Abstract
Knowledge of human gingival cell responses to dental monomers is critical for the development of new dental materials. Testing standards have been developed to provide guidelines to evaluate biological functionality of dental materials and devices. However, one shortcoming of the traditional testing platforms is that they do not recapitulate the multi-layered configuration of gingiva, and thus cannot evaluate the layer-specific cellular responses. An oral mucosa-chip with two cell layers was previously developed as an alternative platform to assess the oral mucosa responses to dental biomaterials. The mucosa-chip consists of an apical keratinocyte layer attached to a fibroblast-embedded collagen hydrogel through interconnecting pores in a three-microchannel network. Here, cell responses in the mucosa-chip were evaluated against 2-hydroxyethyl methacrylate (HEMA), a common monomer used in restorative and aesthetic dentistry. The response of mucosal cell viability was evaluated by exposing the chip to HEMA of concentrations ranging from 1.56 to 25 mM and compared to cells in conventional well-plate monoculture. The co-cultured cells were then stained and imaged with epifluorescence and confocal microscopy to determine the layer-specific responses to the treatment. Mucosa-chips were demonstrated to be more sensitive to assess HEMA-altered cell viability than well-plate cultures, especially at lower doses (1.56 and 6.25 mM). The findings suggest that the mucosa-chip is a promising alternative to traditional platforms or assays to test a variety of biomaterials by offering a multi-layered tissue geometry, accessible layer-specific information, and higher sensitivity in detecting cellular responses.
Keywords: Mucosa-on-a-chip, Gingiva, Dental materials testing, 2-hydroxyethyl methacrylate, Cell viability
1. Introduction
The oral mucosa is a mucous membrane consisting of three distinct layers. The superficial layer is stratified squamous epithelium, which may be keratinized or non-keratinized depending on the location within the oral cavity. A thin basement membrane lies basal to the epithelium, consisting of collagen, laminins, and other proteoglycans. Beneath the basement membrane is a fibrous connective tissue layer, the lamina propria, which provides support and nourishment to the epithelium (Izumi et al. 2015; Keyvan Moharamzadeh 2017). Oral epithelial cells participate in inflammatory processes and act as a protective barrier against bacterial infection, and are also the cells most proximal to many dental materials (Cho et al. 2013; Keyvan Moharamzadeh 2017; K. Moharamzadeh et al. 2007). Mucosal cells cultured in vitro are used to study the biology and pathologies of oral mucosal cells, as well as to test mucosal responses to new dental treatments. It is important that these in vitro models closely resemble native oral mucosa (Tra et al. 2012).
Testing standards have been developed to evaluate the bio-compatibility and cytotoxicity of dental materials. Existing testing models include individual cell lines cultured on a solid substrate (Caldas et al. 2019), the in vitro pulp chamber (Hanks et al. 1988), and decellularized mucosal tissue (Hildebrand et al. 2002). These conventional cell cultures have been commonly employed to study cellular functions and molecular mechanisms owing to their simplicity and scalability. However, the lack of a multi-layered configuration of gingival cells in these traditional testing platforms prevents easy assessment of gingival cell-specific responses to dental materials. Over the past two decades, microfluidic integrated platforms have been intensively implemented in biomedical and tissue engineering studies owing to their low reagent consumption, rapid fabrication, high sensitivity and controllability, and cost-effectiveness (Bilitewski et al. 2003; Lignos et al. 2017; Wu et al. 2016). Although not exactly like in vivo models, these tissue-on-a-chips are more clinically relevant than monocultures mostly in their ability to reconstruct cell layer geometry on a biological scale and monitor cellular interactions in metabolic time course studies.
In terms of dental research, microfluidic platforms have been implemented to test dental biofilm growth (Lam et al. 2016), to study the growth and differentiation of odontoblasts (Niu et al. 2019), and to assess the response of pulp cells to dental material through a tooth-on-a-chip (Franca et al. 2020). To increase the clinical relevance of gingiva on a chip construct, some design requirements deserve close attention without compromising the general advantages of microfluidic platforms. These highly desired features include, but are not limited to, a layered apical-basal cellular configuration mimicking natural tissue, the capability to modulate flow dynamics and biological activities, and the ability to test cellular-specific response of physiological relevance to dental materials and dental- pathogen interactions. In our previous study, we developed an oral mucosa-on-a-chip (mucosa-chip) on a microfluidic platform with luminal, epithelial, and subepithelial compartments arranged in a histology section-like configuration (Rahimi et al. 2018). The mucosa-chip platform consisted of an apical layer of keratinocytes attached to a fibroblast-embedded collagen hydrogel through interconnecting pores in a three-microchannel network. This mucosal equivalent has been optimized to be stable for up to 7 days. In our proof-of-concept experiments, we also demonstrated the usability of mucosa-chips in assessing oral mucosa responses to 2-hydroxyethyl methacrylate (HEMA) of 25 mM and Streptococcus mutans, in which HEMA exposure and bacteria lowered the mucosal cell viability and the transepithelial electrical resistance, respectively. HEMA is a commonly used monomer in restorative and aesthetic dentistry. The adverse effects of HEMA have been well-documented in relation to tissue inflammation, apoptosis, genotoxic effects, inhibition of DNA and protein synthesis, and altering cell division and activity (Bouillaguet et al. 1998; Hanks et al. 1991; Szczepanska et al. 2012). Although the dose-response of gingival cells with respect to HEMA have been well-investigated, only a few studies have explored the integrated mucosal response towards HEMA at low concentrations, in vitro (Falconi et al. 2007).
Taking advantage of a single field-of-view observation of the keratinocyte and fibroblast layer morphology in the previously developed mucosa-chip, the aim of this study was to further assess the responses of specific mucosal cells in oral mucosa to HEMA at varied concentrations, in particular sensitivity to physiologically relevant doses that are typically less than 25 mM. We then compared the sensitivity of cellular responses to HEMA treatment in mucosa-chips with that in well-plate monoculture, a conventional testing platform. We demonstrated that the mucosa-chip reflected more sensitive mucosal cell responses to HEMA treatment, especially in a lower dose scenario, as compared to well-plate monoculture. These findings suggest that the mucosa-chip can be a promising alternative to traditional platforms or assays to test a variety of biomaterials.
2. Materials and methods
2.1. Materials
Sylgard 184 and its curing agent for polydimethylsiloxane (PDMS) device fabrication was purchased from Ellsworth Adhesives (Germantown, WI). Polytetrafluoroethylene (PTFE) tubing of 0.022″ ID (interior diameter), 0.042″ OD (outside diameter) was purchased from Cole-Parmer (Vernon Hills, IL). Stainless steel catheter plugs and hollow metal couplers of 22-gauge size were purchased from Instech Laboratory Inc. (Plymouth, PA). Disposable syringes of 1 ml volume were purchased from Becton, Dickinson and Company (Franklin Lakes, NJ). Phosphate buffered saline (PBS, pH 7.4) was purchased from Sigma-Aldrich (St. Louis, MO). The keratinocyte Gie-No3B11 (Gie or keratinocyte) (Gröger et al. 2008) and human gingival fibroblast (HGF or fibroblast) (Illeperuma et al. 2012; Vardar-Sengul et al. 2009) cell line, Prigrow III and IV media (supplemented with 10% fetal bovine serum (ATCC, Manassas, VA), 500 U/mL penicillin and 500 μg/mL streptomycin (Corning, Bedford, MA)) used to culture fibroblasts and keratinocytes, respectively, were purchased from Applied Biological Materials, Inc. (Richmond, British Columbia). Collagen type I (rat tail, 8.86 mg/mL) was purchased from Corning (Bedford, MA). HEMA was donated from Esstech (Essington, PA). Bovine serum albumin (BSA), AlexaFluor 546-conjured phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) and LIVE/DEAD® Viability/Cytotoxicity kit were obtained from ThermoFisher Scientific (Waltham, MA). All other chemicals can be purchased from major suppliers.
