Abstract
PURPOSE
To compare clinical outcomes in a cohort of patients with advanced non–small-cell lung cancer (NSCLC) with targetable genomic alterations detected using plasma-based circulating tumor DNA (ctDNA) or tumor-based next-generation sequencing (NGS) assays treated with US Food and Drug Administration–approved therapies at a large academic research cancer center.
METHODS
A retrospective review from our MD Anderson GEMINI database identified 2,224 blood samples sent for ctDNA NGS testing from 1971 consecutive patients with a diagnosis of advanced NSCLC. Clinical, treatment, and outcome information were collected, reviewed, and analyzed.
RESULTS
Overall, 27% of the ctDNA tests identified at least one targetable mutation and 73% of targetable mutations were EGFR-sensitizing mutations. Among patients treated with first-line epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) therapies, there were no significant differences in progression-free survival of 379 days and 352 days (P value = .41) with treatment based on tissue (n = 40) or ctDNA (n = 40), respectively. Additionally, there were no differences in progression-free survival or objective response rate among those with low (n = 8, 0.01%-0.99%) versus high (n = 16, ≥ 1%) levels of ctDNA of the targetable mutation as measured by variant allele frequency (VAF). Overall, there was excellent testing concordance (n = 217 tests) of > 97%, sensitivity of 91.7%, and specificity of 99.7% between blood-based ctDNA NGS and tissue-based NGS assays.
CONCLUSION
There were no significant differences in clinical outcomes among patients treated with approved EGFR-TKIs whose mutations were identified using either tumor- or plasma-based comprehensive profiling and those with very low VAF as compared with high VAF, supporting the use of plasma-based profiling to guide initial TKI use in patients with metastatic EGFR-mutant NSCLC.
INTRODUCTION
The availability of tumor genomic information from simple, minimally invasive blood collection has the potential to significantly affect patient care. Next-generation sequencing (NGS) of circulating tumor DNA (ctDNA) testing is available in Clinical Laboratory Improvement Amendments (CLIA)–approved laboratories, has recently gained US Food and Drug Administration (FDA) approval, and is being used for genomic profiling of human cancers. We report on the clinical utility of a comprehensive ctDNA NGS blood test in patients with advanced non–small-cell lung cancer (NSCLC) and the outcome of treatments with targeted therapies.
CONTEXT
Key Objective
We analyzed a cohort of patients with advanced non–small-cell lung cancer (NSCLC) with targetable genomic alterations detected using either tumor- or plasma-based genomic assays and investigated clinical outcomes in those receiving US Food and Drug Administration–approved targeted therapies including epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors in frontline setting.
Knowledge Generated
Patients with NSCLC found to have classical sensitizing EGFR mutations (exon 19 deletion or L858R) diagnosed via either plasma- or tumor-based profiling had similar clinical outcomes when treated with standard-of-care EGFR-tyrosine kinase inhibitors in the frontline setting. Additionally, patients with very low variant allele frequency for their EGFR mutation had similar clinical benefits as those with high variant allele frequency.
Relevance
The use of plasma-based genomic profiling to identify targetable genomic alterations, and guide treatment decisions, is growing rapidly for patients with NSCLC and other tumor types. We found that patients with EGFR-mutant NSCLC had similar outcomes when mutations were identified using plasma- or tumor-based profiling, providing support that appropriate treatment choices can be guided by plasma-based profiling.
Since the development and regulatory approval of highly effective molecularly targeted therapeutics such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) inhibitors, tumor genotyping is considered the standard of care for the optimal management of advanced nonsquamous NSCLC. Obtaining this tumor genomic information from tissue specimens has been the standard practice for identifying these actionable alterations. However, cell-free DNA, specifically the ctDNA fraction, is a blood-based biomarker with demonstrated clinical utility in multiple solid-tumor types for molecular profiling.1,2 In advanced-stage NSCLC, ctDNA is increasingly used as a liquid biopsy to inform therapies for EGFR, ALK, and other molecular subtypes.3,4
Comprehensive plasma-based genotyping with ctDNA in advanced NSCLC is highly concordant with tissue-based genotyping5,6 and increases the rate of targetable alteration detection, compared with tissue testing alone.5,7 Additionally, a prospective cohort study showed that 86% of patients who received targeted therapy on the basis of ctDNA testing achieved at least a complete or a partial response or stable disease.5 A recent retrospective review of 66 patients with NSCLC reported the median time on first-line targeted therapy with treatment on the basis of ctDNA NGS assay results was similar to that of historical reports with tissue-based assays.8 However, long-term clinical outcomes have not been directly compared in patients with NSCLC who were treated with standard-of-care targeted agents on the basis of actionable mutations identified using tissue- and plasma-based profiling, particularly in the frontline setting with EGFR-tyrosine kinase inhibitors (TKIs).
We hypothesized that patients with advanced NSCLC and actionable oncogenic driver mutations identified by ctDNA have similar objective response rate and progression-free survival (PFS) as those with tissue-based testing when treated with appropriate targeted therapy. Additionally, we hypothesized that there are no differences in tumor response and clinical outcome from identified targeted mutations from ctDNA in terms of variant allele frequency (VAF) of the oncogenic driver, between low versus high. In this retrospective study, we analyzed these clinical outcomes in patients who were tested with CLIA-certified tissue-based and plasma-based NGS to comprehensively evaluate the clinical utility of liquid biopsies for the treatment of advanced NSCLC.
