SUMMARY
Proper lung function relies on the precise balance of specialized epithelial cells that coordinate to maintain homeostasis. Herein, we describe essential roles for the transcriptional regulators YAP/TAZ in maintaining lung epithelial homeostasis, reporting that conditional deletion of Yap and Wwtr1/Taz in the lung epithelium of adult mice results in severe defects, including alveolar disorganization and the development of airway mucin hypersecretion. Through in vivo lineage tracing and in vitro molecular experiments, we reveal that reduced YAP/TAZ activity promotes intrinsic goblet transdifferentiation of secretory airway epithelial cells. Global gene expression and chromatin immunoprecipitation sequencing (ChIP-seq) analyses suggest that YAP/TAZ act cooperatively with TEA domain (TEAD) transcription factors and the NuRD complex to suppress the goblet cell fate program, directly repressing the SPDEF gene. Collectively, our study identifies YAP/TAZ as critical factors in lung epithelial homeostasis and offers molecular insight into the mechanisms promoting goblet cell differentiation, which is a hallmark of many lung diseases.
Graphical Abstract

In brief
Hicks-Berthet et al. report that deletion of YAP/TAZ in adult lung epithelium results in defects that include widespread goblet cell metaplasia. YAP/TAZ restrict goblet cell differentiation through a transcriptional network that includes direct SPDEF repression. These findings offer insight into signals that direct goblet cell fate in the lung.
INTRODUCTION
The lung epithelium is made up of specialized cells that coordinate to perform the essential functions of gas exchange and barrier host defense. Alveolar type I and type II (ATI and ATII) cells act in concert to maintain the alveolar structures required for efficient oxygenation. Airway secretory club cells, goblet cells, multiciliated cells, neuroendocrine cells, basal cells, and chemosensory tuft cells collectively function to regulate the process of mucociliary clearance and maintain balanced cellular turnover that controls bronchial homeostasis. Goblet cells, in particular, play notable roles in airway homeostasis via the production and secretion of mucins (Whitsett, 2018). Normally maintained at relatively low numbers during homeostasis, goblet cells are specified upon exposure to environmental insults or inflammatory signals. Uncontrolled specification of goblet cells leads to increased production and secretion of mucins within the airways, with accumulation of excess mucins being a hallmark feature of numerous airway diseases, including asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis (Fahy and Dickey, 2010).
YAP and TAZ are transcriptional effectors that have emerged as key regulators of epithelial injury responses in a variety of organs, including the lung (LaCanna et al., 2019; Lange et al., 2015; Liu et al., 2016). Aberrant activation of YAP/TAZ nuclear activity has been shown to initiate injury-related stem cell expansion in the lung epithelium (Gokey et al., 2018; Volckaert et al., 2017; Zhao et al., 2014), and deletion of YAP and/or TAZ limits the regenerative potential of the lung epithelium in response to damage (LaCanna et al., 2019; Liu et al., 2016). However, studies of YAP/TAZ activity in lung homeostasis are limited, and given the interest in therapeutically targeting these factors, understanding such roles remains important.
In this study we probed YAP and TAZ functions in lung homeostasis and found that conditional deletion of YAP/TAZ within the mouse lung epithelium results in severe lung damage, increased inflammation, and matrix remodeling. We further discovered that YAP/TAZ repress goblet cell differentiation, as deletion of Yap/Taz results in widespread goblet metaplasia. Our experiments revealed that YAP/TAZ act through the TEA domain (TEAD) family (TEAD1–4) transcription factors and cooperate with the nucleosome remodeling and deacetylase (NuRD) complex to directly repress SPDEF expression and downstream goblet cell fate programs. We provide evidence that Hippo pathway kinases, which restrict YAP/TAZ activity, are required for cytokine-stimulated goblet cell gene expression. These data demonstrate a requirement for YAP/TAZ in lung epithelial homeostasis and highlight molecular mechanisms controlling goblet cell differentiation, offering important insight into a cell type commonly dysregulated in lung disease.
RESULTS
Yap and Taz are essential for maintaining adult lung epithelial homeostasis
Given the importance of Yap and Taz in lung epithelial development and injury responses (Isago et al., 2020; LaCanna et al., 2019; Lange et al., 2015; Liu et al., 2016; Mahoney et al., 2014; Zhao et al., 2014), we sought to test the function of these factors in adult lung homeostasis. For this we tested the consequences of conditionally deleting Yap, Taz, or both Yap and Taz (Yap/Taz) in the mouse lung epithelium by crossing Yap- and Taz-floxed mice (Reginensi et al., 2013) with mice expressing tamoxifen-inducible Cre recombinase from the Nkx2.1 promoter (Nkx2.1-CreERT2) (Taniguchi et al., 2011) (herein referred to as YapNxk-KO, TazNxk-KO, and Yap/TazNxk-KO, respectively) (Figure 1A). Consistent with the widespread expression of Nkx2.1 in the lung epithelium, we observed loss of Yap and Taz protein throughout the alveolar and intrapulmonary airway epithelium following tamoxifen treatment (Figure S1A). Individual deletion of Yap or Taz resulted in no overt phenotypes, but co-deletion of Yap/Taz yielded rapid and severe morbidity with a mean survival of 12 days after tamoxifen exposure (Figure 1B). Gross analysis of the Yap/TazNxk-KO lungs showed regions of severe lung collapse and necrosis, with evident alterations in epithelial morphology and increased cellularity, which were not evident in control, YapNxk-KO, or TazNxk-KO lungs (Figures 1C and S1B). Histological examination of other Nxk2.1-expressing tissues, such as the thyroid, similarly showed altered morphologies in Yap/Taz null mice (Figures S1C and S1D).
Figure 1. Yap/Taz deletion leads to lung injury, epithelial remodeling, and goblet cell metaplasia.
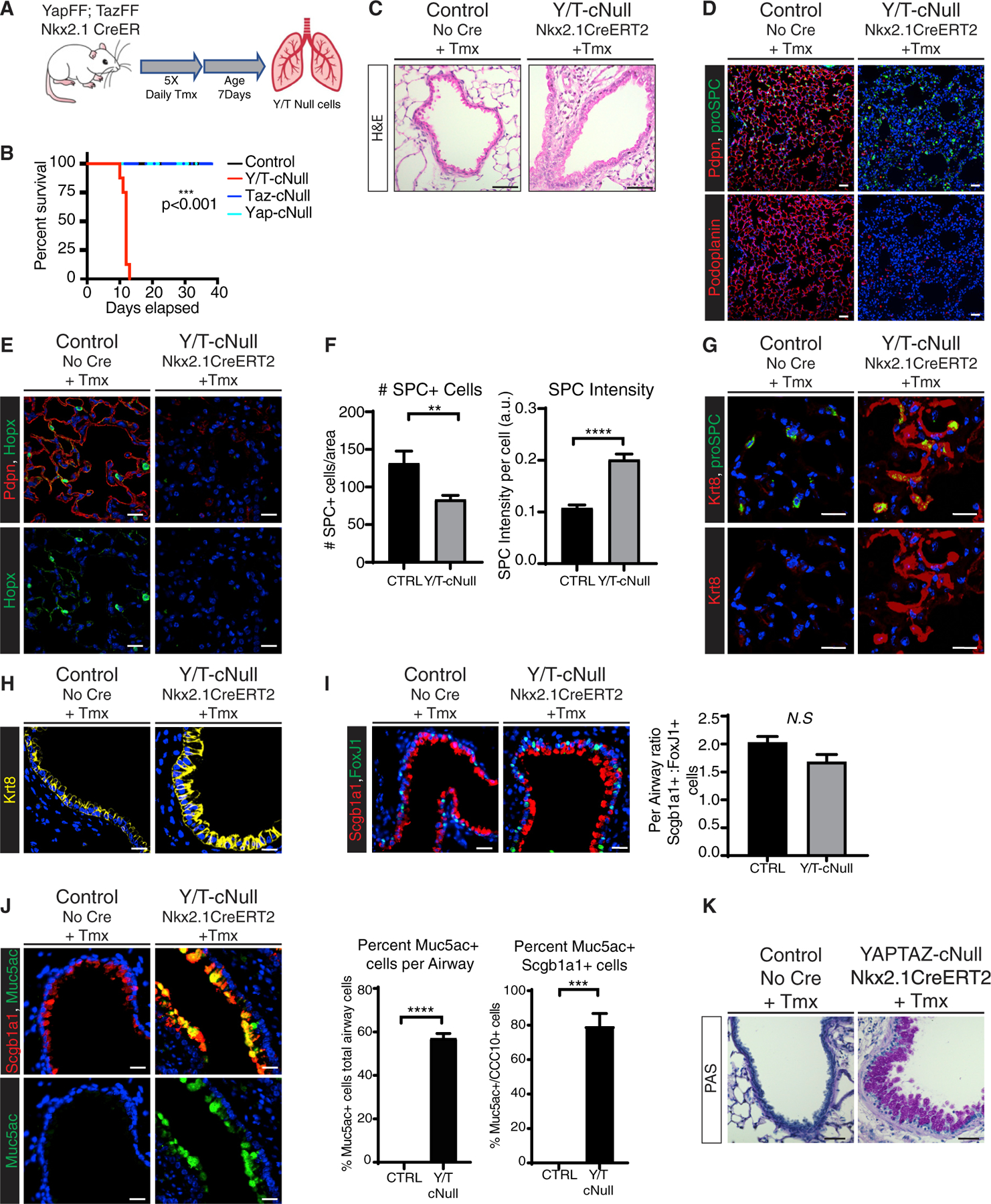
(A) Outline of tamoxifen (Tmx) injection scheme for inducible knockout of Yap and Taz using the Nxk2.1-CreERT2 model. Lung tissues for all experiments described below were collected 12 days following tamoxifen treatment.
(B) Survival curve for Yap/TazNxk-KO (n = 8), YapNxk-KO (n = 4), TazNxk-KO (n = 3), and control (n = 7) mice (log-rank [Mantel-Cox] test, ***p < 0.0001).
(C) H&E staining of representative airway sections from control and Yap/TazNxk-KO lungs (scale bars, 50 μm).
(D–I) Immunostaining of representative tissue sections from control and Yap/TazNxk-KO lungs. DAPI stained nuclei are in blue.
(D) Pdpn and pro-SPC immunostaining (scale bars, 50 μm).
(E) Immunostaining for Hopx with Pdpn co-staining in representative lungs.
(F) The number and intensity of pro-SPC+ cells were calculated per image area (from n = 3 images per mouse, from n = 3 control, and n = 5 knockout mice; unpaired t test, **p < 0.01, ****p < 0.0001).
(G) Immunostaining for Krt8 and pro-SPC reveals elevated Krt8 expression in SPC+ alveolar cells in Yap/TazNxk-KO lungs.
(H) Krt8 immunostaining in representative airways.
(I) Scgb1a1 and Foxj1 immunostaining reveals similar ratios of ciliated and club cells in control and Yap/TazNxk-KO lungs. For quantitation, the numbers of FoxJ1+ cells and Scgb1a1+ cells were counted per airway and the ratio was compared for n = 5 airways for each genotype (unpaired t test, p = 0.0763).
(J) Scgb1a1 and Muc5ac immunostaining in control and Yap/TazNxk-KO lungs. Quantification of percent Muc5ac+ cells/total cells per airway. The total numbers of Muc5ac cells were counted per airway and the percentage as a fraction of total nuclei per airway was plotted; n = 5 per genotype (unpaired t test, ****p < 0.0001). The percentage of double-positive cells was calculated from a minimum of n = 3 images per genotype (unpaired t test, ***p = 0.0003).
(K) Periodic acid-Schiff staining of secreted mucins in control and Yap/TazNxk-KO airways (scale bars, 50 μm).
Scale bars, 20 μm (unless otherwise noted). In all plots, data are represented as mean ± SEM. See also Figure S1.
