Abstract
With polygenic risk score (PRS) for autoimmune type 1 diabetes (T1D), this study identified T1D cases with low T1D PRS and searched for susceptibility loci in these cases. Our hypothesis is that genetic effects (likely mediated by relatively rare genetic variants) of non-mainstream (or non-autoimmune) T1D might have been diluted in the previous studies on T1D cases in general. Two cohorts for the PRS modeling and testing respectively were included. The first cohort consisted of 3302 T1D cases and 6181 controls, and the independent second cohort consisted of 3297 T1D cases and 6169 controls. Cases with low T1D PRS were identified using PRSice-2 and compared to controls with low T1D PRS by genome-wide association (GWA) test. Thirteen novel genetic loci with high imputation quality (Quality Score r2 > 0.91) were identified of SNPs/SNVs associated with low PRS T1D at genome-wide significance (P ≤ 5.0 × E−08), in addition to 4 established T1D loci, 3 reported loci by our previous study, as well as 9 potential novel loci represented by rare SNVs, but with relatively low imputation quality (Quality Score r2 < 0.90). For the 13 novel loci, 9 regions have been reported of association with obesity related traits by previous GWA studies. Three loci encoding long intergenic non-protein coding RNAs (lncRNA), and 2 loci involved in N-linked glycosylation are also highlighted in this study.
Subject terms: Genetic association study, Endocrinology
Introduction
Type 1 diabetes (T1D) is caused by T-cell mediated autoimmune destruction of pancreatic β-cells1. There is no cure for T1D to date. The molecular mechanisms underlying T1D are complex and not completely understood. Human genetic studies have uncovered multiple T1D genes that contribute to our understanding of the pathogenesis ofT1D2–7. With the rapid advances in human genomics technology in recent years, over 70 T1D loci have been identified8 (https://www.ebi.ac.uk/gwas/). While these discoveries of T1D-associated genes have greatly increased our knowledge of T1D, our current genetic knowledge on T1D is far from complete, and a large number of T1D genes remain uncovered9. A key bottleneck for the GWAS approach is limitation of sample size even with the presence of collaborative international consortia10. The phenotype of type 1 diabetes has been regarded as heterogeneous. While the majority of T1D patients have autoimmune disease, 5–10% of Caucasian diabetic subjects with recent-onset T1D do not have islet cell antibodies, often referred to as T1bD11. Due to different pathogenesis, T1bD cases may be associated with different genetic loci from autoimmune T1D, or T1aD. However, the smaller proportion of T1bD cases suggests that T1bD-related genetic effects have been diluted in the previous studies with T1D cases studied in general. Besides T1bD, the non-autoimmune and monogenic form of pediatric diabetes, maturity-onset diabetes of the young (MODY) cases, may be misdiagnosed as T1D12, which further contributes to the heterogeneity of the T1D phenotype.
With numerous genetic loci for many human complex diseases identified to date, polygenic risk scores (PRS) aggregate the effects of many genetic variants across the human genome into a single score, an approach that has been shown of improve disease prediction and differential diagnosis13. The T1D loci identified by the GWAS studies to date are mainly associated with the genetic susceptibility of the major component of the heterogeneous T1D phenotype, i.e. T1aD, while the genetic susceptibility of the minor non-autoimmune components (e.g. T1bD and misdiagnosed MODY) are under-represented in those results likely as a result of being diluted. In this study, we propose that a high T1D PRS score predicts or suggests a T1aD case, whereas a low T1D PRS score in a T1D case suggests the opposite and represents our major interest in this study. Our aim in this study is to identify low PRS T1D cases and to run a separate GWAS in an attempt to uncover genetic loci associated with T1bD patients. Our approach effectively concentrates the dilution of non-mainstream T1D by excluding high PRS T1D cases, to uncover novel genetic loci associated with non-mainstream T1D. Therefore, the dilution of low PRS T1D by misdiagnosed MODY is not a concern. On the other hand, although the low PRS cases may include MODY patients, there are no MODY mutation identified with genome-wide significance in this GWAS study, which is as expected while next generation sequencing, e.g. whole exome sequencing, is the more proper approach.