2.2. Fabrication of microfluidic platform
The mold for the microfluidic platform was fabricated using conventional photolithographic techniques with negative photoresist SU-83035 on a 4″ silicon wafer. The PDMS microfluidic device as shown in Fig. 1a was fabricated using a conventional soft-lithography method as previously reported (Hu et al. 2020; Ly et al. 2020a). Briefly, PDMS in liquid form was prepared by mixing Sylgard 184 and its curing agent at 10:1 ratio, degassed, poured on top of the molds sitting in a house-made aluminum foil container, and cured at 65 °C on a hotplate for 4 h. The solidified PDMS was then delaminated from the molds and cut into the desired pieces. The microfluidic networks were punched for input and output connections. Oxygen plasma (200 mTorr, 10 psi gas source from an oxygen tank, 30 s, medium RF level) was used to bind the punched PDMS slabs to glass slides that were cleaned with a series of acetone, methanol, and isopropyl using a Plasma Cleaner PDC-32G (Harrick Plasma). The bonded devices were then put in an oven at 120 °C and left at least for overnight to restore the hydrophobicity of PDMS. The microfluidic platform used in this study is comprised of three parallel channels. The height of the microchannel networks was 50 μm. The microchannels were 400 μm in width near the aperture area. The fourteen apertures and twelve PDMS pillars between the apertures were 50 and 100 μm in width, respectively.
Fig. 1.
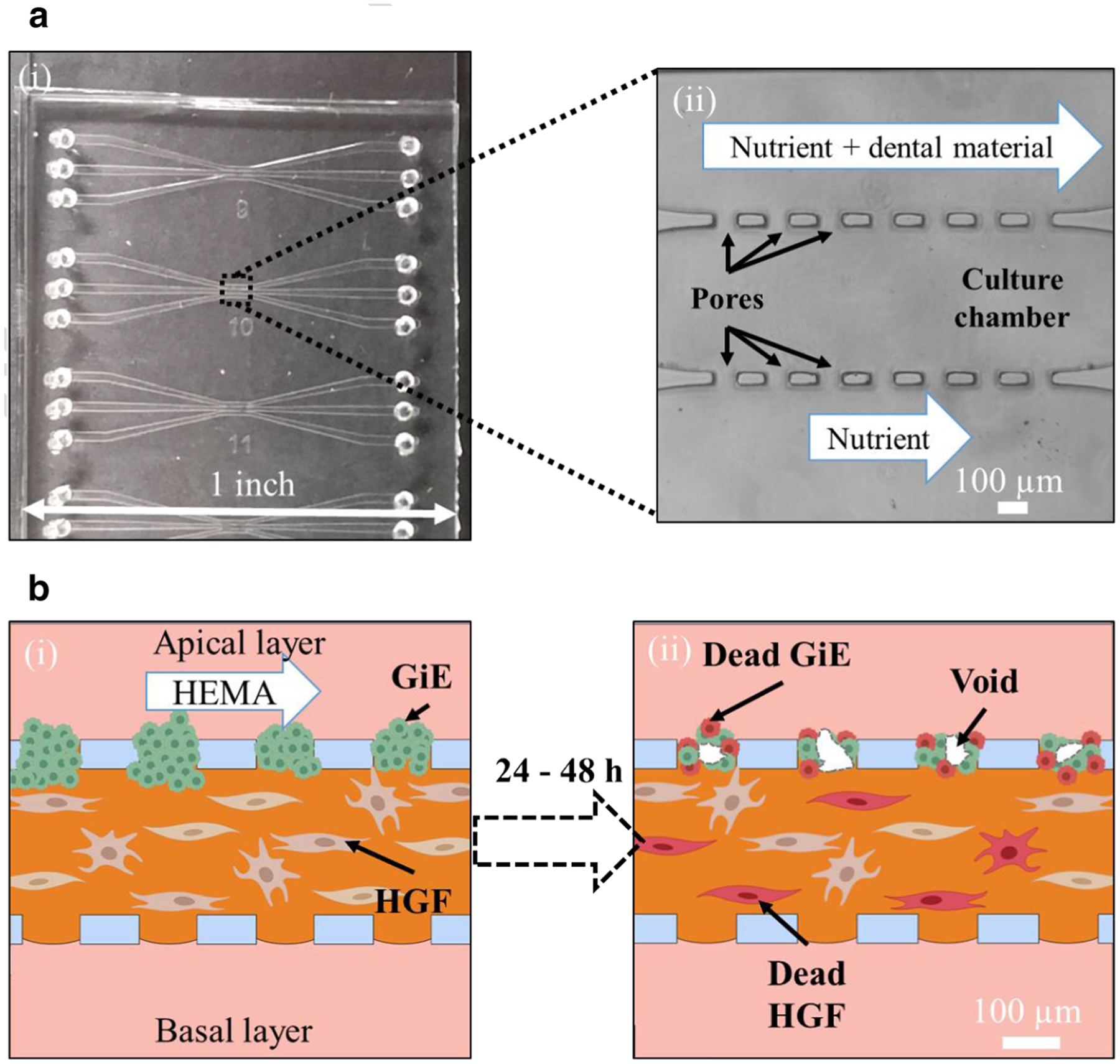
Mucosa-chip to evaluate layer-specific cellular responses to dental monomer HEMA. (a) The mucosal-chip device: (i) a photograph of the PDMS chip attached to glass slide containing the three-channel devices, and (ii) the apertures region under transmitted light microscopy. (b) The cartoon representatives of the mucosa-chip (i) being exposed to HEMA and (ii) post-HEMA exposure
2.3. Cell culture
Two keratinocytes and fibroblasts cell lines were maintained in 25 cm2 polystyrene tissue culture flasks coated with type I collagen (Advanced Biological Materials, Inc.). Nutrient medium was replenished every other day. The two cell lines were extracted from culture flasks by trypsinization. Then the cell suspensions were centrifuged (252 × g in 3 min for Gie and 350 × g in 3 min for HGF), resuspended in 1 mL of media, and counted using a hemocytometer. The mucosal construct in microfluidic chips was developed in a previous study with the optimal densities of fibroblasts and keratinocytes determined to be 5000 cells/μL and 2000 cells/μL, respectively (Ly et al. 2020b; Rahimi et al. 2018). The HGF density of 5000 cells/μL induced minimal gel contraction and allowed the gel to last for over a week while the Gie density of 2000 cells/μL allowed around 97 ± 9% keratinocytes filling to the pores. Furthermore, collagen at 4 mg/mL was the best available concentration to reduce matrix contraction and keratinocyte invasion in the mucosa chip, leading to construct stability over one week of culture. Briefly, the mixture of 4 mg/mL collagen type I and fibroblasts with density of 5000 cells/μL was gently injected to the central channel of the device using micropipette with the flow rates estimated to be approximate 1–4 μL/min, and the chips were incubated at 37 °C for 30 min in order for collagen to polymerize. Next, keratinocytes at a density of 2000 cells/μL were seeded to one side channel by micropipette injection, followed by the vertical tilting of the device to allow gravity to settle keratinocytes into the interconnecting pores. The device was further incubated at 37 °C for 30 min, before the unattached keratinocytes in the side channel were washed away. During the fabrication of the mucosa-chip construct, extra care is taken to not generate high pressure that disturb the 400-μm-wide collagen gel layer. The mucosa-chip constructs are checked under a phase contrast microscope, and chips with ripped gels are not used. The chips were then fed daily with 50/50 Prigrow III/IV media. Fresh media droplets of unequal volume (2 and 10 μL) were placed on top of four ports connecting to the apical and basal channels to allow the fresh media to be introduced by surface tension-driven flow. The central collagen-containing channel was not subjected to media droplets to avoid gel damage. To prevent the drying of media, the chips were enclosed in Petri dishes with sterile water droplets pipetted around the dish edges to maintain the humidity. Figure 1b-(i) shows the illustrative layout of the mucosa-chip after incubation for 48 h, in which an apical layer of keratinocyte attached to a fibroblast-embedded collagen hydrogel in the central channel through interconnecting pores in a three-microchannel chip.