METHODS
From May 2015 to February 2018, a retrospective review (UTMDACC IRB-approved protocol PA16-0061) from our Lung Cancer Moon Shot GEMINI database identified 2,224 blood samples from 1971 consecutive patients with a diagnosis of advanced NSCLC collected and sent for molecular profiling by NGS at a CLIA-certified laboratory (Guardant360; Guardant Health, Redwood City, CA [assay detailed in the Data Supplement]) with a reported 80% sensitivity to detect any guideline-recommended biomarker.7,9
Tumor tissue from either the initial diagnostic specimen or newly biopsied sample was submitted to our institution's molecular diagnostic laboratory (MDL) for genomic profiling. The MDL NGS platform changed over the period of this retrospective study, initially with an NGS assay targeting hotspot regions of 50 genes as detailed in the Data Supplement.10,11 In April 2016, an expanded NGS platform was used called Solid Tumor Genomic Assay V1 (OncoMine Comprehensive Assay V1; Thermo Fisher Scientific, Waltham, MA). This NGS-based assay was used for the detection of 148 single-nucleotide variants, 49 insertions or deletions, 40 copy number aberrations, and a subset of gene fusions as reported.12 All these assays have been validated with CLIA certifications at our institution for routine clinical patient care.
From our GEMINI database, we further identified patients with any of the targetable genes listed above and all available clinical information were included in our genotyping cohort for complete review and validation manually from their electronic medical records. Those patients with long-term follow-up in our clinic with or without available radiologic imaging scans were included. Radiologic imaging scans were completed following standard-of-care practice guidelines or at the discretion of treating physicians. Review and analysis of target lesions were completed by our diagnostic imaging collaborators to provide tumor measurements and tumor responses (RECIST 1.1 guidelines) with last follow-up reviewed on March 12, 2020. Kaplan-Meier methodology was used to calculate median PFS with log-rank test assessment at a significance level of 5%.
For genomic concordance analysis between tissue and ctDNA, a period of 6 months between the collection of tissue and blood without any targeted therapy was applied because during this time period many of our patients are coming from outside oncology facilities were treated with standard chemotherapies with either incomplete tissue genotype or not done. The patients with tissue-based only genomic testing for the frontline EGFR-TKI analysis were identified consecutively by reverse dating from our GEMINI database, with a starting date immediately before the availability of the ctDNA assay in May 2015.
During our initial experience of using this new ctDNA assay, there were many questions from our clinicians regarding the percent of variant allele frequency (%VAF) listed with any gene reported from the Guardant360 assay. There are no cutoff values or parameters in the traditional sense with clinical laboratory tests to help guide clinicians whether to initiate treatment on the basis of these reported %VAFs. In particular when ctDNA reports show very low %VAF of < 1%, the concern was that the calculated %VAF is at or near the limit of assay detection and may not be representative of true tumor DNA (ie, false positive), furthermore not allowing the patient to gain any potential clinical benefit from standard chemotherapy regimen plus potential for harm with TKI treatment–associated toxicities. This particular ctDNA assay (Guardant360) sensitivity is reported to be < 0.1%, whereas other NGS assays’ sensitivities can range from 0.01% to 1%.2 On the basis of these information, we evaluated the clinical outcome from a group of treatment-naive patients receiving EGFR-TKI on the basis of detection of EGFR mutation with %VAF available from ctDNA reports. We separated these patients into two groups: high-ctDNA group are those with VAF ≥ 1% and those with VAF 0.01%-0.99% were designated in the low-ctDNA group. Our hypothesis is that there will be no differences in clinical benefit between the two groups and that the reported %VAF represents true tumor burden and/or tumor shedding.13,14
RESULTS
A summary of patient characteristics and identified mutations is listed in the Data Supplement. The majority of the testings were completed at progression of disease at 53.5%, with 28.4% and 18% of testings after initial diagnosis and during treatment, respectively. Some patients had more than one test done, with 162 patients with two tests and 33 patients with three or more tests. Overall, 87.3% (1,942 of 2,224) of the tests identified at least one molecular alteration, with 26.7% (594 of 2,224) of tests identifying at least one targetable molecular mutation.
A summary chart details the identified targetable mutations (Data Supplement); the most common targetable gene identified was EGFR accounting for more than 80%, with detailed alterations charted in the Data Supplement (right pie chart). The remaining included BRAF (6.7%), MET exon 14 skipping (2.4%), and gene fusion groups ALK (6.1%), RET (2.9%), and ROS1 (1%). Each patient was treated with appropriate therapy on the basis of FDA-approved labeling, National Comprehensive Cancer Network guidelines for targeted therapies, or emerging targets. Additionally, patients with targetable mutations but without approved treatment indications were evaluated for potential enrollment onto available clinical research trials.
A group of 100 patients with diverse actionable mutations identified by ctDNA to have received targeted therapies with long-term follow-up and appropriate radiologic imaging over the treatment period were evaluated (Data Supplement). Of the 14 patients who had a second ctDNA test completed, a change in therapy from erlotinib to osimertinib occurred in 6 with identification of EGFR exon 20 T790M at the time of disease progression. The remainder showed no changes with mutation status; however, in two patients with dual EGFR L858R plus T790M, therapy was switched from erlotinib to osimertinib. The median PFS for the three main mutation groups were as follows: ALK mutations (n = 17, 321 [91-701] days), EGFR-sensitive mutations (n = 54, 242 [27-848] days), and EGFR exon 20 T790M (n = 37, 215 [54-894] days) (Data Supplement, Results).