Methods
Subjects
6599 European T1D cases and 12,350 European controls were included in this study. The T1D cases were from the Children's Hospital of Philadelphia (CHOP)14, The Montreal Children's Hospital14, The Diabetes Control and Complications Trial—Epidemiology of Diabetes Interventions and Complications (DCCT-EDIC) cohort (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000086.v2.p1), the Type 1 Diabetes Genetics Consortium (T1DGC, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000180.v1.p1), and later recruited subjects at CHOP, respectively. The T1D cases were mainly recruited by clinical diagnosis, i.e. insulin dependent for at least 6 months, and diagnosed under the age of 18 for the subjects recruited at CHOP and Montreal. The non-mainstream T1D cases in this study were defined by low T1D PRS scores, with the cut-off value of PRC determined by Receiver Operating Characteristic (ROC) curve analysis. The included cases were all confirmed of European ancestry by principal component analysis (PCA) with genome-wide SNP markers, with individuals from other populations or with admixed ancestries excluded. The genotyping was done by the Illumina Human Hap550 Genotyping BeadChip or a newer version of Illumina Genotyping BeadChip. Other demographic, phenotypic and genotypic details about these individuals were described in our previous publication15. Imputation of single nucleotide polymorphisms (SNP) on auto-chromosomes was done using the TOPMed Imputation Server (https://imputation.biodatacatalyst.nhlbi.nih.gov) with the TOPMed (Version R2 on GRC38) Reference Panel, with the quality filters of R2 ≥ 0.3. Altogether, 104,689,647 autosomal single nucleotide variants (SNV) with quality R2 ≥ 0.3 were included in this study. Population stratification was assessed by PCA analysis, and genetic association tests conditioned on sex were corrected by the first 10 principal components (PC). The association test was done using PLINK1.9 software16.
Polygenic risk scores (PRS)
To avoid the issue of overfitting for PRS scoring, the subjects were randomly split into two independent cohorts without duplication, i.e. the PRS training cohort (Cohort A) including 3302 T1D cases (1739 males, 1560 females, and 3 cases with undetermined sex) and 6181 controls(3326 males, 2840 females, and 15 cases with undetermined sex), and the PRS testing cohort (Cohort B) including 3297 T1D cases (1744 males, 1549 females, and 4 cases with undetermined sex) and 6169 controls (3339 males, 2818 females, and 12 cases with undetermined sex). PRSs of the test cohort were calculated using the Polygenic Risk Score software (PRSice-2)17, based on the statistics of the training group. The performance of a series of cutoff of T1D association P values (including 10–10, 10–9 , 10–8, 10–7, 10–6, 10–5, 10–4, 0.001, 0.01, 0.05, 0.1, 0.2, and 1) for selection of SNP markers was assessed by the Area Under the ROC Curve (AUC). The P value cutoff with the largest AUC was adopted.
GWAS of T1D patients with low PRS
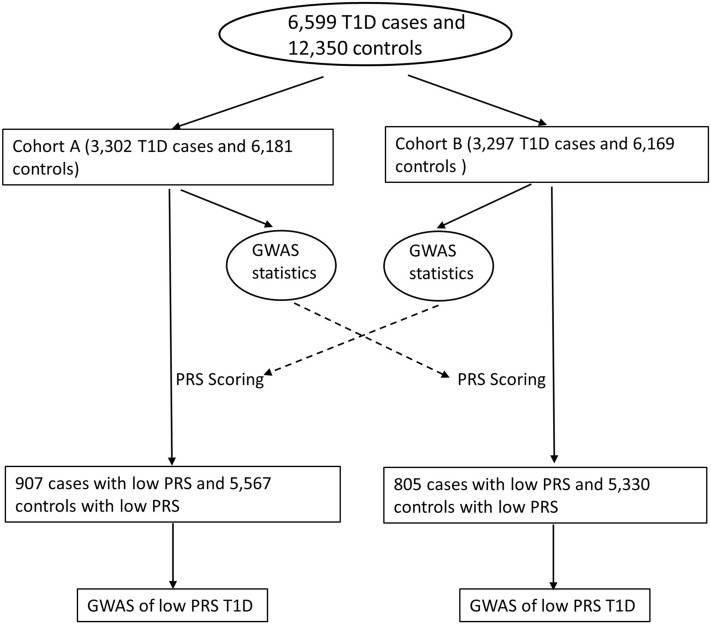
The flow chart of the study approach is shown in Fig. 1. According to the PRS values, the T1D patients were separated into two groups, i.e. a low PRS group and a high PRS group. The PRS cutoff was determined by the maximum Matthews correlation coefficient (MCC). Using the same PRS cutoff, health controls with low T1D PRS were identified. The GWAS of T1D patients with low PRS was performed by comparing to health controls with low T1D PRS. The Manhattan plots were done using the SNPEVG software18. Genetic association signals within each locus were plotted by LocusZoom19.
Figure 1.
The flow chart of the study approach.
Cohort switch
Consequently, we switched the two cohorts, i.e. using Cohort B for the statistics of PRS modelling, then we tested the PRS models in Cohort A. GWAS of T1D patients with low PRS was done using the same approach as described above.
Data and resource availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
All methods were carried out in accordance with relevant guidelines and regulations. The study was approved by the Institutional Review Boards of Children’s Hospital of Philadelphia (CHOP). Written informed consent was obtained from each participating subject or, if subjects are under 18, their parent/guardian.
Significance statement
Type 1 diabetes (T1D) is a highly heterogeneous genetic disease. Human genetic and genomic study on T1D has gained us significant knowledge on the molecular basis of autoimmunity in T1D. However, it has been recognized for long that a small number of T1D cases present without autoantibodies and are considered non-autoimmune. Human genetic approach has not been helpful for the study of these patients, as genetic effects of these non-mainstream (or non-autoimmune) T1D have been diluted in the previous studies on T1D cases in general. For the first, we identified non-mainstream T1D cases represented by low T1D polygenic risk score (PRS), and identified 13 novel loci represented by rare SNVs. This study presents a brand-new genomic landscape of pediatric T1D.