Characterization of the increased cellularity in Yap/TazNxk-KO lungs revealed extensive infiltration of CD45+ leukocytes (Figure S1E). Yap/TazNxk-KO lungs also exhibited increased alveolar and airway associated matrix deposition, but no obvious changes in α-smooth muscle actin (α-SMA) expression (Figures S1F and S1G). Assessment of alveolar epithelial markers showed that Yap/TazNxk-KO lungs exhibited a dramatic loss of podoplanin (Pdpn) (Figure 1D) and homeodomain-only protein (HopX) (Figure 1E), both markers of ATI cells, suggesting that Yap/Taz are required to maintain ATI cell homeostasis. Examination of the ATII marker pro-surfactant protein C (pro-SPC) revealed a significant increase in per-cell pro-SPC intensity, although the overall number of ATII cells declined slightly (Figure 1F). We observed a striking increase in the levels of Krt8 in the alveolar epithelium of Yap/TazNxk-KO lungs (Figure 1G), including in pro-SPC+ cells, reminiscent of observations in re-generating lung epithelium (Choi et al., 2020; Strunz et al., 2020). Notably, SPC+ cells with high Krt8 levels were marked by a distinctive squamous morphology (Figure 1G), suggesting that loss of Yap/Taz alters ATII-related fate in the alveolar epithelium.
In addition to the alveolar defects, dramatic architectural differences were apparent throughout the airway of Yap/TazNxk-KO mice. Cells exhibited a more columnar morphology and increased levels of Krt8 (Figure 1H), which are associated with luminal cell maturation. Profiling of bronchial epithelial cell markers by immunostaining revealed that ratios of Scgb1a1+ club cells and FoxJ1+ multiciliated cells remained unaffected (Figure 1I). However, Yap/TazNxk-KO airway epithelial cells exhibited a striking increase in cells expressing the mucin Muc5ac as well as elevated periodic acid-Schiff (PAS) staining (Figures 1J, 1K, and S1H), indicating increased mucus production throughout the mutant lungs. Muc5ac was nearly undetectable in the lungs of control mice, whereas approximately 58% of the epithelial cells within Yap/TazNxk-KO airways expressed Muc5ac. Interestingly, upon co-staining, we observed that 79% of Scgb1a1+ cells in Yap/TazNxk-KO airways were positive for Muc5ac (Figure 1J), suggesting an acquisition of goblet cell features within club cells. Collectively, our observations indicated essential functions for YAP/TAZ in lung epithelial homeostasis and suggested pleiotropic effects across the different epithelial lineages.
Yap and Taz are required in club cells to prevent goblet cell metaplasia
Scgb1a1+ club cells serve as local stem cells within the conducting airways, with the capacity for both self-renewal and differentiation (Rawlins et al., 2009). Given the coordinate co-staining of Scgb1a1 and Muc5ac within cells following Yap/Taz deletion (Figure 1J), we hypothesized that mucin-producing cells arose through transdifferentiation of club cells toward a goblet cell fate in response to Yap/Taz loss. To test the role of Yap and Taz in club cells, we crossed Yap/Taz-floxed mice with mice expressing tamoxifen-inducible Cre recombinase from the Scgb1a1 locus along with a lox-STOP-lox-EYFP lineage trace (herein referred to as Yap/TazYFP-Scgb-KO). Yap/TazYFP-Scgb-KO mice survived without any outward morbidity following tamoxifen treatment (Figures 2A and 2B). However, Yap/Taz-deleted lungs displayed dramatically increased mucin secretion measured by PAS staining (Figure 2C), along with abundant Muc5ac levels throughout the intrapulmonary and tracheal airways detected by immunofluorescence microscopy (Figures 2D, S2A, and S2B). 93% of yellow fluorescent protein (YFP) lineage-traced cells within Yap/TazYFP-Scgb-KO lungs that were marked by enhanced YFP (EYFP) expression were also Muc5ac-positive (Figure 2E), indicating that Scgb1a1-expresing cells were an origin of the goblet-like cells observed following Yap/Taz deletion. Flow cytometry purified lineage-traced Scgb1a1+ cells from Yap/TazYFP-Scgb-KO lungs also expressed significantly higher levels of Muc5ac transcript compared to lineage-traced control cells, as measured by qPCR (Figure 2F).
Figure 2. Deletion of Yap/Taz in Scgb1a1+ club cells drives goblet cell differentiation.

(A) Schematic of tamoxifen injection regimen for inducible knockout of Yap and Taz using the Scgb1a1-CreERT2 model in vivo. Lung tissues from the experiments described below were collected for analysis 12 days following tamoxifen treatment.
(B) Survival curve for Yap/TazScgb1a1-KO mice (n = 5) following tamoxifen injection (log-rank [Mantel-Cox] test, not significant).
(C) Representative images of Periodic acid-Schiff staining of secreted mucins in control and Yap/TazScgb1a1-KO airways (scale bars, 50 μm).
(D) Immunostaining for Scgb1a1 Muc5ac, and YFP lineage trace in large airways.
(E) Quantification of the percent YFP+ cells co-expressing Muc5ac. Total YFP+ and Muc5ac/YFP double-positive cells were counted per airway from n = 4 images per genotype (data are represented as mean ± SEM; unpaired t test, ****p < 0.0001).
(F) Quantification of relative Muc5ac expression by qPCR of isolated Scgb1a1 lineage-traced cells. Cells were purified by flow cytometry from Yap/TazYFP-Scgb-KO (n = 5) and controlYFP-Scgb (n = 3) mice. (Data are represented as mean ± SEM; unpaired t test, *p < 0.05.)
(G and H) Wild-type pups were sensitized with ovalbumin postnatally and subsequently challenged with ovalbumin or PBS to model allergic asthma.
(G) Yap/Taz immunostaining (using a dual antibody recognizing both proteins) and Muc5ac in P21 lung.
(H) Yap/Taz fluorescence intensity was measured for all individual nuclei within field of view (n = 49, n = 46) (unpaired t test, ****p < 0.0001).
(I) Representative images of Muc5ac and CD45 immunostaining on control and Yap/TazYFP-Scgb-KO lungs revealing elevated infiltration of CD45+ cells proximal to airways expressing high levels of mucin (scale bars, 50 μm).
(J) The number of CD45+ cells was counted within representative sections from control and Yap/Taz-cNull lungs (n = 5 images per genotype; unpaired t test, *p < 0.05). The total number of non-airway cells was obtained by counting DAPI-stained nuclei. The percentage was calculated as the number of CD45+ cells divided by total nuclei.
In all immunostaining images DAPI-stained nuclei are in blue. Scale bars, 20 μm (unless otherwise noted). In all plots, data are represented as mean ± SEM. See also Figure S2.
As in Yap/TazNxk-KO lungs, the epithelium of Yap/TazScgb-KO mice was marked by elevated Krt8 expression (Figure S2C), and the relative numbers of Scgb1a1+ and FoxJ1+ cells appeared unaffected (Figure S2D). Analysis of the proliferative marker Ki67 indicated that Yap/TazYFP-Scgb-KO airway epithelial cells did not exhibit increased proliferation compared to wild-type controls, suggesting that the increase in Muc5ac+ cells did not arise from hyperplastic expansion of goblet cells but rather from the transdifferentiation of individual club cells (Figure S2E).
The dramatic increase in mucin levels within airways following Yap/Taz deletion resembles that observed in a number of lung diseases, such as bronchitis, COPD, and asthma. Data in mouse models of asthma, such as the ovalbumin sensitization model of allergic asthma, indicate that the Scgb1a1-expressing club cells act as a cell of origin for goblet cell metaplasia (Chen et al., 2009). We reasoned that endogenous YAP/TAZ protein may be downregulated in the aberrant goblet cells observed in such diseases. Indeed, we observed that the Muc5ac-expressing epithelium of ovalbumin-challenged mice following ovalbumin sensitization displayed significantly lower total and nuclear levels of Yap/Taz compared to PBS-challenged controls (Figures 2G and 2H).
Yap and Taz intrinsically repress epithelial goblet cell fate
Goblet cells are rarely observed in adult mouse lungs under homeostatic conditions, and their presence is generally accompanied by local inflammation and cytokine stimulation (Kuperman et al., 2002; Wills-Karp et al., 1998). Our in vivo observations following epithelial deletion of Yap/Taz suggested that the induction of mucin hypersecretion was intrinsic to the epithelium; however, we did observe significant local infiltration of CD45+ leukocytes proximal to the airways displaying high mucin expression (Figures 2I and 2J). In order to rule out the influence of extrinsic signals in vivo on goblet cell differentiation, we generated a mouse model that allowed for robust Yap/Taz conditional deletion in vitro (Yap/Taz-floxed mice crossed to mice expressing tamoxifen-inducible Cre from the Rosa26 locus; Yap/TazR26-KO) and used this model to grow tracheal explant cultures with induced deletion of Yap/Taz in vitro (Figure S3A). We found that in vitro tracheal deletion of Yap/Taz consistently led to elevated Muc5ac levels (Figure S3B). To further test the cell-intrinsic roles of YAP/TAZ, we isolated primary mouse airway epithelial cells (mAECs) and expanded and differentiated these cells as three-dimensional airway organoid “bronchosphere”’ cultures or two-dimensional air-liquid interface (ALI) cultures (Figure 3A) (Rock et al., 2009; You and Brody, 2013). We found that deletion of Yap/Taz in differentiated bronchospheres led to a dramatic increase in Muc5ac levels, filling the developed lumens (Figure 3B). Similarly, deletion of Yap/Taz in differentiated ALI cultures led to a loss of YAP/TAZ protein and transcripts (Figures S3C–S3E) along with increased Muc5ac expression (Figure 3C). Morphological changes in Yap/Taz-deleted epithelial cells were observable in ALI cultures, resembling in vivo observations, including elevated levels of the luminal marker Krt8 (Figure S3F). Analysis of airway epithelial markers by qPCR showed reduced Yap and Taz transcripts in Yap/Taz-deleted cultures compared to controls (Figure S3E), accompanied by increased Muc5ac and Scgb1a1expression and reduced expression of the basal cell marker Krt5 (Figure 3D).
Figure 3. Loss of Yap/Taz in the airway epithelium promotes Muc5ac production in vitro.
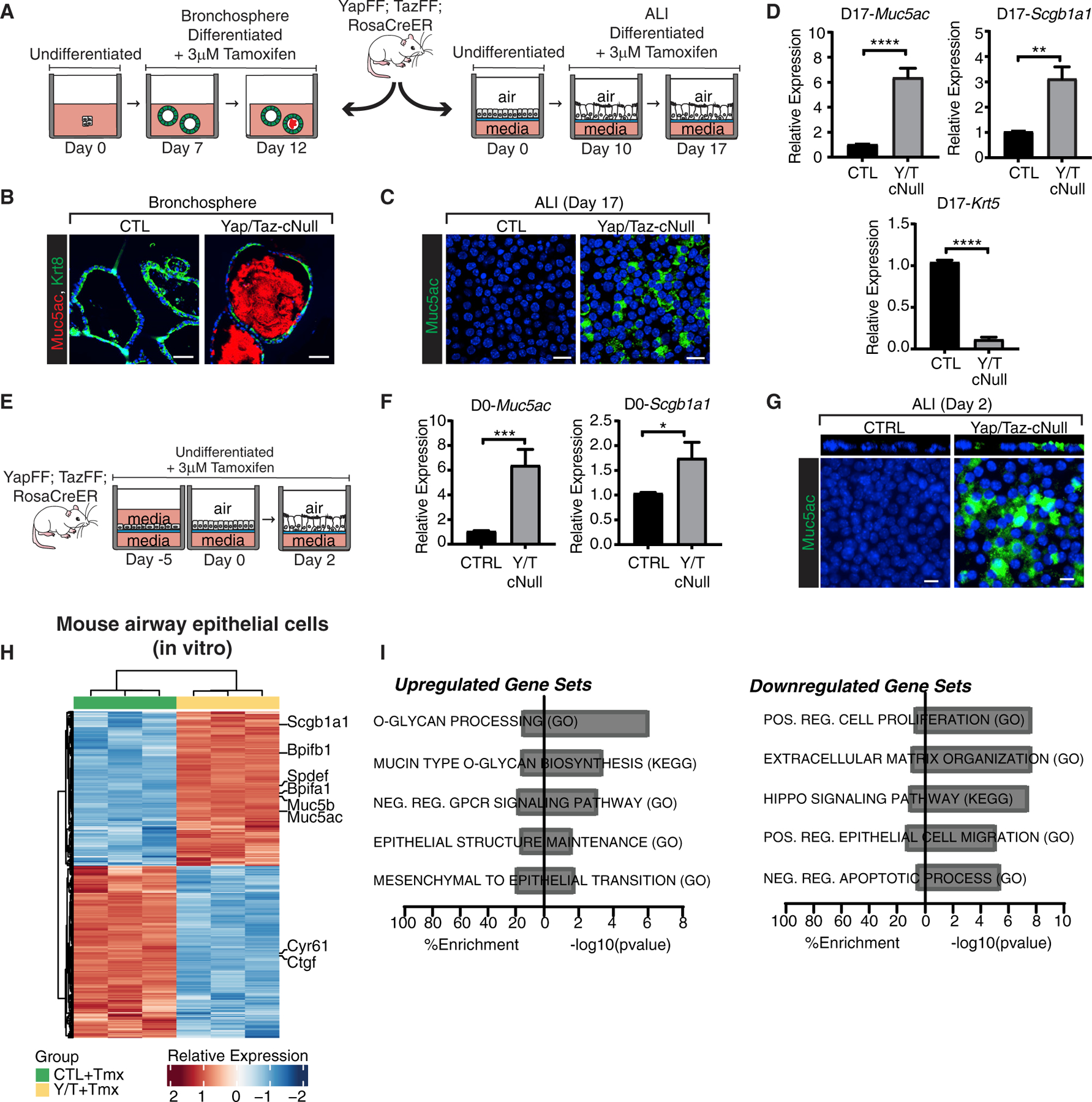
(A) Schematic outlining Yap/Taz deletion in differentiated airway epithelial cells using air-liquid interface (ALI) and bronchosphere cultures in vitro. For the ALI, basal cells isolated from mouse tracheas were grown to confluence (day 0), differentiated at the ALI for 10 days, then treated with 4-hydroxytamoxifen for 7 days to induce Yap/Taz deletion. For bronchosphere cultures (illustration on the left), basal cells isolated from mouse tracheas were differentiated for 7 days in Matrigel, before treatment with 4-hydroxytamoxifen for 5 days.
