Feedback mechanisms ensure the exclusive expression of olfactory receptors in Drosophila.
Abstract
In olfactory systems across phyla, most sensory neurons express a single olfactory receptor gene selected from a large genomic repertoire. We describe previously unknown receptor gene–dependent mechanisms that ensure singular expression of receptors encoded by a tandem gene array [Ionotropic receptor 75c (Ir75c), Ir75b, and Ir75a, organized 5′ to 3′] in Drosophila melanogaster. Transcription from upstream genes in the cluster runs through the coding region of downstream loci and inhibits their expression in cis, most likely via transcriptional interference. Moreover, Ir75c blocks accumulation of other receptor proteins in trans through a protein-dependent, posttranscriptional mechanism. These repression mechanisms operate in endogenous neurons, in conjunction with cell type–specific gene regulatory networks, to ensure unique receptor expression. Our data provide evidence for inter-olfactory receptor regulation in invertebrates and highlight unprecedented, but potentially widespread, mechanisms for ensuring exclusive expression of chemosensory receptors, and other protein families, encoded by tandemly arranged genes.
INTRODUCTION
The selective neuronal expression of olfactory receptors is fundamental to the sensory representation of odors in the brain. All current models explaining singular receptor expression in vertebrates and invertebrates invoke the binding of transcriptional activators to one receptor locus while other receptor genes are silenced through transcriptional repressors and/or repressive chromatin structure (1–3). In invertebrates, this process is thought to rely only on deterministic, transcriptional “codes,” where combinations of trans-acting factors function through modular, cis-regulatory elements of receptor genes to achieve specificity of neuronal expression (3–5). By contrast, mammalian olfactory receptor regulation also incorporates a feedback pathway, in which an expressed receptor protein prevents activation of other receptor genes via induction of the unfolded protein response that, ultimately, stabilizes the choice of the expressed receptor gene and precludes desilencing of other receptor loci (1, 2). In all animals, many olfactory receptor genes are found in tandem arrays, as a result of their duplication by nonallelic homologous recombination (6). In some cases in vertebrates, such clustered receptors share enhancer elements (1, 2), but whether this genomic organization has other consequences is unclear.
Here, we investigated the mechanisms ensuring the exclusive expression of members of a “model” cluster of three Drosophila melanogaster Ionotropic receptors (Irs), a gene family that encodes chemosensory receptors distantly related to ionotropic glutamate receptors (7–9). This gene array, Ir75c, Ir75b, and Ir75a (organized 5′ to 3′, spanning <10 kb) (Fig. 1A), is thought to have arisen through duplication of an Ir75a-like ancestral gene in the last common Drosophilidae ancestor (10). These genes encode volatile carboxylic acid–sensing “tuning” receptors that are expressed, together with the co-receptor Ir8a (11), in distinct, spatially stereotyped populations of olfactory sensory neurons (OSNs) housed in different sensory hairs (sensilla) in the main olfactory organ, the antenna (Fig. 1A) (7, 10, 12).
Fig. 1. RNAi screens identify roles for E93 in repression and activation of Ir expression.
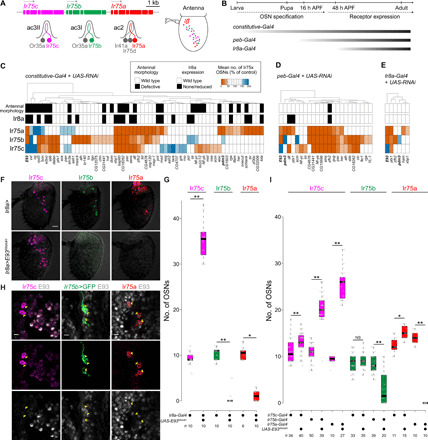
(A) Schematics of Ir75c, Ir75b, and Ir75a (arrows indicate the direction of transcription), the corresponding antennal coeloconic (ac) sensillar neurons, and their antennal distribution (10). (B) Schematics of the phases of OSN development and the temporal expression patterns of the RNA interference (RNAi) screen Gal4 drivers. (C) Hits from the constitutive-Gal4 RNAi screen. At least three antennae were scored for each line [see tables S1 to S7 for data for (C) to (E)]. For Ir75a/b/c neuron numbers, an RNAi line was considered to give a phenotype if there were less than half, or more than 1.5×, the numbers of cells in controls (as illustrated by the heatmap). Euclidian distance was used for the hierarchical clustering in (C) to (E). Control genotype: ey-FLP,Act5c(FRT-CD2)-Gal4/+. (D) Hits from the peb-Gal4 RNAi screen. Control genotype: peb-Gal4,UAS-Dcr-2/+. (E) Hits from the Ir8a-Gal4 RNAi screen. Control genotype: Ir8a-Gal4/+. (F) Representative images of Ir75c, Ir75b, and Ir75a immunofluorescence (on a bright-field background) in control (Ir8a-Gal4/+) and E93RNAi (Ir8a-Gal4/UAS-E93RNAi#1) antennae. Scale bar, 20 μm. (G) Quantification of neuron numbers in the genotypes shown in (F). Boxplots show the median, first, and third quartile of the data, overlaid with individual data points. Pairwise Wilcoxon rank sum test: **P < 0.001 and *P < 0.05. The sample size is shown beneath the plot. (H) Representative images of immunofluorescence on antennal sections for E93 and Ir75c or Ir75a in w1118 flies, or E93 and green fluorescent protein (GFP) in Ir75b-Gal4/+;UAS-myr:GFP/+ animals. Yellow arrowheads indicate the soma. Scale bars, 5 μm. (I) Quantification of neuron numbers in control and Ir OSN-targeted E93RNAi antennae. Genotypes: UAS-myr:GFP/+;Ir75c-Gal4/+, UAS-myr:GFP/+;Ir75c-Gal4/UAS-E93RNAi#1, Ir75b-Gal4/+;UAS-myr:GFP/+, Ir75b-Gal4/+;UAS-myr:GFP/UAS-E93RNAi#1, Ir75a-Gal4/+;[UAS-myr:GFP or +]/+, and Ir75a-Gal4/+;[UAS-myr:GFP or +]/UAS-E93RNAi#1 (GFP signal was not used in any quantifications). Pairwise Wilcoxon rank sum test: **P < 0.001 and *P < 0.05; NS, P > 0.05.