Results
AUC of different cutoffs of T1D association P values for SNP selection and PRS
The AUCs of different cutoffs of T1D association P values for selection of SNP sets are shown in Table 1a. The best AUC (0.8607) is seen at the cutoff of P value ≤ 1E−05, which suggests that stricter cutoff may cause the missing of informative SNPs, while looser may introduce noise by including SNPs with spurious T1D association. Based on the SNP markers with T1D association P value ≤ 1E−05, a PRS score was acquired for each individual in the independent test cohort. By the maximum MCC (Supplementary Table 1), a PRS cutoff of 1.11E−03 has the maximum MCC (0.6294). A PRS ≤ 1.11E−03 was defined as low risk, and a PRS > 1.11E−03 was defined as high risk. With this threshold, the sensitivity (True positive rate, TPR) for T1D prediction is 75.9%, and the specificity (True negative rate, TFR) for T1D prediction is 86.4%. By PRS ≤ 1.11E−03, 805 (24.4%, including 407 males, 396 females, and 2 cases with undetermined sex) out of 3297 T1D cases had low PRS; and 5330 (86.4%, including 2882 males, 2436 females, and 12 cases with undetermined sex) out of 6169 controls had low PRS.
Table 1.
The AUCs of different cutoffs of T1D association P values.
| P value* | AUC** |
|---|---|
| a. First cohort | |
| ≤ 1.00E−10 | 0.8462 |
| ≤ 1.00E−09 | 0.8487 |
| ≤ 1.00E−08 | 0.8518 |
| ≤ 1.00E−07 | 0.8565 |
| ≤ 1.00E−06 | 0.8604 |
| ≤ 1.00E−05 | 0.8607 |
| ≤ 1.00E−04 | 0.8590 |
| ≤ 0.001 | 0.8561 |
| ≤ 0.01 | 0.8546 |
| ≤ 0.05 | 0.8502 |
| ≤ 0.1 | 0.8508 |
| ≤ 0.2 | 0.8530 |
| ≤ 0.5 | 0.8563 |
| ≤ 1 | 0.8579 |
| b. Switched cohort | |
| ≤ 1.00E−10 | 0.8576 |
| ≤ 1.00E−09 | 0.8589 |
| ≤ 1.00E−08 | 0.8588 |
| ≤ 1.00E−07 | 0.8609 |
| ≤ 1.00E−06 | 0.8633 |
| ≤ 1.00E−05 | 0.8654 |
| ≤ 1.00E−04 | 0.8618 |
| ≤ 0.001 | 0.8555 |
| ≤ 0.01 | 0.8470 |
| ≤ 0.05 | 0.8441 |
| ≤ 0.1 | 0.8446 |
| ≤ 0.2 | 0.8467 |
| ≤ 0.5 | 0.8521 |
| ≤ 1 | 0.8533 |
*The P values are based on the statistics of the PRS training cohort.
**The AUCs are the PRS performances in the independent testing cohort.
GWAS of T1D patients with low PRS
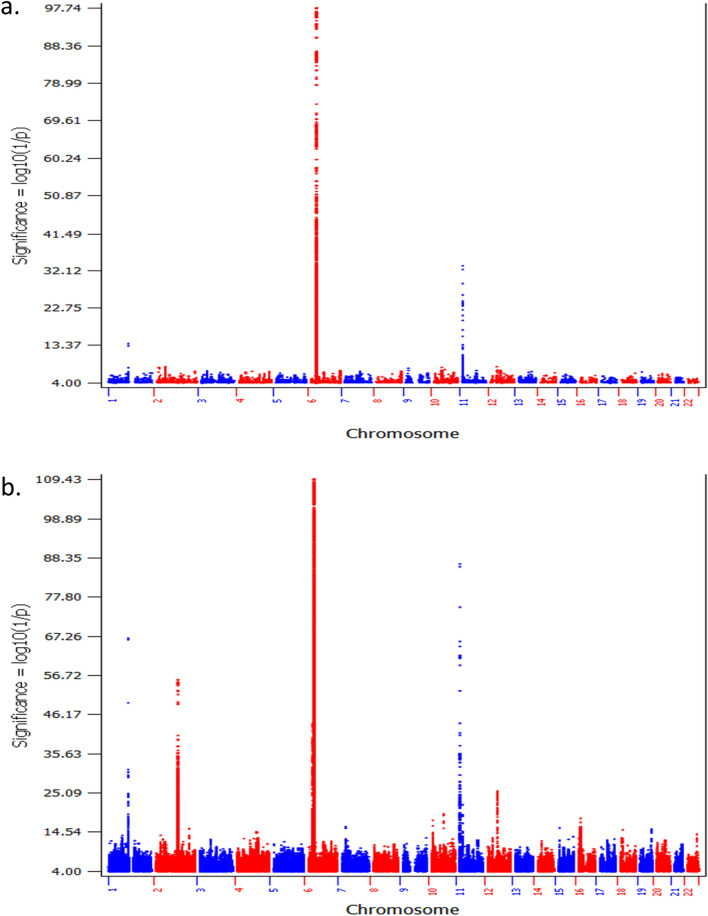
The GWAS of T1D patients with low T1D PRS compared to controls with low T1D PRS identified a large number of SNPs associated with T1D with genome-wide significance (P ≤ 5.0 × E−08), from 10 genetic loci (Supplementary Table 2, Fig. 2). Among these 10 genetic loci, 3 loci have been established of T1D association by previous studies, including HLA, INS, and PTPN22 (Table 2a). By looking at the established leading T1D signal of each locus, the frequencies of the predisposing alleles of HLA and PTPN22 were lower in the low T1D PRS cohort, while the protective allele of INS were higher in the low T1D PRS