(B) Muc5ac and Keratin 8 immunostaining of control and Yap/Taz null bronchospheres (scale bars, 50 μm).
(C) Muc5ac immunostaining of control and Yap/Taz null differentiated mAEC ALI cultures shows enhanced Muc5ac levels following Yap/Taz deletion.
(D) Yap/Taz deletion in differentiated mAEC ALI cultures increases Muc5ac and Scgb1a1 expression by qPCR (n = 5; unpaired t test, **p < 0.01, ****p < 0.0001). Reduced Krt5 expression by qPCR was observable in Yap/Taz null mAECs (n = 3; unpaired t test, ****p < 0.0001).
(E) Schematic of Yap/Taz deletion in undifferentiated basal cell cultures using the Rosa26-CreERT2 model. Briefly, basal cells were isolated from mouse tracheas and cultured submerged in media on transwell filters in the presence of 4-hydroxytamoxifen for 5 days to reach confluence (day 0) and then shifted to an ALI for 2 days (day 2).
(F) Analysis of Muc5ac and Scgb1a1 expression by qPCR in undifferentiated Y/T null mAECs collected on day 0 of ALI (n = 7; unpaired t test, *p < 0.05, **p < 0.01).
(G) Muc5ac immunostaining of Yap/Taz-deleted mouse basal cells collected on day 2 of the ALI.
(H) Heatmap displaying gene expression changes in Yap/Taz-deleted mAECs compared to controls analyzed by microarray (n = 3 per condition, 2-fold change cutoff, FDR = 0.05). Cells were cultured to density and treated with 4-hydroxytamoxifen for 5 days before analysis.
(I) Pathway enrichment of significantly upregulated and downregulated genes following Yap/Taz depletion in mouse ALI cultures identified by GSEA. Both the percentage representation and the −log10 p value within each gene set are displayed.
For all immunostaining images DAPI-stained nuclei are in blue. Scale bars, 10 μm (unless otherwise noted). In all bar plots data are represented as mean ± SEM. See also Figure S3 and Tables S1 and S2.
Evidence suggests that airway basal cells also have the capacity for differentiation to the goblet cell lineage (Guseh et al., 2009). We therefore sought to test the consequence of Yap/Taz deletion in undifferentiated mAECs, which are primarily composed of airway basal cells (Figures 3E and S3G–S3I). Indeed, primary cell cultures of Krt5+ cells submerged in media increased Muc5ac and Scgb1a1 transcripts following Yap/Taz deletion (Figure 3F). A loss of the basal cell markers Tp63 and Krt5 was also observed following Yap/Taz deletion (Figures S3H and S3I) with evidence of only slight cell apoptosis (Figure S3J), suggesting differentiation of the large majority of the cells. After only 2 subsequent days of differentiation at the ALI, Yap/Taz-deleted cultures exhibited an accumulation of secreted mucins (Figure 3G), which was striking in this time frame given the complete absence in control cultures even after prolonged differentiation at the ALI.
Transcriptional profiling of Yap/Taz-deleted airway epithelial cells reveals a goblet cell gene expression program
Given the well-characterized roles for YAP and TAZ as transcriptional effectors, we carefully examined the nuclear localization of these factors throughout the adult mouse airway epithelium. In mouse lung tissues we observed that the mature luminal cells of the bronchial epithelium, delineated by Krt8, displayed a pan-cellular localization of YAP/TAZ with many cells exhibiting abundant nuclear staining (Figure S3K), similar to previous reports (Lange et al., 2015). To gain insight into airway epithelial transcriptional programs controlled by Yap/Taz, we examined global RNA alterations in mouse airway cultures following deletion in culture when grown at high confluence on collagencoated transwell filters. Differential gene expression analysis of YAP/TAZ-deleted cells compared to controls identified a signature of 885 differentially expressed genes in mAECs (2-fold change cutoff, false discovery rate [FDR] = 0.05) (Figure 3H; Table S1) with 466 downregulated genes, including Yap and Wwtr1 (Taz) transcripts, as well as the canonical Yap/Taz target genes Ctgf and Cyr61. Pathway enrichment analysis of the downregulated genes indicated expected roles for Yap/Taz in promoting matrix remodeling and cell proliferation (Figure 3I; Table S2). Conversely, 419 genes were significantly upregulated in response to Yap/Taz knockout, and gene set enrichment analysis (GSEA) of this genetic signature identified a number of pathways involved in mucin production and glycosylation (Figure 3I; Table S2). Several lung-associated mucin genes (Rose and Voynow, 2006) were induced in Yap/Taz null mAECs, including the gel-forming mucins Muc5ac and Muc5b, as well as the membrane-associated mucins Muc4, Muc13, and Muc16. Yap/Taz null mAECs expressed significantly higher levels of UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl-transferases such as Galnt4, t5, t6, t7, and t12, which catalyze the initial step in protein glycosylation (Bennett et al., 2012). Overall, these data demonstrate that the transcriptional program in Yap/Taz-depleted cells outlines a global shift in cellular function toward the production, glycosylation, and secretion of mucins, consistent with differentiation toward the goblet cell lineage.
Yap/Taz restrict mucin production in human cells
Given the importance of Yap/Taz in limiting mucin expression in mAECs, we sought to understand how YAP/TAZ might be regulated in human airways where goblet cells are broadly observable. Human lung tissues showed a pan-cellular YAP/TAZ distribution within airway epithelial cells, including evident nuclear localization similar to mouse airway epithelium (Figure 4A). Notably, all cells expressing Muc5ac showed low levels of YAP/TAZ, including very low nuclear YAP/TAZ (Figure 4B). Primary human bronchial epithelial cells (HBECs) differentiated in vitro in ALI cultures showed similar YAP/TAZ localization patterns, with all Muc5ac-expressing cells exhibiting very low total and nuclear YAP/TAZ levels (Figure 4C).
Figure 4. YAP/TAZ depletion induces MUC5AC expression in human primary epithelial cells.

(A) Representative YAP/TAZ and MUC5AC immunostaining of human airway sections (performed using an antibody recognizing YAP and TAZ). DAPI-stained nuclei are in blue. Nuclei within MUC5AC+ cells are highlighted with a yellow dotted line and with a blue dotted line in MUC5AC− cells. A white dotted line marks the basal surface of the epithelium (scale bars, 10 μm).
(B) Quantification of nuclear YAP/TAZ intensity in airway epithelial cells across multiple sections from two patient donors. Cells were scored as either MUC5AC-positive or -negative and the intensity of YAP/TAZ staining was measured within the nuclear area outlined by DAPI staining (minimum of n = 14; unpaired t test, ****p < 0.0001).
(C) YAP/MUC5AC immunostaining of human ALI cultures imaged by confocal microscopy. A z stack view is shown in the top panels (scale bars,10 μm).
(D) HBECs were transfected with control siRNA (siCTL) or siRNA targeting YAP/TAZ (siY/T). MUC5AC, SCGB1A1, YAP, and WWTR1/TAZ qPCR analysis of lysates collected 72 h after knockdown (n = 6; unpaired t test, **p = 0.001, ****p < 0.0001).
(E) Heatmap of gene expression changes resulting from YAP/TAZ knockdown in HBECs analyzed by RNA sequencing (RNA-seq). 2 distinct patient isolates were treated with three independent siCTLs or siRNA targeting YAP/TAZ (siY/T), and global gene expression changes were examined by RNA-seq after 48 h of culture (n = 3 per condition, 2-fold change cutoff, FDR = 0.05).
(F) Pathway enrichment of significantly upregulated and downregulated genes following YAP/TAZ depletion in human airway epithelial cells identified by GSEA. Both the −log10 p value and the percentage representation within each gene set are displayed.
In all bar plots data are represented as mean ± SEM. See also Figure S4 and Tables S2 and S3.
In HBECs, depletion of YAP and TAZ transcripts by RNAi resulted in a striking increase in MUC5AC and SCGB1A1 expression and reduced levels of the canonical YAP/TAZ target gene CTGF (Figures 4D, S4A, and S4B). These observations demonstrate a conserved function for YAP/TAZ in the repressing goblet fate of airway epithelial cells and suggest that nuclear YAP/TAZ activity may be facilitating this function.
To further characterize the transcriptional activity of YAP and TAZ in human cells, we performed bulk RNA sequencing of YAP/TAZ-deleted HBECs from two independent donors. Differential expression analysis identified 989 significantly downregulated and 871 significantly upregulated genes in response to Yap/Taz deletion (2-fold change cutoff, FDR = 0.05) (Figure 4E; Table S3). As expected, significantly downregulated transcripts included YAP and TAZ mRNAs as well as canonical YAP/TAZ target genes CTGF and CYR61. Pathway enrichment analysis of these downregulated genes indicated conserved roles for YAP/TAZ in promoting matrix remodeling and cell junction organization (Figure 4F). Notably, the lung-associated mucins MUC5AC and MUC5B transcripts were induced in YAP/TAZ-depleted cells as well as SPDEF, which encodes a transcription factor necessary and sufficient for airway goblet cell differentiation (Chen et al., 2009; Park et al., 2007; Rajavelu et al., 2015). BPIFA1 and BPIFA2, which encode innate immune proteins expressed in airway goblet cells (De Smet et al., 2017), were similarly upregulated. GSEA of the upregulated gene signature revealed pathways involved in asthma, cytokine-mediated signaling, as well as the cellular response to type 1 interferon, all of which have been linked to goblet cell differentiation (Figure 4F). Enrichment analysis of the YAP/TAZ-regulated mouse signature among the human signature displayed significant overlap between the datasets, indicating a high degree of conservation (Figures S4C and S4D).
YAP/TAZ suppress mucin expression via the TEAD family of transcription factors in HBECs
As transcriptional co-factors, YAP and TAZ rely on binding to transcription factors to exert control over downstream gene expression, with the TEAD family of transcription factors (TEAD1–4) being the best characterized effectors of YAP/TAZ function (Dey et al., 2020). We therefore examined TEAD function in the control of HBEC differentiation. Simultaneous knockdown of TEAD1–4 yielded an induction of MUC5AC expression, as we observed with YAP/TAZ depletion (Figure 5A), suggesting that YAP/TAZ-TEAD complexes function together to control goblet cell fate.
Figure 5. ChIP-Seq analysis of TEAD activity in HBECs.

(A) HBECs were plated at high density and transfected with a pool of siRNAs targeting TEAD1–4 or a non-targeting siCTL, and lysates were collected 48 h later. Samples were examined by qPCR analysis for TEAD1–4, CTGF, MUC5AC, and SCGB1A1 expression (n = 3; data are represented as mean ± SEM; unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001).
(B and C) ChIP-seq was performed in triplicate using a pan-TEAD antibody from low-passage, near-confluent HBECs cultured in PneumaCult-EX Plus. The data displayed represent the overlapping peaks from the replicate ChIP-seq datasets.