RESULTS
RNAi screening for trans-acting regulators of Ir75c, Ir75b, and Ir75a
To identify trans-acting factors that regulate the expression of Ir75c, Ir75b, and Ir75a, we performed a transgenic RNA interference (RNAi) screen of 121 genes encoding transcription factors and chromatin regulators that were previously implicated in peripheral olfactory system development (13). We induced RNAi with a constitutive Gal4 driver that is active broadly throughout antennal development (Fig. 1B) (13) and examined expression of Ir75c, Ir75b, Ir75a, and Ir8a with antibodies, as well as overall antennal morphology.
Sixty-two genes exhibited RNAi phenotypes (Fig. 1C; fig. S1, A and B; and tables S1 to S3): Most of the phenotypes reflect reductions in the number of OSNs expressing these tuning receptors, but increases in OSN population size were occasionally observed. RNAi of individual genes could affect the expression all three tuning receptors (and often also Ir8a), but many affected only one OSN type or different combinations of two neuron classes. These results revealed a complex and unique gene regulatory network for the three OSN populations.
Because RNAi is induced early in OSN development, many of the identified genes are likely to have upstream roles in antennal development and/or OSN fate specification; many lines produced morphological defects of the antenna (Fig. 1C, fig. S1B, and table S1). To identify candidate molecules that directly regulate receptor expression, we performed RNAi using two additional drivers, pebbled (peb)–Gal4 (14) and Ir8a-Gal4 (11), which are expressed ~16 hours after puparium formation (APF) and ~48 hours APF, respectively (Fig. 1B). As expected, only a subset of genes showed phenotypes with these drivers (Fig. 1, D and E, and tables S4 to S7). Notably, coexpression of Ir75c and Ir75b (which were visualized simultaneously) was extremely rare with any driver (table S1), suggesting that redundant mechanisms exist to prevent these receptors from being expressed in the same neuron.
E93 has a dual role in inhibition and activation of Ir expression
From these screens, we first focused on Ecdysone-induced protein 93F (E93) whose depletion produced a similar phenotype with all drivers: E93RNAi animals display drastic loss of Ir75a and Ir75b expression, while Ir75c is expressed in many additional neurons (Fig. 1, C to G). This phenotype was confirmed in two independent RNAi lines as well as in E93 mutant animals (fig. S2, A to C). While loss of Ir75a and Ir75b was consistently observed regardless of the driver, the number of Ir75c-expressing cells varied depending on the timing of knockdown (fig. S2A).
Previous studies showed that E93 is expressed broadly in the antenna and is necessary for the expression of several Odorant receptor (Or) genes (4). Colabeling with E93 and Ir antibodies [or an Ir transgenic reporter (10)] revealed that E93 is present in Ir75b and Ir75a neurons but not in Ir75c neurons (Fig. 1H). Ir8a-Gal4–driven E93RNAi (hereafter, Ir8a>E93RNAi) occurs at the time when tuning receptor transcription begins (15), suggesting that E93 has a very proximal (if not direct) role in regulating expression of these receptors. The number and spatial distribution of additional Ir75c-positive cells in Ir8a>E93RNAi antennae were consistent with Ir75b and Ir75a neurons expressing Ir75c instead of their normal receptors (Fig. 1, A, F, and G). We tested this possibility by inducing E93RNAi with drivers for these individual neuron populations (Fig. 1I). Ir75a>E93RNAi led to loss of Ir75a expression and an increase in Ir75c OSN numbers. Ir75b>E93RNAi led to reduction (but not complete loss) of Ir75b expression and an increase in Ir75c OSN numbers. Ir75c>E93RNAi has little phenotypic consequence, consistent with the lack of E93 expression in this neuron population.
Together, these observations suggested an initial simple model: In Ir75a and Ir75b neurons, E93 promotes expression of Ir75a or Ir75b (presumably together with OSN subtype–specific transcription factors) and inhibits expression of Ir75c. In Ir75c neurons, E93 is absent, which permits expression of Ir75c (induced by other transcriptional activators).
Ir75c-dependent repression of Ir75b and Ir75a expression
In other tissues, E93 regulates local chromatin structure and/or directly controls gene transcription (16), raising the question of whether expression of Ir75c, Ir75b, and Ir75a is affected at the transcriptional level by the absence of this protein. The boundaries of transcription units of these receptor genes were previously unclear, confounded by the detection of partial transcripts containing exons from adjacent members of this cluster (10). To clarify this issue and determine the effect of loss of E93 on transcription from these loci, we performed bulk RNA sequencing (RNA-seq) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) of control and E93RNAi antennae.
RNA-seq analysis of control tissue provided evidence for the existence of seven distinct transcripts encoded by these three receptor genes (Fig. 2A). Notably, many transcripts initiating from Ir75c do not terminate at the 3′ end of this gene but rather run through the Ir75b and Ir75a exons. However, these transcripts are very unlikely to encode Ir75b or Ir75a proteins because they lack the first exon (containing the start codon) of the downstream genes (Fig. 2A). Similarly, most transcripts initiating from Ir75b incorporate exons 2 to 7 of Ir75a (Fig. 2A). These extended RNAs appear to reflect a failure in transcription termination at the 3′ end of Ir75c and Ir75b, consistent with the absence of canonical transcription termination/polyadenylation sequences downstream of their coding regions; such a sequence is only present 3′ of Ir75a (Fig. 2A). We confirmed the presence of such transcripts by analysis of Expressed Sequence Tag datasets and Sanger sequencing, as well as by RNA fluorescence in situ hybridization (FISH; see below and fig. S3).
Fig. 2. Ir75c-dependent repression of Ir75b and Ir75a expression.