cohort. The effect sizes of HLA (P = 6.67E−08) and PTPN22 (P = 0.052) were smaller in the low PRS cases. Besides these 3 established T1D loci, 7 loci associated with low PRS T1D were identified (Table 3a). LocusZoom plots for genetic association signals within each locus are shown in Supplementary Figures 1–7. The association signals of these loci are only seen in low PRS T1D cases, but not in the T1D cases overall, and were missed previously due to diluted genetic effects. Among the 7 loci, 6 loci are novel, while the ankyrin 3 (ANK3) locus, related to neural control of the endocrine pancreas20, has been identified of genome-wide significance in our study on low T1D genetic risk scores (GRS) patients21,22.
Figure 2.
The Manhattan plots of cohort B. (a) The plot of the GWAS of T1D patients with low T1D PRS compared to controls with low T1D PRS (805 cases vs. 5330 controls); (b) the plot of the GWAS of all T1D patients compared to all controls (3297 cases vs. 6169 controls).
Table 2.
Leading SNPs at three loci have been established of T1D association.
| CHR | dbSNP | BP (hg38) | Gene.refGene | A1 | Quality Score (r2) | Genotyped | Low PRS cases versus low PRS controls | U95 | P | All cases versus all controls in cohort B | U95 | P | OR heterogeneity P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | n | OR | L95 | MAF | n | OR | L95 | ||||||||||||
| a. First cohort | |||||||||||||||||||
| 1 | rs2476601 | 11,38,34,946 | PTPN22 | A | 0.99897 | Genotyped | 0.08676 | 6118 | 1.87 | 1.594 | 2.194 | 1.55E−14 | 0.1112 | 9450 | 2.244 | 2.048 | 2.458 | 1.92E−67 | 0.052 |
| 6 | rs9273368 | 3,26,58,698 | HLA-DQB1 | A | 0.97908 | Imputed | 0.2586 | 6118 | 4.073 | 3.616 | 4.587 | 1.24E−118 | 0.3841 | 9450 | 6.018 | 5.572 | 6.5 | < 1E−350 | 6.67E−08 |
| 11 | rs689 | 21,60,994 | INS | A | 0.89726 | Imputed | 0.2572 | 6118 | 0.391 | 0.3354 | 0.4559 | 4.02E−33 | 0.2331 | 9450 | 0.4442 | 0.4098 | 0.4815 | 1.03E−86 | 0.149 |
| b. Switched cohort | |||||||||||||||||||
| 1 | rs2476601 | 11,38,34,946 | PTPN22 | A | 0.99897 | Genotyped | 0.09515 | 6460 | 2.216 | 1.921 | 2.557 | 1.03E−27 | 0.1152 | 9465 | 2.268 | 2.07 | 2.484 | 3.53E−69 | 0.789 |
| 6 | rs9273368 | 3,26,58,698 | HLA-DQB1 | A | 0.97908 | Imputed | 0.2584 | 6460 | 3.809 | 3.416 | 4.247 | 6.19E−128 | 0.376 | 9465 | 5.937 | 5.503 | 6.404 | < 1E−350 | 5.49E−11 |
| 11 | rs689 | 21,60,994 | INS | A | 0.89726 | Imputed | 0.259 | 6460 | 0.4553 | 0.3972 | 0.5218 | 1.23E−29 | 0.2366 | 9465 | 0.4883 | 0.4517 | 0.5279 | 1.54E−72 | 0.383 |
| 12 | rs1702877 | 5,60,34,024 | IKZF4 | T | 0.98704 | Imputed | 0.3276 | 6460 | 1.353 | 1.22 | 1.501 | 1.03E−08 | 0.3427 | 9465 | 1.366 | 1.282 | 1.455 | 3.59E−22 | 0.877 |
Italic values indicate the smallest P value.
Table 3.
