Abstract
Purpose.
While abundant myeloid cell populations in the pancreatic ductal adenocarcinoma (PDAC) microenvironment have been postulated to suppress anti-tumor immunity, the composition of these populations, their spatial locations, and how they relate to patient outcomes are poorly understood.
Experimental Design.
To generate spatially-resolved tumor and immune cell data at single cell resolution, we developed two quantitative multiplex immunofluorescence assays to interrogate myeloid cells (CD15, CD14, ARG1, CD33, HLA-DR) and macrophages [CD68, CD163, CD86, interferon regulatory factor 5 (IRF5), MRC1 (CD206)] in the PDAC tumor microenvironment. Spatial point pattern analyses were conducted to assess the degree of co-localization between tumor cells and immune cells. Multivariable-adjusted Cox proportional hazards regression was used to assess associations with patient outcomes.
Results.
In a multi-institutional cohort of 305 primary PDAC resection specimens, myeloid cells were abundant, enriched within stromal regions, highly heterogeneous across tumors, and differed by somatic genotype. High densities of CD15+ARG1+ immunosuppressive granulocytic cells and M2-polarized macrophages were associated with worse patient survival. Moreover, beyond cell density, closer proximity of M2-polarized macrophages to tumor cells was strongly associated with disease-free survival, revealing the clinical significance and biologic importance of immune cell localization within tumor areas.
Conclusions.
A diverse set of myeloid cells are present within the PDAC tumor microenvironment and are distributed heterogeneously across patient tumors. Not only the densities but also the spatial locations of myeloid immune cells are associated with patient outcomes, highlighting the potential role of spatially-resolved myeloid cell subtypes as quantitative biomarkers for PDAC prognosis and therapy.
Translational Relevance.
Using multiplex immunofluorescence combined with digital image analysis and supervised machine learning, we comprehensively evaluated the composition, spatial characteristics and prognostic significance of myeloid cells in the PDAC microenvironment in a multi-institutional cohort of >300 patients with resected pancreatic ductal adenocarcinoma (PDAC). Myeloid cells were highly abundant in the human PDAC microenvironment with large inter-tumor heterogeneity. Not only the densities but also the spatial locations of myeloid immune cells were associated with patient outcomes, revealing PDAC microenvironmental complexity with important implications for design and interpretation of immunotherapy clinical trials.
Keywords: macrophage, myeloid immune cell, pancreatic cancer, spatial analysis, tumor microenvironment
INTRODUCTION
Pancreatic cancer is currently the fourth leading cause of cancer-related death in the western world (1,2) and has a 5-year survival rate of less than 9% (1). Over the past several decades, improvements in patient outcomes have been modest (3), reflecting both the lack of effective tools for early detection and a paucity of treatment options, especially for patients with advanced disease (3). While recent advances in immunotherapy have been important to the treatment of a broad variety of cancers, pancreatic ductal adenocarcinoma (PDAC) has proven resistant to treatment with immune checkpoint blockade (ICB), with efficacy shown only in the small subset of patients with microsatellite instability-high (MSI-H) tumors, representing less than 1% of patients (4). One of the presumed contributors to ICB therapy resistance in PDAC is an immunosuppressive tumor microenvironment (5).
Myeloid-derived suppressor cells (MDSCs) are a heterogeneous population of immature myeloid cells that can be divided into two subpopulations, polymorphonuclear MDSCs (PMN-MDSC) and monocytic MDSCs (M-MDSC), based upon their phenotypic and morphologic resemblance to neutrophils and monocytes, respectively (6). The common characteristic of these cells is their ability to suppress adaptive Tcell immunity by blocking the proliferation and activity of T lymphocytes in the tumor microenvironment, thereby attenuating anti-tumor immunity (7). Additionally, preclinical studies have shown that MDSCs may play a role in tumor angiogenesis and in formation of the pre-metastatic niche (8,9). A growing body of evidence indicates that higher levels of circulating MDSCs are associated with advanced disease stage, poor prognosis, and treatment failure across tumor types (5).
Tumor-associated macrophages (TAMs) form an important subpopulation of mature myeloid immune cells. Previous studies in several tumor types have reported an association between higher macrophage density and worse clinical outcome, although contradictory results have been reported (10). Traditionally, macrophages are divided into a pro-inflammatory and anti-tumorigenic M1-phenotype, and an anti-inflammatory and pro-tumorigenic M2-phenotype (10). However, this traditional categorization is increasingly recognized as an over-simplification, as macrophages exist on a spectrum of polarization states between the M1 and M2 phenotypic extremes and exhibit functional plasticity within the tumor microenvironment (11,12).
Despite a potentially important role for pro- and anti-inflammatory myeloid populations in the PDAC microenvironment, the lack of single protein markers to identify these populations has precluded detailed histologic analysis. However, recent advances in multiplex immunofluorescence (mIF) and digital image analysis have enabled simultaneous examination of multiple protein targets in a single tissue section, providing unique insights into cell phenotypes (13). Furthermore, improved spatial point pattern analysis methods allow for better characterization of cellular proximity in the tumor microenvironment and its potential clinical implications (14). We leveraged these advances to develop novel mIF panels and interrogated over 300 surgically resected PDACs with dense clinical and genomic annotation to define the landscape, spatial distribution, and prognostic implications of myeloid cells in the PDAC microenvironment.
MATERIALS AND METHODS
Patients
The study population consisted of patients with PDAC who underwent up-front pancreatectomy with curative intent at the Dana-Farber/Brigham and Women’s Cancer Center (DF/BWCC) between 10/26/2002 and 05/21/2012, at the University of Rochester Medical Center (URMC) between 03/01/2006 and 11/01/2013, and at the Stanford Cancer Institute (SCI) between 09/26/1995 and 05/22/2013 (Supplementary Table S1). Tumor tissue blocks were collected for histologic and molecular evaluation and for construction of tissue microarrays (TMAs). The TMAs were constructed by collecting two 1 mm2 cores from each formalin-fixed paraffin-embedded (FFPE) tumor block. The cores were chosen from tumor areas that exhibited representative overall morphology and immune cell infiltration. Clinicopathological data such as sex, age at surgery, type of pancreatic resection, perioperative chemotherapy/radiotherapy, tumor location, AJCC (7th ed.) pT and pN stages, histologic grade, lymphovascular invasion, and resection margin status were collected from the medical records and by histology slide review, as previously described (15). No patients who had received chemotherapy and/or radiotherapy before surgery were included in the study population. The status of KRAS, CDKN2A, SMAD4 and TP53 was determined using a combination of sequencing and immunohistochemistry, as previously described (15). Patients treated at DF/BWCC signed written informed consent for participation in the study, whereas, at URMC and SCI, informed consent was waived as patients were identified retrospectively, according to institutional review board (IRB)-exempt protocols. The study design was approved by the IRB at each institution. The study was conducted in accordance with the U.S. Common Rule.
OS was defined as time between surgery and death for any reason. Patients with metastatic disease at resection and with 30-day or in-hospital mortality were excluded from the OS analyses (n=18). DFS was defined as time between surgery and disease recurrence, and four additional patients with R2 resection margin status were excluded from these analyses (Supplementary Figure S1).
Multiplex immunofluorescence
Two mIF panels were developed to characterize myeloid cells in the PDAC immune microenvironment. The first panel includes: (a) two myeloid lineage markers: CD14 for the monocytic lineage and CD15 for the granulocytic lineage (6), (b) a suppressive marker ARG1 (16), and (c) two myeloid differentiation markers: CD33, a transmembrane sialic acid-binding receptor highly expressed on myeloid cells in their early stages of maturation (6,17), and HLA-DR, the MHC class II cell-surface receptor that presents antigens and is commonly highly expressed on differentiated macrophages (18) (Supplementary Figure S2). The second panel was designed to identify macrophages and characterize their M1-M2 polarization state. We employed CD68 and CD163 as macrophage markers, defining cells expressing either or both of these markers as macrophages, but inadequate sensitivity as single markers (19,20). The polarization state of each macrophage was then assigned by a comparative calculation of the intensities of M1 polarization (CD86 and IRF5) (10) and M2 polarization (scavenger receptors CD163 and MRC1) markers (10), thereby allowing placement of each identified macrophage at a point on a polarization spectrum. Both panels included 2-(4-amidinophenyl)-1H -indole-6-carboxamidine (DAPI) as a nuclear marker and cytokeratin to identify epithelial cells.