2.4. HEMA exposure
The HEMA solution of 25 mM was prepared by dissolving a precisely weighed amount of HEMA powder in 50/50 Prigrow III/IV media. Other concentrations (1.56 and 6.25 mM) were obtained by serial dilution with 50/50 Prigrow III/IV media. HEMA of various concentrations (1.56, 6.25, and 25 mM) were gently injected to the apical layers of the mucosa-chips using micropipette. Figure 2 shows the timeline for chip culture including exposure to HEMA at 0 h. The time from the fabrication of mucosal construct to HEMA exposure was around 48 h. After an additional 24–48 h of culture time, the effects of HEMA on mucosal construct inside the chips were evaluated with immunofluorescence and live/dead staining.
Fig. 2.
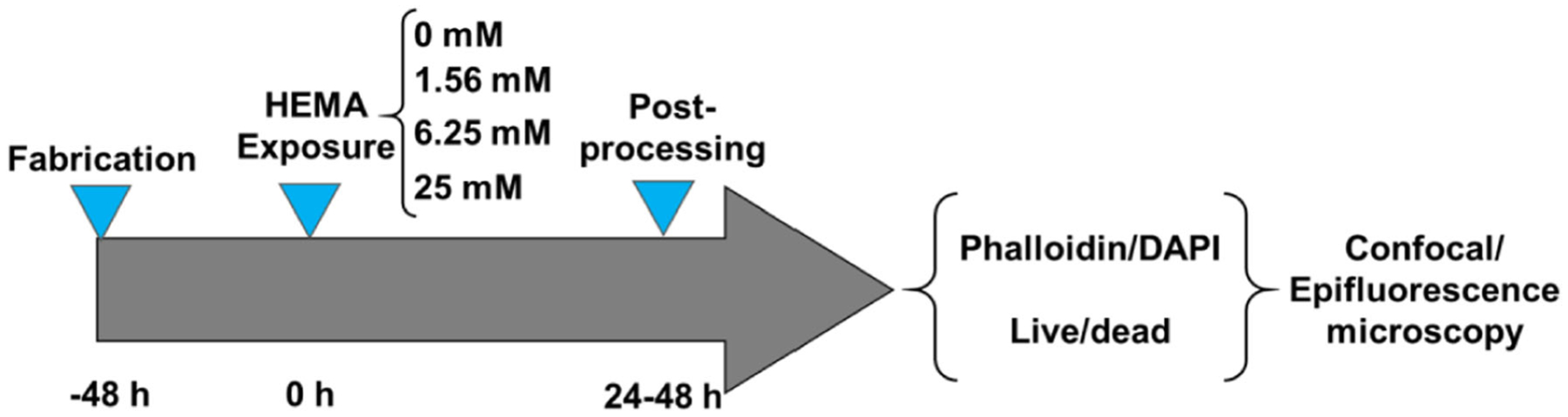
The experimental timeline, in which the time the mucosa-chips exposed to HEMA was set as 0 time point. Post-HEMA exposure, the chips were stained and characterized with confocal and epifluorescence microscopy
2.5. Fluorescence staining
Twenty-four-hour post-HEMA exposure, the mucosa-chips treated with different HEMA concentrations were fixed in 4% formaldehyde for 10 min at room temperature, followed by rinsing with PBS 1X twice. Then the chips were permeabilized with 0.1% Triton-X solution for 30 min before blocking with a solution of 1% BSA in 0.1% Triton-X for 24 h. AlexaFluor 546-conjured phalloidin and DAPI were used to stain the F-actin cytoskeleton and the nuclei of cells in the mucosal constructs, respectively.
2.6. Live/dead assay
The cell viability of keratinocytes and fibroblasts on mucosa-chips and traditional well-plates was investigated and compared using the Live/Dead assay. The staining procedures were as follows:
Mucosa-chip Mucosa-chips post-HEMA exposure (24 and 48 h) were stained using LIVE/DEAD® Viability/Cytotoxicity Kit followed manufacturer guidelines. Briefly, channels were rinsed with serum-free media before adding a mixture of 8X ethidium homodimer-1 (8 μM) and 4X calcein acetoxymethyl ester (2.67 μM) reagent in PBS 1X, and incubating for one hour prior to epifluorescence imaging. Dead cells were counted by visual inspection of the epifluorescence images, spanning the entire working culture area of the microfluidic chip, in comparison to co-registered phase contrast images to delineate cell borders. Three rectangular regions-of-interest (ROI) of 15,000 μm2 within fibroblasts and keratinocytes region were randomly selected and the area fraction of live signal within the ROI was measured using ImageJ by thresholding the ROI at 43 ± 4 of the background in the ROI. The live area was calculated by multiplying the area by the area fraction. Experiments were conducted in three separate trials, one chip per trial containing 5–8 mucosal constructs per chip and data was represented as mean ± standard deviation (SD) (for dead cells count) or standard error of the means (SEM) (for live area fraction).
Well-plate The cell viability of keratinocytes and fibroblasts were examined separately on 96-well plate. For experiments, two cell lines were obtained from a sub-confluent stock culture. Direct contact cytotoxicity testing was conducted as described in previous studies (Bienek et al. 2018a; Bienek et al. 2018b). Adherent cells (~104 per well) were exposed to twofold serial dilutions of HEMA starting from 25 mM to the final concentration of 0.195 mM for 24 h. Thereafter, cells were assessed for cell viability using the LIVE/DEAD® Viability/Cytotoxicity kit. After HEMA exposure, cells were washed with phosphate-buffered saline (without calcium and magnesium) and then incubated with 1X ethidium homodimer-1 (1 μM) and 1X calcein acetoxymethyl ester (0.658 μM) for 20 min. A Spark plate reader (Tecan, Morrisville, NC) was used to assess the fluorescence of ethidium homodimer (excitation 530 nm, emission 645 nm) and calcein acetoxymethyl ester (excitation 485 nm, emission 530 nm). The live/dead signal of the control group was set as 100 and the live/dead signals of HEMA-treated groups were normalized based on that. Means were obtained from 5 independent trials, with each trial tested in triplicate.
2.7. Microscopy
Epifluorescence microscopy was performed with an inverted microscope (Accu-Scope EXI-310, Ludesco, LLC) and a fluorescence light source (LM-75 PhotoFluor, 89 North). The same specimens were also imaged with a confocal microscope (LSM 510, Zeiss) with standard laser wavelengths for DAPI, AlexaFluor 546 excitation, and emission filters of 408–462 and 552–642 nm, acquired in two separate channels. Confocal images were 512 × 512 pixels, spanning an (800 × 800) μm2 field-of view.
2.8. Statistical analysis
All experiments were performed in triplicate, unless specified otherwise. Results are reported in the text as mean ± SD, except live area fraction data are reported as mean ± SEM. The effects on live and dead-stained keratinocytes and fibroblasts by concentration and duration of HEMA exposure were evaluated with two-factor ANOVAs, for factors of concentration (0, 1.56, 6.25, and 25 mM) and duration in culture with HEMA (24 and 48 h), with Tukey’s post hoc tests to compare between pairs of groups. Linear regression was performed on live and dead cell parameters versus HEMA concentration, with the slopes compared to 0 by t-tests. All tests were performed using Systat (version 13, Systat Software, Inc.) with the level of significance set at p < 0.05.
3. Results
3.1. Mucosal layer-specific organization is disrupted by HEMA
Cell morphology and organization closely resemble the native gingiva in the co-cultured mucosa-on-a-chip not exposed to HEMA, after 24 h culture (Fig. 3a). At 0 mM HEMA, the keratinocyte cytoskeleton was distributed cortically in polygonal cells forming a layer up to four cells thick. Exposure to HEMA disrupted the F-actin network, causing cytoplasmic shrinkage and appearance of voids in the keratinocyte layer, in which as HEMA concentration increased, the void spaces became larger (Fig. 3b-(ii)). The area of the void was greater for higher HEMA concentrations (Fig. 3c). In particular, the void area of construct post-exposure to 1.56, 6.25, and 25 mM HEMA was 7.3 ± 3.2, 35.0 ± 7.1, and 72.4 ± 17.5 (×1000) μm2, respectively. Confocal microscopy revealed the 3D cytoskeletal architecture of the mucosal constructs with and without HEMA treatment throughout the 50-μm-tall channels as shown in Fig. 4. Without HEMA exposure, the keratinocytes and fibroblasts were tightly connected and formed contiguous cytoskeleton networks (Fig. 4a). Constructs exposed to 25 mM HEMA had less F-actin signal, despite the presence of cell nuclei (Fig. 4b).