(B) The percent distribution of TEAD peaks across genomic features is shown as a statistical circular graph.
(C) The distribution of mapped TEAD peak locations is shown within 5 kb upstream and downstream of transcriptional start sites (TSSs) at genes with TEAD-bound promoters.
(D) GSEA of all genes identified with TEAD peaks mapped to the promoter region (within 5 kb of TSSs).
(E) Representation of the association of human YAP/TAZ-regulated genes (signature shown in Figure 4E) with TEAD peaks identified by ChIP-seq analysis.
(F) Tables of genes identified with TEAD binding at the promoter that were also found to be differentially expressed in both mouse and human YAP/TAZ-depleted airway epithelial cells (i.e., direct gene targets conserved across mouse and human airway cells). See also Figure S5 and Tables S4 and S5.
To gain insight into the transcriptional events directly regulated by YAP/TAZ-TEAD complexes we performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis of HBECs using antibodies specific to TEADs. A total of 16,306 annotated genome-binding peaks were identified for the TEAD HBECs. Although a plurality (~36%) of all peaks localized to distal intergenic regions, consistent with previous reports in cancer cells (Zanconato et al., 2015), a large number of peaks (~25%) were located in the promoter region of genes, 0–5 kb around the transcriptional start site (TSS), with such peaks exhibiting a high density around the TSS (Figures 5B and 5C). Canonical target genes of YAP/TAZ-TEAD complexes such as CTGF, ANKRD1, and AJUBA all showed promoter-associated peaks. Motif enrichment of peaks associated with TEAD binding confirmed the enrichment of predicted TEAD motifs (Figure S5A). Intriguingly, pathway enrichment analysis of the genes associated with TEAD binding at the promoter revealed significant association with pathways such as tumor necrosis factor (TNF)-α/nuclear factor κB (NF-κB) signaling and interleukin (IL)-1R signaling, which have been implicated in goblet cell specification in vivo (Figure 5D; Table S4) (Broide et al., 2005; Chen et al., 2019; Poynter et al., 2004).
To interpret the relationship between TEADs and the YAP/TAZ-regulated gene expression program, we identified TEAD peaks that reside within 5 kb of the TSS of genes that we defined as YAP/TAZ targets in HBEC and mAEC transcriptional analysis. In HBECs we found that 18.25% of the upregulated and 34.68% of the downregulated genes were associated with direct binding of TEADs at gene promoters, suggesting that the expression of these genes is directly mediated by YAP/TAZ-TEAD complexes (Figure 5E). Similar frequency of TEAD binding at the promoter of target genes was also observed within the mouse dataset (Figure S5B). We found that collectively, 43.74% of the upregulated gene set and 62.08% of the downregulated gene set aligned with mapped peaks in HBECs (Figure 5E), and similarly that 51.59% of the upregulated gene set and 63.59% of the downregulated genes in mAECs aligned with mapped peaks (Figure S5B). Notable genes implicated in basal airway homeostasis, such as KRT5 and ITGB4, were identified as positively regulated direct YAP/TAZ-TEAD targets in both mouse and human datasets (Figure 5F; Table S5). Several interesting negatively regulated YAP/TAZ-TEAD targets were identified with TEAD promoter peaks across both datasets, including the goblet cell regulating transcription factor SPDEF (Figure 5F).
YAP/TAZ-TEAD repression of SPDEF control a goblet cell gene expression program
We notably observed a TEAD binding peak at the SPDEF promoter near the TSS in our ChIP-seq analysis of HBECs (Figure 6A) and confirmed this binding by ChIP-qPCR using a TEAD antibody. We additionally performed ChIP-qPCR using a YAP antibody and identified YAP binding at the same site, suggesting direct gene regulation by YAP-TEAD complexes (Figure 6B). YAP and TEAD binding were similarly observed at the promoter of the canonical YAP/TAZ-TEAD target CTGF in our ChIP-qPCR analysis (Figure S6A).
Figure 6. YAP/TAZ and TEADs regulate SPDEF expression and correlate with SPDEF in human lung tissue.

(A) TEAD binding at the SPDEF promoter identified by ChIP-seq performed in HBECs using a pan-TEAD antibody.
(B) ChIP-qPCR of YAP and TEAD binding to the same site at the SPDEF promoter in HBECs (representative of a minimum of three independent experiments, n = 3; unpaired t test, *p < 0.05, ***p < 0.001).
(C) qPCR for SPDEF in HBECs treated with siCTL or siRNA targeting YAP/TAZ (siY/T) (n = 6; unpaired t test, **p < 0.01).
(D) qPCR analysis of Spdef transcripts in Y/T-deleted mAECs (n = 3; unpaired t test, *p < 0.05).
(E) qPCR analysis of HBECs treated with a pool of siRNAs targeting TEAD1–4 (siTEAD) or a non-targeting siCTL. Samples were collected from HBECs grown in submerged culture for 48 h following siRNA transfection (n = 3; unpaired t test, **p < 0.01).
(F) qPCR analysis of Scgb1a1 lineage-traced cells, purified by flow cytometry from Yap/TazYFP-Scgb-KO (n = 5) and controlYFP-Scgb (n = 3) mice. The relative expression of Spdef transcripts was analyzed by qPCR revealing increased Spdef expression in Yap/Taz-deleted cells (unpaired t test, *p < 0.05).
(G) In situ RNAScope for Spdef mRNA in control and Yap/TazScgb1a1-KO lungs collected 12 days after tamoxifen treatment reveals extensive increase in SPDEF transcripts as a consequence of Yap/Taz deletion in vivo (scale bars, 20 μm).
(H) YFP+ cells purified by flow cytometry from Yap/TazYFP-Scgb-KO and controlYFP-Scgb were analyzed by RNA-seq. Heatmap displays the relative expression for a subset of these genes important for mucin production, previously found to be regulated by Spdef expression in vivo (Chen et al., 2009).
(I) qPCR analysis of HBECs that were transfected with siRNA targeting YAP/TAZ and SPDEF to induce YAP/TAZ depletion (siY/T) or YAP/TAZ/SPDEF (siY/T/SPDEF) co-depletion (n = 6; unpaired t test, ***p < 0.001, ****p < 0.0001).
(J) Enrichment plots for YAP/TAZ-regulated genes among GTEX Lung database genes ranked by correlation with SPDEF expression (Kolmogorov-Smirnov test (KS test), p < 2.2e−16). In both plots, all genes in the database are ranked by correlation with SPDEF. Each plot shows the enrichment of the upregulated gene set or the downregulated gene set as a consequence of YAP/TAZ depletion.
(K) Model of Sobel mediation testing within the GTEX lung dataset and a plot of the mediation p value for each gene in the designated signature (i.e., testing for how YAP/TAZ-regulated genes depend on SPDEF expression) (Fisher’s exact test, non-parametric bootstrap p < 1e−5). Genes within the YAP/TAZ-regulated signature exhibit significantly smaller mediation p values indicating a pattern of mediation by SPDEF within the signature.
In all bar plots data are represented as mean ± SEM. See also Figure S6 and Table S6.
Our global gene expression profiling of Yap/Taz-depleted mAECs and HBECs identified increased expression of SPDEF transcripts, which we confirmed by qPCR (Figures 6C and 6D). Correspondingly, we found that SPDEF expression was significantly upregulated as a consequence of TEAD1–4 knockdown in primary human airway cells (Figure 6E). Given the identification of SPDEF as a Yap/Taz-Tead target gene in mouse and human in vitro models, we sought to confirm this in vivo. qPCR analysis of Spdef transcripts in sorted CC10+ lineage-traced cells from control and Yap/TazYFP-Scgb-KO lungs revealed Spdef upregulation as a consequence of Yap/Taz deletion (Figure 6F). We further validated the increased Spdef expression by RNAScope in situ hybridization in the airways of Yap/TazScgb-KO lungs (Figure 6G).
Given the upregulation of Spdef, we were curious whether the overall transcriptional program of Yap/Taz null epithelial cells in vivo reflected increased Spdef transcriptional activity. We performed bulk RNA sequencing of sorted CC10+ lineage-traced cells from controlYFP-Scgb and Yap/TazYFP-Scgb-KO lungs, identifying 1,110 differentially expressed genes in response to Yap/Taz deletion (fold change > 2, FDR < 0.05) (Figure S6B; Table S6). Prior studies have described a transcriptional signature linked to increased Spdef activity (Chen et al., 2009), and we found that key genes induced by Spdef expression were similarly upregulated in response to club cell-specific deletion of Yap/Taz in vivo (Figure 6H). Conversely, genes found to be repressed in response to induced Spdef expression were similarly downregulated in our in vivo Yap/TazYFP-Scgb-KO cells (Figure 6H). Comparison of our in vivo derived Yap/Taz-deleted gene expression signature revealed significant enrichment of the in vitro-derived Yap/Taz-regulated expression signature, with both upregulated and downregulated gene sets (p = 0.0004 and p = 0.026, respectively) (Figures S6C, S6D, and 3H), indicating a conserved in vitro and in vivo Yap/Taz-regulated transcriptional program that impacts goblet cell-related genes.
The conserved upregulation of Spdef across all knockout models tested in the present study suggested that SPDEF transcriptional activity may drive goblet cell specification under these conditions. To test SPDEF dependence in this context, we co-depleted SPDEF and YAP/TAZ in primary HBECs, and observed that loss of SPDEF blocked the induction of MUC5AC expression following YAP/TAZ depletion (Figures 6I and S6E). To test this on a broader level, we analyzed gene expression patterns in the GTEX database, containing gene expression data from 427 human lung tissue samples (GTEx Consortium, 2015). We ranked all genes by their correlation with SPDEF expression and performed enrichment analysis of our YAP/TAZ gene signature, finding that genes upregulated as a result of YAP/TAZ depletion were significantly enriched among the positively correlated genes (p < 2.2e−16). Conversely, the genes downregulated by YAP/TAZ loss were significantly enriched among genes negatively correlated with SPDEF expression in human lung tissue (p < 2.2e−16) (Figure 6J). To evaluate whether regulation of the identified YAP/TAZ signature genes is mediated by SPDEF, we performed a Sobel test of mediation in human lung tissue from the GTEX dataset. YAP/TAZ signature genes displayed strong evidence of mediation by SPDEF, with p values for mediation testing that were significantly smaller compared to the p values of the remaining control genes in the dataset (Fisher’s exact test, non-parametric bootstrap, p < 1e−5) (Figure 6K). This statistical modeling thus suggests a dependence of the YAP/TAZ signature genes on SPDEF that is distinct from direct YAP/TAZ control. Collectively, our in vitro observations and statistical analyses of human gene expression data suggest that the increased expression of SPDEF due to YAP/TAZ loss mediates a gene expression program that promotes the goblet cell fate.
The Hippo pathway mediates cytokine-induced SPDEF expression
The specification of goblet cells has been linked to signals induced by extrinsic inflammatory cytokines, such as IL-13, IL-1α, and IL-1β (Chen et al., 2019; Kuperman et al., 2002). We therefore asked how YAP/TAZ modulate signals induced by goblet cell-stimulating cytokines. Depletion of YAP and TAZ individually in HBECs supplemented with a cocktail of IL-13, IL-1α, and IL-1β did not alter the expression of SPDEF or the response to cytokine stimulation (Figures 7A, 7B, and S7A). However, co-depletion of YAP and TAZ resulted in increased SPDEF expression along with a strong sensitization to cytokine-induced signals, with a greater increase in SPDEF transcripts following cytokine treatment compared to controls.
Figure 7. Hippo pathway inhibition restricts cytokine-driven goblet cell specification, and the NuRD complex regulates SPDEF expression.

(A) Schematic of HBEC culture and siRNA-mediated knockdown together with cytokine media (CM) supplementation (PnemaCult-EX plus with and without IL-1α, IL-1β, and IL-13).
(B) HBECs were cultured with and without CM and YAP and/or TAZ were depleted by siRNA treatment. qPCR analysis reveals that SPDEF transcripts were minimally affected by individual knockdown of YAP or TAZ. Simultaneous knockdown of YAP/TAZ significantly increased SPDEF transcripts (minimum of n = 3; unpaired t test, ****p < 0.0001).
(C) LATS1/2 were depleted by siRNA. qPCR analysis demonstrates significant increases in SPDEF and MUC5AC with cytokine treatment and significant repression following knockdown (n = 4; unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.001).
