [Google Scholar]
- 16. Gudbjartsson DF, Helgason A, Jonsson H. et al. Spread of SARS-CoV-2 in the Icelandic Population. N Engl J Med 2020;382:2302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferretti L, Wymant C, Kendall M. et al. Quantifying SARS-CoV-2 transmission suggests epidemic control with digital contact tracing. Science 2020;368:eabb6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Streeck H, Schulte B, Kümmerer BM, et al. Infection fatality rate of SARS-CoV-2 infection in a German community with a super-spreading event. Nat Commun 2020;11(1):5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S, Kim T, Lee E. et al. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med 2020;180:1447. 10.1001/jamainternmed.2020.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corman VM, Landt O, Kaiser M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wikramaratna P, Paton RS, Ghafari M, Lourenco J.. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. Euro Surveill 2020;25(50):2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kucirka L, Lauer S, Laeyendecker O, Boon D, Lessler J.. Variation in false negative rate of RT-PCR based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020;173(4):262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singapore MOH . MOH Updates on COVID-19. 2020. https://www.moh.gov.sg/covid-19 (September 2020, date last accessed).
- 24. Singapore COVID-19 Dashboard. Dashboard of the COVID-19 Virus Outbreak in Singapore. 2020. https://web.archive.org/web/20210330201133/https://www.againstcovid19.com/singapore/dashboard (September 2020, date last accessed).
- 25. van der Hoek W, Backer JA, Bodewes R. et al. The role of children in the transmission of SARS-CoV-2. Ned Tijdschr Geneeskd 2020;164:D5140. [PubMed] [Google Scholar]
- 26. Yu H-J, Hu Y-F, Liu X-X. et al. Household infection: the predominant risk factor for close contacts of patients with COVID-19. Travel Med Infect Dis 2020;36:101809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X, Zhou Q, He Y. et al. Nosocomial outbreak of COVID-19 pneumonia in Wuhan, China. Eur Respir J 2020;55:2000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Son H, Lee H, Lee M. et al. Epidemiological characteristics of and containment measures for COVID-19 in Busan, Korea. Epidemiol Health 2020;42:e2020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun WW, Ling F, Pan JR. et al. Epidemiological characteristics of COVID-19 family clustering in Zhejiang Province. Zhonghua Yu Fang Yi Xue Za Zhi 2020;54:625–29. [DOI] [PubMed] [Google Scholar]
- 30. Wu J, Huang Y, Tu C. et al. Household transmission of SARS-CoV-2, Zhuhai, China, 2020. Clin Infect Dis 2020;71:2099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kwok KO, Wong VWY, Wei WI, Wong SYS, Tang JW-T.. Epidemiological characteristics of the first 53 laboratory-confirmed cases of COVID-19 epidemic in Hong Kong, 13 February 2020. Euro Surveill 2020;25(16):2000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li WZhang BLu Jet al. . Characteristics of Household Transmission of COVID-19. Clin Infect Dis 2020;71:1943–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jing Q-L, Liu M-J, Zhang Z-B. et al. Household secondary attack rate of COVID-19 and associated determinants in Guangzhou, China: a retrospective cohort study. Lancet Infect Dis 2020;20:1141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park SY, Kim Y-M, Yi S. et al. Coronavirus disease outbreak in call center, South Korea. Emerg Infect Dis 2020;26:1666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Z, Ma W, Zheng X, Wu G, Zhang R. Household transmission of SARS-CoV-2. J Infect 2020;81:179–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boscolo-Rizzo P, Borsetto D, Spinato G. et al. New onset of loss of smell or taste in household contacts of home-isolated SARS-CoV-2-positive subjects. Eur Arch Otorhinolaryngol 2020;277:2637–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosenberg ES, Dufort EM, Blog DS. et al. ; New York State Coronavirus 2019 Response Team. COVID-19 testing, epidemic features, hospital outcomes, and household prevalence, New York State-March 2020. Clin Infect Dis 2020;71:1953–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawson P, Rabold EM, Laws RL. et al. Loss of taste and smell as distinguishing symptoms of coronavirus disease 2019. Clin Infect Dis 2021;72:682–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Tian H, Zhang L. et al. Reduction of secondary transmission of SARS-CoV-2 in households by face mask use, disinfection and social distancing: a cohort study in Beijing, China. BMJ Glob Health 2020;5:e002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xin H, Jiang F, Xue A. et al. Risk factors associated with occurrence of COVID-19 among household persons exposed to patients with confirmed COVID-19 in Qingdao Municipal. Transbound Emerg Dis 2021;68:782–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laxminarayan R, Wahl B, Dudala SR. et al. Epidemiology and transmission dynamics of COVID-19 in two Indian states. Science 2020;370:691–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koh D. Occupational risks for COVID-19 infection. Occup Med (Lond) 2020;70:3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Google. COVID-19 Community Mobility Reports. 2020. https://www.google.com/covid19/mobility/ (September 2020, date last accessed).
- 45. Jing Q-L, Liu M-J, Yuan J. et al. Household secondary attack rate of COVID-19 and associated determinants. Lancet Infect Dis 2020;20(10):1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davies NG, Kucharski AJ, Eggo RM. et al. ; Centre for the Mathematical Modelling of Infectious Diseases COVID-19 working group. Effects of non-pharmaceutical interventions on COVID-19 cases, deaths, and demand for hospital services in the UK: a modelling study. Lancet Public Health 2020;5:e375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rothwell J. Americans’ Social Contacts During the COVID-19 Pandemic. 2020. https://news.gallup.com/opinion/gallup/308444/americans-social-contacts-during-covid-pandemic.aspx (September 2020, date last accessed).
- 48. The Lancet. The plight of essential workers during the COVID-19 pandemic. The Lancet 2020;395:1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kucharski AJ, Russell TW, Diamond C. et al. ; Centre for Mathematical Modelling of Infectious Diseases COVID-19 working group. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis 2020;20:553–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Firth JA, Hellewell J, Klepac P. et al. ; CMMID COVID-19 Working Group. Using a real-world network to model localized COVID-19 control strategies. Nat Med 2020;26:1616–22. [DOI] [PubMed] [Google Scholar]
- 51. Aleta A, Martin-Corral D, Piontti A. y, et al. Modeling the Impact of Social Distancing, Testing, Contact Tracing and Household Quarantine on Second-Wave Scenarios of the COVID-19 Epidemic. Nat Hum Behav. 2020 Sep;4(9):964–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hinch R, Probert W, Nurtay A, et al. Effective Configurations of a Digital Contact Tracing App: A Report to NHSX. En In:(Apr 2020). https://Github Com/BDI-Pathogens/Covid-19_instant_tracing/Blob/Master/Report2020 (September 2020, date last accessed).
- 53. Center for Disease Control. Prevent Getting Sick. 2020. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/index.html (September 2020, date last accessed).
- 54. Block P, Hoffman M, Raabe IJ. et al. Social network-based distancing strategies to flatten the COVID-19 curve in a post-lockdown world. Nat Hum Behav 2020;4:588–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Outside of what was directly downloaded from cited works, the data for this work (namely formatted contact-tracing data from Singapore) and scripts for reproducing the results are available at https://github.com/Andrewilyas/covid-household-transmission. The data were scraped and processed from the following web dashboard: https://www.againstcovid19.com/singapore/dashboard.