Fig. 3.
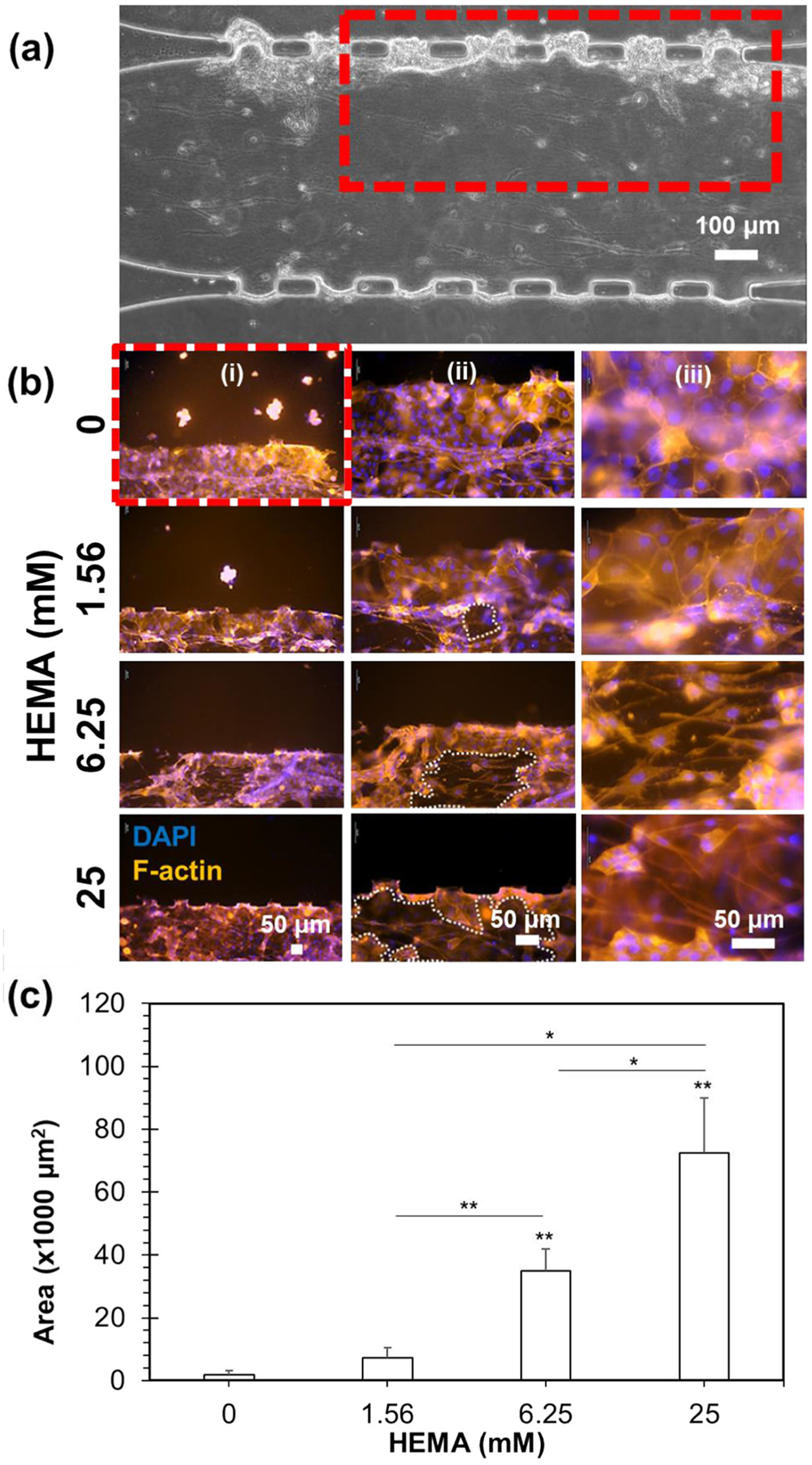
Epifluorescence micrographs of mucosa-chip chamber post-HEMA exposure. (a) The mucosal construct under phase contrast microscopy. The red rectangle indicates the zoom-in image in B(i). (b) The mucosal construct treated with different HEMA concentration under epifluorescence microscopy, in which the cells were fixed and stained with phalloidin (red) and DAPI (blue) to reveal the epithelial and sub-epithelial layers. The images were taken at different magnification with scale bars as indicated. The white-dashed regions in (ii) indicate the voids emerged post-HEMA exposure. (c) The area of voids within mucosa-chips post-exposure to different HEMA concentration. Data = mean ± SD, (*) indicates p < 0.05 and (**) indicates p < 0.01
Fig. 4.
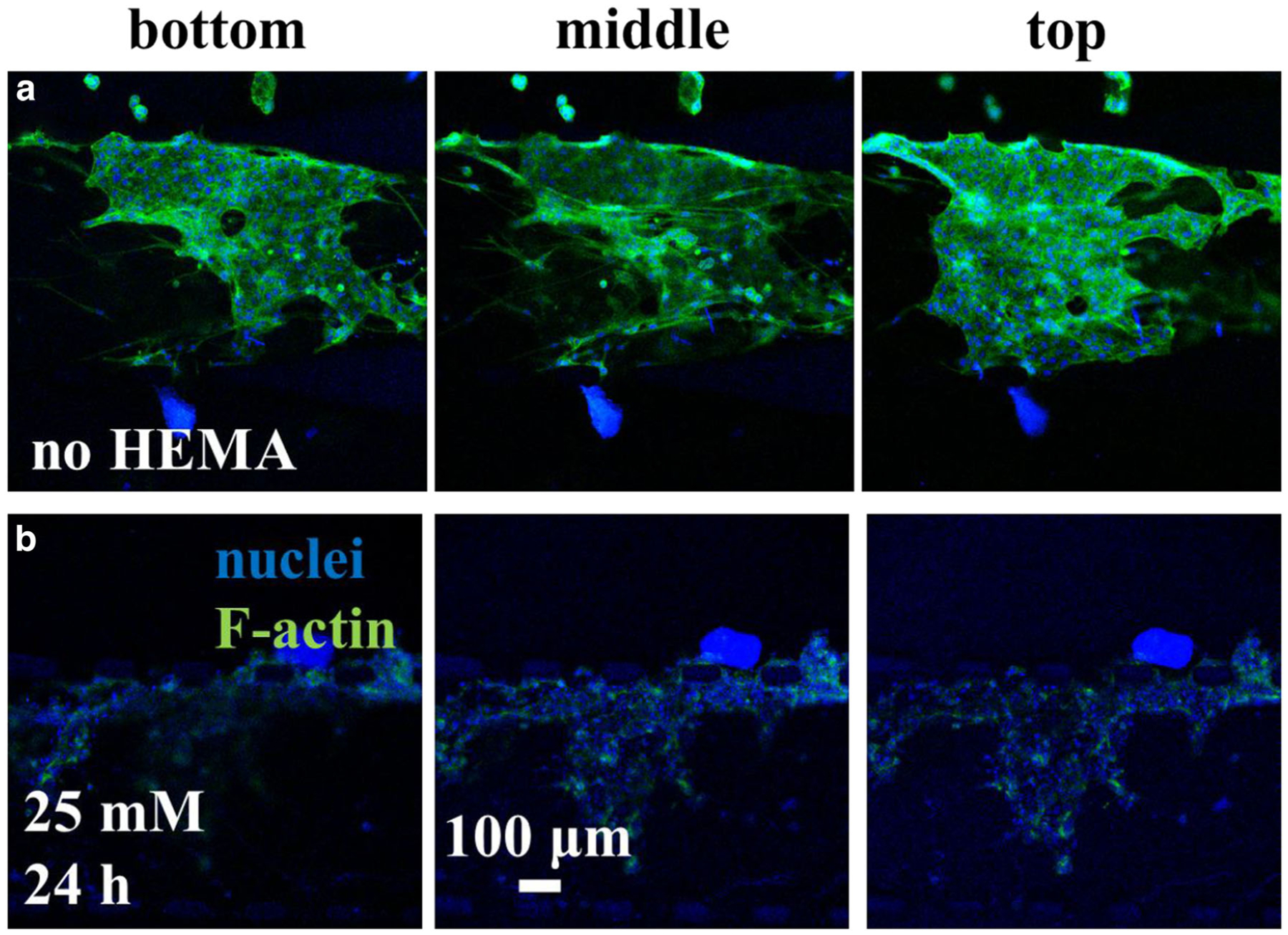
Confocal micrographs of mucosa-chip chamber (a) without and (b) with HEMA exposure at 24-h time point. The nuclei were stained in blue while F-actin in green
3.2. HEMA affects cell viability in mucosa-chip platform
Exposure to HEMA affected live cells in a time and concentration-dependent manner. In particular, less viable cells were present at higher HEMA concentrations, though viable cell area was larger at longer exposure times (Fig. 5a). The keratinocyte layer live area fraction depended on HEMA concentration (F = 14.2, p < 0.001) and duration of exposure (F = 11.5, p < 0.01, 2-factor ANOVA). Specifically, keratinocyte layer live area fraction was 41 ± 23% after 24 h exposure and 59 ± 21% after 48 h exposure. After 24 and 48 h exposure, keratinocyte layer live area fraction was lower with higher concentration of HEMA, from 62 ± 12% and 76 ± 5% for controls not exposed to HEMA at 24 and 48 h, respectively, to 13 ± 7% and 25 ± 3% for mucosa chips exposed to 25 mM HEMA at 24 and 48 h, respectively (Fig. 5b). Similarly, fibroblast layer live area fraction depended on HEMA concentration (F = 22.7, p < 0.001) but not duration of exposure (2-factor ANOVA). Specifically, fibroblast layer live area fraction was 16 ± 9% after 24 h exposure and 20 ± 10% after 48 h exposure. Fibroblast layer live area fraction was lower with higher concentration of HEMA, from 26 ± 8% for controls not exposed to HEMA to 4 ± 2% for mucosa chips exposed to 25 mM HEMA (Fig. 5b).