All antibodies were first optimized via chromogenic IHC in PDAC or normal lymph node tissue samples to ensure contextual specificity. The antibodies were then tested in IF, and fluorophore pairings and concentrations were iteratively modifed to maximize the signal-to-noise ratio. Antibodies were then combined into two seven-marker mIF panels in a manner that minimized spectral and spatial overlap and produced results that matched single-plex IHC (Supplementary Figure S3). The mIF staining protocol, consisting of sequential rounds of antigen retrieval, antigen detection, and fluorescent labeling using tyramide signal amplification, was performed on a Leica BOND RX Research Stainer (Leica Biosystems, Buffalo, IL) using 4-μm-thick sections of formalin-fixed paraffin-embedded (FFPE) PDAC TMAs (n=7) (Supplementary Figure S4, Supplementary Tables S2 and S3). All slides for each mIF panel were processed in a single batch, yielding fluorescence signal intensities for each marker that were similar across TMAs (Supplementary Figure S5).
Myeloid cell detection
Stained slides were scanned at 200x magnification and spectrally unmixed using an automated multispectral imaging system (Vectra 3.0, Akoya Biosciences, Hopkinton, MA). Only tumor cores containing invasive adenocarcinoma in at least 5% of total tissue area and which were free of artifacts were selected for further analysis, resulting in an exclusion of 40 tumors from the myeloid cell panel data and 35 tumors from the macrophage panel data. Supervised machine learning algorithms (InForm 2.4.1, Akoya Biosciences) were sequentially employed to perform tissue segmentation, cell segmentation, and cell phenotyping (Supplementary Figure S2), as previously described (13). For the myeloid cell panel, the phenotyping algorithm was trained to identify four cell phenotypes (granulocytic lineage cells: CD15+CD14−CK−, monocytic lineage cells: CD15−CD14+CK−, tumor epithelial cells: CD15−CD14−CK+ and other cells: CD15−CD14CK− ), while the phenotyping algorithm for the macrophage panel recognized three phenotypes (macrophages: CD68+ and/or CD163+, tumor cells: CD68−CD163−CK+ and other cells: CD68−CD163−CK−).
The single cell level data exported from InForm was further processed with the R statistical programing language (version 3.5.3, R Foundation for Statistical Computing, Vienna, Austria). In the myeloid panel, expression levels of HLA-DR (cytoplasm), CD33 (cytoplasm), ARG1 (cytoplasm) and MRC1 (cytoplasm) were used to further classify myeloid cells according to their maturation state and potential ARG1-dependent suppressive activity (Figure 1, Supplementary Figure S2).
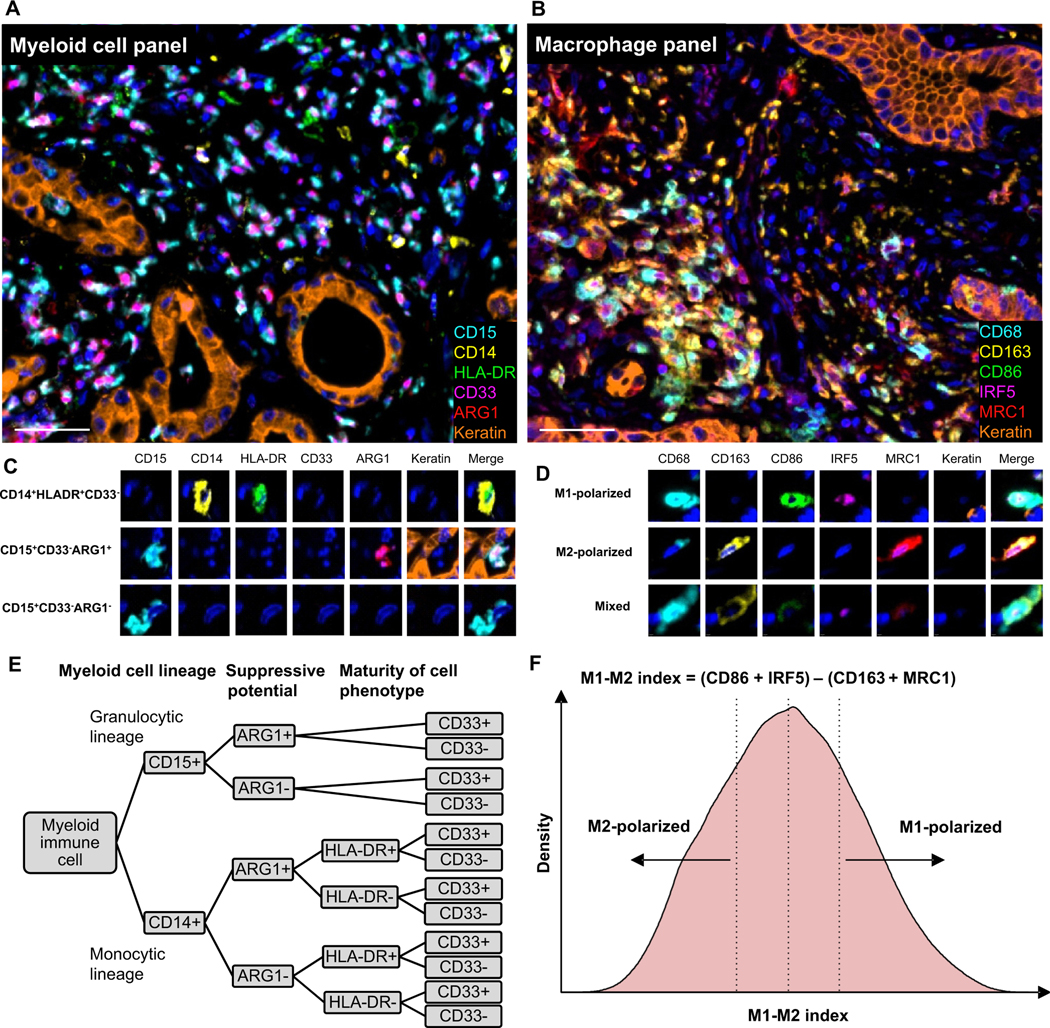
Figure 1. Multiplex analysis of myeloid immune cells in the pancreatic cancer microenvironment.
Examples of multiplex immunofluorescence (mIF) images from the (A) myeloid cell panel and (B) macrophage panel, and (C, D) corresponding cell phenotypes. Gating strategy to classify myeloid cell lineage (CD15, CD14), suppressive potential (ARG1) and cell phenotype maturity (HLA-DR, CD33) (E). Evaluation of macrophage polarization (F). The distribution plot is based on all macrophages in the study cohort (n=296); M1-polarized macrophages were defined as macrophages within the 4th quartile and M2-polarized macrophages as macrophages within the 1st quartile of the M1-M2 index distribution. The scale bar is 50 μm.
To evaluate the polarization state of each macrophage, we first standardized the scale of each polarization marker by calculating signal intensity z-scores within each TMA starting from the mean intensity from each marker in the relevant subcellular compartment of every cell (CD86: cytoplasm, IRF5: nucleus, CD163: cytoplasm, MRC1: cytoplasm). Next, we calculated an M1-M2 index, using percentiles of the scaled intensity z-scores, as “(CD86+IRF5) - (CD163+MRC1)” (Figure 1). In this approach, a high M1-M2 index indicates polarization toward an M1-like phenotype, while a low M1-M2 index indicates polarization toward an M2-like phenotype. For our main analysis, we evaluated macrophages within the most extreme quartiles (1st and 4th) of the polarization spectrum, thereby focusing on more highly polarized phenotypes (Figure 1).
Evaluation of myeloid cell spatial distribution
To evaluate spatial relationships between myeloid cells and tumor cells beyond tissue compartments (i.e. tumor and stroma), we conducted spatial analyses using the “nearest neighbor distance” (NND) and Gcross functions implemented in the spatstat package in R. NND analysis was conducted by calculating the distance (μm) from each point i (i.e. immune cell) to its nearest neighbor point j (i.e. tumor cell) and Gcross analysis was conducted to estimate the probability of finding at least one point j (i.e. immune cell) within a specified radius (μm) of any point i (i.e. tumor cell) (Supplementary Figure S6). The Kaplan-Meier (km) method was employed to correct for edge effects due to unobservable points in Gcross analysis. We selected radii of 10μm, 20 μm, and 30 μm for study, as it has been previously suggested that these distances represent physiologically plausible distances for direct cell-cell interactions (14).