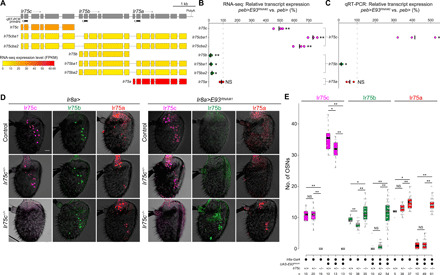
(A) Top: Detailed structure of the Ir75c, Ir75b, and Ir75a loci, indicating qRT-PCR primers and the predicted polyadenylation (polyA) site. Bottom: Heatmap representation of expression levels of seven isoforms encoded by these genes, determined by RNA-seq of antennae from control flies (peb-Gal4,UAS-Dcr-2/+;VIE-260B/+). FKPM, fragments per kilobase million. (B) Relative expression levels (assessed by RNA-seq) of the seven transcript isoforms in antennae of individual biological replicates (n = 3) of peb>E93RNAi flies (peb-Gal4,UAS-Dcr-2/+;UAS-E93RNAi#2/+) normalized to the mean of control samples [genotype as in (A)]. The black bars represent the mean. Two-sample t test (comparing the means of E93RNAi and control samples without normalization): **P < 0.001 and *P < 0.05; NS, P > 0.05. (C) Relative expression (assessed by qRT-PCR) of Ir75c, Ir75b, and Ir75a transcripts in peb>E93RNAi antennae normalized to the mean of the controls [genotypes are as in (A)]. The primer pairs used do not distinguish different isoforms for a given gene. Each dot represents the mean value of three technical replicates. Two-sample t test was performed for ddCt values (comparing the means of E93RNAi and control samples without normalization): *P < 0.05; NS, P > 0.05 (n = 3 biological replicates). (D) Representative images of Ir75c, Ir75b, and Ir75a immunofluorescence in antennae of control (left) and E93RNAi (right) animals in Ir75c+/+, Ir75c+/−, or Ir75c−/− backgrounds. Genotypes: Ir8a-Gal4/+, Ir8a-Gal4/+;Ir75cMB08510/+, Ir8a-Gal4/+;Ir75cMB08510/Ir75cMB08510, Ir8a-Gal4/UAS-E93RNAi#1, Ir8a-Gal4/UAS-E93RNAi#1;Ir75cMB08510/+, and Ir8a-Gal4/UAS-E93RNAi#1;Ir75cMB08510/Ir75cMB08510. Scale bar, 20 μm. (E) Quantification of neuron numbers in the genotypes shown in (D). Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Benjamini and Hochberg method: **P < 0.001 and *P < 0.05; NS, P > 0.05.
We next examined changes in transcript levels for the three Ir genes in E93RNAi antennae. Transcripts initiating from Ir75c are highly up-regulated in the absence of E93 (Fig. 2, B and C), consistent with the ectopic expression of receptor protein (Fig. 1, F and G). Coding transcripts for Ir75b and Ir75a were identified as those containing the first exon of these genes: Transcription initiating from Ir75b is strongly diminished (Fig. 2, B and C), concordant with the loss of Ir75b expression (Fig. 1, F and G). By contrast, Ir75a transcript levels were only slightly, but nonsignificantly, reduced in E93RNAi antennae (Fig. 2, B and C), despite the loss of detectable Ir75a protein (Fig. 1, F and G). These observations refined our model as they indicate that E93 inhibits Ir75c transcription and promotes Ir75b transcription but has only an indirect role in promoting Ir75a expression.
We were unable to identify an E93 binding motif (16) within the minimal promoter element of Ir75b (10). Given the increase in Ir75c expression in Ir75b neurons (Fig. 1I) and the existence of transcriptional read-through across these loci (Fig. 2A), we hypothesized that the decrease in Ir75b transcription is an indirect consequence of the ectopic expression of Ir75c through a process of transcriptional interference (17), i.e., where transcription from one gene (here, Ir75c) inhibits, in cis, the expression of a downstream gene (here, Ir75b).
To test this possibility, we obtained an Ir75c mutant (Ir75cMB08510) bearing a Minos insertion in the 5′ regulatory region (fig. S3A), which we found leads to essentially complete loss of Ir75c RNA (fig. S3, B to D). By contrast, transcripts of Ir75b and Ir75a were still detected (fig. S3C). Moreover, analysis of the number of neurons detected by RNA probes for these receptor genes in control and Ir75c mutants provided further evidence of read-through of Ir75c transcripts through these downstream loci (fig. S3, C and D). We noticed that numbers of Ir75b (and Ir75a) neurons were slightly elevated in Ir75c mutant antennae compared to controls (Fig. 2, D and E); we return to this phenomenon in the last section of the results. Most notably, combination of the Ir75c mutant with E93RNAi was sufficient to restore Ir75b expression (Fig. 2, D and E). This result has two implications: first, that E93 is not directly required for transcription of Ir75b and, second, that expression of Ir75c can inhibit transcription of Ir75b.
Loss of Ir75c in an E93RNAi background also led to restoration of Ir75a expression (Fig. 2, D and E). As Ir75a is still robustly transcribed in E93RNAi antennae (Fig. 2, B and C), this observation indicates that Ir75c inhibits Ir75a expression predominantly posttranscriptionally (although the existence of Ir75cba transcripts raises the possibility of a minor, cis-acting effect of Ir75c on Ir75a transcription). We also noted that Ir75b expression is only very slightly increased in E93RNAi antennae in a heterozygous Ir75c mutant, where one allele of Ir75b should not be subject to transcriptional interference by Ir75c (Fig. 2E). We found no evidence that Ir75c inhibits Ir75b transcription through interchromosomal regulation (i.e., transvection) (fig. S4) (18), suggesting that Ir75b must be additionally repressed posttranscriptionally by Ir75c.