Novel loci associated with low PRS T1D.
| CHR | BP (hg38) | SNP | dbSNP | allele | allele_Frq | Quality Score (r2) | Genotyped | n | OR | L95 | U95 | P | n | OR | L95 | U95 | P | Func.refGene | Gene.refGene |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low PRS cases versus low PRS controls | All cases versus all controls in the cohort | ||||||||||||||||||
| a. First cohort | |||||||||||||||||||
| 2 | 1,07,27,877 | chr2:10727877:C:T | rs147458998 | T | 0.00203 | 0.718 | Imputed | 6118 | 11.06 | 4.818 | 25.4 | 1.45E−08 | 9450 | 3.201 | 1.508 | 6.795 | 0.002454 | Intronic | ATP6V1C2 |
| 2 | 1,71,26,012 | chr2:17126012:C:T | rs56806432 | T | 0.03445 | 0.97878 | Imputed | 6118 | 1.936 | 1.539 | 2.437 | 1.74E−08 | 9450 | 1.149 | 0.976 | 1.352 | 0.09531 | Intergenic | FAM49A;RAD51AP2 |
| 2 | 1,71,59,712 | chr2:17159712:C:A | rs16983255 | A | 0.03454 | 0.97987 | Imputed | 6118 | 1.916 | 1.523 | 2.411 | 2.85E−08 | 9450 | 1.142 | 0.9704 | 1.343 | 0.1101 | Intergenic | FAM49A;RAD51AP2 |
| 2 | 1,71,59,927 | chr2:17159927:C:T | rs60975145 | T | 0.03448 | 0.9799 | Imputed | 6118 | 1.927 | 1.532 | 2.425 | 2.17E−08 | 9450 | 1.148 | 0.9755 | 1.351 | 0.09667 | Intergenic | FAM49A;RAD51AP2 |
| 2 | 5,15,13,429 | chr2:51513429:G:A | rs28958299 | A | 0.00609 | 0.98423 | Imputed | 6118 | 3.4 | 2.194 | 5.269 | 4.39E−08 | 9450 | 1.527 | 1.05 | 2.223 | 0.02693 | ncRNA_intronic | LOC730100 |
| 2 | 5,14,87,374 | chr2:51487374:C:CA | CA | 0.0063 | 0.98739 | Imputed | 6118 | 3.415 | 2.201 | 5.298 | 4.25E−08 | 9450 | 1.566 | 1.079 | 2.275 | 0.0184 | |||
| 2 | 5,15,04,453 | chr2:51504453:C:T | rs57623361 | T | 0.00667 | 0.98795 | Imputed | 6118 | 3.251 | 2.129 | 4.964 | 4.75E−08 | 9450 | 1.532 | 1.07 | 2.192 | 0.0198 | ncRNA_intronic | LOC730100 |
| 2 | 5,15,31,113 | chr2:51531113:T:A | rs28957091 | A | 0.00679 | 0.98915 | Imputed | 6118 | 3.408 | 2.239 | 5.188 | 1.07E−08 | 9450 | 1.611 | 1.124 | 2.308 | 0.009372 | ncRNA_intronic | LOC730100 |
| 2 | 5,15,28,593 | chr2:51528593:T:C | rs1406418 | C | 0.00716 | 0.99126 | Imputed | 6118 | 3.166 | 2.094 | 4.789 | 4.74E−08 | 9450 | 1.531 | 1.079 | 2.172 | 0.01703 | ncRNA_intronic | LOC730100 |
| 2 | 5,15,30,016 | chr2:51530016:T:G | rs28958318 | G | 0.0073 | 0.99201 | Imputed | 6118 | 3.205 | 2.122 | 4.84 | 3.10E−08 | 9450 | 1.579 | 1.113 | 2.24 | 0.01039 | ncRNA_intronic | LOC730100 |
| 2 | 5,15,20,595 | chr2:51520595:C:T | rs28957085 | T | 0.00601 | 0.99376 | Imputed | 6118 | 3.575 | 2.31 | 5.533 | 1.09E−08 | 9450 | 1.616 | 1.113 | 2.346 | 0.01167 | ncRNA_intronic | LOC730100 |
| 2 | 5,14,86,782 | chr2:51486782:AG:A | A | 0.00593 | 0.99402 | Imputed | 6118 | 3.445 | 2.213 | 5.364 | 4.30E−08 | 9450 | 1.476 | 1.01 | 2.156 | 0.04423 | ncRNA_intronic | LOC730100 | |
| 2 | 5,15,25,890 | chr2:51525890:A:G | rs1528792 | G | 0.00684 | 0.99484 | Imputed | 6118 | 3.27 | 2.159 | 4.951 | 2.18E−08 | 9450 | 1.529 | 1.075 | 2.174 | 0.01814 | ncRNA_intronic | LOC730100 |
| 2 | 5,15,28,054 | chr2:51528054:T:C | rs28957087 | C | 0.00735 | 0.99609 | Imputed | 6118 | 3.306 | 2.195 | 4.979 | 1.04E−08 | 9450 | 1.607 | 1.135 | 2.276 | 0.007525 | ncRNA_intronic | LOC730100 |
| 9 | 1,42,29,050 | chr9:14229050:G:C | rs10961435 | C | 0.00389 | 0.95755 | Imputed | 6118 | 4.762 | 2.753 | 8.237 | 2.39E−08 | 9450 | 1.806 | 1.103 | 2.957 | 0.01885 | Intronic | NFIB |