Statistical analyses
Myeloid cell densities and spatial measurements were categorized into quartiles by institution. If a cell population had zero density in more than 25% of patient tumors, then the first quartile was defined as all patient tumors with no detected cells of that phenotype and the remaining patients were divided into three equal groups based on cell population density to form quartiles 2, 3, and 4. Cox proportional hazards regression was used to compute HRs and 95% CIs for DFS and OS. The proportional hazards assumption was satisfied by evaluating a time-dependent variable, which was the product of the exposure variable and time (all P > 0.05). A trend test was performed by including quartile-specific medians as a continuous variable in the Cox regression models and evaluating the Wald Chi-Squared test. In survival analyses, we performed univariable analyses and adjusted for potential prognostic factors in multivariable models as done previously in this patient cohort. Multivariable-adjusted models included age, sex, pathologic N stage (N0, N1, Nx), tumor grade (well/moderately differentiated, poorly differentiated, unknown), lymphovascular invasion (negative, positive, unknown), resection margin status (R0, R1, R2, unknown), and receipt of perioperative treatment (15). To evaluate whether Gtumor:M2 could predict DFS independent of M2-polarized macrophage densities, we used a residual method to account for the high correlation between the two variables (r=0.8). We calculated the residual from a linear regression model in which M2-polarized macrophage density was an independent variable and Gtumor:M2 was the dependent variable. Thus, the residual is an estimate of spatial proximity uncorrelated with cell density. Then, we assessed the association between the residual, as categorized by quartiles, and DFS. Kaplan-Meier curves were constructed to graphically demonstrate survival for time to event analyses and to provide median survival times based upon immune cell exposures. The log-rank test was used to evaluate statistical significance. The Wilcoxon rank-sum test was used to evaluate the association of myeloid cell densities with tumor and patient characteristics. No statistically significant heterogeneity by institutional cohort was identified for major immune cell types and DFS (all P-heterogeneity >0.05), including CD15+ARG1+ cell density, M2-polarized macrophage density, and M2-polarized macrophage spatial localization by Gcross function. Two-sided P values of ≤0.05 were considered statistically significant. All statistical analyses were performed with SAS 9.4 software (SAS institute, Cary, NC, USA).
RESULTS
Myeloid cell densities in resected pancreatic cancer
Two mIF panels were developed to characterize myeloid cells in the PDAC immune microenvironment (Figure 1). The first panel was designed to identify CD14+ monocytic lineage cells and CD15+ granulocytic lineage cells and assess their ARG1-dependent suppressive potential (ARG1; arginase-1) and maturity (CD33, HLA-DR). The second panel identified macrophages and evaluated macrophage polarization by a comparative calculation of the intensities of M1 polarization (CD86 and IRF5, interferon regulatory factor 5) (10) and M2 polarization [scavenger receptors CD163 and MRC1 (CD206)] markers (10), thereby allowing placement of each identified macrophage at a point on a polarization spectrum.
Resected PDACs were analyzed from 305 patients treated at three cancer centers in the United States (Supplementary Table S1). All patients underwent up-front surgical resection, yielding 121 (40%) patients with poorly differentiated tumors and 211 (69%) with lymph node positive disease. Of the 305 patients, the median age was 67 (range, 59–73) years, and 138 (45%) were women. Germline and somatic alterations, such as KRAS, CDKN2A, SMAD4 and TP53 mutations, for these patients have been described previously (15,21) and are similar to those seen in other large cohorts of patients with PDACs (22,23). In total, 159,033 CD15+ granulocytic cells, 219,379 CD14+ monocytic cells, 462,481 macrophages, and 812,758 (myeloid panel)/973,761 (macrophage panel) tumor cells were phenotyped using our myeloid cell and macrophage-focused mIF panels. Cell densities were obtained from 496 tumor cores for the myeloid cell and 513 tumor cores for the macrophage panel, yielding data for 290 (average 1.7 cores/tumor) and 296 tumors (average 1.7 cores/tumor), respectively. To evaluate the representativeness of tissue cores for the original tumor, whole-slide sections were also evaluated for 27 cases with the myeloid cell panel and 29 cases with the macrophage panel. Spearman correlation coefficients between the cell densities in TMA cores and whole-slide sections were 0.83 for CD15+ myeloid cells, 0.58 for CD14+ myeloid cells and 0.46 for macrophages (Supplementary Figure S7).
Overall, myeloid cells were present in high abundance (on average, one myeloid cell for every 2.1 tumor cells) and myeloid cell subtype densities varied widely across tumors (8.8-fold difference between the 10th and 90th total myeloid immune cell density percentiles) (Figure 2, Supplementary Table S4, Supplementary Figure S8). Examination of specific myeloid cell types revealed that monocytic lineage cells (CD14+) were more common than granulocytic lineage cells (CD15+), and both cell types had higher densities in stromal compared with intraepithelial regions (all P<0.05). ARG1 expression was common in granulocytic cells but rare and dim, if present, in monocytic cells. Strong CD33 expression, suggestive of an immature myeloid cell phenotype, was rare in both monocytic and granulocytic cells, while the maturity marker HLA-DR was commonly expressed on CD14+ cells. Macrophage density (in particular, M2-polarized macrophage density) was substantially higher in stromal compared with intraepithelial areas [median stromal:intraepithelial macrophage ratio 5.8 (for M2-polarized macrophages 7.4), both P <0.05].
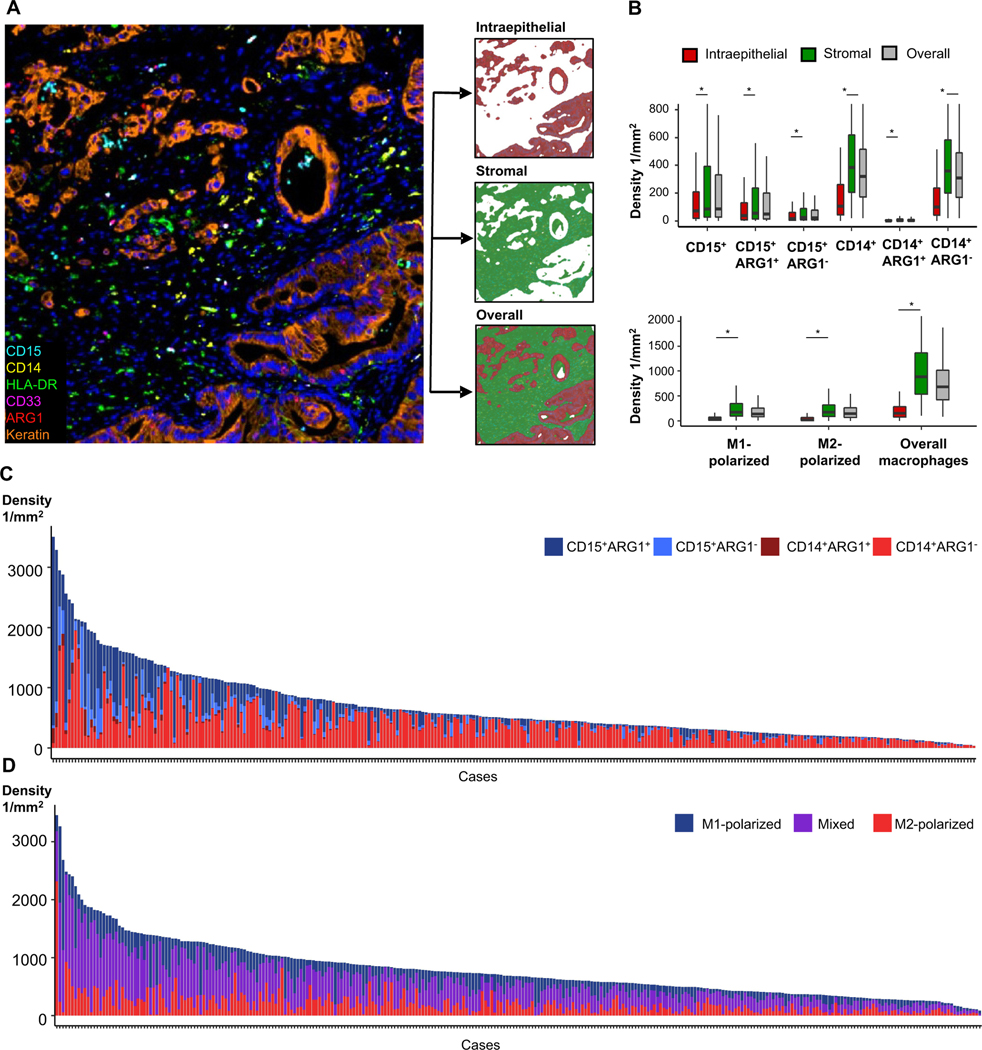
Figure 2. Myeloid immune cell densities within tumor intraepithelial, stromal, and whole tissue areas.