Fig. 5.
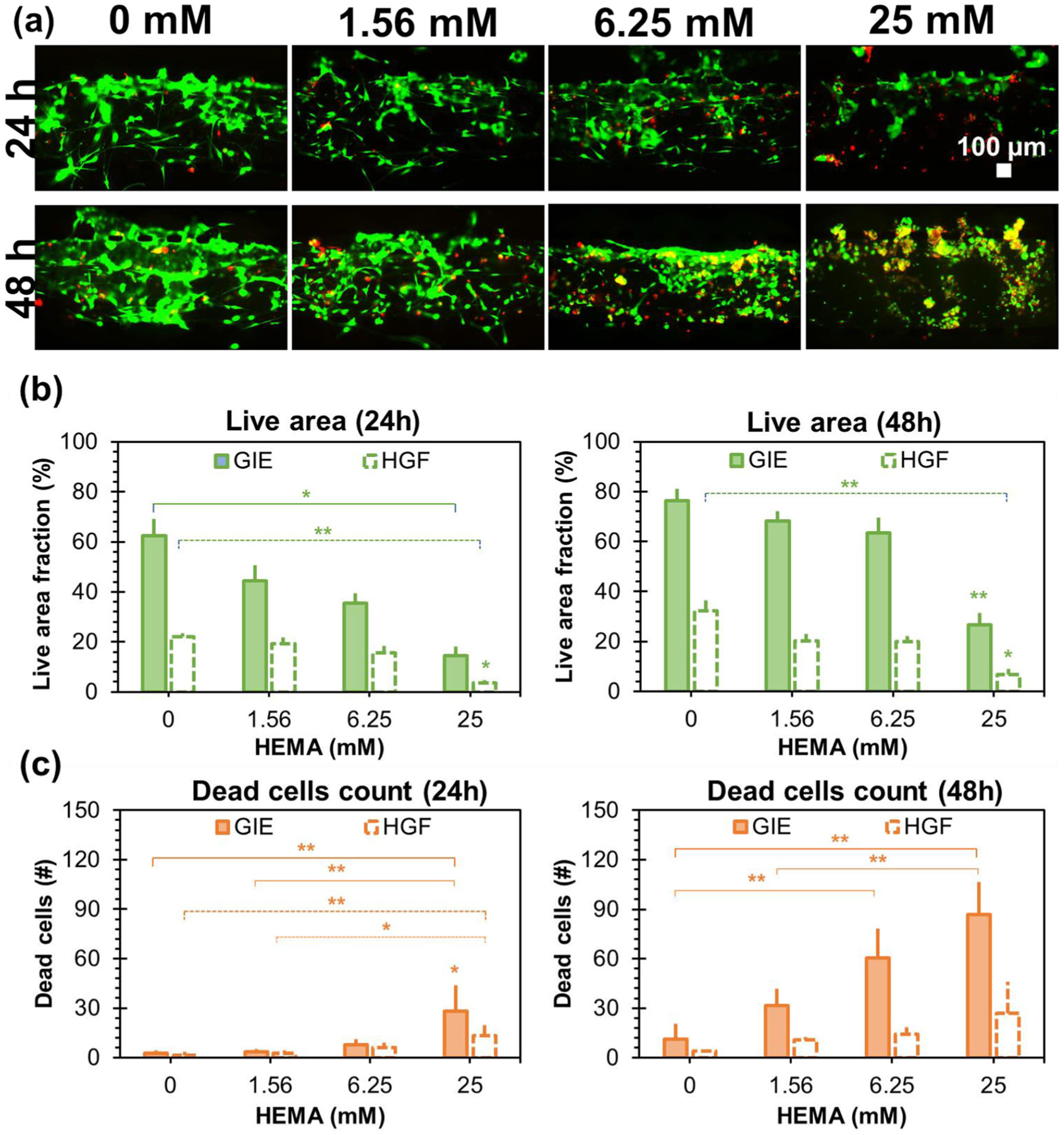
The effects of HEMA exposure on layer viability of the mucosa-chip monitored at 24 and 48 h. (a) The epifluorescence micrographs of live cells stained in green and dead cells stained in red post 24 and 48-h exposure to different HEMA concentration. (b) The live cells area within each condition at 24 and 48 h. (c) The dead cells counted within each condition at 24 and 48 h. Data = mean ± SEM, (*) indicates p < 0.05 and (**) indicates p < 0.01
Exposure to HEMA affected numbers of dead cells in a time and concentration-dependent manner. In particular, more dead cells were present at higher HEMA concentrations and at longer exposure times (Fig. 5a). The number of dead keratinocytes depended on HEMA concentration (F = 15.3, p < 0.001), duration of exposure (F = 120.3, p < 0.001), and the interaction of dose and duration (F = 11.7, p < 0.01, 2-factor ANOVA). Specifically, the number of dead keratinocytes per field was 9 ± 12 after 24 h exposure and 46 ± 32 after 48 h exposure. After 24 and 48-h exposure, numbers of dead keratinocytes per sampled field were higher with higher concentrations of HEMA, from 3 ± 2 and 11 ± 10 for controls not exposed to HEMA at 24 and 48 h, respectively, to 28 ± 16 and 87 ± 19 for mucosa chips exposed to 25 mM HEMA at 24 and 48 h, respectively (Fig. 5c). Similarly, numbers of dead fibroblasts per sampled field depended on HEMA concentration (F = 7.8, p < 0.01) and duration of exposure (F = 7.6, p < 0.05, 2-factor ANOVA). Specifically, the number of dead fibroblasts per sampled field was 5 ± 6 after 24 h exposure and 14 ± 12 after 48 h exposure. After 24 and 48 h exposure, numbers of dead keratinocytes per sampled field were higher with higher concentrations of HEMA, from 2 ± 2 and 4 ± 1 for controls not exposed to HEMA at 24 and 48 h, respectively, to 13 ± 8 and 27 ± 19 for mucosa chips exposed to 25 mM HEMA at 24 and 48 h, respectively (Fig. 5c).