Posttranscriptional repression of Ir75a and Ir75b by Ir75c protein
We examined the hypothesized posttranscriptional inhibitory activity of Ir75c by misexpressing the gene from a transgenic cDNA construct (i.e., unlinked to the endogenous Ir75c locus) using Ir8a-Gal4. We observed a mild but significant reduction in the number of Ir75b-expressing cells and more substantial loss of Ir75a (Fig. 3, A and B). Many of the detectable Ir75b and Ir75a neurons had only weak immunofluorescence signals (Fig. 3C). As a control, we misexpressed Ir75a cDNA using the same driver, but this had no obvious impact on Ir75c expression, and led to a slight but variable increase in Ir75b (Fig. 3, A to C).
Fig. 3. Posttranscriptional inhibition of Ir75a and Ir75b by Ir75c protein.
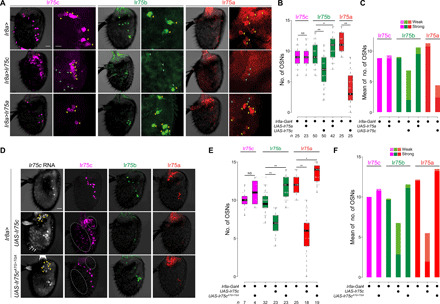
(A) Representative images of Ir75c, Ir75b, and Ir75a immunofluorescence in antennae of control (Ir8a-Gal4/+), Ir75c misexpression (Ir8a-Gal4/UAS-Ir75c), and Ir75a misexpression (Ir8a-Gal4/UAS-Ir75a) animals. Scale bar, 20 μm. A region of interest is shown at higher magnification to the right of each image. Scale bar, 10 μm. Yellow and cyan arrowheads indicate neurons with strong and weak receptor expression, respectively. (B) Quantification of neuron numbers in the genotypes shown in (A) (Ir75a quantification in Ir8a>Ir75a antennae and Ir75c quantification in Ir8a>Ir75c antennae are not shown, as neuron density, comprising the entire Ir8a population, is too high for unambiguous scoring). Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Bonferroni method: **P < 0.001 and *P < 0.05; NS, P > 0.05. (C) Quantification of the mean number of neurons with strong and weak immunofluorescence signals [see arrowheads in (A)] in the genotypes shown in (A). (D) Representative images of Ir75c RNA FISH and Ir75c, Ir75b, and Ir75a immunofluorescence in antennae of control (Ir8a-Gal4/+), Ir75c misexpression (Ir8a-Gal4/UAS-Ir75c), and Ir75cATG>TGA misexpression (Ir8a-Gal4/UAS-Ir75cATG>TGA) animals. Yellow arrowheads indicate examples of neurons expressing Ir75c RNA ectopically. Ectopic Ir75c protein signal is detected only in antennae expressing UAS-Ir75c but not UAS-Ir75cATG>TGA (e.g., in regions highlighted with dashed ovals). Scale bar, 20 μm. (E) Quantification of neuron numbers in the genotypes shown in (D) (Ir75c quantification for Ir8a>Ir75c antennae is not shown, as neuron density, comprising the entire Ir8a population, is too high for unambiguous scoring). Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Bonferroni method: **P < 0.001 and *P < 0.05; NS, P > 0.05. It is unknown why Ir75b and Ir75a neuron numbers increase slightly upon expression of UAS-Ir75cATG>TGA. (F) Quantification of the number of neurons with strong and weak immunofluorescence signals in the genotypes shown in (D).
To test whether the inhibitory effect of Ir75c expression was at the RNA or protein level, we replaced the start codon in the Ir75c cDNA with a stop codon (Ir75cATG>TGA). Ir8a-Gal4–driven expression of this transgene led to ectopic Ir75c RNA, but not ectopic Ir75c protein (Fig. 3D). In these antennae, no reduction in Ir75b or Ir75a expression was observed, contrasting with the impact of the protein-coding transgene (Fig. 3, D to F). These data, together with the analysis of Ir transcript levels (Fig. 2, B and C), suggest a model in which transcription from the endogenous Ir75c gene inhibits Ir75b at the transcriptional level, and Ir75c protein impairs Ir75b and Ir75a expression posttranscriptionally.
Ir75b-dependent, cis-mediated repression of Ir75a
The failure of Ir75b transcripts to terminate efficiently at the 3′ end of this gene (Fig. 2A) raised the possibility that Ir75b can also suppress expression of Ir75a by transcriptional interference, analogous to the inhibition of Ir75b transcription by Ir75c. From our RNAi screen, we noted that both constitutive-Gal4– and peb-Gal4–driven RNAi of pou domain motif 3 (pdm3) led to increased numbers of Ir75b-expressing cells and greatly diminished the number of Ir75a-expressing cells (Ir75c expression is unchanged) (Fig. 1, C and D). An independent study of Pdm3 (19) indicated that this transcription factor is expressed in Ir75a neurons, but not Ir75b or Ir75c OSNs; here, Pdm3 promotes Ir75a neuron fate and suppresses Ir75b neuron fate, encompassing receptor protein expression and axon targeting properties (19). We wondered, however, whether loss of Ir75a in pdm3RNAi antennae might be an indirect consequence of ectopic Ir75b expression.
We first examined whether loss of pdm3 affects transcription of these genes. Results from qRT-PCR analysis corresponded well with changes in the number of neurons expressing Ir75b and Ir75a proteins: peb>pdm3RNAi antennae displayed an increase in Ir75b transcription and decreased Ir75a transcription (fig. S5A). Furthermore, we confirmed by RNA FISH the increase in number of cells expressing Ir75b transcripts upon loss of Pdm3 (fig. S5, B and C). The number of neurons detected by our Ir75a probe was not different between genotypes, presumably because this reagent detects transcripts initiating from all three genes in the cluster; the decrease in Ir75a-expressing cells is therefore counterbalanced by the increase in the size of the Ir75b neuron population (fig. S5, B and C).
To test whether Pdm3’s role in promoting Ir75a transcription was direct or indirect, we combined peb>pdm3RNAi with an Ir75b mutant (Ir75bDsRed), which bears a 1.3-kb insertion in the middle of the gene that disrupts Ir75b transcription and abolishes Ir75b protein expression (fig. S3, A, E, and F, and Fig. 4, A and B). Ir75a expression is partially restored through loss of one copy of Ir75b and fully restored in homozygous Ir75b mutants (Fig. 4, A and B). Together, these data support a model in which Pdm3 functions in Ir75a neurons to repress Ir75b expression, which would otherwise inhibit transcription of Ir75a in cis. We tested whether Ir75b might also affect Ir75a expression posttranscriptionally but found that heterologous expression of a UAS-Ir75b transgene yielded barely detectable ectopic Ir75b, precluding conclusive insights.