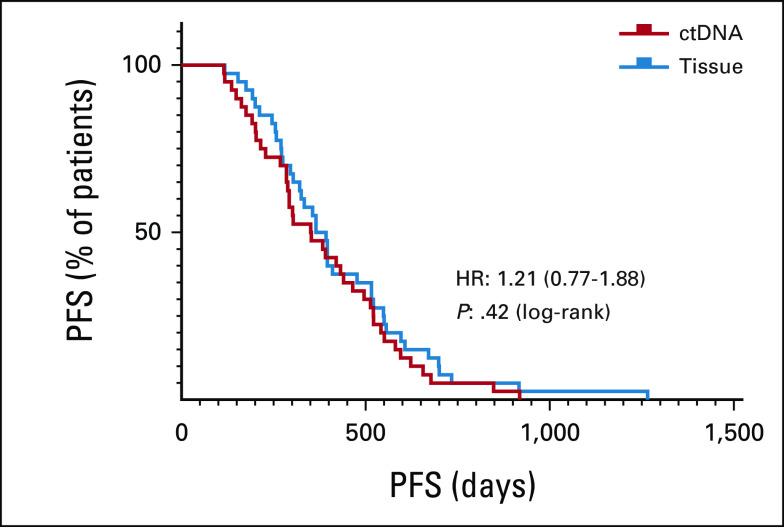
Next, we compared clinical outcomes of advanced NSCLC treatment-naïve patients identified with sensitive EGFR mutations from either tissue-based or ctDNA, receiving front-line FDA-approved EGFR-TKI therapy in 40 consecutive patients for each group, with complete clinical standard of care follow-up visits. Assessment of progression-free survival (PFS) was from date of therapy initiation until clinical progression of disease. Results are summarized in Table 1. Both groups had similar characteristics in terms of sex, age, and types of EGFR mutations. There were no statistical differences (P value = .42) in the median PFS between the two groups (Fig 1), tissue-based and ctDNA-based, 379 (118-1266) days and 353 (115-919) days, respectively.
TABLE 1.
Clinical Outcomes in Patients With Advanced Non-–Small-Cell Lung Cancer Treated With US Food and Drug Administration–Approved Frontline EGFR-Tyrosine Kinase Inhibitor Directed by Genomic Profiling From Either Tissue- or ctDNA-Based Testing
FIG 1.
PFS in patients with advanced non–small-cell lung cancer treated with US Food and Drug Administration–approved frontline epidermal growth factor receptor-tyrosine kinase inhibitor directed by genomic profiling from tissue-based (blue) versus ctDNA-based (red) testing, log-rank test. ctDNA, circulating tumor DNA; HR, hazard ratio; PFS, progression-free survival.
In the frontline EGFR-sensitive ctDNA cohort mentioned above, we assessed whether outcomes differed on the basis of high or low percent VAF of the targetable alteration detected in plasma. VAF has been reported to be associated with tumor burden13,14 and, conceivably, a lower VAF may be associated with a lower diagnostic accuracy. Among this cohort of 40 patients, only 24 had the additional complete radiographic imaging for tumor response assessments. We compared patients in low-VAF group (n = 8), with VAF of 0.01%-0.99%, and those in high-VAF group (n = 16), with VAF of 1% or greater. Figure 2A shows no significant differences in tumor responses (RECIST V1.1) between the low- versus high-VAF group, with mean tumor reduction of –47.25% and –26.11%, respectively (P value = .152). Comparing between the two groups (Fig 2B), there was a trend in low-VAF group having greater clinical benefit than high VAF; with median PFS of 424.5 and 293 days, respectively. The difference was not statistically significant (P value = .062).
FIG 2.
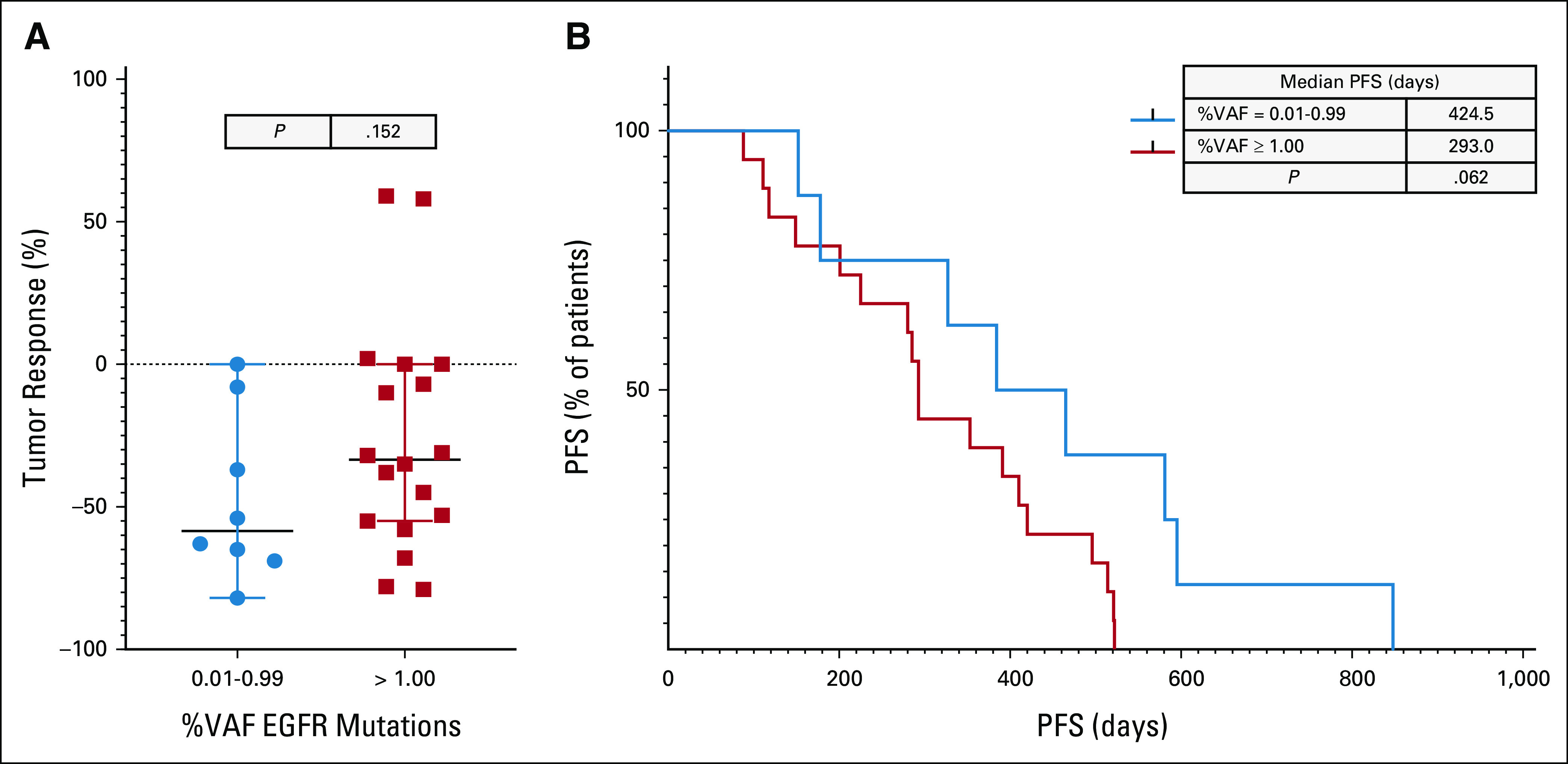
Comparison of clinical responses from patients with advanced NSCLC treated with FDA-approved EGFR-TKI in first line on the basis of ctDNA-identified molecular alterations. (A) Tumor responses (RECIST V1.1) on the basis of low (0.01%-0.99%, blue circle) versus high (≥ 1.00%, red box) VAF, unpaired t test. (B) PFS of low VAF (blue) and high VAF (red), log-rank test. ctDNA, circulating tumor DNA; EGFR, epidermal growth factor receptor; FDA, US Food and Drug Administration; NSCLC, non–small-cell lung cancer; PFS, progression-free survival; TKI, tyrosine kinase inhibitor; VAF, variant allele frequency.
We next tested the concordance between tissue and plasma profiling for targetable mutations. We analyzed the cases for which data were available for the same patient, sufficient tumor was available for complete genotype analysis, and the tests were done within a 6-month period with no new treatment in the intervening period. With this consideration, a total of 217 matched test results were compared between plasma ctDNA and tissue, with complete genomic analysis summarized in Table 2. For the cohort with sensitizing EGFR (exon 19del and exon 21 L858R) mutation groups (n = 109), there was 98% concordance between ctDNA and tissue with