Tumors were segmented into tumor intraepithelial and stromal compartments, enabling compartment-specific density measurements (A). Intraepithelial, stromal, and overall densities of CD15+ granulocytic cells, CD14+ monocytic cells and M1-polarized, M2-polarized and overall macrophages (B). All myeloid cell populations were present at higher density in stromal compared to intraepithelial compartments. P values were calculated with Wilcoxon signed-rank test. *P <0.001. Overall granulocytic and monocytic myeloid cell densities according to ARG1 status (C) and M1-polarized, M2-polarized and mixed phenotype macrophage densities (D) demonstrate significant inter-tumoral variability (n=290 and n=296).
Myeloid cell subsets, clinicopathologic characteristics, and PDAC driver gene alterations
Given the high abundance and substantial interpatient variability of myeloid cells in the PDAC microenvironment, we tested whether myeloid cell densities were associated with clinicopathologic characteristics or alterations in the four main PDAC driver genes (KRAS, CDKN2A, SMAD4, and TP53) (15). No statistically significant associations were found between myeloid cell densities and clinicopathological characteristics such as age, sex, pathologic T and N stage, tumor grade, perineural invasion, or lymphovascular invasion, with the exception of the associations between higher density of overall macrophages and M1-polarized macrophages with nodal metastases (P=0.02 and P=0.01, respectively) ( Supplementary Table S5). We next examined PDAC driver gene alterations and myeloid cell densities (Supplementary Table S6). Higher CD15+ granulocytic cell density was associated with TP53 alterations (P=0.04), due to higher densities of both CD15+ARG1+ and CD15+ARG1− cells in TP53-altered tumors. Lower CD14+ monocytic cell density, mostly representing CD14+ARG1− cells, was associated with SMAD4 inactivation (P=0.04). In the macrophage panel, higher overall macrophage density was associated with TP53 alterations (P=0.04). Thus, the densities of myeloid immune cell populations were not associated with clinicopathologic characteristics in our patient cohort, but these populations did vary according to driver gene status.
Myeloid cell densities and patient outcomes
Patients with resected PDAC have a high risk for disease recurrence and many have short overall survival times (24–26). The likelihood of long-term survival has been associated with several pathologic factors, such as lack of lymph node involvement and tumor differentiation (27,28), but also T cell infiltration and quantity and quality of tumor neoantigens (14,29,30). We hypothesized that outcomes in patients with resected PDAC would be associated with myeloid immune cell population densities, and particularly that poor survival would be associated with greater tumor infiltration by immunosuppressive myeloid cell populations. Given that several of the detected cell phenotypes were rare in the PDAC microenvironment (Supplementary Table S4), we focused subsequent analyses on the main myeloid cell lineages further classified by ARG1 expression (i.e. CD15+ARG1+ and CD15+ARG1−; CD14+ARG1+ and CD14+ARG1−) as well as M1- and M2-polarized macrophages. Among these cell types, ARG1+ granulocytic and monocytic myeloid cells, along with M2-polarized macrophages, were hypothesized to have the highest potential for orchestrating an immunosuppressive microenvironment and limiting disease control.
Univariable Cox regression models and Kaplan-Meier curves revealed associations between higher overall CD15+ARG1+ granulocytic cell density and shorter disease-free survival (DFS; median DFS time for the top vs. bottom quartile, 9.9 months vs. 17.1 months; Table 1, Supplementary Figure S9). The median CD15+ARG1+ cell density in the bottom quartile was 6.6 cells/mm2, compared with 545.2 cells/mm2 in the top quartile, again demonstrating the high degree of variability in myeloid cell infiltration across patient tumors. In a multivariable-adjusted Cox regression model, the association between higher overall CD15+ARG1+ cell density and worse DFS was independent of other covariates (top versus bottom quartile: hazard ratio (HR) 1.82, 95% confidence interval (CI) 1.19–2.77, Ptrend = 0.002) (Table 1), whereas no significant association was observed between this population and OS (Supplementary Table S7). To further investigate this cell population, CD15+ARG1+ cells were classified based upon their location in either stromal or intraepithelial regions. The association of stromal CD15+ARG1+ cell density was strongly associated with worse DFS when comparing the top to the bottom quartile (HR 1.95; 95%CI, 1.25–3.02; Ptrend=0.002; Supplementary Table S8). Similar results were seen in analyses of overall survival (OS), with patients in the top quartile of CD15+ARG1+ granulocytic cell density having shorter median OS time than those in the bottom quartile (16.9 versus 26.7 months, Supplementary Table S7, Supplementary Figure S9).
Table 1.
Univariable and multivariable Cox regression models for disease-free survival according to ARG1+ and ARG1− monocytic (CD14+) and granulocytic (CD15+) cells.
| Myeloid immune cell subseta | No. of patients | Cell Density, Median (IQR) (1/mm2) | Median survival (months) | Univariable HR (95% CI) | Multivariable HR (95% CI)b |
|---|---|---|---|---|---|
| CD15+ARG1+ granulocytic cells | |||||
| Q1 | 72 | 6.6 (3.2–9.9) | 17.1 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 71 | 27.5 (22.8–38.3) | 13.4 | 1.17 (0.79–1.75) | 0.98 (0.65–1.48) |
| Q3 | 73 | 119.2 (79.6–164.4) | 10.1 | 1.64 (1.10–2.42) | 1.32 (0.87–1.99) |
| Q4 | 70 | 545.2 (341.8–771.3) | 9.9 | 1.67 (1.13–2.48) | 1.82 (1.19–2.77) |
| Ptrend | 0.019 | 0.002 | |||
|
CD15+ARG1− granulocytic cells | |||||
| Q1 | 71 | 3.1 (1.5–6.4) | 14.8 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 72 | 14.8 (11.4–20.1) | 11.1 | 1.45 (0.97–2.16) | 1.16 (0.77–1.75) |
| Q3 | 72 | 49.3(34.9–64.1) | 10.6 | 1.56 (1.05–2.32) | 1.37 (0.90–2.09) |
| Q4 | 71 | 170.2 (129.2–225.5) | 10.9 | 1.23 (0.83–1.85) | 0.98 (0.64–1.51) |
| Ptrend | 0.882 | 0.500 | |||
|
CD14+ARG1+
monocytic cells | |||||
| Q1 | 114 | 0.0 (0.0–0.0) | 14.3 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 58 | 3.1 (2.2–3.9) | 11.6 | 1.00 (reference) | 1.34 (0.92–1.95) |
| Q3 | 59 | 8.7 (5.7–14.2) | 12.0 | 1.25 (0.87–1.80) | 1.06 (0.70–1.61) |
| Q4 | 56 | 34.7 (21.3–71.8) | 9.7 | 1.12 (0.76–1.66) | 1.38 (0.91–2.09) |
| Ptrend | 0.156 | 0.197 | |||
|
CD14+ARG1− monocytic cells | |||||
| Q1 | 70 | 108.7 (68.9–143.9) | 11.2 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 72 | 229.4 (190.1–289.2) | 11.5 | 0.97 (0.66–1.44) | 1.20 (0.79–1.83) |
| Q3 | 73 | 416.5 (339.5–459.7) | 14.1 | 0.67 (0.45–0.99) | 0.74 (0.49–1.13) |
| Q4 | 71 | 666.3 (584.5–870.2) | 10.5 | 1.02 (0.70–1.48) | 0.90 (0.60–1.33) |
| Ptrend | 0.612 | 0.247 | |||
The densities of myeloid immune cell subsets are based on the overall densities in tumor area (intraepithelial + stromal).