(D) Schematic of experimental design for small molecule inhibition of the Hippo pathway MST kinases in differentiated HBECs. Briefly, HBECs were differentiated in the ALI for 10 days and then the MST inhibitor XMU-MP-1 (10 μM) was added on an ongoing basis for an additional 10 days before the cells were processed for analysis.
(E) qPCR analysis of differentiated HBECs treated with XMU-MP-1 (10 μM) demonstrates that MUC5AC and SPDEF were significantly repressed by MST inhibition and CTGF was induced (n = 3; unpaired t test, *p < 0.05, ***p < 0.001).
(F and G) qPCR analysis of HBECs transfected with siRNA targeting NuRD complex components. Knockdown of (F) GATAD2A or (G) CHD4 increased SPDEF and MUC5AC expression (n = 5; unpaired t test, *p < 0.05, ***p < 0.001, ****p < 0.0001).
(H) ChIP-qPCR analysis shows binding of GATAD2A and CHD4 to the SPDEF promoter in HBECs (n = 3; unpaired t test, **p < 0.01). qPCR amplification of bound chromatin was performed using the same primers as for the TEAD ChIP-qPCR shown in Figure 6B (i.e., NURD complex binding to the TEAD binding site).
(I) Schematic outline of the MNase-qPCR sites proximal to the SPDEF start site (indicated by the red lines; A, B, C). Blue line represents the TEAD binding site as identified by ChIP-seq. MNase-qPCR of HBECs transfected with siCTL or YAP/TAZ targeting siRNA (siYap/Taz) collected 72 h following knockdown. Purified DNA was crosslinked and digested with MNase. Amplification by site specific primers revealed a shift of the nucleosomes away from the transcriptional start site (n = 3, *p < 0.05, **p < 0.01).
For all plots, data are represented as mean ± SEM. See also Figure S7.
Nuclear YAP/TAZ activity is known to be inhibited by the Hippo pathway kinase cascade that includes the MST1/MST2 and LATS1/LATS2 kinases (Dey et al., 2020). We examined the consequences of Hippo pathway disruption in the context of cytokine stimulation. Co-depletion of LATS1/2 in HBECs by RNAi reduced MUC5AC and SPDEF transcript levels in the presence or absence of goblet cell-specifying cytokines (Figure 7C). We further examined whether pharmacological inhibition of the MST1/2 kinases with XMU-MP-1 treatment would similarly block goblet cell-related gene expression. We found that XMU-MP-1 treatment of HBECs during differentiation at ALI led to reduced levels of goblet cell transcripts (MUC5AC, SPDEF, SCGB1A1) along with significant upregulation of the canonical YAP/TAZ target gene CTGF (Figures S7B and S7C). While these data indicated a requirement for Hippo pathway function during differentiation, we also were curious to understand whether pathway inhibition was sufficient to reverse established goblet cell gene expression in differentiated cultures. We therefore differentiated HBECs at ALI for 10 days and subsequently treated them with XMU-MP-1 (Figure 7D). We found that differentiated HBECs exhibited robust MUC5AC and SPDEF expression whereas differentiated cells treated with XMU-MP-1 exhibited a dramatic reduction in transcript levels for both genes, indicating a loss of the goblet gene expression.
NuRD complex proteins occupy the SPDEF promoter and repress SPDEF expression
Our observations indicate that YAP/TAZ function as transcriptional gatekeepers in differentiating and differentiated cells to regulate the expression of SPDEF and consequently the expression of a pro-mucin transcriptional program. Given the occupancy of YAP and TEADs at the SPDEF promoter, we hypothesized direct repression of SPDEF expression through the regulation of transcriptional repressor complexes. The NuRD complex has previously been identified as a mediator of YAP/TAZ-TEAD-dependent transcriptional repression (Beyer et al., 2013; Kim et al., 2015), suggesting a relationship between the NuRD complex and SPDEF gene regulation. To test this, we depleted the core NuRD complex effectors, CHD4 and GATAD2A (p66α), using RNAi in HBECs and found that depletion of either promoted SPDEF, MUC5AC, and SCGB1A1 expression (Figures 7F and 7G). To test direct regulation of the SPDEF gene by the NuRD complex we performed ChIP-qPCR in HBECs using antibodies specific for these factors and found that both CDH4 and GATAD2A bind to the same site occupied by YAP and TEAD (Figure 7H).
In order to regulate target gene expression, the NuRD complex has been well characterized for its nucleosome remodeling functions (Bornelöv et al., 2018). We hypothesized that YAP/TAZ interactions with the NuRD complex may alter local nucleosome positioning at the SPDEF promoter to control gene expression. Using micrococcal nuclease (MNase)-qPCR, we examined the relative occupancy of nucleosomes near the SPDEF promoter and found that with YAP/TAZ knockdown nucleosomes were shifted away from the SPDEF TSS (Figure 7I). These data therefore indicate that YAP/TAZ impact nucleosome positioning and that YAP/TAZ, TEADs, and the NuRD complex cooperate to actively repress SPDEF and mucin gene expression in airway epithelial cells.
DISCUSSION
Our study demonstrates essential roles for Yap and Taz in lung epithelial homeostasis, and we report important functions for Yap/Taz in restricting airway goblet cell differentiation. Deletion of Yap and Taz in the adult mouse lung epithelium leads to rapid animal lethality, which is associated with severe defects in lung architecture, loss of ATI alveolar cell markers, airway goblet cell metaplasia, lung inflammation, and fibrosis. These striking lung defects were only observed following co-deletion of Yap and Taz, indicating vital redundancy for YAP/TAZ in lung homeostasis.
A notable phenotype associated with Nkx2.1-CreERT2-mediated Yap/Taz deletion in the lung was the observed loss of markers associated with ATI alveolar cells, which are essential for maintaining alveolar structure and proper gas exchange within the distal lung. A role for YAP/TAZ in ATI cell specification is supported by prior studies, which demonstrate that increased YAP activity induces the expression of ATI cell markers (Nantie et al., 2018; van Soldt et al., 2019) and that YAP/TAZ are required for regeneration of ATI cells from ATII cells in response to lung injury (LaCanna et al., 2019; Liu et al., 2016). The loss of ATI markers combined with the insight from prior studies raises the interesting possibility that YAP/TAZ may function in mature AT1 cells to maintain their identity. Our observations further indicated that the severe lung injury associated with Yap/Taz deletion drives microenvironment alterations resulting in inflammation and extracellular matrix deposition. Correspondingly, conditional deletion of Yap/Taz in SPC-expressing lung cells following pneumonia infection has been shown to similarly result in enhanced inflammation and fibrosis (LaCanna et al., 2019).
Given the dramatic expansion of goblet cells following Nkx2.1-CreERT2-mediated Yap/Taz deletion, we chose to examine the function of Yap/Taz directly in club cells using a Scgb1a1-CreERT2 model. We found that Yap/Taz deletion in club cells was sufficient to drive increased mucin production in targeted cells, consistent with goblet transdifferentiation. We interestingly observed co-expression of Scgb1a1 and Muc5ac within targeted cells, which is a phenotype that has previously been reported in studies of airway epithelium (Okuda et al., 2019). Scgb1a1-expressing club cells have been identified as the origin of goblet cells in antigen-challenged mice, and it has been observed that the levels of Scgb1a1 rise and fall during the period of mucin production (Evans et al., 2004), showing similarities to our observations following Yap/Taz deletion. However, goblet cells that have low expression of Scgb1a1 are also described (Vieira Braga et al., 2019), suggesting that reduced Yap/Taz activity may induce a specific subset or distinct state of goblet cells.
We found that deletion/depletion of YAP/TAZ in isolated in vitro cultures of primary mouse or human airway epithelial cells similarly resulted in the rapid induction of mucin expression and secretion, indicating an intrinsic function for YAP/TAZ in the control of this phenotype. Our ChIP-seq and global gene expression analyses of airway epithelial cells reveal a complex network of signals that are directly regulated by YAP/TAZ. Most notably, the goblet cell transcription factor Spdef was significantly upregulated in vivo and in vitro following YAP/TAZ deletion, as were a cohort of genes linked to mucin secretion, previously found to be regulated by Spdef expression. Evidence from our co-depletion studies in primary HBECs further showed that mucin expression resulting from loss of YAP/TAZ was dependent on the induction of SPDEF. Indeed, SPDEF has been previously shown to be both necessary and sufficient to drive the differentiation of club cells into goblet cells (Chen et al., 2009; Park et al., 2007), indicating that tight transcriptional control of SPDEF is critical in the maintenance of airway homeostasis.
Despite the importance of SPDEF in goblet cell differentiation, the transcriptional control of the SPDEF gene in the lung is poorly understood. The Runx2 and Foxa3 transcription factors have been shown to promote SPDEF expression and goblet cell differentiation in bronchial epithelial cells, and transforming growth factor (TGF)-β-regulated SMAD transcription factors contribute to SPDEF repression in conjunctival epithelium (Chen et al., 2014; Feldman et al., 2019; McCauley et al., 2014; Shi et al., 2017), indicating transcriptional collaboration or potential parallel functions between different factors. Interestingly, YAP/TAZ associate with and direct the activity of these factors in other contexts (Varelas et al., 2008; Zaidi et al., 2004), suggesting that their interplay may influence goblet cell fate in the lung.
Our observations also indicate that the NuRD transcriptional repressor complex participates in the transcriptional node that restricts SPDEF expression under basal conditions. The NuRD complex components GATAD2A and CHD4 have previously been identified as YAP/TAZ-TEAD-associated factors that together play transcriptional repressive roles (Beyer et al., 2013; Kim et al., 2015). The NuRD complex is a key mediator of chromatin remodeling and histone modification (Lai and Wade, 2011), and our data indicate that YAP/TAZ alter histone positioning at the SPDEF promoter. Notably, epigenetic dysregulation of SPDEF expression has been implicated in lung diseases, such as COPD (Song et al., 2017), and thus identification of YAP/TAZ-TEAD and NuRD as direct regulators of SPDEF expression may offer molecular insight into aberrant SPDEF expression in such diseases.
A variety of upstream signals have been implicated in airway goblet cell differentiation, and thus YAP/TAZ may function to integrate such signals at the transcriptional level. Indeed, we show that Yap/Taz intersect with cytokine-induced signals to modulate the transcriptional goblet cell differentiation program. We also show that repression of the Hippo pathway MST1/MST2 and LATS1/2 kinases, which leads to increased nuclear Yap/Taz activity, results in a reduction of goblet cell-associated genes even in the presence of goblet cell-inducing cytokines. These observations raise the exciting possibility that inducing Yap/Taz activity may provide a method for repressing goblet cell expansion in lung diseases.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xaralabos Varelas (xvarelas@bu.edu).
Materials availability
All materials used in this study are commercially available.
Data and code availability
Microarray, RNA-seq and ChIP Seq datasets have been deposited to NCBI GEO. Human RNA-seq datasets are listed as GEO: GSE158305 and Chip-Seq as GEO: GSE158306 (superseries GEO: GSE158307). The microarray data is available under GEO: GSE156525. RNA-seq of in vivo purified Scgb1a1+ cells is available under the GEO: GSE171712.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse models
All animal experiments were done in accordance with protocols approved by Boston University School of Medicine (Institutional Animal Care and Use Committee protocol AN-15304). The YAPloxP/loxP mice and TAZloxP/loxP mice were provided by J. Wrana (LTRI institute). All other mice used in this study are outlined below.
| Mouse strain | Alias | Source | Stock Number |
|---|---|---|---|
| B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | R26-LSL-EYFP | The Jackson Laboratory | #006148 |
| B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J | CC10-CreERT2 | The Jackson Laboratory | #016225 |
| Nkx2-1tm1.1(cre/ERT2)Zjh/J | Nkx2.1-CreERT2 | The Jackson Laboratory | #014552 |
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | lsl-tdTomato | The Jackson Laboratory | #007914 |
Tamoxifen administration in vivo
Adult mice (6–10 weeks old) were injected intraperitoneally with Tamoxifen (Sigma, T5648) in corn oil (75mg/kg, 20 mg/ml stock solution) once per day for five consecutive days to induced Cre mediated recombination. The sex of the mice was randomized across experimental groups.