excellent sensitivity (92%-96%) and specificity (99%-100%) and both positive (94%-100%) and negative (98%) predictive values. For the small group of atypical EGFR mutations (n = 10), the concordance was 99.5% with excellent sensitivity (100%) and specificity (99.5%). We also considered dual EGFR mutations with both activating (exon 19 deletion or exon 21 L858R) and resistance (exon 20 T790M) EGFR alterations present from the same sample to be considered a match. Dual EGFR mutations were identified in 24 tumor samples; the sensitivity and specificity of ctDNA were 75% and 99.8%, respectively, with 96.8% concordance. With EML4-ALK fusions testing (n = 17), the sensitivity and specificity were 88.2% and 99.5%, respectively, with a 98.6% concordance. Each of the smaller cohorts consisting of BRAF V600E (n = 9), RET fusions (n = 2), MET exon 14 skipping (n = 3), and ROS1 fusions (n = 2) demonstrated 100% concordance.
TABLE 2.
Testing Agreement of Targeted Mutations Identified From Tissue and ctDNA (n = 217 Matched Cases)
DISCUSSION
As the number of new targetable mutations and effective FDA-approved targeted drugs continue to grow in recent years, particularly for NSCLC, the availability of comprehensive molecular profiling has become critical for routine clinical care. The simplicity and noninvasive nature of acquiring ctDNA from a blood collection enables clinicians an additional tool to assist in the care of their patients. Undergenotyping remains a significant clinical problem among patients with advanced or recurrent NSCLC; investigators have reported only 8%-18% of patients had complete genotype testing from tissue before beginning therapy.7,15
Leighl et al7 compared the results of the molecular profiling from both tissue and plasma ctDNA in 282 newly diagnosed patients with NSCLC, revealing that ctDNA testing identified a similar number of mutation-positive patients as tissue, on the basis of guideline-complete genotype testing. This result is similar to our subcohort of patients with matched tumor molecular testing in which ctDNA sensitivity and positive predictive value were > 90% across most alteration types, including single nucleotide variants, indels, and fusions. The exceptions were the EGFR exon 20 T790M mutation and comutation of activating EGFR mutations, with either exon 19 deletion or L858R, and T790M, for which this ctDNA assay failed to detect T790M in 24% (7 of 29) of cases. This result is not surprising as several reports have shown this disparity,16-18 including a recent meta-analysis of 21 studies evaluating the clinical accuracy of EGFR mutation testing from plasma, showing that > 30% of plasma tests did not detect EGFR T790M as compared with tissue.18,19 In part, this may be explained that some of these EGFR tumors are nonshedders and that the current ctDNA assay is not able to detect T790M at such a low level. In addition, both tissue-based and plasma-based assays had different bioinformatic pipelines and significant changes in their respective NGS assays over the period of this study, which could also have contributed to any disagreement between the two assays. In this analysis, several factors resulted in not having more matched set of plasma and tissue for this concordance analysis including inadequate tissue available for full genotyping, poor tissue quality, and the smaller number of certain targetable mutations, such as MET exon 14 skipping mutation, that were not available in our MDL NGS assay until 2017. In our EGFR frontline cohort, when available, there were no significant differences in clinical benefit when FDA-approved treatment was used on the basis of actionable oncogenic driver mutations identified by ctDNA- or tissue-based testing.