Cox proportional hazards regression model adjusted for age, sex, pathologic N stage (N0, N1, Nx), tumor grade (well/moderately differentiated, poorly differentiated, unknown), lymphovascular invasion (negative, positive, unknown), resection margin status (R0, R1, R2, unknown), receipt of perioperative treatment.
Abbreviations: CI:confidence interval; HR:hazard ratio; IQR:interquartile range; Q:quartile
In contrast to CD15+ARG1+ granulocytic cells, few CD14+ARG1+ monocytic cells were present in the PDAC microenvironment and CD14+ARG1+ cell density was not associated with DFS or OS in univariable or multivariable-adjusted models (Table 1; Supplementary Table S7). Densities of the ARG1− myeloid cells (CD15+ARG1− and CD14+ARG1−) were also not associated with DFS or OS (Table 1, Supplementary Table S7). Myeloid cells undergo a stepwise process of maturation as they acquire their terminally differentiated phenotypes. Given that myeloid cells with varying degrees of maturation could be recruited to the tumor microenvironment, this may result in functional differences for these cell populations based on their cell maturity. In an exploratory analysis by cell maturity, higher overall CD14+HLA-DR+CD33− cell density, representing a mature/differentiated monocytic cell, was associated with better DFS and OS (Supplementary Table S9).
We next examined the association of infiltrating macrophages with clinical outcomes in our patient population with resected PDAC. In univariable analyses, higher macrophage density overall and in stromal regions was associated with modestly worse DFS, but the associations were attenuated with multivariable adjustment and not evident for OS (Supplementary Table S10). When macrophages were classified according to polarization status, higher overall and stromal M2-polarized macrophage densities were also modestly associated with worse DFS, while M1-polarized macrophage densities were not (Table 2, Supplementary Table S11). Given that the relative balance of M1-polarized versus M2-polarized macrophages may be more important than their densities considered separately, we also calculated M1:M2 density ratios but found no associations with survival (data not shown). Although these data somewhat supported the hypothesis that M2-polarized macrophage densities are associated with worse patient outcomes, the associations were modest in magnitude and less pronounced than for CD15+ARG1+ granulocytic cell densities.
Table 2.
Univariable and multivariable Cox regression models for disease-free survival according to M1- and M2-polarized macrophage density and Gcross spatial measurements between tumor cells and macrophages within 20μm radius.
| Myeloid immune cell subseta | No. of patients | Cell Density, Median (IQR) (1/mm2) | Median survival (months) | Univariable HR (95% CI) | Multivariable HR (95% CI)b |
|---|---|---|---|---|---|
| M1-polarized macrophages | |||||
| Cell density | |||||
| Q1 | 69 | 47.8 (33.2–61.0) | 14.2 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 75 | 109.3 (93.6–125.3) | 11.6 | 1.04 (0.70–1.55) | 0.86 (0.56–1.31) |
| Q3 | 75 | 184.1 (159.0–217.1) | 14.4 | 0.99 (0.67–1.47) | 1.02 (0.68–1.52) |
| Q4 | 73 | 378.7 (304.6–488.3) | 10.5 | 1.05 (0.71–1.56) | 0.76 (0.50–1.16) |
| Ptrend | 0.863 | 0.254 | |||
| Gtumor:M1, 20μm radius | |||||
| Q1 | 71 | 0.03 (0.02–0.04) | 14.3 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 74 | 0.06 (0.05–0.07) | 13.8 | 1.29 (0.87–1.92) | 1.22 (0.80–1.84) |
| Q3 | 73 | 0.10 (0.09–0.11) | 11.5 | 1.17 (0.78–1.74) | 1.11 (0.72–1.71) |
| Q4 | 74 | 0.18 (0.15–0.23) | 10.3 | 1.35 (0.91–1.98) | 1.10 (0.72–1.68) |
| Ptrend | 0.222 | 0.939 | |||
| M2-polarized macrophages | |||||
| Cell density | |||||
| Q1 | 71 | 37.3 (20.9–51.3) | 12.0 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 73 | 103.7 (83.9–119.5) | 15.9 | 0.75 (0.50–1.11) | 0.70 (0.46–1.06) |
| Q3 | 75 | 192.5 (167.1–225.5) | 13.5 | 0.90 (0.61–1.32) | 1.05 (0.71–1.56) |
| Q4 | 73 | 353.0 (297.9–479.3) | 9.9 | 1.20 (0.84–1.76) | 1.32 (0.90–1.94) |
| Ptrend | 0.149 | 0.035 | |||
| Gtumor:M2, 20μm radius | |||||
| Q1 | 73 | 0.009 (0.005–0.014) | 15.1 | 1.00 (reference) | 1.00 (reference) |
| Q2 | 73 | 0.03 (0.02–0.03) | 11.1 | 1.41 (0.94–2.11) | 1.23 (0.81–1.88) |
| Q3 | 73 | 0.06 (0.05–0.07) | 10.1 | 1.52 (1.02–2.26) | 1.55 (1.01–2.39) |
| Q4 | 73 | 0.13 (0.11–0.18) | 10.2 | 1.83 (1.25–2.70) | 1.72 (1.14–2.60) |
| Ptrend | 0.004 | 0.009 | |||
The densities of myeloid immune cell subsets are based on the overall densities in tumor area (intraepithelial + stromal). M1-polarized macrophages were defined as macrophages in the 4th and M2-polarized macrophages as macrophages in the 1st quartile of M1-M2-index distribution.
Cox proportional hazards regression model adjusted for age, sex, pathologic N stage (N0, N1, Nx), tumor grade (well/moderately differentiated, poorly-differentiated, unknown), lymphovascular invasion (negative, positive, unknown), resection margin status (R0, R1, R2, unknown), receipt of perioperative treatment.
Abbreviations: CI:confidence interval; HR:hazard ratio; IQR:interquartile range; Q:quartile
Spatial location of myeloid cells and patient outcomes
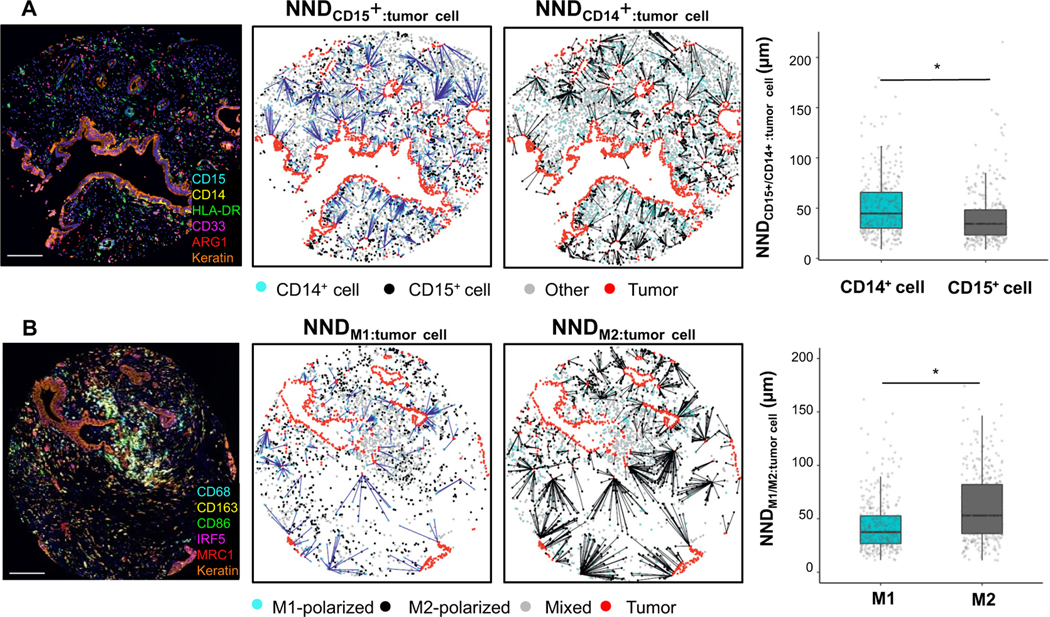
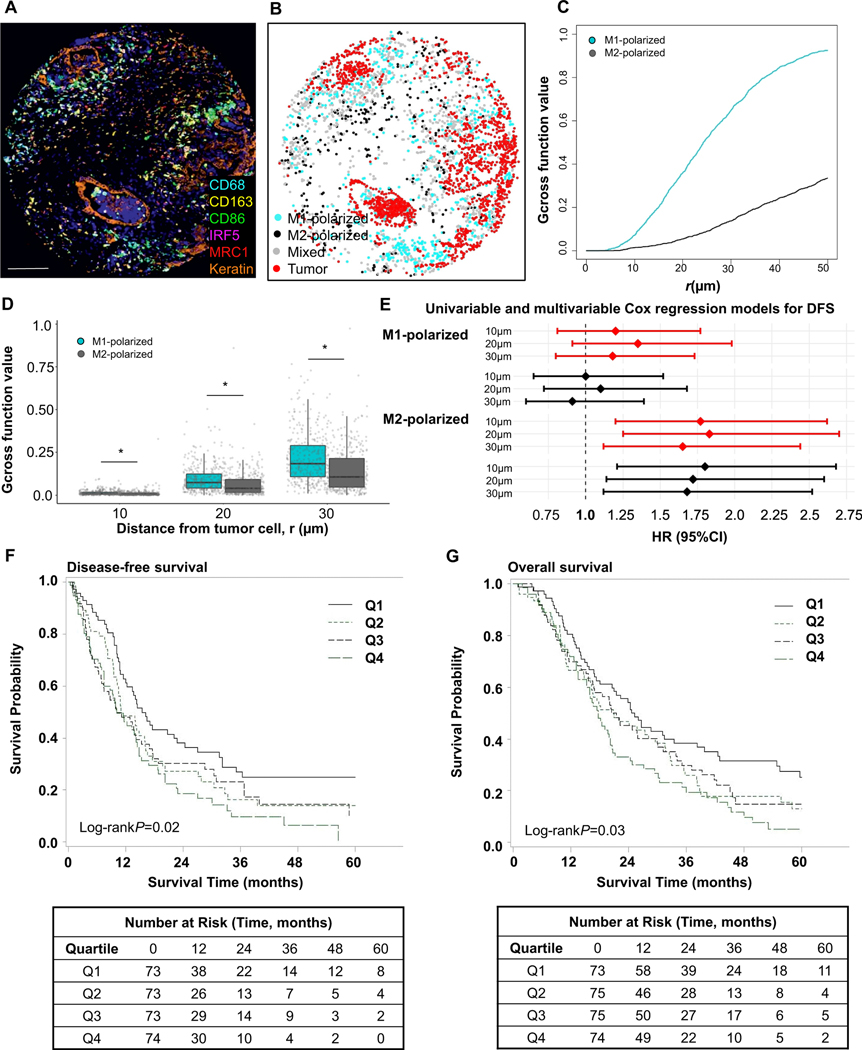