Ovalbumin sensitization model of asthma
Wild-type pups were sensitized to ovalbumin via intraperitoneal injections of 10ug OVA (Sigma Aldrich, A5503) in Imject alum (Thermo Scientific, 77161) at postnatal days 5 and 10. Mice were subsequently challenged with aerosolized 3% OVA or a PBS solution for 20min daily between postnatal day 18–20. Mice were euthanized at postnatal day 21 and lungs were processed for histology.
METHOD DETAILS
ALI and primary cell culture
Primary mouse airway epithelial cells were cultured at ALI as previously described (You et al., 2002). In Yap/Taz knockout experiments, media was supplemented with 3 μM 4-Hydroxytamoxifen (Sigma) to induce Cre recombinase activity or methanol as a vehicle control. Cells were maintained in 4-hydroxytamoxifen for 5–7 days before sample collection. HBECs (Lonza Lot# 269120 and 451973) were cultured in Pneumacult EX Plus media (StemCell Technologies). siRNA transfection was carried out with Lipofectamine RNAi-max (Invitrogen, 13778150) and cells were maintained in submerged culture for 3 days before analysis. For cytokine supplementation HBECS were cultured in Pnemacult media with Il-13 (10ng/mL), Il-1α (5ng/mL), and Il-1β (5ng/mL). For MST inhibition, media was supplemented with 10 μm XMU-MP-1 compound.
Tracheal explant cultures
Tracheas were dissected and cut longitudinally along the dorsal side. Tracheas were then placed on 1cm2 Surgifoam absorbable sterile gelatin sponges (Ethicon), with the epithelium exposed to air. Tracheas were maintained, unsubmerged, in DMEM:F12 media supplemented with Penicillin/Streptomycin and Amphoterocin. For knockout experiments, 3–10 μM 4-Hydroxytamoxifen (Sigma) or methanol as a vehicle control, were added to the media for 7–10 days before fixation in 4% paraformaldehyde.
Histology and immunostaining
Lungs were inflated with 4% paraformaldehyde (PFA) and fixed overnight at 4C, followed by ethanol dehydration and paraffin embedding. Tissues were sectioned at 5um for histology and were prepared according to a standard deparaffinization and rehydration protocol. For immunostaining, samples were exposed to microwave-assisted antigen retrieval in a low pH Citrate buffer (Vector Laboratories, H-3300) followed by blocking, primary, and secondary antibody incubation in 5% Donkey serum (EMD Millipore, S30–100ML). For DAB staining, slides were processed as above and submerged in 3% H2O2 before incubation with HRP conjugated secondary antibodies. Slides were developed in DAB substrate (Cell Signaling, 8059) for 1 to 10minutes and then counterstained with hematoxylin before dehydration and mounting. Periodic Acid Schiff staining was performed according to manufacturer’s protocol (Sigma, 395B-1KT). H&E staining was performed with Harris Hematoxylin (Fisher) and Eosin (Sigma).
ALI inserts were fixed in 4% PFA and then blocked/permeabilized in 0.2% Triton X-100/0.1% BSA/PBS. Cells were subsequently incubated with primary and secondary antibodies in a 0.1% Triton X-100/0.2% BSA/PBS buffer. Nuclear immunofluorescence intensity was measured by FIJI and cell quantification was performed using CellProfiler pipelines (McQuin et al., 2018; Schindelin et al., 2012).
All primary antibodies used are outlined in the key resources table and the dilutions used are as follows: mouse anti-Muc5ac (1:100), rabbit anti-CCSP (1:200), mouse anti-FoxJ1 (1:400), rabbit anti-proSPC (1:1000), hamster anti-podoplanin (1:1000), mouse anti-Hopx (1:100), rabbit anti-Krt5 (1:500), chicken anti-Krt5 (1:200), rat anti-Krt8 (1:300), chicken anti-YFP (1:400), rabbit anti-Ki67 (1:100), mouse anti-αSMA (1:400), rabbit anti-CD45 (1:100), rabbit anti-Nkx2.1 (1:100), goat anti-tdTomato (1:200).
Whole-mount staining
Tracheas for whole mount staining were permeabilized in 0.3% Triton X-100 for 3 hours, followed by blocking for 24 hours in a solution of 0.1% Triton X-100, 5% Donkey Serum in Rodent Block IgG (Biocare). Antibodies were delivered in a solution of 5% Donkey Serum in PBS-T for 48 hours at 4C, according to the table in Supplemental Experimental Procedures. Tracheas were mounted in Prolong Gold antifade reagent (Invitrogen) and imaged by confocal microscopy.
Gene expression profiling
RNA for all experiments was extracted using the RNeasy Mini kit (QIAGEN). For quantitative reverse transcriptase PCR analysis RNA was reverse transcribed using the iScript cDNA Synthesis kit (BioRad, 1708891). Both TaqMan Universal Master Mix II with UNG (Applied Biosystems, 444038) and Fast SYBR Green Master Mix (Applied Biosystems, catalog no. 4385612) enzymes were used for analysis of gene expression in this study. A complete list of primers used for SYBR based qPCR is available in Table S7.
For defining global YAP/TAZ regulated gene expression in mouse airway cells, RNA was extracted from primary tracheal cultures isolated from YAP/TAZ-R26CreERT2 mice as described above. All cells were cultured in the presence of 3 μM 4-hydroxytamoxifen for 5 days in submerged mouse ALI expansion medium. RNA was subject to quality assessment by Bioanalyzer. Mouse Gene 2.0 ST CEL files were normalized to produce gene-level expression values using the implementation of the Robust Multiarray Average (RMA) in the affy package (version 1.36.1) included in the Bioconductor software suite (version 2.11) and an Entrez Gene-specific probeset mapping (17.0.0) from the Molecular and Behavioral Neuroscience Institute (Brainarray) at the University of Michigan. Array quality was assessed by computing Relative Log Expression (RLE) and Normalized Unscaled Standard Error (NUSE) using the affyPLM package (version 1.34.0). Differential expression was assessed using the moderated (empirical Bayesian) t test implemented in the limma package (version 3.14.4) (i.e., creating simple linear models with lmFit, followed by empirical Bayesian adjustment with eBayes). Correction for multiple hypothesis testing was accomplished using the Benjamini-Hochberg false discovery rate (FDR). Human homologs of mouse genes were identified using HomoloGene (version 68). All microarray analyses were performed using the R environment for statistical computing (version 2.15.1). The differentially expressed gene signature was defined by a 2-fold change cutoff with FDR ≤ 0.05.
For generating the YAP/TAZ regulated gene expression signature in human airway cells, HBECS were cultured in Pneumacult EX plus and transfected with control and dual Yap/Taz targeting siRNAs and RNA was extracted for quality assessment and library prep for RNA-sequencing. 3 unique siRNA control sequences were used and all siRNA transfected samples were collected in triplicate, 48hours after transfection.
For gene expression profiling of in vivo knockout epithelial cells, Scgb1a1 lineage-traced cells were isolated by fluorescence activated cell sorting (FACS). Control mice and Yap/Taz floxed mice were injected with tamoxifen for 5 consecutive days and lungs were collected with perfusion on Day 12 after onset of administration. Lungs were dissociated to 10 μm on a McIlwain Tissue Chopper (Ted Pella) and digested with Collagenase A (Sigma) to prepare a single cell suspension for sorting. Cells were sorted directly into Trizol for RNA isolation.
RNA quality for all samples was assessed by BioAnalyzer before proceeding with library preparation for sequencing. Sequencing libraries were prepared from total RNA samples using Illumina TruSeq RNA Sample Preparation Kit v2. The libraries from individual samples were pooled sequencing. HBEC samples were sequenced on the Illumina HiSeq 2500 platform to generate single end 50bp reads. Murine in vivo purified samples were sequenced in the Illumina NextSeq 2000 P2 to generate 50pb paired end reads to depth of ~70million reads per sample.
Demultiplexing and creation of FASTQ files were performed using Illumina BaseSpace. Samples were aligned using hg19 and 2-pass STAR alignment. Gene and transcript level counts were calculated using RSEM using Ensembl v75 annotation. Quality metrics were calculated by STAR and RSeQC. EdgeR was used to compute normalized data (library sizes normalized using TMM, trimmed mean of M-values, and log2 counts per million computed) and genes were excluded that either had an interquartile range equal to zero or a sum across samples equal or less than 1. For the generation of HBEC heatmaps, we calculated residual expression by regressing out the differences in the count between the two HBEC cell lines using limma package. All statistical analyses used R statistical software 3.5.1 and packages: limma v3.38.3, biomaRt 2.38.0, msigdbr 6.2.1, edgeR 3.24.1, nlme 3.1–137, and clusterProfiler 3.11.1. Enrichment analysis for comparison of mouse and human transcriptional signatures was computed using fgsea 1.14.0
Transcriptional GSEA
GSEA (version 2.2.1) was used to identify biological terms, pathways and processes that are coordinately up- or downregulated within each pairwise comparison. The Entrez Gene identifiers of the human homologs of the genes interrogated by the array were ranked according to the Moderated t statistic computed between the knockout and wild-type groups. Mouse genes with multiple human homologs (or vice versa) were removed prior to ranking, so that the ranked list represents only those human genes that match exactly one mouse gene. This ranked list was then used to perform pre-ranked GSEA analyses (default parameters with random seed 1234) using the Entrez Gene versions of the Hallmark, Biocarta, KEGG, Reactome, Gene Ontology (GO), and transcription factor and microRNA motif gene sets obtained from the Molecular Signatures Database (MSigDB), version 6.0.
MNase-qPCR
HBECs were cultured in Pnemucult EX Plus media and transfected with siControl and siYAP/TAZ siRNAs as described above. Samples were collected for analysis 72 hours after transfection. Briefly, cells were crosslinked with 1% formaldehyde, permeabilized, and digested with Micrococcal nuclease. Decrosslinked chromatin was purified, 150bp fragments were gel extracted, and quantified by PicoGreen (ThermoFisher) for qPCR. Primer sequences are available in Table S7.
Chromatin immunoprecipitation
HBECs for ChIP were cultured to density in Pnemucult EX Plus media. Cells were cross-linked in 1mM EGS in PBS for 30min followed by a 1% formaldehyde treatment for 10 min. Fixation was subsequently neutralized with 0.125M glycine in PBS. Harvested chromatin was sonicated using the Bioruptor UCD- 200 for 2 cycles of 10 min (max power 30 s on 30 s off). After sonication, chromatin was incubated with designated amount of antibody at 4 °C overnight. The following antibody amounts were used as follows: Rabbit anti-Yap (Abcam, Cat# ab52771, 3ug), Rabbit anti-Yap (Novus, Cat#NB110–58358, 1ug), Rabbit anti-GATAD2A (Bethyl, Cat# A302–357A, 2ug), Mouse anti-CHD4 (Abcam, Cat#ab70469, 2ug), Rabbit anti-TEAD (AvivaSysBio, Cat# ARP38276, 1ug). Immuno-precipitated complexes were collected by Protein A/G Magnetic beads (Pierce, 8802). Samples were washed with low salt buffer (20mM Tris, 140mM NaCl, 1mM EDTA, 0.1% NaDeoxycholate, 0.1% SDS, 1% Triton X-100), followed by a high salt buffer (20mM Tris, 500mM NaCl, 1mM EDTA, 0.5% NaDeoxycholate, 1% Triton X-100), and a LiCl buffer (20mM Tris, 1mM EDTA, 0.1% NaDeoxycholate, 1% Triton X-100, 250mM LiCl). Chromatin was de-crosslinked overnight at 65C and purified using the Qiaquick PCR purification kit (QIAGEN, 28104). ChIP qPCR experiments were repeated at least three times and representative results are shown as samples mean between technical replicates ± standard deviation. ChIP-qPCR for Tead, Yap, and NuRD proteins was assessed using the same primers targeting the promoter region of SPDEF. Yap and Tead binding was also confirmed at CTGF. Primers for the CTGF and SPDEF locus are available in Table S7. For ChIP-seq, three independent experiments were sequenced. The purified DNA was ligated to specific adaptors and sequenced using DNB-seq, performed by BGI, to a depth of 40 million reads.