3.3. HEMA affects cell viability in monoculture platform
Keratinocyte and fibroblast monolayers were also affected by exposure to HEMA. Table 1 summarizes the normalized live/dead data of HEMA-treated groups to untreated group (control). Only at the highest doses of HEMA (12.5 and 25 mM) was viability of cells in monolayers measured to be less than cells not exposed to HEMA. Fibroblasts were affected more than keratinocytes. For instance, at 12.5 mM HEMA, fibroblasts were less viable (44 ± 39%) than keratinocytes (73 ± 16%). A similar trend existed at 25 mM HEMA. Metrics (i.e., number of viable cells) were altered (2-factor ANOVA, p < 0.05) in keratinocytes and fibroblasts well-plate monocultures after 24 h exposure to 12.5 and 25 mM HEMA, but not lower HEMA concentrations. There was no significant effect of HEMA on the percentage of dead fibroblasts (2-factor ANOVA). Meanwhile, the percentage of dead keratinocytes depended on HEMA concentration (F = 2.4, p < 0.05) and the interaction of dose (F = 2.7, p < 0.05, 2-factor ANOVA). Despite that, there was not real sensitivity to the dead well-plate signals, in which the percentage of dead was close to 100% for nearly all conditions, except 12.5 mM.
Table 1.
Normalized percent of viabilitya of Gie and HGF cells exposed to HEMA for 24 h quantified fluorometrically
| Concentration (mM) | Gie | HGF | |
|---|---|---|---|
| 25 | Live | 46.2 ± 17.5 | 15.6 ± 10.5 |
| Dead | 108.7 ± 9.4 | 114.4 ± 23.2 | |
| 12.5 | Live | 72.9 ± 15.9 | 44.4 ± 39.1 |
| Dead | 125.2 ± 14.2 | 103.8 ± 13.9 | |
| 6.25 | Live | 120.8 ± 9.8 | 99.2 ± 26.0 |
| Dead | 106.6 ± 6.6 | 98.8 ± 9.0 | |
| 3.125 | Live | 124.1 ± 8.4 | 108.1 ± 33.3 |
| Dead | 100.9 ± 6.1 | 100.5 ± 10.5 | |
| 1.563 | Live | 124.3 ± 10.7 | 109.7 ± 27.8 |
| Dead | 105.3 ± 9.4 | 95.0 ± 11.3 | |
| 0.781 | Live | 125.4 ± 9.5 | 112.7 ± 28.8 |
| Dead | 99.8 ± 3.2 | 96.4 ± 9.5 | |
| 0.391 | Live | 122.5 ± 13.5 | 112.4 ± 25.0 |
| Dead | 105.3 ± 9.2 | 97.3 ± 8.7 | |
| 0.195 | Live | 121.6 ± 7.2 | 107.4 ± 25.9 |
| Dead | 101.7 ± 4.6 | 95.0 ± 8.2 | |
| 0 | Live | 100.0 | 100 |
| Dead | 100.0 | 100 |
mean ± SEM for five independent replicates tested in triplicate
3.4. Comparison of HEMA effects on cell viability between mucosa-chip and well-plate platform
Metrics of cell viability and death were more sensitive from the chip than monolayer culture in well-plates. Altered numbers of dead cells occurred at different levels of HEMA in the mucosa-chip (Fig. 6a, particularly compare values at 6.25, 12.5, and 25 mM), while dead signals from well-plate monocultures were not greatly altered (Fig. 6b). After 24 h of HEMA exposure, the mucosa chip live cell area fraction sensitivity to HEMA concentration was −1.7 ± 0.5%/mM and −0.8 ± 0.1%/mM for keratinocytes and fibroblasts, respectively (mean ± standard error; p < 0.01 and p < 0.001, respectively). Similarly, the sensitivity of mucosa chip dead cell number per sampled field to HEMA concentration was 1.0 ± 0.2 dead cells/mM, and 0.5 ± 0.1 dead cells/mM for keratinocytes and fibroblasts, respectively (p < 0.001 for both). In contrast, after 24 h of HEMA exposure, the well-plate monoculture live signal sensitivity to HEMA concentration was −0.1 ± 0.3%/mM and −0.2 ± 0.2%/mM for keratinocytes and fibroblasts, respectively (mean ± standard error; not significantly different from no sensitivity). Similarly, the sensitivity of well-plate dead cell signal was 0.5 ± 0.3%/mM and 0.7 ± 0.1%/mM, respectively (mean ± standard error; p = 0.1 and p < 0.001, respectively).
Fig. 6.
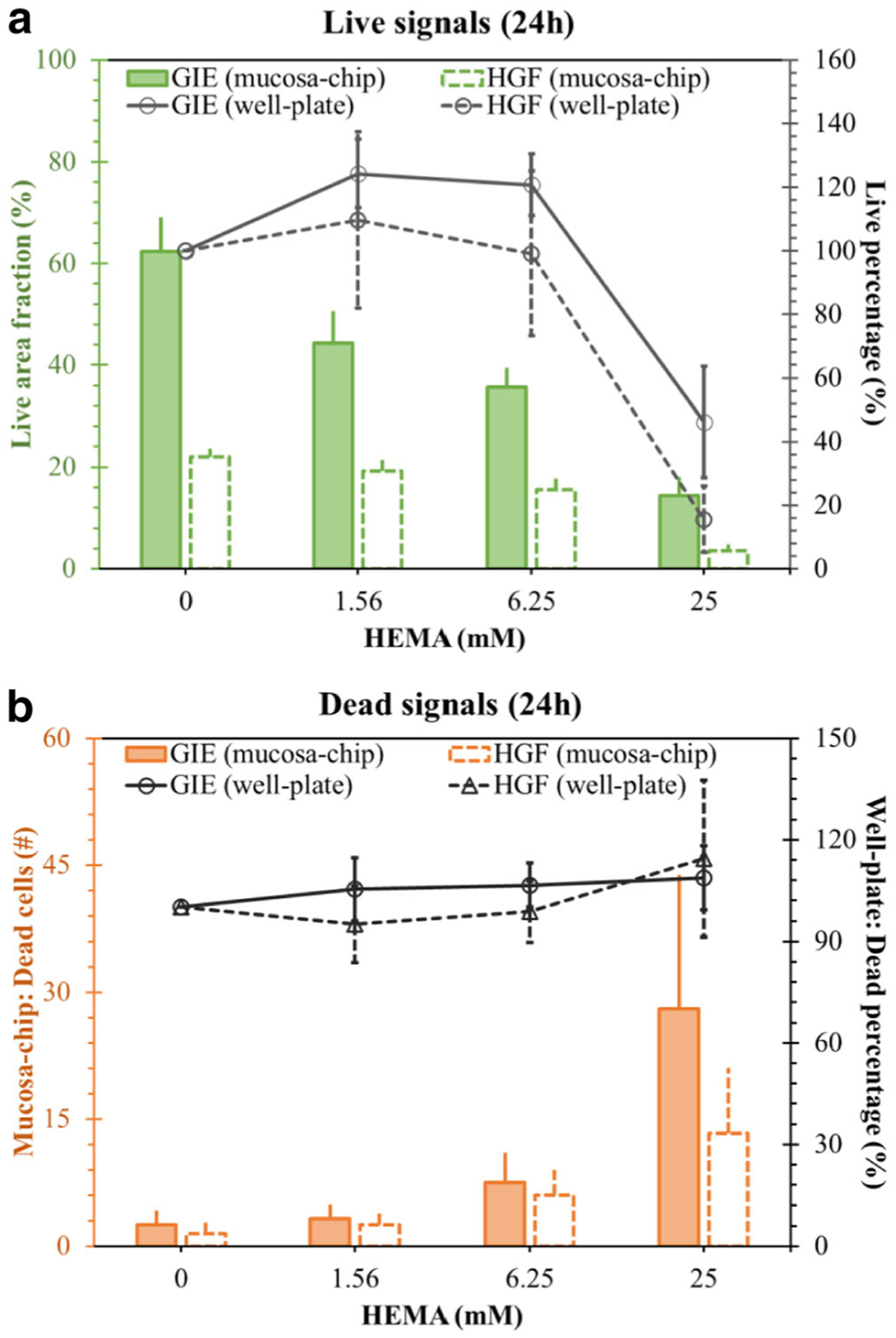
Comparison of (a) live signals and (b) dead signals at 24 h between mucosa-chip (first vertical axis) and well-plate model (secondary vertical axis)
4. Discussion
In this study, we aimed to explore the suitability and sensitivity of the mucosa-chip as a testing platform for cell responses to dental materials in general, using the specific example of HEMA. A major advantage of the mucosal chip in this context was found to be the horizontal layout of the platform which allowed direct observation and assessment of cellular responses to HEMA in a single field-of-view of an inverted microscope. Other advantages of mucosa-chips are the ability to capture keratinocyte and fibroblast layer morphology, their sensitive responses to HEMA in a single-field-of-view. Specifically, the current study goes beyond our previous single-dose, proof-of-concept demonstrations (Rahimi et al. 2018) by assessing the layer-specific responses of the mucosa chip and comparing with well-plate monoculture in a dose-response manner. The results suggest that the mucosa-chip is a promising alternative to traditional platforms or assays to test dental biomaterials by offering a multi-layered tissue geometry, accessible layer-specific information, and higher sensitivity in detecting cellular responses.