During the course of image analysis, we observed that macrophages near tumor cells often expressed high levels of M1 markers (CD86 and IRF5) whereas macrophages with higher M2 marker (CD163 and MRC1) intensities were often located further from tumor cells (Figure 3). To verify this observation, we performed a “nearest neighbor distance” (NND) analysis, measuring the distance from each M1-polarized or M2-polarized macrophage to the nearest tumor cell (Figure 3). This analysis revealed that M1-polarized macrophages were, on average, closer to tumor cells than M2-polarized macrophages. We also expanded this evaluation to include the spatial location of granulocytic CD15+ and monocytic CD14+ cells and found that CD15+ cells lie closer to tumor cells as compared to CD14+ cells. These findings led us to hypothesize that differential spatial locations for M1- and M2- polarized macrophages might represent differences in their biologic function with prognostic significance. To more precisely model the spatial relationships between tumor cells and macrophages, we utilized the Gcross (Gtumor:macrophage) function to evaluate the probability of a tumor cell co-localizing with at least one macrophage within a specified radius. High Gtumor:macrophage values indicate a high density of macrophages located close to tumor cells. Consistent with the NND analysis, Gtumor:M1 was higher than Gtumor:M2 across radii of 10 μm, 20 μm, and 30 μm (Figure 4). To test whether proximity between tumor cells and differentially polarized macrophages was associated with patient outcomes, we conducted a Cox regression analysis using Gcross function values at varying radii. This analysis indicated that high Gtumor:M2 at radii of 10 μm, 20 μm, and 30 μm were associated with worse DFS independent of potential confounders (Table 2, Figure 4, Supplementary Table S12). For example, the univariable- and multivariable-adjusted HRs for DFS were 1.83 (95%CI 1.25–2.70; Ptrend=0.004) and 1.72 (95%CI 1.14–2.60; Ptrend=0.009), comparing the top to the bottom quartile of Gcross function values at a radius of 20 μm for M2-polarized macrophages (Table 2). In contrast, no associations were found between M1-polarized macrophage spatial location and DFS using the Gtumor:M1 metric (Table 2, Figure 4, Supplementary Table S12). Similar findings were identified in analyses of OS (Supplementary Table S12). Thus, the proximity of immunosuppressive M2-polarized macrophages to tumor cells was strongly associated with patient survival.
Figure 3. Nearest neighbor distance (NND) analysis for myeloid immune cells and tumor cells.
Example of a tumor stained with the myeloid cell panel (A), NND calculated from granulocytic cells (CD15+) and monocytic cells (CD14+) to their nearest tumor cells, and boxplot of the average NND between myeloid immune cells and tumor cells. Example of a tumor stained with the macrophage panel (B), NND calculated from M1- and M2-polarized macrophages to their nearest tumor cells, and boxplot of the average NND between macrophages and tumor cells. P values were calculated with Wilcoxon rank-sum test. *P <0.001. The scale bar is 200 μm.
Figure 4. Spatial analyses of M1-polarized and M2-polarized macrophage populations with Gcross function.
Examples of multiplex immunofluorescence image (A), corresponding phenotype map with M1- and M2-polarized macrophages (B), and Gcross (Gtumor:M1 and Gtumor:M2) function plot (C). Boxplot of the comparison between Gtumor:M1 and Gtumor:M2 function values at 10μm, 20μm and 30μm radii. P values were calculated with Wilcoxon sum-rank test. *P <0.001 (D). Higher Gtumor:macrophage values indicate higher densities of macrophages located close to tumor cells. M1-polarized macrophages are located closer to tumor cells than M2-macrophages at all radii evaluated up to 50 μm. Forest plot of univariable (red) and multivariable (black) Cox regression models for disease-free survival (DFS) according to Gcross spatial measurements between tumor cells and M1- and M2-polarized macrophages. HRs are for the top compared to the bottom quartile; closer location of M2-polarized macrophages to tumor cells was associated with worse patient outcomes, while this association was not identified for M1-polarized macrophages (E). Kaplan-Meier disease-free survival (F) and overall survival (G) curves according to Gtumor:M2 function values within 20μm. P values were calculated with log-rank test. The scale bar is 200 μm.
To evaluate the relative contributions of M2-polarized macrophage density and spatial location to patient survival, we categorized patients into four groups based on the M2-polarized macrophage density and Gcross function values. Patients with tumors harboring a higher density of M2-polarized macrophages and a higher Gtumor:M2 function values (i.e. M2-polarized macrophages located closer to tumor cells) had the worst DFS across radii of 10 μm, 20 μm, and 30 μm (Supplementary Table S13), although Gcross values appeared to contribute more strongly than macrophage densities to the associations with patient survival. Thus, we examined the independent contributions to patient survival of M2-polarized macrophage density and proximity to tumor cells. Notably, the association between high Gtumor:M2 function values and shorter DFS was independent of cell density (Supplementary Table S14), emphasizing the importance of considering immune cell location relative to tumor cells.