From our cohort of 2,224 ctDNA NGS tests from 1971 patients, 26.7% of the tests identified at least one targetable mutation in EGFR, ROS1, BRAF, RET, ALK, MET exon 14, or ERBB2, with many of these patients treated with targeted therapies as part of standard-of-care practice. This detection rate is lower than the 35% detection rate reported by Aggarwal et al5 in their smaller cohort of 323 patients with NSCLC, which may be due, in part, to the higher number of first-line patients included in their cohort (51.4% v 28.4% in our cohort). With our retrospective analysis, the total number of patients in each of our cohort is limited to those patients with a complement of full clinical follow-up evaluations. As patients are referred for evaluation and treatment recommendation, they return to their primary oncologists to receive their treatment and management.
In our cohort of frontline EGFR-TKI, there were no differences in clinical outcomes (PFS) for patients receiving FDA-approved treatment on the basis of either tissue-based or ctDNA blood-based NGS with median PFS of 379 days and 352 days, respectively. These median PFS results are similar to those reported in multiple clinical trials with either erlotinib or gefitinib for frontline EGFR-TKI therapies on the basis of either tissue- or ctDNA-targeted EGFR-TKIs.20-29 With our small cohort of patients receiving first-line EGFR-TKI for sensitive EGFR mutations on the basis of ctDNA genomic testing, we did not find any significant differences in tumor responses and PFS when comparing low (0.01%-0.99%) versus high (≥ 1.00%) percent VAF. However, a trend of greater clinical benefit was observed in those with a low %VAF before initiation of EGFR-TKI. This has been reported by other investigators among patients with NSCLC treated with EGFR-TKIs.30,31 With more advancement in testing platforms and increasing sensitivity reaching as low as 0.001% VAF, stratifying patients on the basis of ctDNA levels with high ctDNA levels getting more intensive therapy versus a less intensive regimen for those with low ctDNA has been proposed.32 For those patients with ctDNA %VAF at the very low end of the ultrasensitive ctDNA assay, a treatment break may be instituted with close ctDNA monitoring with resuming of treatment if and when ctDNA rises to above a certain threshold, which needs to be determined and validated with a prospective clinical study. Additionally, in this analysis, we did not account for presence of comutations such as TP53 and others that may play a role in a patient's tumor response to EGFR-TKI and clinical outcome. Increasing evidence supports ctDNA results as both a prognostic and predictive marker.30,33-35
Our retrospective study provides additional evidence supporting the clinical utility of ctDNA NGS to identify targetable mutations with clinical outcomes comparable with tissue-directed therapy in patients with advanced NSCLC.36 In our limited cohort of patients, ctDNA testing at initial diagnosis identifies similar molecular alterations as tissue-based testing and treatment on the basis of ctDNA results leads to similar tumor responses and clinical benefits as compared with treatment directed by tissue-based results. Additionally, there were no significant differences in the outcome in our EGFR cohort with high or low ctDNA (%VAF) treated with FDA-approved TKIs on the basis of ctDNA NGS results. Molecular testing of ctDNA is a simple, minimally invasive test. With high concordance and equivalent clinical outcomes, ctDNA-guided results can be used in the first-line advanced NSCLC setting, especially when tissue is not available, adequate for complete genotyping, or possible to obtain through biopsy. In addition to tissue requirements for genomic screening for clinical trial entry, liquid biopsy is also becoming more acceptable and may even replace tissue-based, especially at initial or subsequent disease progression as reported and seen in our center.37
With the current and newer generation of assays, ctDNA can also be useful as a long-term management tool not only in detecting emergent resistance or identifying rare molecular alterations at disease progression but also in providing a more precise test for minimal residual disease and molecular response, illustrating the advantages of broad profiling.38-41
ACKNOWLEDGMENT
The authors would like to acknowledge the thoracic clinical team for their research support. We would also like to acknowledge the generous philanthropic contributions to the University of Texas MD Anderson Cancer Center Moon Shots Program and Guardant Health for their support and collaboration on this project, especially Richard B. Lanman and Victoria M. Raymond.