Fig. 4. Ir75b-dependent repression of Ir75a.
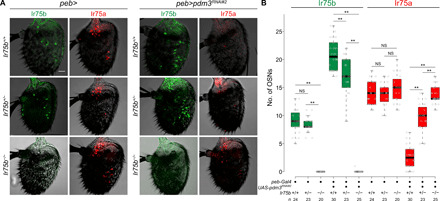
(A) Representative images of Ir75b and Ir75a immunofluorescence in control and pdm3RNAi antennae in Ir75b+/+, Ir75b+/−, and Ir75b−/− backgrounds. Genotypes (top to bottom): peb-Gal4,UAS-Dcr-2/+, peb-Gal4,UAS-Dcr-2/+;;Ir75bDsRed/+, peb-Gal4,UAS-Dcr-2/+;;Ir75bDsRed/Ir75bDsRed (left), peb-Gal4,UAS-Dcr-2/+;UAS-pdm3RNAi#2/+, peb-Gal4,UAS-Dcr-2/+;UAS-pdm3RNAi#2/+;Ir75bDsRed/+, and peb-Gal4,UAS-Dcr-2/+;UAS-pdm3RNAi#2/+;Ir75bDsRed/Ir75bDsRed (right). Scale bar, 20 μm. (B) Quantification of neuron numbers in the genotypes shown in (A). Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Bonferroni method: **P < 0.001; NS, P > 0.05.
Ir75c suppresses expression of Ir75b and Ir75a in its own neurons
Last, we asked whether Ir75c and Ir75b exert a repressive effect on other genes in the cluster in their endogenous neurons and not only when expressed ectopically in other neurons (i.e., in E93RNAi or pdm3RNAi antennae or through transgenic misexpression of Ir75c). In Ir75c mutant antennae, we observed a small and variable increase in the number of neurons expressing Ir75b and Ir75a, compared to controls (Fig. 5, A and B; see also Fig. 2E). Similarly, in Ir75b mutants, we observed an increase in numbers of neurons expressing Ir75a, but no effect on those expressing Ir75c, consistent with a contribution of Ir75b in suppressing expression of Ir75a in its own neurons (Fig. 5, A and B). This latter effect was sensitive to the genetic background, however, as we did not observe such an increase in all genotypes (e.g., Fig. 4B).
Fig. 5. Ectopic expression of Ir75b and Ir75a in Ir75c mutant neurons.
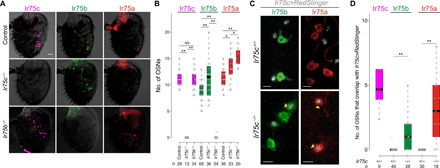
(A) Representative images of Ir75c, Ir75b, and Ir75a immunofluorescence in control (w1118), Ir75c−/− (Ir75cMB08510/Ir75cMB08510), and Ir75b−/− (Ir75bDsRed/Ir75bDsRed) antennae. Scale bar, 20 μm. (B) Quantification of neuron numbers in the genotypes shown in (A). Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Benjamini and Hochberg method: **P < 0.001 and *P < 0.05; NS, P > 0.05. (C) Representative images of Ir75b and Ir75a immunofluorescence for a subset of Ir75c neurons [labeled with RedStinger (gray)] in Ir75c+/− and Ir75c−/− antennae. Yellow arrowheads mark Ir75c−/− neurons that ectopically express Ir75b or Ir75a. Genotypes: UAS-RedStinger/+;Ir75c-Gal4,Ir75cMB08510/+ and UAS-RedStinger/+;Ir75c-Gal4,Ir75cMB08510/Ir75cMB08510. Scale bars, 5 μm. (D) Quantification of Ir75c neurons that ectopically express Ir75b or Ir75a in Ir75c+/− and Ir75c−/− flies [genotypes as in (C)]. Pairwise Wilcoxon rank sum test and P values adjusted for multiple comparisons with the Bonferroni method: **P < 0.001. In the control Ir75c immunofluorescence samples, all neurons [81 of 81 (100%)] express both Ir75c and Ir75c>RedStinger. Ectopic expression of Ir75b and Ir75a in Ir75c−/− neurons appeared to be mutually exclusive [only 1 of 18 (5.5%) neurons expressed both of these receptors, presumably, in part, due to the repressive effect of Ir75b expression on Ir75a (Fig. 4)].
We therefore focused on the Ir75c mutant and examined whether the additional expression of Ir75b and Ir75a is in Ir75c neurons, by labeling these cells transgenically with Ir75c>RedStinger. In control tissue, we detected, as expected, no expression of Ir75a or Ir75b in Ir75c neurons (with the exception of one cell) (Fig. 5, C and D). By contrast, in Ir75c mutants, several Ir75c>RedStinger-labeled neurons expressed one or other of these Irs (Fig. 5, C and D), indicating that Ir75c represses these receptors in its own neural population. It is not unexpected that the ectopic expression of Ir75b and Ir75a is limited to only a small subset of Ir75c mutant neurons as these cells presumably only partially resemble the gene regulatory environment of endogenous Ir75b or Ir75a neurons, as illustrated by our original RNAi screens (Fig. 1, C to E).
DISCUSSION
Through a screen for trans-acting regulators controlling the expression of a model tandem cluster of olfactory receptor genes, we have found, serendipitously, novel types of feedback pathway to ensure exclusive receptor expression. These findings provide evidence for inter-olfactory receptor regulation in invertebrates (Fig. 6).
Fig. 6. Model of the regulatory interactions between Ir75c, Ir75b, and Ir75a.