DISCUSSION
The PDAC microenvironment is characterized by a dense, fibrotic stroma and prominent myeloid cell infiltration, which is thought to contribute to localized immunosuppression that impedes adaptive Tcell immunity (4). Nevertheless, PDAC patients with high T cell infiltration in the tumor microenvironment and neoantigen qualities promoting T cell responses have improved survival (29,30). These results suggest that therapies combining immune checkpoint blockade and inhibitors of immunosuppressive myeloid cells could potentially serve as an effective therapeutic strategy for patients with PDAC (31–33). While such strategies have shown promise in pre-clinical studies and early clinical trials (31,34), such approaches remain limited by our incomplete understanding of myeloid cell populations in human PDAC. In the current study, we analyzed the composition, spatial characteristics, and prognostic significance of myeloid cells in a multi-institutional cohort of over 300 resected PDAC specimens. We found that the PDAC microenvironment was characterized by high numbers of granulocytic and monocytic myeloid cells, but with large inter-patient variability in density and location of these cell types. Notably, composition and spatial locations of myeloid cells were strong, independent predictors of patient outcomes. In particular, high density of CD15+ARG1+ granulocytes was associated with significantly shorter survival time, but a similar relationship was not identified for macrophages. However, patients with closer proximity of M2-polarized macrophages to tumor cells had significantly worse survival after cancer resection, suggesting that not only cell densities but also their spatial location have important clinical implications. Furthermore, myeloid cell densities were associated with alterations in PDAC driver genes, suggesting that tumor intrinsic factors may influence the PDAC microenvironment and hold importance in understanding therapeutic efficacy for ongoing immune-oncology clinic trials. While a previous study reported an association between SMAD4 inactivation and high CD15+ myeloid cell density in PDAC (35), our study could not replicate this finding. However, we observed that higher CD15+ granulocytic cell density was associated with TP53 alterations, and lower CD14+ monocytic cell density was associated with SMAD4 inactivation.
Many of the previously published studies focused on myeloid cells in PDAC have investigated mouse models (36,37) or circulating myeloid cells in peripheral blood (38–42). However, murine myeloid cell populations are not fully analogous to human myeloid cell populations, and myeloid cells in the peripheral circulation are often different from their tissue-resident counterparts (6,43,44). Previous studies with human tissue cohorts have suggested associations between immunosuppressive myeloid cells and worse patient outcomes, but these studies have been limited by relatively small sample size or limited marker detection strategies (41,45–48). Furthermore, these prior studies have primarily focused on overall myeloid cell densities, without assessing cell subtypes or evaluating spatial relationships with tumor cells. In the current study, we utilized multiplex IF panels to characterize the expression of multiple markers in myeloid cells at single cell resolution. We found that myeloid cell densities were significantly higher in stromal areas than intraepithelial areas, and that monocytic cells were, on average, more common than granulocytic cells. Despite their lower abundance, granulocytic cells expressed ARG1 at much higher levels than monocytic cells, consistent with prior studies in other tumor types that identified PMN-MDCSs frequently employing ARG1 as a mechanism for Tcell suppression (41,46). Similarly, while monocytic cells have been reported to suppress Tcell function via an ARG1-dependent mechanism in some infectious diseases, such as with hepatitis C and human deficiency virus (HIV) infections, the role of the ARG1 pathway in tumor-associated monocytic cells has remained controversial (49). We also found that higher CD15+ARG1+ granulocytic cell density was associated with worse clinical outcomes. Although granulocytic cells may use other mechanisms, such as reactive oxygen species (ROS) production, to suppress Tcell activity (6), our findings suggest that ARG1 expression may represent one of the key characteristics defining prognostically divergent populations of CD15+ cells in the PDAC microenvironment.
Prior studies have reported increased levels of MDSCs, especially PMN-MDSCs, in PDAC tissue relative to healthy control pancreas and also in PDAC tissue upon tumor progression (39–41). Our study suggested that both immature granulocytic (CD15+CD33+) and monocytic cells (CD14+HLA-DR−CD33+), regardless of ARG1 expression, were uncommon in the PDAC microenvironment. While our resection cohort precludes evaluation of myeloid cell dynamics over time, these results suggest a high degree of plasticity within myeloid cell populations and raise the possibility that myeloid cells rapidly mature and differentiate when they exit the circulation and enter the tumor microenvironment. Nevertheless, a continuum is likely present between immature myeloid cells and their differentiated progeny, and the precise relationships between PMN-MDSCs and neutrophils, as well as M-MDSCs and TAMs, in the PDAC microenvironment requires further study (18,43).
Higher density of macrophages in the PDAC microenvironment has been associated with worse prognosis (10). However, estimates of M1- and M2-polarized macrophages in PDAC have varied greatly, likely due to variability in marker selection and methods used for their detection (12,48,50,51). Since no known protein markers are exclusive for M1 or M2 subpopulations, accurate assignment of macrophage polarization states necessitates combinatorial, multi-marker approaches. We investigated the macrophage polarization spectrum at the single cell level based on the simultaneous measurement of four polarization markers with construction of a polarization index. Furthermore, recent advances in spatial analysis methodologies have enabled more sophisticated exploration of the spatial relationship between immune and tumor cells (14,52). We found that M1-polarized macrophages were located, on average, closer to tumor cells than M2-polarized macrophages. This result is similar to that from a recent study of gastric cancer (53), and suggests an interplay between tumor cells and mechanisms that govern macrophage polarization. By combining a novel, multi-marker, macrophage polarization index and single cell localization approaches, we identified a strong association between proximity of M2-polarized macrophages to tumor cells and patient outcomes. These results highlight the importance of considering spatial relationships of immune and tumor cells, in addition to the presence or absence of specific immune cell subtypes in the tumor microenvironment.
Our study has several limitations that require consideration. Although our mIF approach simultaneously incorporated more myeloid cell markers than possible using traditional IHC and enabled us to investigate the overall landscape of immunosuppressive cells in the PDAC microenvironment, a consensus set of protein markers for MDSCs has not yet been defined and therefore differences in marker selection exist across studies (6,49). Thus, future studies incorporating alternative myeloid cell markers could reveal additional insights into the significance of myeloid cells in PDAC. Our study concentrated on immunosuppressive cells in the myeloid lineage but did not include markers for other immune cell types or non-immune cells such as fibroblasts, necessitating future studies to evaluate the importance of these other cell types in the PDAC microenvironment. Thus, future studies will be needed to assess which immunosuppressive cells influence T cell exclusion and exhaustion in the PDAC microenvironment. Our study was performed using TMAs that contain cores from representative areas of each tumor. To maximize the generalizability of the findings to whole tumor mass, we examined multiple cores from each tumor and combined the results to better estimate immune cell densities for patients with resected PDAC. Furthermore, we compared tissue cores to whole slide sections in a subset of cases and identified moderate to high correlations. Our study tested several hypotheses, increasing the risk of type I statistical error. However, our study was hypothesis-driven and based on a strong biologic rationale for the importance of immunosuppressive myeloid cell types in the tumor microenvironment. Furthermore, immune cell subsets can be related to one another, as can occur when daughter cells with distinct phenotypes are derived from the same ancestor cell. Nevertheless, independent studies are needed to confirm the described findings.
Our study also has important strengths. Our cohort of resected, previously untreated tumors was drawn from multiple institutions across the United States, included a large sample size, and had extensive clinicopathologic, immunohistochemical and molecular annotation. Our customized mIF methods were optimized specifically to detect myeloid cell markers present in PDAC and allowed for co-detection of multiple markers at the single cell level, thereby enabling high-confidence cell phenotyping and accurate quantification of true cell densities rather than just marker positivity. These densities, reported as cell number per mm2, stand in contrast to most prior PDAC immune cell studies that provide only semi-quantitative measurements, and can serve as reference points for future studies. Our approach using four polarization markers to calculate a continuous M1-M2 polarization index enables better characterization of macrophage phenotypes than prior tissue-based approaches that relied upon binarized expression of a single marker. Finally, our tissue-based analysis revealed unsuspected spatial organization for myeloid cells and macrophages that could not be identified using dissociative techniques such as flow cytometry or single cell RNA sequencing.
In conclusion, the current study shows that a diverse set of myeloid cells are present within the PDAC tumor microenvironment and are distributed heterogeneously across patient tumors. Not only the densities but also the spatial locations of immune cells are associated with patient outcomes, revealing complexity to the PDAC microenvironment that will be important to design and interpretation of immunotherapy clinical trials for patients with PDAC.
Supplementary Material
ACKNOWLEDGEMENTS
S.A.V. was supported by the Finnish Cultural Foundation and Orion Research Foundation sr. S.O. was supported in part by NIH grant R35 CA197735. B.M.W. was supported by the Hale Family Center for Pancreatic Cancer Research, Lustgarten Foundation Dedicated Laboratory program, NIH grant U01 CA210171, NIH grant P50 CA127003, Stand Up to Cancer, Pancreatic Cancer Action Network, Noble Effort Fund, Wexler Family Fund, Promises for Purple and McCarthy Strong.