Hai T. Tran
Research Funding: Bayer, Guardant Health, ZIOPHARM Oncology, Bristol Myers Squibb
Vincent K. Lam
Consulting or Advisory Role: Takeda, Seattle Genetics, Bristol Myers Squibb
Research Funding: Guardant Health, Takeda, GlaxoSmithKline, Bristol Myers Squibb
Yasir Y. Elamin
Consulting or Advisory Role: Lilly, AstraZeneca, Turning Point Therapeutics
Research Funding: Spectrum Pharmaceuticals, AstraZeneca, Takeda, Xcovery, Lilly, Elevation Oncology, Turning Point Therapeutics
Travel, Accommodations, Expenses: Lilly
Islam S. A. Hassan
Employment: Agios, Servier
Stock and Other Ownership Interests: Agios
Mehmet Altan
Consulting or Advisory Role: GlaxoSmithKline, Shattuck Labs
Research Funding: Lilly, Bristol Myers Squibb, Novartis, GlaxoSmithKline, Jounce Therapeutics, Adaptimmune, Merck, Genentech, Nektar, Shattuck Labs
George R. Blumenschein
Employment: Janssen, Johnson & Johnson
Stock and Other Ownership Interests: Virogin Biotech
Consulting or Advisory Role: Bristol Myers Squibb, Bayer, Celgene, Clovis Oncology, AbbVie, ARIAD, Merck, Genentech, Novartis, Xcovery, Adicet Bio, Amgen, AstraZeneca, Roche, MedImmune, Maverick Therapeutics, Johnson & Johnson, Virogin Biotech, Gilead Sciences, Daiichi Sankyo Inc
Research Funding: Merck, Celgene, Genentech, Xcovery, Novartis, Bristol Myers Squibb, GlaxoSmithKline, Adaptimmune, Macrogenics, Kite, a Gilead Company, Immatics, Torque, Incyte, MedImmune, Exelixis, Immunocore, Roche, AstraZeneca, Bayer, Tmunity Therapeutics Inc, Regeneron, BeiGene, Repertoire Immune Medicines, Daiichi Sankyo Inc
Lauren E. Byers
Consulting or Advisory Role: AbbVie, AstraZeneca, PharmaMar, Genmab, Bristol Myers Squibb, Sierra Oncology, Pfizer, Merck, Jazz Pharmaceuticals, Genentech, Alethia, Debiopharm Group, Tolero Pharmaceuticals
Research Funding: Genmab, AstraZeneca, Sierra Oncology, Tolero Pharmaceuticals
Anne S. Tsao
Consulting or Advisory Role: Novartis, Boehringer Ingelheim, Genentech/Roche, Lilly, Bristol Myers Squibb, Epizyme, AstraZeneca/MedImmune, ARIAD, EMD Serono, Takeda, Heron
Research Funding: Merck, Genentech/Roche, Seattle Genetics, Millennium, Bristol Myers Squibb, Boehringer Ingelheim, Polaris, EMD Serono, Seattle Genetics, Takeda
Patents, Royalties, Other Intellectual Property: UptoDate
Don L. Gibbons
Stock and Other Ownership Interests: Exact Sciences, Nektar
Consulting or Advisory Role: Sanofi, GlaxoSmithKline, Janssen Research & Development, Ribon Therapeutics, Mitobridge, Lilly, Menarini
Research Funding: Janssen Research & Development, Takeda, AstraZeneca, Mitobridge, Ribon Therapeutics, Boehringer Ingelheim, Mirati Therapeutics
Travel, Accommodations, Expenses: AstraZeneca/MedImmune, BerGenBio, Takeda
Bonnie S. Glisson
Research Funding: Pfizer, Medimmune, ISA Pharmaceuticals, CUE Biopharma
Travel, Accommodations, Expenses: AbbVie/Stemcentrx, ISA Pharmaceuticals
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, Origimed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital, Geneplus
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, Johnson & Johnson/Janssen
Research Funding: Merck
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Lilly, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Catalyst Biotech, Foundation Medicine, Novartis, Mirati Therapeutics, BrightPath Biotheraputics, Janssen, Nexus Health Systems, Pneuma Respiratory, Kairos Ventures, Roche, Leads Biolabs
Research Funding: AstraZeneca, Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Preliminary results from this study were presented at IASLC 19th (September 23-26, 2018, Toronto, Canada) and 20th (September 7-10, 2019, Barcelona, Spain) World Conference on Lung Cancer.
SUPPORT
Supported by the University of Texas MD Anderson Lung Moon Shot Initiative, UT MD Anderson Cancer Center NIH CCSG CA016672 and Guardant Health Inc.
AUTHOR CONTRIBUTIONS
Conception and design: Hai T. Tran, Vincent K. Lam, Nabil A. Elshafeey, Jianjun Zhang, John V. Heymach
Financial support: Nabil A. Elshafeey
Administrative support: Nabil A. Elshafeey, Waree Rinsurongkawong, Anne S. Tsao
Provision of study materials or patients: Yasir Y. Elamin, Nabil A. Elshafeey, Anne S. Tsao, Don L. Gibbons
Collection and assembly of data: Hai T. Tran, Vincent K. Lam, Yasir Y. Elamin, Lingzhi Hong, Rivka Colen, Islam S. A. Hassan, George R. Blumenschein, Waree Rinsurongkawong, Melvin J. Rivera, Mayra E. Vasquez, Brett W. Carter, Lauren E. Byers, Don L. Gibbons, Frank Fossella, Jianjun Zhang, John V. Heymach