Schematics of the predicted interactions between Ir75c, Ir75b, and Ir75a gene products in each neuron class in wild-type, E93RNAi, pdm3RNAi, and Ir75c−/− genetic backgrounds. RNA polymerase II complexes (RNA polII) are shaded yellow. Protein-coding transcripts and long 3′ untranslated region extensions (lacking the first exons of the downstream genes) are shown as unbroken and broken lines, respectively. It is unknown whether E93 and Pdm3 inhibit transcription of Ir75c and Ir75b, respectively, directly or indirectly (indicated by dashed lines). Pdm3 is selectively expressed in Ir75a neurons (19).
The cis-mediated transcriptional repression of Ir75b by Ir75c, and of Ir75a by Ir75b, is most simply explained by transcriptional interference (Fig. 6) (17). This model is supported by the existence of transcripts containing exons from adjacent genes within this array and the presence of a canonical transcription terminator sequence downstream of only Ir75a. Our previous mapping of RNA polymerase II occupancy in these neurons by Targeted DamID (19) provides further evidence for the failure in efficient termination of the transcription machinery at the 3′ end of Ir75c and Ir75b. The biochemical mechanism of transcriptional interference is not fully understood but is thought to result from steric competition on the DNA, where RNA polymerase II complexes transcribing an upstream gene prevent assembly of the transcription machinery at the promoter of a downstream gene (17, 20). Testing this model explicitly would require the ability to terminate RNA polymerase II transcription at the 3′ end of Ir75c and Ir75b. As such, we cannot currently exclude alternative (or additional) mechanisms to explain the reduced transcription of the downstream gene when the upstream gene is active, such as competition during promoter selection; investigating this possibility will depend on much better knowledge of the transcriptional regulatory sequences of each gene (10). However, we do not find any evidence for interchromosomal regulation (transvection) between these loci.
Similar to the Irs analyzed in this work, several Gustatory receptor (Gr) genes are arranged in clusters in Drosophila, with transcription termination signals only apparent after the last gene in the array (21). Analysis of transcripts encoded by the tandem sugar-sensing Gr genes revealed that they encompass exons of several individual loci (21–23). While polycistronic transcription of these genes might underlie the neuronal coexpression observed for some of these receptors (21–23), an alternative (or additional) possibility is that such a pattern of transcription reflects a mechanism by which Gr genes inhibit each other’s expression through transcriptional interference. Future analysis of this and other tandem arrays of chemosensory receptor genes will reveal how widespread the phenomenon is. In an evolutionary context, the existence of cis-mediated transcriptional repression in recently duplicated chemosensory genes may provide a substrate for natural selection to favor rapid acquisition of distinct expression patterns (or otherwise pseudogenization of one duplicate). Such a possibility was supported by genome-wide expression analysis of nested (nonchemosensory) genes in drosophilids (24). For the Irs studied here, recruitment of E93 and Pdm3 to their gene regulatory networks may have introduced important inhibitory connections: By preventing transcription of upstream genes in the cluster, these transcription factors permit expression of downstream genes (Fig. 6).
The mechanism of posttranscriptional (trans) suppression by Ir75c protein of Ir75b and, in particular, Ir75a remains unclear. As Ir75a transcription is not substantially diminished by Ir75c misexpression, it seems unlikely that this inhibition operates via the same feedback mechanism between mammalian olfactory receptor loci, which ensures the silenced transcriptional state of nonchosen receptor genes (2). The repression of Ir75a by Ir75c also appears to be distinct from the exclusion mechanism between Rhodopsin 5 (Rh5) and Rh6 in the Drosophila visual system, in which an Rh6 protein-dependent signal represses rh5 promoter activity (25), as well as the broad suppression of Or transcription resulting from transgenic overexpression of one Or in Anopheles gambiae (26). One possibility is that Ir protein subunits compete with each other for the binding to the common Ir8a co-receptor, which is essential for their stabilization in OSNs (11, 27). Precise details of the repression mechanism(s) will require future development of methods to quantify the processes controlling the translation of receptor transcripts and regulating the turnover of proteins within specific OSN populations.
Given the widespread occurrence of chemosensory receptor gene families in arrays in the genomes of most or all animals (6), our discoveries may have broad implications for the evolution of their distinct expression patterns. Moreover, the dominance of expression of 5′-located Ir genes over 3′-located genes is reminiscent of the “posterior dominance” and “posterior prevalence” of transcripts and proteins encoded by clustered Hox transcription factor genes (28, 29). Study of the regulatory interactions between arrays of olfactory receptor genes may reveal general insights into how other types of proteins encoded by tandemly duplicated genes evolve distinct expression patterns.
MATERIALS AND METHODS
Drosophila strains and culture
Flies were maintained at 25°C in 12-hour light:12-hour dark conditions, except where noted. D. melanogaster strains were maintained on a standard wheat flour–yeast–fruit juice medium. Published mutant and transgenic D. melanogaster are described in table S8. For histological experiments, flies were 3 to 8 days old. To increase the efficiency of RNAi, flies were moved to 27°C after 2 to 3 days. As peb>E93RNAi#1 flies were not viable at 27°C, this genotype was raised at 19°C.
New Drosophila strains
UAS-Ir75cATG>TGA transgenic line
Site-directed mutagenesis of the Ir75c start codon in pUAST-Ir75c attB (10) was performed using a primer pair (CGATTTCCACACCTGAACTAGTTGGCCATTG/CAATGGCCAACTAGTTCAGGTGTGGAAATCG; the introduced stop codon is underlined) using standard methods. The presence of the mutation was confirmed by sequencing, and the mutated Ir75c cDNA was subcloned into a new pUAST attB vector backbone to avoid other, unintended mutations in the construct, before transgenesis via the phiC31 site-specific integration system (landing site attP40) by BestGene Inc.