Conflict of interests: R.F.D. receives honoraria for consulting from Exelixis Inc. O.R. has research support from Merck, is a speaker for activities supported by educational grants from BMS and Merck and is a consultant for Merck, Celgene, Five Prime, GSK, GFK, Bayer, Roche/Genentech, Puretech, Imvax and Sobi. In addition, he has patent “Methods of using pembrolizumab and trebananib” pending. A.J.A. has consulted for Oncorus, Inc., Arrakis Therapeutics, and Merck & Co., Inc., and has research funding from Mirati Therapeutics and Deerfield, Inc. that is unrelated to this project. A.C.K is a stockholder and advisor for Aravive Inc. B.M.W. has research funding from Celgene and Eli Lilly and consulting for BioLineRx, Celgene, G1 Therapeutics, and GRAIL.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;49:1374–403. [DOI] [PubMed] [Google Scholar]
- 3.Nevala-Plagemann C, Hidalgo M, Garrido-Laguna I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat Rev Clin Oncol. 2020;17:108–23. [DOI] [PubMed] [Google Scholar]
- 4.Morrison AH, Byrne KT, Vonderheide RH. Immunotherapy and Prevention of Pancreatic Cancer. Trends in Cancer. 2018;4:418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabrilovich DI. Myeloid-Derived Suppressor Cells. Cancer Immunol Res. 2017;5:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Mitri D, Toso A, Alimonti A. Molecular Pathways: Targeting Tumor-Infiltrating Myeloid-Derived Suppressor Cells for Cancer Therapy. Clin Cancer Res. 2015;21:3108–12. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Ding Y, Guo N, Wang S. MDSCs: Key Criminals of Tumor Pre-metastatic Niche Formation. Front Immunol. 2019;10:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541–66. [DOI] [PubMed] [Google Scholar]
- 11.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. [DOI] [PubMed] [Google Scholar]
- 12.Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, Ungefroren H, Vogel I, et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int J cancer. 2014;135:843–61. [DOI] [PubMed] [Google Scholar]
- 13.Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget M-A, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep. 2017;7:13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian ZR, Rubinson DA, Nowak JA, Morales-Oyarvide V, Dunne RF, Kozak MM, et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol. 2018;4:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munder M Arginase: an emerging key player in the mammalian immune system. Br J Pharmacol. 2009;158:638–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons D, Seed B. Isolation of a cDNA encoding CD33, a differentiation antigen of myeloid progenitor cells. J Immunol. 1988;141:2797–800. [PubMed] [Google Scholar]
- 18.Elliott LA, Doherty GA, Sheahan K, Ryan EJ. Human Tumor-Infiltrating Myeloid Cells: Phenotypic and Functional Diversity. Front Immunol. 2017;8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev Y V. CD68/macrosialin: not just a histochemical marker. Lab Invest. 2017;97:4–13. [DOI] [PubMed] [Google Scholar]
- 20.Tremble LF, McCabe M, Walker SP, McCarthy S, Tynan RF, Beecher S, et al. Differential association of CD68+ and CD163+ macrophages with macrophage enzymes, whole tumour gene expression and overall survival in advanced melanoma. Br J Cancer. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurgelun MB, Chittenden AB, Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet Med. 2018;0:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA. 2018;319:2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K, et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: The CONKO-001 randomized trial. JAMA. 2013;310:1473–81. [DOI] [PubMed] [Google Scholar]
- 25.Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389:1011–24. [DOI] [PubMed] [Google Scholar]
- 26.Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379:2395–406. [DOI] [PubMed] [Google Scholar]
- 27.Allen PJ, Kuk D, Castillo CF, Basturk O, Wolfgang CL, Cameron JL, et al. Multi-institutional Validation Study of the American Joint Commission on Cancer (8th Edition) Changes for T and N Staging in Patients With Pancreatic Adenocarcinoma. Ann Surg. 2017;265:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morales-Oyarvide V, Rubinson DA, Dunne RF, Kozak MM, Bui JL, Yuan C, et al. Lymph node metastases in resected pancreatic ductal adenocarcinoma: predictors of disease recurrence and survival. Br J Cancer. 2017;117:1874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orhan A, Vogelsang RP, Andersen MB, Madsen MT, Hölmich ER, Raskov H, et al. The prognostic value of tumour-infiltrating lymphocytes in pancreatic cancer: a systematic review and meta-analysis. Eur J Cancer. 2020;132:71–84. [DOI] [PubMed] [Google Scholar]
- 31.Osipov A, Saung MT, Zheng L, Murphy AG. Small molecule immunomodulation: the tumor microenvironment and overcoming immune escape. J Immunother cancer. 2019;7:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017;19:203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsujikawa T, Crocenzi T, Durham JN, Sugar EA, Wu AA, Onners B, et al. Evaluation of Cyclophosphamide/GVAX Pancreas Followed by Listeria-mesothelin (CRS-207) With or Without Nivolumab in Patients with Pancreatic Cancer. Clin Cancer Res. 2020;clincanres.3978.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneda MM, Cappello P, Nguyen A V, Ralainirina N, Hardamon CR, Foubert P, et al. Macrophage PI3Kγ Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016;6:870–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W-Q, Liu L, Xu H-X, Wu C-T, Xiang J-F, Xu J, et al. Infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br J Surg. 2016;103:1189–99. [DOI] [PubMed] [Google Scholar]
- 36.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stromnes IM, Brockenbrough JS, Izeradjene K, Carlson MA, Cuevas C, Simmons RM, et al. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut. 2014;63:1769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma P, Beatty PL, McKolanis J, Brand R, Schoen RE, Finn OJ. Circulating Myeloid Derived Suppressor Cells (MDSC) That Accumulate in Premalignancy Share Phenotypic and Functional Characteristics With MDSC in Cancer. Front Immunol. 2019;10:1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markowitz J, Brooks TR, Duggan MC, Paul BK, Pan X, Wei L, et al. Patients with pancreatic adenocarcinoma exhibit elevated levels of myeloid-derived suppressor cells upon progression of disease. Cancer Immunol Immunother. 2015;64:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60:1419–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khaled YS, Ammori BJ, Elkord E. Increased levels of granulocytic myeloid-derived suppressor cells in peripheral blood and tumour tissue of pancreatic cancer patients. J Immunol Res. 2014;2014:879897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javeed N, Gustafson MP, Dutta SK, Lin Y, Bamlet WR, Oberg AL, et al. Immunosuppressive CD14+HLA-DRlo/neg monocytes are elevated in pancreatic cancer and “primed” by tumor-derived exosomes. Oncoimmunology. 2017;6:e1252013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tcyganov E, Mastio J, Chen E, Gabrilovich DI. Plasticity of myeloid-derived suppressor cells in cancer. Curr Opin Immunol. 2018;51:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjornson-Hooper ZB, Fragiadakis GK, Spitzer MH, Madhireddy D, McIlwain D, Nolan GP. A comprehensive atlas of immunological differences between humans, mice and non-human primates. bioRxiv. 2019;574160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trovato R, Fiore A, Sartori S, Canè S, Giugno R, Cascione L, et al. Immunosuppression by monocytic myeloid-derived suppressor cells in patients with pancreatic ductal carcinoma is orchestrated by STAT3. J Immunother cancer. 2019;7:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Xu X, Shi M, Chen Y, Yu D, Zhao C, et al. CD13hi Neutrophil-like myeloid-derived suppressor cells exert immune suppression through Arginase 1 expression in pancreatic ductal adenocarcinoma. Oncoimmunology. 2017;6:e1258504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helm O, Mennrich R, Petrick D, Goebel L, Freitag-Wolf S, Röder C, et al. Comparative characterization of stroma cells and ductal epithelium in chronic pancreatitis and pancreatic ductal adenocarcinoma. PLoS One. 2014;9:e94357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wartenberg M, Zlobec I, Perren A, Koelzer VH, Gloor B, Lugli A, et al. Accumulation of FOXP3+T-cells in the tumor microenvironment is associated with an epithelial-mesenchymal-transition-type tumor budding phenotype and is an independent prognostic factor in surgically resected pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:4190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, et al. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. J Surg Res. Elsevier Ltd; 2011;167:e211–9. [DOI] [PubMed] [Google Scholar]
- 52.Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020; [DOI] [PubMed] [Google Scholar]
- 53.Huang Y-K, Wang M, Sun Y, Di Costanzo N, Mitchell C, Achuthan A, et al. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun. 2019;10:3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.