ChIP-Seq data pre-processing
Single end 50bp reads were checked for quality, trimmed with Trim Galore (v0.4.1), and aligned to the human reference genome (hg19) with Bowtie2 (v2.3.4.1) retaining only uniquely mapped reads, both with default parameters. Redundant reads were removed and the resulting SAM files were converted to BAM format with samtools (v1.7). Narrow peak calling was performed in individual replicates with MACS2 (Model-based Analysis for ChIP-Seq, v2.1.2) using a minimum q-value cutoff of 0.05 for peak detection. Peaks overlapped between replicates were identified using the findOverlapsOfPeaks from the ChIPpeakAnno package (v3.22.2) and were used for downstream analysis. Peaks were annotated and summary plots of feature annotations were generated with the R package ChIPseeker (v1.20.0) where promoter regions were defined as 5000 bases upstream and downstream of the transcription start site (TSS) of known genes. De novo motif enrichment analysis was done within the TEAD peak regions using findMotifsGenome.pl program from HOMER (v4.10) with the default parameters.
Pathway enrichment
Pathway enrichment was performed using the hypergeometric test via the R package hypeR (v1.2.0) using the Molecular Signatures Database (MsigDB) from the Broad Institute.
Mediation testing
To investigate the SPDEF mediation of YAP, we tested gene signatures from a YAP/TAZ knockout experiment against SPDEF expression in the publicly available GTEx Lung datasets (GTEx Consortium, 2015). The Genotype-Tissue Expression (GTEx) Lung RNA-Seq count matrix and available metadata was downloaded through the GTEx Portal. We normalized the data with the R package edgeR (v3.24.1) using the TMM (Trimmed Mean of M-values) method. We first tested the correlation of YAP/TAZ signature genes against SPDEF in GTEx-LUNG. We calculated and ranked the Pearson correlation of all genes and performed a two-sided Kolmo-gorov- Smirnov comparing the positions of up and down gene signatures to a uniform distribution. Additionally, we used Sobel’s test to measure the SPDEF-mediation of YAP’s effect on YAP/TAZ signature genes compared to the background population of genes. We use the Sobel implementation from the R package multilevel (v2.6). We compared the p values of SPDEF-mediation of YAP with YAP/TAZ signature genes against YAP with background genes using a Wilcoxon rank-sum test.
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of statistical analyses, sample size, and tests used are listed in the relevant figure legend and results sections. For all qPCR and cell counting data plots are displayed as a mean ± SEM from a minimum of 3 replicates, as noted in the figure legends. For these data Student’s t tests were performed using Prism 9.0. Survival curve data was also analyzed using Prism 9. P values are noted in the figures and figure legends.
All images for quantification were obtained on a Zeiss Axio Observer fluorescence microscope or Zeiss LSM 710 confocal microscope. FoxJ1+ and Scgb1a1+ cells were quantified manually. Images were processed in ImageJ for nuclear intensity measurements. Cell Profiler was used for quantification of CD45+ and SPC+ cells in lung tissue sections.
Statistical analysis of global gene expression and ChIP seq datasets is described in detail in the appropriate methods sections. For transcriptional profiling of YAP/TAZ depleted NHBECS by RNA seq, 2 independent donor lines were used. For each line, cells were transfected in triplicate with 3 separate control siRNAs and a dual siRNA targeting YAP and TAZ.Tead ChIP in NHBECS was performed in three independent replicates for sequencing. In vivo lineage traced cells were isolated from 3 separate ControlYFP-Scgb and Yap/TazYFP-Scgb-KO mice for RNA sequencing.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Muc5ac | Novus | Cat# NBP2–15196 |
| Rabbit anti-CCSP | EMD Millipore | Cat# 07–623; RRID:AB_310759 |
| Mouse anti-FoxJ1 | ThermoFisherScientific | Cat# 14-9965-82; RRID:AB_1548835 |
| Rabbit anti-ProSFTPC | Seven Hills Bioreagents | Cat# WRAB-9337; RRID:AB_2335890 |
| Hamster anti-Podoplanin | DSHB | Cat# 8.1.1; RRID:AB_531893 |
| Mouse anti-Hopx | Santa Cruz | Cat# SC398703; RRID: AB_2687966 |
| Rabbit anti-Krt5 | BioLegend | Cat# 905501; RRID:AB_2565050 |
| Chicken anti-Krt5 | BioLegend | Cat# 905901; RRID:AB_2565054 |
| Rat anti-Krt8 | DSHB | Cat# TROMA-I; RRID:AB_531826 |
| Chicken anti-YFP | Aves Labs | Cat# GFP-1020; RRID:AB_10000240 |
| Rabbit anti-Ki67 | Abcam | Cat# ab16667; RRID:AB_302459 |
| Mouse anti-aSmooth Muscle Actin | Sigma | Cat# A2547; RRID:AB_476701 |
| Rabbit anti-CD45 | Abcam | Cat# ab10558; RRID: AB_442810 |
| Rabbit anti-Nkx2.1 | Abcam | Cat# ab76013; RRID:AB_1310784 |
| Goat anti-tdTomato | Sicgen | Cat# AB8181–200; RRID:AB_2722750 |
| Rabbit anti-Yap/Taz (D24E4) | Cell Signaling | Cat# 8418; RRID:AB_10950494 |
| Rabbit anti-Yap XP (D8H1X) | Cell Signaling | Cat# 14074; RRID:AB_2650491 |
| Rabbit anti-Taz | Sigma | Cat# HPA007415; RRID: AB_1080602 |
| Rabbit anti-Yap | Abcam | Cat# ab52771; RRID:AB_2219141 |
| Rabbit anti-Yap | Novus | Cat# NB110–58358; RRID:AB_922796 |
| Rabbit anti-Gatad2a | Bethyl | Cat# A302–357A; RRID:AB_1907235 |
| Mouse anti-CHD4 | Abcam | Cat# ab70469; RRID:AB_2229454 |
| Rabbit anti-TEAD | AvivaSystemsBiology | Cat# ARP38276_P050; RRID:AB_614654 |
| Donkey Anti-Mouse; Alexa Fluor 488 | Jackson | Cat# 715-546-151; RRID:AB_2340850 |
| Donkey Anti-Mouse; Cy3 | Jackson | Cat# 715-166-151; RRID:AB_2340817 |
| Donkey Anti-Mouse; Alexa Fluor 647 | Jackson | Cat# 715-606-151; RRID:AB_2340866 |
| Donkey Anti-Chicken; AlexaFluor488 | Jackson | Cat# 703-546-155; RRID:AB_2340376 |
| Donkey Anti-Chicken; AlexaFluor594 | Jackson | Cat# 703-585-155; RRID:AB_2340377 |
| Goat Anti-Chicken; AlexaFluor 647 | ThermoFisher | Cat# A-21449; RRID:AB_2535866 |
| Donkey Anti-Rat; AlexaFluor 488 | Jackson | Cat# 712-546-153; RRID:AB_2340686 |
| Donkey Anti-Rat; AlexaFluor 594 | Jackson | Cat# 712-586-153; RRID:AB_2340691 |
| Goat Anti-Rat; AlexaFluor 647 | Molecular Probes | Cat# A-21247; RRID:AB_141778 |
| Donkey Anti-Rabbit; AlexaFluor 488 | Molecular Probes | Cat# A-21206; RRID:AB_2535792 |
| Donkey Anti-Rabbit; Cy3 | Jackson | Cat# 711-166-152; RRID:AB_2313568 |
| Donkey Anti-Rabbit; AlexaFluor 647 | Molecular Probes | Cat# A-31573; RRID:AB_2536183 |
| Goat Anti-Syrian Hamster; AlexaFluor 647 | Jackson | Cat# 107-605-142; RRID:AB_2337482 |
| Biological samples | ||
| NHBE-Bronchial Epi Cells w/RA, BEGM, cryo | Lonza | CC-2540 |
| Chemicals, peptides, and recombinant proteins | ||
| Harris hematoxylin | Fisher Scientific | 6765003 |
| Eosin Y Solution | Sigma | HT110316 |
| 4-Hydroxytamoxifen | Sigma | H7904 |
| Tamoxifen | Sigma | T5648– 5 g |
| Corn oil | Sigma | C8267–2.5L |
| Pneumacult EX Plus | Lonza | 5040 |
| Pneumacult ALI Medium | Lonza | 5001 |
| DMEM | Corning | 10–013-CV |
| F12 | Corning | 10–080-CM |
| Pen/Strep | GIBCO | 15140–122 |
| Amphoterocin | GIBCO | 15290–018 |
| Insulin | Sigma | I6634–50MG |
| Transferrin | Sigma | T1147–100MG |
| Cholera Toxin | Sigma Aldrich | C-8052 |
| EGF | Fisher (Corning) | CB-40001 |
| Retinoic Acid | Sigma | R2625–50mg |
| XMU-MP-1 | Medchemexpress LLC | HY100526–10mg |
| IL-13 | R&D | 21–3ILB-100CF |
| Il1-a | R&D | 200-LA-002 |
| IL-1b | R&D | 200-LA-002 |
| Collagen I, Rat | Fisher | CB-40236 |
| EGS (ethylene glycol bis(succinimidyl succinate)) | Life Tech | 21565 |
| Collagenase A | Sigma | 11088793001 |
| ACK Lysis Buffer | GIBCO | A1049201 |
| Critical assays | ||
| Molecular Probes Click-iT Plus TUNEL Assay, Alexa Fluor 647 dye | Fisher | C10619 |
| mm-Spdef RNAScope Probe | ACDBio | 544421 |
| RNAscope 2.5 HD Reagent Kit-RED | ACDBio | 322350 |
| RNAscope Positive Control Probe- Mm-Ppib | ACDBio | 313911 |
| RNAscope Negative Control Probe- DapB | ACDBio | 310043 |
| PAS | Sigma | 395B-1KT |
| Trichrome Stain (Masson) | Sigma | HT15–1KT |
| Invitrogen Lipofectamine RNAiMAX Transfection Reagent | Fisher Scientific | 13-778-150 |
| QIAquick PCR Purification Kit (250) | QIAGEN | 28106 |
| RNeasy Mini | QIAGEN | 74106 |
| iScript cDNA Synthesis Kit | BioRad | 1708891 |
| Fast SYBR Green Master Mix | Life Technologies | 43-856-18 |
| Applied Biosystems TaqMan Universal Master Mix II, With UNG | Fisher Scientific | 4440038 |
| Mouse GAPDH Taqman probe Mm99999915_g1 | ThermoFisher | 4331182 |
| Mouse Muc5ac Taqman probe Mm01276718_m1 | ThermoFisher | 4331182 |
| Mouse Scgb1a1 Taqman probe Mm00442046_m1 | ThermoFisher | 4331182 |
| Mouse FoxJ1 Taqman probe Mm01267279_m1 | ThermoFisher | 4331182 |
| Mouse trp63 Taqman probe Mm00495788_m1 | ThermoFisher | 4331182 |
| Mouse Krt5 Taqman probe Mm01305291_g1 | ThermoFisher | 4331182 |
| Mouse Yap1 Taqman probe Mm01143263_m1 | ThermoFisher | 4331182 |
| Mouse Wwtr1 Taqman probe Mm01289583_m1 | ThermoFisher | 4331182 |
| Mouse Ctgf Taqman probe Mm01192933_g1 | ThermoFisher | 4331182 |
| Mouse Spdef Taqman probe Mm00600221_m1 | ThermoFisher | 4331182 |
| Human CTGF Taqman probe Hs00170014_m1 | ThermoFisher | 4331182 |
| Human YAP Taqman probe Hs00902712_g1 | ThermoFisher | 4331182 |
| Human WWTR1 Taqman probe Hs01086149_m1 | ThermoFisher | 4331182 |
| Human MUC5AC Taqman probe Hs01365616_m1 | ThermoFisher | 4331182 |
| Human SCGB1A1 Taqman probe Hs00171092_m1 | ThermoFisher | 4331182 |
| Human GATAD2A Taqman Probe Hs00214293_m1 | ThermoFisher | 4331182 |
| Human CHD4 Taqman probe Hs00172349_m1 | ThermoFisher | 4331182 |
| Human SPDEF Taqman probe Hs00171942_m1 | ThermoFisher | 4331182 |
| Human PPIA Taqman probe Hs04194521_s1 | ThermoFisher | 4331182 |
| Deposited data | ||
| Mouse Yap/Taz knockout ALI Microarray | This paper | GEO: GSE156525 |
| Human Yap/Taz Knockdown RNA-Seq | This paper | GEO: GSE158305 |
| Human (HBEC) ChiP-Seq | This paper | GEO: GSE158306 |
| Mouse Scgb1a1+ Control and Yap/Taz Knockout RNA-Seq | This paper | GEO: GSE171712 |
| Experimental models: Organisms/strains | ||
| Mus musculus B6.129X1-Gt(ROSA) 26Sortm1(EYFP)Cos/J | The Jackson Laboratory | 006148 |
| Mus musculus B6N.129S6(Cg)-Scgb1a1tm1(cre/ERT)Blh/J | The Jackson Laboratory | 016225 |
| Mus musculus Nkx2-1tm1.1(cre/ERT2)Zjh/J | The Jackson Laboratory | 014552 |
| Mus musculus B6.Cg-Gt(ROSA) 26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | 007914 |
| Oligonucleotides | ||
| Negative Control siRNA (20 nmol) | QIAGEN | 1027310 |
| ON-TARGETplus Non-targeting siRNA #1 | Dharmacon | D-001810–01 |
| ON-TARGETplus Non-targeting siRNA #2 | Dharmacon | D-001810–02 |
| SMARTpool: siGENOME SPDEF siRNA | Dharmacon | L-020199-00-0005 |
| SMARTpool: siGENOME Lats1 siRNA | Dharmacon | L-004632-00-0005 |
| SMARTpool: siGENOME Lats2 siRNA | Dharmacon | L-003865-00-0005 |
| SMARTpool: siGENOME GATAD2A siRNA | Dharmacon | M-015311-00-0005 |
| SMARTpool: siGENOME CHD4 siRNA | Dharmacon | M-009774-01-0005 |
| siRNA Targeting Yap/Taz UGUGGAUGAGAUGGAUACA | Hiemer et al., 2014 | N/A |
| siRNA Targeting Tead1/3/4 UGAUCAACUUCAUCCACAA | Tilston-Lunel et al., 2021 | N/A |
| siRNA Targeting Tead2 CCUGGUGAAUUUCUUGCACAA | Tilston-Lunel et al., 2021 | N/A |
| Software and algorithms | ||
| Cell Profiler | McQuin et al., 2018 | https://cellprofiler.org/ |
| ImageJ/Fiji | Schindelin et al., 2012 | https://imagej.net/software/Fiji |
| Basespace | Illumina | https://www.illumina.com/products/by-type/informatics-products/basespace-sequence-hub.html |
| STAR 2.7.1a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| MACS2 2.2.7.1 | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| ChIPSeeker 1.24.0 | Yu et al., 2015 | https://guangchuangyu.github.io/software/ChIPseeker/ |
| limma 3.44.3 | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/edgeR.html/ |
| biomaRt 2.44.1 | Durinck et al., 2009 | https://github.com/grimbough/biomaRt |
| msigdbr 7.4.1 | msigdbr package | https://github.com/igordot/msigdbr |
| edgeR 3.30.3 | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html/ |
| nlme 3.1 | Pinheiro et al., 2021 | https://cran.r-project.org/web/packages/nlme/index.html |
| clusterProfiler 3.16.1 | Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| fgsea 1.14.0 | Korotkevich et al., 2021 | https://github.com/ctlab/fgsea/ |
| Other | ||
| 0.4um Transwell Inserts (polyester) | Corning | 3470 |
| Protein A/G Beads | Fisher Scientific | 88803 |
| Surgifoam Gelatin Sponges | Ethicon | MS0005 |
| ProLong Gold Antifade reagent with DAPI | Invitrogen | P36931 |
Highlights.