Ir75b mutant
The sgRNA expression vector was generated by cloning an annealed primer pair (GTCGCCCTCAGTGGCTGACGCGCC/AAACGGCGCGTCAGCCACTGAGGG) into Bbs I–digested pCFD3-dU6-3gRNA (Addgene, no. 49410), as previously described (30). The donor vector was constructed by amplifying Ir75b-specific homology arms (1.1 to 1.4 kb) from genomic DNA of {Act5C-Cas9.P.RFP-}ZH-2A DNAlig4[169] flies (31) [homology arm 1 (5′ to 3′): GATCCACCTGCGATCTCGCCACCTCGAAGCAGAGCCGGT/GATCCACCTGCGATCCTACCGGCCTGGTAACCCAATCTA; homology arm 2: GATCGCTCTTCGTATGCGTCAGCCACTAAGGGATATGTT/GATCGCTCTTCGGACCACGGCCACCGTGTGATTATGCTG] and inserting the resultant products into pHD-DsRed-attP (Addgene, no. 51019) (32) via Aar I/Sap I restriction enzyme–based cloning.
Transgenesis of D. melanogaster was performed in-house following standard protocols (http://gompel.org/methods). For CRISPR-Cas9–mediated homologous recombination, we injected a mix of the sgRNA expression (final concentration of 150 ng μl−1) and donor vector (final concentration of 400 ng μl−1) into {Act5C-Cas9.P.RFP-}ZH-2A DNAlig4[169] flies. Ir75b mutant animals were selected on the basis of DsRed expression, and correct integration was confirmed by PCR. The Ir75bDsRed allele was outcrossed to w1118 for three generations before establishment of a homozygous mutant stock.
Histology and image analysis
Immunofluorescence and RNA FISH on whole-mount antennae or antennal cryosections were performed essentially as previously described (33). Primary and secondary antibodies used are listed in table S9. Guinea pig anti-Ir75c antibodies were raised against the same peptide epitope (KEYLSELHLRPRLQHRMD) as used to generate the rabbit anti-Ir75c antibodies (10) and affinity-purified by Proteintech Group Inc. RNA FISH probes complementary to sequences of Ir75a, Ir75b, and Ir75c were as previously described (10). Imaging was performed on a Zeiss confocal microscope LSM710 or, for RNA FISH samples, a Zeiss confocal microscope 880 Airyscan, using 40× or 63× oil immersion objectives. All images were processed in Fiji, and the cell counter plugin was used to quantify neuron numbers.
RNA-seq and analysis
Antennal RNA was extracted from three biological replicates of control [peb-Gal4,UAS-Dcr-2/+;VIE-260B/+ (VIE-260B is an empty landing site used for RNAi transgene insertion)] and E93RNAi (peb-Gal4,UAS-Dcr-2/+;UAS-E93RNAi#2/+) animals. For each pair of biological replicates, ~350 females were grown under identical conditions and antennae were harvested and RNA was extracted from 2- to 5-day-old flies in parallel, as previously described (13). RNA quality was assessed on a Fragment Analyzer (Advanced Analytical Technologies Inc.); all RNAs had an RNA quality number of 9 to 10. mRNA isolation, RNA-seq library preparation, and sequencing were performed as previously described (34).
Reads were aligned against D. melanogaster.BDGP6.92 genome using STAR (Spliced Transcripts Alignment to a Reference) v2.5.3a (35). The reads mapped to the region 3L:17,815,000-17,823,000 containing the Ir75c/Ir75b/Ir75a genes were extracted using SAMtools v1.8 (36), and transcripts were assembled using StringTie v1.3.3b (37) with parameters for stranded library and a minimum transcript length of 1000 bases. Transcript abundance was quantified with StringTie using the D. melanogaster.BDGP6.92 reference annotation complemented with the four novel isoforms (options: -G, -m 1000, -e, and -B).
Quantitative reverse transcription polymerase chain reaction
For analysis of E93RNAi animals, the same RNA samples were used as for the RNA-seq experiment. For other experiments, RNA was isolated from 2- to 5-day-old flies (~50 antennae) with the QIAGEN RNeasy Mini Kit (catalog no. 74104), and cDNA was prepared using Invitrogen SuperScript II Reverse Transcriptase (catalog no. 18064022).
qRT-PCR was performed on the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) using iQ SYBR Green Supermix from Bio-Rad. Primer pairs (5′ to 3′) were as follows: Ir75c (TGCTGGTCCATAAAGGAGGC/GGCTTTGTCGCAACCCAAAT), Ir75b (ATTTTGCACTGCTCACTTTCGC/AACTGTACCACTTTTGTCGCA), Ir75a (GCTGGCAGAGTGATGAAAGTCT/TCCTGAGTCTGATCGCACTTT), and control (tubulin: αTub84B isoform) (TGTCGCGTGTGAAACACTTC/AGCAGGCGTTTCCAATCTG). qRT-PCR was performed with three technical replicates for each of three biological replicates for each primer pair. The mean of the three technical replicates was used for each biological replicate. ddCt values were calculated with normalization to tubulin values.
Statistical analysis
Statistical analyses and plotting were performed in RStudio v3.5.2 (R Foundation for Statistical Computing, Vienna, Austria, 2005; R-project-org). For multiple comparisons, a two-tailed Wilcoxon rank sum test was performed followed by the Benjamini and Hochberg method or the Bonferroni method for P value adjustment. For fig. S5A, P values were calculated by one-way analysis of variance (ANOVA) test of ddCt values followed by a Tukey’s post hoc test for multiple comparisons. For other qRT-PCR experiments and for the RNA-seq analysis, P values were calculated by a two-sample t test.