Data analysis and interpretation: Hai T. Tran, Vincent K. Lam, Lingzhi Hong, Rivka Colen, Islam S. A. Hassan, Mehmet Altan, George R. Blumenschein, Melvin J. Rivera, Brett W. Carter, Lauren E. Byers, Anne S. Tsao, Don L. Gibbons, Frank Fossella, Bonnie S. Glisson, Jianjun Zhang, John V. Heymach
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hai T. Tran
Research Funding: Bayer, Guardant Health, ZIOPHARM Oncology, Bristol Myers Squibb
Vincent K. Lam
Consulting or Advisory Role: Takeda, Seattle Genetics, Bristol Myers Squibb
Research Funding: Guardant Health, Takeda, GlaxoSmithKline, Bristol Myers Squibb
Yasir Y. Elamin
Consulting or Advisory Role: Lilly, AstraZeneca, Turning Point Therapeutics
Research Funding: Spectrum Pharmaceuticals, AstraZeneca, Takeda, Xcovery, Lilly, Elevation Oncology, Turning Point Therapeutics
Travel, Accommodations, Expenses: Lilly
Islam S. A. Hassan
Employment: Agios, Servier
Stock and Other Ownership Interests: Agios
Mehmet Altan
Consulting or Advisory Role: GlaxoSmithKline, Shattuck Labs
Research Funding: Lilly, Bristol Myers Squibb, Novartis, GlaxoSmithKline, Jounce Therapeutics, Adaptimmune, Merck, Genentech, Nektar, Shattuck Labs
George R. Blumenschein
Employment: Janssen, Johnson & Johnson
Stock and Other Ownership Interests: Virogin Biotech
Consulting or Advisory Role: Bristol Myers Squibb, Bayer, Celgene, Clovis Oncology, AbbVie, ARIAD, Merck, Genentech, Novartis, Xcovery, Adicet Bio, Amgen, AstraZeneca, Roche, MedImmune, Maverick Therapeutics, Johnson & Johnson, Virogin Biotech, Gilead Sciences, Daiichi Sankyo Inc
Research Funding: Merck, Celgene, Genentech, Xcovery, Novartis, Bristol Myers Squibb, GlaxoSmithKline, Adaptimmune, Macrogenics, Kite, a Gilead Company, Immatics, Torque, Incyte, MedImmune, Exelixis, Immunocore, Roche, AstraZeneca, Bayer, Tmunity Therapeutics Inc, Regeneron, BeiGene, Repertoire Immune Medicines, Daiichi Sankyo Inc
Lauren E. Byers
Consulting or Advisory Role: AbbVie, AstraZeneca, PharmaMar, Genmab, Bristol Myers Squibb, Sierra Oncology, Pfizer, Merck, Jazz Pharmaceuticals, Genentech, Alethia, Debiopharm Group, Tolero Pharmaceuticals
Research Funding: Genmab, AstraZeneca, Sierra Oncology, Tolero Pharmaceuticals
Anne S. Tsao
Consulting or Advisory Role: Novartis, Boehringer Ingelheim, Genentech/Roche, Lilly, Bristol Myers Squibb, Epizyme, AstraZeneca/MedImmune, ARIAD, EMD Serono, Takeda, Heron
Research Funding: Merck, Genentech/Roche, Seattle Genetics, Millennium, Bristol Myers Squibb, Boehringer Ingelheim, Polaris, EMD Serono, Seattle Genetics, Takeda
Patents, Royalties, Other Intellectual Property: UptoDate
Don L. Gibbons
Stock and Other Ownership Interests: Exact Sciences, Nektar
Consulting or Advisory Role: Sanofi, GlaxoSmithKline, Janssen Research & Development, Ribon Therapeutics, Mitobridge, Lilly, Menarini
Research Funding: Janssen Research & Development, Takeda, AstraZeneca, Mitobridge, Ribon Therapeutics, Boehringer Ingelheim, Mirati Therapeutics
Travel, Accommodations, Expenses: AstraZeneca/MedImmune, BerGenBio, Takeda
Bonnie S. Glisson
Research Funding: Pfizer, Medimmune, ISA Pharmaceuticals, CUE Biopharma
Travel, Accommodations, Expenses: AbbVie/Stemcentrx, ISA Pharmaceuticals
Jianjun Zhang
Honoraria: Roche, Sino-USA Biomedical Platform, Geneplus, Origimed, Innovent Biologics, CancerNet, Zhejiang Cancer Hospital, Geneplus
Consulting or Advisory Role: AstraZeneca, Geneplus, Capital Medical University, Johnson & Johnson/Janssen
Research Funding: Merck
Travel, Accommodations, Expenses: Innovent Biologics, Zhejiang Cancer Hospital
John V. Heymach
Stock and Other Ownership Interests: Cardinal Spine, Bio-Tree
Consulting or Advisory Role: AstraZeneca, Bristol Myers Squibb, Spectrum Pharmaceuticals, Guardant Health, Hengrui Pharmaceutical, GlaxoSmithKline, EMD Serono, Lilly, Takeda, Sanofi/Aventis, Genentech/Roche, Boehringer Ingelheim, Catalyst Biotech, Foundation Medicine, Novartis, Mirati Therapeutics, BrightPath Biotheraputics, Janssen, Nexus Health Systems, Pneuma Respiratory, Kairos Ventures, Roche, Leads Biolabs
Research Funding: AstraZeneca, Spectrum Pharmaceuticals, GlaxoSmithKline
Patents, Royalties, Other Intellectual Property: Licensing agreement between Spectrum and MD Anderson (including myself) regarding intellectual property for treatment of EGFR and HER2 exon 20 mutations
No other potential conflicts of interest were reported.