| 10 | 4,42,60,591 | chr10:44260591:C:T | rs746298 | T | 0.00446 | 0.98941 | Imputed | 6118 | 4.175 | 2.552 | 6.83 | 1.26E−08 | 9450 | 2.304 | 1.485 | 3.576 | 0.000197 | Intergenic | LINC00841;C10orf142 |
| 10 | 6,06,64,647 | chr10:60664647:T:C | rs1816797 | C | 0.00183 | 0.93498 | Imputed | 6118 | 8.974 | 4.087 | 19.7 | 4.55E−08 | 9450 | 4.188 | 2.04 | 8.599 | 9.52E−05 | Intronic | ANK3 |
| 12 | 3,46,99,338 | chr12:34699338:G:T | rs12424461 | T | 0.00231 | 0.85309 | Imputed | 6118 | 8.528 | 4.101 | 17.73 | 9.54E−09 | 9450 | 2.127 | 1.105 | 4.094 | 0.02396 | Intergenic | ALG10;NONE |
| b. Switched cohort | |||||||||||||||||||
| 2 | 6,24,48,472 | chr2:62448472:C:T | rs78389245 | T | 0.00223 | 0.93017 | Imputed | 6460 | 6.453 | 3.353 | 12.42 | 2.37E−08 | 9465 | 2.246 | 1.248 | 4.041 | 0.006953 | Intergenic | B3GNT2;TMEM17 |
| 2 | 6,24,42,411 | chr2:62442411:A:G | rs75610843 | G | 0.00349 | 0.96936 | Imputed | 6460 | 5.023 | 2.83 | 8.917 | 3.54E−08 | 9465 | 2.114 | 1.278 | 3.498 | 0.003576 | Intergenic | B3GNT2;TMEM17 |
| 2 | 6,24,46,036 | chr2:62446036:C:T | rs76505469 | T | 0.0031 | 0.96962 | Imputed | 6460 | 5.655 | 3.117 | 10.26 | 1.20E−08 | 9465 | 1.959 | 1.156 | 3.319 | 0.01245 | Intergenic | B3GNT2;TMEM17 |
| 2 | 6,24,40,366 | chr2:62440366:C:T | rs75921605 | T | 0.00346 | 0.97057 | Imputed | 6460 | 5.023 | 2.83 | 8.917 | 3.54E−08 | 9465 | 2.114 | 1.278 | 3.498 | 0.003576 | Intergenic | B3GNT2;TMEM17 |
| 2 | 7,03,52,234 | chr2:70352234:A:G | rs116081627 | G | 0.002 | 0.91477 | Imputed | 6460 | 7.679 | 3.755 | 15.7 | 2.34E−08 | 9465 | 1.911 | 1 | 3.651 | 0.05001 | Intergenic | FAM136A;TGFA |
| 3 | 4,89,51,134 | chr3:48951134:T:C | rs143836109 | C | 0.00397 | 0.58298 | Imputed | 6460 | 4.986 | 2.801 | 8.875 | 4.77E−08 | 9465 | 1.782 | 1.059 | 2.999 | 0.02969 | Intronic | ARIH2 |
| 4 | 8,83,57,077 | chr4:88357077:T:C | rs76377119 | C | 0.002 | 0.71507 | Imputed | 6460 | 9.545 | 4.389 | 20.76 | 1.26E−08 | 9465 | 4.076 | 1.927 | 8.622 | 0.000237 | Intergenic | LOC105369192;HERC6 |
| 4 | 14,15,56,946 | chr4:141556946:C:T | rs72615940 | T | 0.00275 | 0.95975 | Imputed | 6460 | 6.361 | 3.301 | 12.26 | 3.25E−08 | 9465 | 2.651 | 1.469 | 4.784 | 0.001204 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,792 | chr4:141557792:T:C | rs115762557 | C | 0.00279 | 0.96694 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,58,035 | chr4:141558035:A:C | rs72615945 | C | 0.00278 | 0.96769 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,58,497 | chr4:141558497:G:GATTTTCA | GATTTTCA | 0.00278 | 0.96769 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | |||
| 4 | 14,15,58,842 | chr4:141558842:C:T | rs72615950 | T | 0.00278 | 0.96771 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,59,253 | chr4:141559253:G:A | rs72615951 | A | 0.00278 | 0.96771 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,59,260 | chr4:141559260:G:A | rs72615952 | A | 0.00278 | 0.96771 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,845 | chr4:141557845:T:A | rs77083164 | A | 0.00278 | 0.96773 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,678 | chr4:141557678:A:G | rs75736694 | G | 0.00278 | 0.96774 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,58,749 | chr4:141558749:G:A | rs72615948 | A | 0.00278 | 0.96774 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,600 | chr4:141557600:A:G | rs72615942 | G | 0.00278 | 0.96775 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,58,193 | chr4:141558193:G:C | rs72615946 | C | 0.00278 | 0.96775 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,58,199 | chr4:141558199:C:T | rs72615947 | T | 0.00278 | 0.96775 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,291 | chr4:141557291:T:C | rs72615941 | C | 0.00278 | 0.96776 | Imputed | 6460 | 6.046 | 3.166 | 11.54 | 4.96E−08 | 9465 | 2.474 | 1.368 | 4.475 | 0.002725 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,56,501 | chr4:141556501:C:T | rs17007424 | T | 0.00256 | 0.97308 | Imputed | 6460 | 6.361 | 3.301 | 12.26 | 3.25E−08 | 9465 | 2.454 | 1.35 | 4.462 | 0.003252 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,56,393 | chr4:141556393:A:G | rs72615939 | G | 0.00257 | 0.97583 | Imputed | 6460 | 6.361 | 3.301 | 12.26 | 3.25E−08 | 9465 | 2.454 | 1.35 | 4.462 | 0.003252 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,56,881 | chr4:141556881:C:T | rs200157898 | T | 0.00257 | 0.97584 | Imputed | 6460 | 6.361 | 3.301 | 12.26 | 3.25E−08 | 9465 | 2.454 | 1.35 | 4.462 | 0.003252 | Intergenic | LINC02432;IL15 |
| 4 | 14,15,57,024 | chr4:141557024:TACTC:T | T | 0.00257 | 0.97591 | Imputed | 6460 | 6.361 | 3.301 | 12.26 | 3.25E−08 | 9465 | 2.454 | 1.35 | 4.462 | 0.003252 | Intergenic | LINC02432;IL15 | |
| 5 | 15,94,80,234 | chr5:159480234:C:T | rs117952033 | T | 0.00138 | 0.83707 | Imputed | 6460 | 14.31 | 5.602 | 36.55 | 2.68E−08 | 9465 | 4.707 | 1.862 | 11.9 | 0.001062 | Intergenic | LINC01845;LINC01847 |
| 5 | 15,94,99,811 | chr5:159499811:C:T | rs10515798 | T | 0.00148 | 0.84382 | Imputed | 6460 | 14.31 | 5.602 | 36.55 | 2.68E−08 | 9465 | 4.707 | 1.862 | 11.9 | 0.001062 | Intergenic | LINC01845;LINC01847 |
| 6 | 17,02,83,857 | chr6:170283857:C:T | rs3734776 | T | 0.00695 | 0.93794 | Imputed | 6460 | 3.602 | 2.351 | 5.518 | 3.87E−09 | 9465 | 1.912 | 1.334 | 2.741 | 0.000422 | Exonic | DLL1 |
| 6 | 17,02,88,217 | chr6:170288217:C:T | rs3818115 | T | 0.00634 | 0.93938 | Imputed | 6460 | 3.633 | 2.336 | 5.649 | 1.02E−08 | 9465 | 1.819 | 1.255 | 2.638 | 0.001589 | Intronic | DLL1 |
| 6 | 17,02,84,744 | chr6:170284744:C:T | rs2273214 | T | 0.00632 | 0.94094 | Imputed | 6460 | 3.633 | 2.336 | 5.649 | 1.02E−08 | 9465 | 1.819 | 1.255 | 2.638 | 0.001589 | Intronic | DLL1 |
| 6 | 17,02,90,297 | chr6:170290297:C:T | rs3823301 | T | 0.00644 | 0.94127 | Imputed | 6460 | 3.639 | 2.356 | 5.619 | 5.74E−09 | 9465 | 1.791 | 1.241 | 2.585 | 0.001843 | UTR5 | DLL1 |
| 6 | 17,02,87,112 | chr6:170287112:A:C | rs3800238 | C | 0.00632 | 0.94131 | Imputed | 6460 | 3.633 | 2.336 | 5.649 | 1.02E−08 | 9465 | 1.819 | 1.255 | 2.638 | 0.001589 | Intronic | DLL1 |
| 6 | 17,02,87,178 | chr6:170287178:C:A | rs3800237 | A | 0.00861 | 0.94352 | Imputed | 6460 | 3.085 | 2.12 | 4.49 | 3.98E−09 | 9465 | 1.543 | 1.126 | 2.115 | 0.007003 | Intronic | DLL1 |
| 7 | 7,20,10,707 | chr7:72010707:G:C | rs118182411 | C | 0.00376 | 0.93903 | Imputed | 6460 | 4.074 | 2.474 | 6.71 | 3.42E−08 | 9465 | 1.513 | 0.9562 | 2.393 | 0.07696 | Intronic | CALN1 |
| 7 | 8,93,18,777 | chr7:89318777:C:T | rs77205087 | T | 0.00582 | 0.9747 | Imputed | 6460 | 3.447 | 2.238 | 5.308 | 1.94E−08 | 9465 | 1.321 | 0.8943 | 1.951 | 0.1619 | Intronic | ZNF804B |