Acknowledgments
We thank E. Baehrecke and C. Doe for reagents, C. Dumayne for help with qRT-PCR experiments, and M. C. Gambetta and members of the Benton laboratory for discussions and comments on the manuscript. Funding: L.L.P.-G. was supported by a FEBS Long-Term Fellowship. T.O.A. was supported by a Human Frontier Science Program Long-Term Fellowship (LT000461/2015-L) and by a Swiss National Science Foundation Ambizione Grant (PZ00P3 185743). Research in R.B.’s laboratory was supported by the University of Lausanne, European Research Council Consolidator and Advanced Grants (615094 and 833548, respectively), and the Swiss National Science Foundation (310030B_185377). Author contributions: All authors contributed to the experimental design, analysis, and interpretation of results. K.M. performed all experiments, except for those in Fig. 3D and figs. S3 (C to H) and S5 (B and C), as well as RNA isolation for RNA-seq and qRT-PCR experiments, which were performed by S.C. P.C.C. provided the candidate gene list for the RNAi screen and performed initial analysis of RNA-seq data. L.L.P.-G. supervised K.M. during the initial stages of the project. T.O.A. generated the Ir75b mutant. S.P. performed RNA-seq data analysis. K.M. and R.B. wrote the paper with input from all other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data are available in GEO (accession GSE150296). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/32/eabe3745/DC1
REFERENCES AND NOTES
- 1.Nagai M. H., Armelin-Correa L. M., Malnic B., Monogenic and monoallelic expression of odorant receptors. Mol. Pharmacol. 90, 633–639 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Dalton R. P., Lomvardas S., Chemosensory receptor specificity and regulation. Annu. Rev. Neurosci. 38, 331–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barish S., Volkan P. C., Mechanisms of olfactory receptor neuron specification in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 4, 609–621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jafari S., Alkhori L., Schleiffer A., Brochtrup A., Hummel T., Alenius M., Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 10, e1001280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuss S. H., Ray A., Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol. Cell. Neurosci. 41, 101–112 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Nei M., Niimura Y., Nozawa M., The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 9, 951–963 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B., Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rytz R., Croset V., Benton R., Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Ni L., The structure and function of ionotropic receptors in Drosophila. Front. Mol. Neurosci. 13, 638839 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prieto-Godino L. L., Rytz R., Cruchet S., Bargeton B., Abuin L., Silbering A. F., Ruta V., Dal Peraro M., Benton R., Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93, 661–676.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Abuin L., Bargeton B., Ulbrich M. H., Isacoff E. Y., Kellenberger S., Benton R., Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prieto-Godino L. L., Rytz R., Bargeton B., Abuin L., Arguello J. R., Peraro M. D., Benton R., Olfactory receptor pseudo-pseudogenes. Nature 539, 93–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chai P. C., Cruchet S., Wigger L., Benton R., Sensory neuron lineage mapping and manipulation in the Drosophila olfactory system. Nat. Commun. 10, 643 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komiyama T., Sweeney L. B., Schuldiner O., Garcia K. C., Luo L., Graded expression of semaphorin-1a cell-autonomously directs dendritic targeting of olfactory projection neurons. Cell 128, 399–410 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Li H., Li T., Horns F., Li J., Xie Q., Xu C., Wu B., Kebschull J. M., McLaughlin C. N., Kolluru S. S., Jones R. C., Vacek D., Xie A., Luginbuhl D. J., Quake S. R., Luo L., Single-cell transcriptomes reveal diverse regulatory strategies for olfactory receptor expression and axon targeting. Curr. Biol. 30, 1189–1198.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uyehara C. M., Nystrom S. L., Niederhuber M. J., Leatham-Jensen M., Ma Y., Buttitta L. A., McKay D. J., Hormone-dependent control of developmental timing through regulation of chromatin accessibility. Genes Dev. 31, 862–875 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shearwin K. E., Callen B. P., Egan J. B., Transcriptional interference—A crash course. Trends Genet. 21, 339–345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukaya T., Levine M., Transvection. Curr. Biol. 27, R1047–R1049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arguello J. R., Abuin L., Armida J., Mika K., Chai P. C., Benton R., Targeted molecular profiling of rare olfactory sensory neurons identifies fate, wiring, and functional determinants. eLife 10, e63036 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Proudfoot N. J., Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slone J., Daniels J., Amrein H., Sugar receptors in Drosophila. Curr. Biol. 17, 1809–1816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahanukar A., Lei Y. T., Kwon J. Y., Carlson J. R., Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii S., Yavuz A., Slone J., Jagge C., Song X., Amrein H., Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 25, 621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assis R., Transcriptional interference promotes rapid expression divergence of Drosophila nested genes. Genome Biol. Evol. 8, 3149–3158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasiliauskas D., Mazzoni E. O., Sprecher S. G., Brodetskiy K., Johnston R. J. Jr., Lidder P., Vogt N., Celik A., Desplan C., Feedback from rhodopsin controls rhodopsin exclusion in Drosophila photoreceptors. Nature 479, 108–112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maguire S. E., Afify A., Goff L. A., Potter C. J., A feedback mechanism regulates odorant receptor expression in the malaria mosquito, Anopheles gambiae. bioRxiv, 2020.07.23.218586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ai M., Blais S., Park J. Y., Min S., Neubert T. A., Suh G. S., Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J. Neurosci. 33, 10741–10749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gummalla M., Galetti S., Maeda R. K., Karch F., Hox gene regulation in the central nervous system of Drosophila. Front. Cell. Neurosci. 8, 96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durston A. J., Global posterior prevalence is unique to vertebrates: A dance to the music of time? Dev. Dyn. 241, 1799–1807 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, –E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Koolhaas W. H., Schnorrer F., A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 4, 2409–2418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., O’Connor-Giles K. M., Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saina M., Benton R., Visualizing olfactory receptor expression and localization in Drosophila. Methods Mol. Biol. 1003, 211 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Prieto-Godino L. L., Silbering A. F., Khallaf M. A., Cruchet S., Bojkowska K., Pradervand S., Hansson B. S., Knaden M., Benton R., Functional integration of "undead" neurons in the olfactory system. Sci. Adv. 6, eaaz7238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L., StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mou X., Duncan D. M., Baehrecke E. H., Duncan I., Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc. Natl. Acad. Sci. U.S.A. 109, 2949–2954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.S. Chyb, N. Gompel, Atlas of Drosophila Morphology: Wild-Type and Classical Mutants (Academic Press, 2013). [Google Scholar]
- 40.Bing X., Rzezniczak T. Z., Bateman J. R., Merritt T. J. S., Transvection-based gene regulation in Drosophila is a complex and plastic trait. G3 4, 2175–2187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/32/eabe3745/DC1