With the expansion in demand for restorative and aesthetic dentistry, resin-based dental restorative materials have been developed as implants with conservative, esthetic, and long-lasting restorative properties (Gabriella Teti et al. 2015). Despite improvements in restorative design, HEMA, one of the common monomers in resin-based materials, has been found to be responsible for major clinical failures and to have adverse effects on various vital cellular functions (Krifka et al. 2013; Schweikl et al. 2006a, b). For instance, HEMA has been identified as the cause of persistent inflammatory responses, leading to cellular stress via generation of reactive oxygen species (ROS) (Schmalz et al. 2011). Sustained, elevated levels of ROS are likely to lead to excessive oxidative stress, induce apoptosis, delay cell proliferation and impair the healing process (Loan Khanh et al. 2019; Tran et al. 2019). In this study, the native oral mucosa in mucosa-chips was exposed to varied HEMA concentrations. Cell death post-HEMA treatment in either mucosa-chips or well-plate models was similar to previous reports. However, cells inside the constructs but not in well-plates responded sensitively to HEMA at low concentrations (1.56 and 6.25 mM) (Fig. 6). The dead cells observed in both keratinocyte and fibroblast regions indicates that HEMA was distributed over the whole construct and strongly affected both cell types. A higher live area in keratinocytes than fibroblasts is partly due to the intrinsic nature of keratinocytes that form stratified epithelial layers while fibroblasts tend to stay as individual cells surrounded by extracellular matrix. On the other hand, the higher number of dead keratinocytes might be due to the fact that HEMA had been in direct contact with the apical layer of the mucosal construct, thus causing more damage to keratinocytes. In clinical trials, HEMA leaching from adhesive materials was determined to be at around 1.5–8 mM, close to the lower levels investigated in this study (Schweikl et al. 2006a, b).
The sensitivity comparison of Fig. 6 suggests that mucosa-chips are a more sensitive platform than well-plate culture to test gingival cell responses to dental materials for pre-clinical purposes. Furthermore, the results reveal that HEMA disrupted the cell-cell networks, resulting in a large void in the keratinocyte layer, which would likely lead to compromised barrier function (Figs. 3 and 4). These data are in agreement with studies reporting that HEMA down-regulates several extracellular matrix proteins and disturbs reparative dentinogenesis (About et al. 2002; G. Teti et al. 2009). In future work, the real-time assessment of cellular responses should be performed to assess mucosal responses to HEMA (Franca et al. 2020). Adding an air-liquid interface as found in human oral mucosa, and evaluation of gingival cell differentiation markers would also increase clinical relevancy of the model. Furthermore, the long-term culture of mucosa-chip should be improved to evaluate the long-term effects of dental materials on oral cell responses in mucosa-chip. Finally, future work should test chip-based responses to other dental materials such as triethylene glycol dimethacrylate (TEGDMA) (Kaufman and Skrtic 2019), another commonly used monomer that can leach out from resin composites and have the potential to penetrate into the tissues, to demonstrate the feasibility of this platform as a potential standard testing approach.
5. Conclusions
The mucosa-chip is a promising platform that possesses multi-layered tissue configuration of oral mucosa, mimics the physiological conditions of oral mucosa interface, and allows direct observation of mucosal cells’ responses to dental materials. Compared to a traditional well-plate platform, the mucosa chip presents higher sensitivity in assessing cell layer-specific responses in dose-dependent manner to the dental monomer HEMA in a physiologically relevant range.
Acknowledgements
This effort was supported in part by the American Dental Association (ADA), ADA Foundation, the National Institute of Dental and Craniofacial Research (R01-DE26122-02), and the National Institute of General Medical Science (1R15GM129766-01). Authors gratefully acknowledge donation of HEMA from Esstech, Essington, PA.
Footnotes
Data availability The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest The authors have no conflicts of interest to declare.
References
- About I, Camps J, Mitsiadis TA, Bottero MJ, Butler W, Franquin JC, Influence of resinous monomers on the differentiation in vitro of human pulp cells into odontoblasts. J. Biomed. Mater. Res 63(4), 418–423 (2002). 10.1002/jbm.10253 [DOI] [PubMed] [Google Scholar]
- Bienek DR, Frukhtbeyn SA, Giuseppetti AA, Okeke UC, Pires RM, Antonucci JM, Skrtic D, Ionic dimethacrylates for antimicrobial and remineralizing dental composites. Ann. Dent. Oral Disor 1, 108 (2018a) [PMC free article] [PubMed] [Google Scholar]
- Bienek DR, Frukhtbeyn SA, Giuseppetti AA, Okeke UC, Skrtic D, Antimicrobial monomers for polymeric dental restoratives: Cytotoxicity and physicochemical properties. J Funct Biomat 9(1), 20 (2018b). 10.3390/jfb9010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilitewski U, Genrich M, Kadow S, Mersal G, Biochemical analysis with microfluidic systems. Anal. Bioanal. Chem 377(3), 556–569 (2003). 10.1007/s00216-003-2179-4 [DOI] [PubMed] [Google Scholar]
- Bouillaguet S, Virgillito M, Jc W, Ciucchi B, Holz J, The influence of dentine permeability on cytotoxicity of four dentine bonding systems, in vitro. J. Oral Rehabil 25, 45–51 (1998). 10.1046/j.1365-2842.1998.00205.x [DOI] [PubMed] [Google Scholar]
- Caldas IP, Alves GG, Barbosa IB, Scelza P, de Noronha F, Scelza MZ, In vitro cytotoxicity of dental adhesives: A systematic review. Dent. Mater 35(2), 195–205 (2019). 10.1016/j.dental.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Cho K-H, Yu S-K, Lee M-H, Lee D-S, Kim H-J, Histological assessment of the palatal mucosa and greater palatine artery with reference to subepithelial connective tissue grafting. Anat. cell biol 46(3), 171–176 (2013). 10.5115/acb.2013.46.3.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi M, Teti G, Zago M, Pelotti S, Breschi L, Mazzotti G, Effects of HEMA on type I collagen protein in human gingival fibroblasts. Cell Biol. Toxicol 23(5), 313–322 (2007). 10.1007/s10565-006-0148-3 [DOI] [PubMed] [Google Scholar]
- Franca CM, Tahayeri A, Rodrigues NS, Ferdosian S, Puppin Rontani RM, Sereda G, et al. , The tooth on-a-chip: A microphysiologic model system mimicking the biologic interface of the tooth with biomaterials. Lab Chip 20(2), 405–413 (2020). 10.1039/c9lc00915a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröger S, Michel J, Meyle J, Establishment and characterization of immortalized human gingival keratinocyte cell lines. J. Periodontal Res 43(6), 604–614 (2008). 10.1111/j.1600-0765.2007.01019.x [DOI] [PubMed] [Google Scholar]