YAP/TAZ maintain alveolar and airway integrity in adult mice
Nuclear YAP/TAZ restrict goblet cell differentiation and mucin hypersecretion
YAP/TAZ, TEAD, and the NuRD complex cooperate to repress SPDEF expression
Hippo-regulated YAP/TAZ intersect with goblet cell-specifying cytokine cues
ACKNOWLEDGMENTS
This work was supported by the Boston University Sequencing Core and Flow Cytometry Core and by BU-CTSI grant UL1TR001430. We thank Jeffrey Wrana for sharing the Yap-loxP/loxP and Wwtr1-loxP/loxP mice. X.V. was funded by NIH/NHLBI R01HL124392 and NIH/NICHD R21HD094012 and by an ACS Research Scholar Grant (RSG-17-138-01-CSM). J.H.-B. was funded by NIH/NHLBI grants F31HL132506 and T32 HL007035. S.M. was in part funded by Find the Cause Breast Cancer Foundation. A.F. and B.N. were funded by Moorman-Simon Fellowships in Computational Biomedicine.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109347.
DECLARATION OF INTERESTS
X.V and J.B. were supported in part by a Janssen Pharmaceuticals Sponsored Research Agreement. The remaining authors declare no competing interests.
REFERENCES
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, and Tabak LA (2012). Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, and Wrana JL (2013). Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624. [DOI] [PubMed] [Google Scholar]
- Bornelöv S, Reynolds N, Xenophontos M, Gharbi S, Johnstone E, Floyd R, Ralser M, Signolet J, Loos R, Dietmann S, et al. (2018). The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell 71, 56–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broide DH, Lawrence T, Doherty T, Cho JY, Miller M, McElwain K, McElwain S, and Karin M (2005). Allergen-induced peribronchial fibrosis and mucus production mediated by IκB kinase β-dependent genes in airway epithelium. Proc. Natl. Acad. Sci. USA 102, 17723–17728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, and Whitsett JA (2009). SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Invest 119, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Karp CL, Impey S, Xu Y, Randell SH, Kitzmiller J, Maeda Y, Haitchi HM, Sridharan A, et al. (2014). Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am. J. Respir. Crit. Care Med 189, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Sun L, Kato T, Okuda K, Martino MB, Abzhanova A, Lin JM, Gilmore RC, Batson BD, O’Neal YK, et al. (2019). IL-1β dominates the promucin secretory cytokine profile in cystic fibrosis. J. Clin. Invest 129, 4433–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, and Lee JH (2020). Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet EG, Seys LJ, Verhamme FM, Vanaudenaerde BM, Brusselle GG, Bingle CD, and Bracke KR (2017). Association of innate defense proteins BPIFA1 and BPIFB1 with disease severity in COPD. Int. J. Chron. Obstruct. Pulmon. Dis 13, 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Varelas X, and Guan KL (2020). Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov 19, 480–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, DeMayo FJ, Burns AR, Smith C, Reynolds SD, et al. (2004). Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am. J. Respir. Cell Mol. Biol 31, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV, and Dickey BF (2010). Airway mucus function and dysfunction. N. Engl. J. Med 363, 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MB, Wood M, Lapey A, and Mou H (2019). SMAD signaling restricts mucous cell differentiation in human airway epithelium. Am. J. Respir. Cell Mol. Biol 61, 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokey JJ, Sridharan A, Xu Y, Green J, Carraro G, Stripp BR, Perl AT, and Whitsett JA (2018). Active epithelial Hippo signaling in idiopathic pulmonary fibrosis. JCI Insight 3, e98738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, and Rajagopal J (2009). Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemer SE, Szymaniak AD, and Varelas X (2014). The transcriptional regulators TAZ and YAP direct transforming growth factor β-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem 289, 13461–13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isago H, Mitani A, Mikami Y, Horie M, Urushiyama H, Hamamoto R, Terasaki Y, and Nagase T (2020). Epithelial expression of YAP and TAZ is sequentially required in lung development. Am. J. Respir. Cell Mol. Biol 62, 256–266. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim T, Johnson RL, and Lim DS (2015). Transcriptional co-repressor function of the Hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282. [DOI] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI] [Google Scholar]
- Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, and Erle DJ (2002). Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med 8, 885–889. [DOI] [PubMed] [Google Scholar]
- LaCanna R, Liccardo D, Zhang P, Tragesser L, Wang Y, Cao T, Chapman HA, Morrisey EE, Shen H, Koch WJ, et al. (2019). Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J. Clin. Invest 129, 2107–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai AY, and Wade PA (2011). Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange AW, Sridharan A, Xu Y, Stripp BR, Perl AK, and Whitsett JA (2015). Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J. Mol. Cell Biol 7, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu Q, Li J, Huang H, Cai T, Ji H, et al. (2016). MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 16, 1810–1819. [DOI] [PubMed] [Google Scholar]
- Mahoney JE, Mori M, Szymaniak AD, Varelas X, and Cardoso WV (2014). The Hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev. Cell 30, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley HA, Liu CY, Attia AC, Wikenheiser-Brokamp KA, Zhang Y, Whitsett JA, and Guasch G (2014). TGFβ signaling inhibits goblet cell differentiation via SPDEF in conjunctival epithelium. Development 141, 4628–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantie LB, Young RE, Paltzer WG, Zhang Y, Johnson RL, Verheyden JM, and Sun X (2018). Lats1/2 inactivation reveals Hippo function in alveolar type I cell differentiation during lung transition to air breathing. Development 145, dev163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, Radicioni G, Kesimer M, Chua M, Dang H, et al. (2019). Localization of secretory mucins MUC5AC and MUC5B in normal/healthy human airways. Am. J. Respir. Crit. Care Med 199, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, Khurana Hershey GK, Chen G, and Whitsett JA (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Invest 117, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, and Sarkar D; R Core Team (2021). nlme: Linear and nonlinear mixed effects models. R package version 3.1–152. https://CRAN.R-project.org/package=nlme. [Google Scholar]
- Poynter ME, Cloots R, van Woerkom T, Butnor KJ, Vacek P, Taatjes DJ, Irvin CG, and Janssen-Heininger YM (2004). NF-κB activation in airways modulates allergic inflammation but not hyperresponsiveness. J. Immunol 173, 7003–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavelu P, Chen G, Xu Y, Kitzmiller JA, Korfhagen TR, and Whitsett JA (2015). Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J. Clin. Invest 125, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BL (2009). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, and McNeill H (2013). Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 9, e1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MC, and Voynow JA (2006). Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev 86, 245–278. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi N, Zhang J, and Chen SY (2017). Runx2, a novel regulator for goblet cell differentiation and asthma development. FASEB J. 31, 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Heijink IH, Kistemaker LEM, Reinders-Luinge M, Kooistra W, Noordhoek JA, Gosens R, Brandsma CA, Timens W, Hiemstra PS, et al. (2017). Aberrant DNA methylation and expression of SPDEF and FOXA2 in airway epithelium of patients with COPD. Clin. Epigenetics 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, Tsidiridis G, Lange M, Mattner LF, Yee M, et al. (2020). Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun 11, 3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilston-Lunel A, Mazzilli S, Kingston NM, Szymaniak AD, Hicks-Berthet J, Kern JG, Abo K, Reid ME, Perdomo C, Wilson AA, et al. (2021). Aberrant epithelial polarity cues drive the development of precancerous airway lesions. Proc. Natl. Acad. Sci. USA 118, e2019282118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soldt BJ, Qian J, Li J, Tang N, Lu J, and Cardoso WV (2019). Yap and its subcellular localization have distinct compartment-specific roles in the developing lung. Development 146, dev175810.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, and Wrana JL (2008). TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol 10, 837–848. [DOI] [PubMed] [Google Scholar]
- Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, Brouwer S, Gomes T, Hesse L, Jiang J, et al. (2019). A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med 25, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Volckaert T, Yuan T, Chao CM, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, et al. (2017). Fgf10-Hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48–59.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA (2018). Airway epithelial differentiation and mucociliary clearance. Ann. Am. Thorac. Soc 15 (Suppl 3), S143–S148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, and Donaldson DD (1998). Interleukin-13: Central mediator of allergic asthma. Science 282, 2258–2261. [DOI] [PubMed] [Google Scholar]
- You Y, and Brody SL (2013). Culture and differentiation of mouse tracheal epithelial cells. Methods Mol. Biol 945, 123–143. [DOI] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, and Brody SL (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. American J. Physiology-Lung Cellular and Molecular Physiology 283, 1315–1321. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, and He Q-Y (2015). ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, and Stein GS (2004). Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 23, 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, and Piccolo S (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol 17, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Fallon TR, Saladi SV, Pardo-Saganta A, Villoria J, Mou H, Vinarsky V, Gonzalez-Celeiro M, Nunna N, Hariri LP, et al. (2014). Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev. Cell 30, 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray, RNA-seq and ChIP Seq datasets have been deposited to NCBI GEO. Human RNA-seq datasets are listed as GEO: GSE158305 and Chip-Seq as GEO: GSE158306 (superseries GEO: GSE158307). The microarray data is available under GEO: GSE156525. RNA-seq of in vivo purified Scgb1a1+ cells is available under the GEO: GSE171712.