REFERENCES
- 1.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA Nat Rev Cancer 17223–2382017 [DOI] [PubMed] [Google Scholar]
- 2.Zill OA, Banks KC, Fairclough SR, et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients Clin Cancer Res 243528–35382018 [DOI] [PubMed] [Google Scholar]
- 3.US Food and Drug Administration . cobas EGFR Mutation Test V2. 2016. https://www.accessdata.fda.gov/cdrh_docs/pdf12/P120019S007c.pdf [Google Scholar]
- 4.Ettinger DS, Wood DE, Aisner D, et al: National Comprehensive Cancer Network Non–Small Cell Lung Cancer, Version 2.2018 J Natl Compr Canc Netw; 15504–5352017 [DOI] [PubMed] [Google Scholar]
- 5.Aggarwal C, Thompson JC, Black TA, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer JAMA Oncol 5173–1802018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA Clin Cancer Res 225772–57822016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leighl NB, Page RD, Raymond VM, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer Clin Cancer Res 254691–47002019 [DOI] [PubMed] [Google Scholar]
- 8.Reckamp KL, Patil T, Kirtane K, et al. Duration of targeted therapy in patients with advanced non-small-cell lung cancer identified by circulating tumor DNA analysis Clin Lung Cancer 21545–552.e12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies Clin Cancer Res 243539–35492018 [DOI] [PubMed] [Google Scholar]
- 10.Mehrotra M, Luthra R, Abraham R, et al. Validation of quantitative PCR-based assays for detection of gene copy number aberrations in formalin-fixed, paraffin embedded solid tumor samples Cancer Genet 212-21324–312017 [DOI] [PubMed] [Google Scholar]
- 11.Goswami RS, Luthra R, Singh RR, et al. Identification of factors affecting the success of next-generation sequencing testing in solid tumors Am J Clin Pathol 145222–2372016 [DOI] [PubMed] [Google Scholar]
- 12.Luthra R, Patel KP, Routbort MJ, et al. A targeted high-throughput next-generation sequencing panel for clinical screening of mutations, gene amplifications, and fusions in solid tumors J Mol Diagn 19255–2642017 [DOI] [PubMed] [Google Scholar]
- 13.Strijker M, Soer EC, de Pastena M, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma Int J Cancer 1461445–14562020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chemi F, Rothwell DG, McGranahan N, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse Nat Med 251534–15392019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: Gaps and opportunities Clin Lung Cancer 18651–6592017 [DOI] [PubMed] [Google Scholar]
- 16.Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non-small cell lung cancer: A meta-analysis Cancer Epidemiol Biomarkers Prev 24206–2122015 [DOI] [PubMed] [Google Scholar]
- 17.Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer J Thorac Oncol 121061–10702017 [DOI] [PubMed] [Google Scholar]
- 18.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer J Clin Oncol 343375–33822016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passiglia F, Rizzo S, Di Maio M, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: A systematic review and meta-analysis. Sci Rep. 2018;8:13379. doi: 10.1038/s41598-018-30780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: Analyses from the phase III, randomized, open-label, ENSURE study Ann Oncol 261883–18892015 [DOI] [PubMed] [Google Scholar]
- 21.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations J Clin Oncol 313327–33342013 [DOI] [PubMed] [Google Scholar]
- 22.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): An open-label, randomised phase 3 trial Lancet Oncol 15213–2222014 [DOI] [PubMed] [Google Scholar]
- 23.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial Lancet Oncol 13239–2462012 [DOI] [PubMed] [Google Scholar]
- 24.Schuler M, Yang JC, Park K, et al. Afatinib beyond progression in patients with non-small-cell lung cancer following chemotherapy, erlotinib/gefitinib and afatinib: Phase III randomized LUX-lung 5 trial Ann Oncol 27417–4232016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma N Engl J Med 361947–9572009 [DOI] [PubMed] [Google Scholar]
- 26.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial Lancet Oncol 11121–1282010 [DOI] [PubMed] [Google Scholar]
- 27.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR N Engl J Med 3622380–23882010 [DOI] [PubMed] [Google Scholar]
- 28.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study Lancet Oncol 12735–7422011 [DOI] [PubMed] [Google Scholar]
- 29.Mayo-de-Las-Casas C, Jordana-Ariza N, Garzon-Ibanez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions Ann Oncol 282248–22552017 [DOI] [PubMed] [Google Scholar]
- 30.O'Kane GM, Liu G, Stockley TL, et al. The presence and variant allele fraction of EGFR mutations in ctDNA and development of resistance Lung Cancer 13186–892019 [DOI] [PubMed] [Google Scholar]
- 31.Beagan JJ, Bach S, van Boerdonk RA, et al. Circulating tumor DNA analysis of EGFR-mutant non-small cell lung cancer patients receiving osimertinib following previous tyrosine kinase inhibitor treatment Lung Cancer 145173–1802020 [DOI] [PubMed] [Google Scholar]
- 32.Cheng ML, Pectasides E, Hanna GJ, et al. Circulating tumor DNA in advanced solid tumors: Clinical relevance and future directions CA Cancer J Clin 71176–1902021 [DOI] [PubMed] [Google Scholar]
- 33.Pairawan S, Hess KR, Janku F, et al. Cell-free circulating tumor DNA variant allele frequency associates with survival in metastatic cancer Clin Cancer Res 261924–19312020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients Eur J Cancer 881–92018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madison R, Schrock AB, Castellanos E, et al. Retrospective analysis of real-world data to determine clinical outcomes of patients with advanced non-small cell lung cancer following cell-free circulating tumor DNA genomic profiling Lung Cancer 14869–782020 [DOI] [PubMed] [Google Scholar]
- 37.Corcoran RB.Liquid biopsy versus tumor biopsy for clinical-trial recruitment Nat Med 261815–18162020 [DOI] [PubMed] [Google Scholar]
- 38.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC Clin Cancer Res 246195–62032018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion Cancer Discov 81529–15392018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis AA, Iams WT, Chan D, et al. Early assessment of molecular progression and response by whole-genome circulating tumor DNA in advanced solid tumors Mol Cancer Ther 191486–14962020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling Cancer Discov 71394–14032017 [DOI] [PMC free article] [PubMed] [Google Scholar]