- Hanks CT, Craig RG, Diehl ML, Pashley DH, Cytotoxicity of dental composites and other materials in a new in vitro device. J. Oral. Pathol 17(8), 396–403 (1988). 10.1111/j.1600-0714.1988.tb01304.x [DOI] [PubMed] [Google Scholar]
- Hanks CT, Strawn SE, Wataha JC, Craig RG, Cytotoxic effects of resin components on cultured mammalian fibroblasts. J. Dent. Res 70(11), 1450–1455 (1991). 10.1177/00220345910700111201 [DOI] [PubMed] [Google Scholar]
- Hildebrand HC, Hakkinen L, Wiebe CB, Larjava HS, Characterization of organotypic keratinocyte cultures on deepithelialized bovine tongue mucosa. Histol Histopathol 17(1), 151–163 (2002). 10.14670/hh-17.151 [DOI] [PubMed] [Google Scholar]
- Hu P, Rooholghodos SA, Pham LH, Ly KL, Luo X, Interfacial Electrofabrication of freestanding biopolymer membranes with distal electrodes. Langmuir 36(37), 11034–11043 (2020). 10.1021/acs.langmuir.0c01894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illeperuma RP, Park YJ, Kim JM, Bae JY, Che ZM, Son HK, et al. , Immortalized gingival fibroblasts as a cytotoxicity test model for dental materials. J. Mater. Sci. Mater. Med 23(3), 753–762 (2012). 10.1007/s10856-011-4473-6 [DOI] [PubMed] [Google Scholar]
- Izumi K, Kato H, & Feinberg S (2015). Tissue engineered Oral mucosa. In (pp. 721–731)
- Kaufman G, Skrtic D, Morphological and kinetic study of oral keratinocytes assembly on reconstituted basement membrane: Effect of TEGDMA. Arch. Oral Biol 104, 103–111 (2019). 10.1016/j.archoralbio.2019.05.019 [DOI] [PubMed] [Google Scholar]
- Krifka S, Spagnuolo G, Schmalz G, Schweikl H, A review of adaptive mechanisms in cell responses towards oxidative stress caused by dental resin monomers. Biomaterials 34(19), 4555–4563 (2013). 10.1016/j.biomaterials.2013.03.019 [DOI] [PubMed] [Google Scholar]
- Lam RHW, Cui X, Guo W, Thorsen T, High-throughput dental biofilm growth analysis for multiparametric microenvironmental biochemical conditions using microfluidics. Lab Chip 16(9), 1652–1662 (2016). 10.1039/C6LC00072J [DOI] [PubMed] [Google Scholar]
- Lignos I, Maceiczyk R, deMello AJ, Microfluidic technology: Uncovering the mechanisms of Nanocrystal nucleation and growth. Acc. Chem. Res 50(5), 1248–1257 (2017). 10.1021/acs.accounts.7b00088 [DOI] [PubMed] [Google Scholar]
- Loan Khanh L, Thanh Truc N, Tan Dat N, Thi Phuong Nghi N, van Toi V, Thi Thu Hoai N, et al. , Gelatin-stabilized composites of silver nanoparticles and curcumin: Characterization, antibacterial and antioxidant study. Sci. Technol. Adv. Mater 20(1), 276–290 (2019). 10.1080/14686996.2019.1585131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly K, Raub CB, & Luo X (2020a). Tuning the porosity of biofabricated chitosan membranes in microfluidics with co-assembled nanoparticles as template. Mater. Adv, 34–44. doi: 10.1039/D0MA00073F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly K, Rooholghodos S, Rahimi C, Rahimi B, Bienek DR, Kaufman G, … Luo X (2020b, October 4–9, 2020). Oral mucosa-chip as an alternative platform to evaluate the impacts of dental monomers. Paper presented at the 24th international conference on miniaturized Systems for Chemistry and Life Sciences [Google Scholar]
- Moharamzadeh K (2017). Oral mucosa tissue engineering. In Tayebi L & Moharamzadeh K (Eds.), Biomaterials for Oral and Dental Tissue Engineering (pp. 223–244): Woodhead publishing [Google Scholar]
- Moharamzadeh K, Brook IM, Van Noort R, Scutt AM, Thornhill MH, Tissue-engineered oral mucosa: A review of the scientific literature. J. Dent. Res 86(2), 115–124 (2007). 10.1177/154405910708600203 [DOI] [PubMed] [Google Scholar]
- Niu L, Zhang H, Liu Y, Wang Y, Li A, Liu R, et al. , Microfluidic Chip for Odontoblasts in vitro. ACS Biomater. Sci. Eng 5(9), 4844–4851 (2019). 10.1021/acsbiomaterials.9b00743 [DOI] [PubMed] [Google Scholar]
- Rahimi C, Rahimi B, Padova D, Rooholghodos SA, Bienek DR, Luo X, et al. , Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 12(5), 054106 (2018). 10.1063/1.5048938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalz G, Krifka S, Schweikl H, Toll-like receptors, LPS, and dental monomers. Adv. Dent. Res 23(3), 302–306 (2011). 10.1177/0022034511405391 [DOI] [PubMed] [Google Scholar]
- Schweikl H, Spagnuolo G, Schmalz G, Genetic and cellular toxicology of dental resin monomers. J. Dent. Res 85, 870–877 (2006a). 10.1177/154405910608501001 [DOI] [PubMed] [Google Scholar]
- Schweikl H, Spagnuolo G, Schmalz G, Genetic and cellular toxicology of dental resin monomers. J. Dent. Res 85(10), 870–877 (2006b). 10.1177/154405910608501001 [DOI] [PubMed] [Google Scholar]
- Szczepanska J, Poplawski T, Synowiec E, Pawlowska E, Chojnacki CJ, Chojnacki J, Blasiak J, 2-hydroxylethyl methacrylate (HEMA), a tooth restoration component, exerts its genotoxic effects in human gingival fibroblasts trough methacrylic acid, an immediate product of its degradation. Mol. Biol. Rep 39(2), 1561–1574 (2012). 10.1007/s11033-011-0895-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teti G, Mazzotti G, Zago M, Ortolani M, Breschi L, Pelotti S, et al. , HEMA down-regulates procollagen alpha1 type I in human gingival fibroblasts. J. Biomed. Mater. Res. A 90(1), 256–262 (2009). 10.1002/jbm.a.32082 [DOI] [PubMed] [Google Scholar]
- Teti G, Orsini G, Salvatore V, Focaroli S, Mazzotti MC, Ruggeri A, … Falconi M (2015). HEMA but not TEGDMA induces autophagy in human gingival fibroblasts. 6(275). doi: 10.3389/fphys.2015.00275, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tra WMW, van Neck JW, Hovius SER, van Osch GJVM, Perez-Amodio S, Characterization of a three-dimensional mucosal equivalent: Similarities and differences with native Oral mucosa. Cells Tissues Organs 195(3), 185–196 (2012). 10.1159/000324918 [DOI] [PubMed] [Google Scholar]
- Tran HA, Ly KL, Fox KE, Tran PA, Nguyen TH, Immobilization of antimicrobial silver and antioxidant flavonoid as a coating for wound dressing materials. Int. J. Nanomedicine 14, 9929–9939 (2019). 10.2147/ijn.S230214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardar-Sengul S, Arora S, Baylas H, & Mercola D (2009). Expression Profile of Human Gingival Fibroblasts Induced by Interleukin-1β Reveals Central Role of Nuclear Factor-Kappa B in Stabilizing Human Gingival Fibroblasts During Inflammation. 80(5), 833–849. doi: 10.1902/jop.2009.080483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, He Z, Chen Q, Lin J-M, Biochemical analysis on microfluidic chips. TrAC Trends Anal. Chem 80, 213–231 (2016). 10.1016/j.trac.2016.03.013 [DOI] [Google Scholar]