| 7 | 8,93,09,079 | chr7:89309079:C:T | rs76060515 | T | 0.00578 | 0.97724 | Imputed | 6460 | 3.447 | 2.238 | 5.308 | 1.94E−08 | 9465 | 1.321 | 0.8943 | 1.951 | 0.1619 | Intronic | ZNF804B |
| 7 | 10,08,14,253 | chr7:100814253:G:A | rs3890144 | A | 0.00757 | 0.95297 | Imputed | 6460 | 3.061 | 2.066 | 4.537 | 2.47E−08 | 9465 | 1.352 | 0.959 | 1.906 | 0.0853 | Intronic | EPHB4 |
| 7 | 10,07,99,808 | chr7:100799808:C:T | rs60224425 | T | 0.00756 | 0.95354 | Imputed | 6460 | 3.021 | 2.043 | 4.467 | 3.01E−08 | 9465 | 1.403 | 1 | 1.966 | 0.04969 | Intergenic | ZAN;EPHB4 |
| 9 | 11,02,69,080 | chr9:110269080:T:G | rs10816957 | G | 0.00712 | 0.94616 | Imputed | 6460 | 3.054 | 2.063 | 4.52 | 2.44E−08 | 9465 | 1.306 | 0.9282 | 1.838 | 0.1254 | Intergenic | TXN;TXNDC8 |
| 12 | 3,36,97,680 | chr12:33697680:T:C | rs11052850 | C | 0.00691 | 0.99026 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,97,996 | chr12:33697996:G:A | rs1352395 | A | 0.0069 | 0.9906 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,95,324 | chr12:33695324:G:A | rs12228218 | A | 0.00689 | 0.9914 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,80,534 | chr12:33680534:G:A | rs4142676 | A | 0.00691 | 0.99149 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,93,014 | chr12:33693014:T:C | rs11052843 | C | 0.00689 | 0.99193 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,97,208 | chr12:33697208:T:C | rs2087269 | C | 0.00689 | 0.992 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 3,36,95,992 | chr12:33695992:G:T | rs11052847 | T | 0.00689 | 0.99201 | Imputed | 6460 | 2.758 | 1.918 | 3.966 | 4.37E−08 | 9465 | 1.131 | 0.8173 | 1.565 | 0.4581 | Intergenic | SYT10;ALG10 |
| 12 | 13,29,15,197 | chr12:132915197:G:A | rs11147161 | A | 0.00503 | 0.96369 | Imputed | 6460 | 3.768 | 2.344 | 6.059 | 4.36E−08 | 9465 | 1.366 | 0.8825 | 2.115 | 0.1617 | Intergenic | LOC101928530;ZNF605 |
| 12 | 13,28,98,032 | chr12:132898032:A:C | rs12230138 | C | 0.00501 | 0.96567 | Imputed | 6460 | 3.899 | 2.436 | 6.239 | 1.41E−08 | 9465 | 1.411 | 0.9143 | 2.177 | 0.1199 | Intergenic | LOC101928530;ZNF605 |
| 15 | 6,97,78,062 | chr15:69778062:T:C | rs75516437 | C | 0.00621 | 0.64954 | Imputed | 6460 | 3.937 | 2.498 | 6.207 | 3.60E−09 | 9465 | 1.559 | 1.033 | 2.353 | 0.03452 | Intergenic | PCAT29;LINC00593 |
| 15 | 9,23,50,924 | chr15:92350924:T:C | T | 0.02086 | 0.30905 | Imputed | 6460 | 3.36 | 2.177 | 5.186 | 4.45E−08 | 9465 | 1.432 | 0.9822 | 2.089 | 0.06196 | Intergenic | SLCO3A1;ST8SIA2 | |
| 20 | 5,50,80,179 | chr20:55080179:G:A | rs144851111 | A | 0.00386 | 0.30246 | Imputed | 6460 | 8.283 | 4.084 | 16.8 | 4.62E−09 | 9465 | 2.649 | 1.352 | 5.191 | 0.00452 | Intergenic | DOK5;LINC01441 |
| 21 | 2,81,70,508 | chr21:28170508:A:C | rs2831598 | C | 0.00548 | 0.85895 | Imputed | 6460 | 3.812 | 2.421 | 6.001 | 7.48E−09 | 9465 | 1.972 | 1.34 | 2.903 | 0.000575 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,64,783 | chr21:28164783:C:T | rs139600098 | T | 0.00601 | 0.86242 | Imputed | 6460 | 3.641 | 2.37 | 5.594 | 3.69E−09 | 9465 | 1.825 | 1.265 | 2.631 | 0.001282 | ncRNA_intronic | LINC01695 |
| 21 | 2,84,29,678 | chr21:28429678:G:A | rs145901638 | A | 0.0053 | 0.8642 | Imputed | 6460 | 4.115 | 2.594 | 6.527 | 1.86E−09 | 9465 | 1.829 | 1.229 | 2.722 | 0.002898 | Intergenic | LINC01695;LINC00161 |
| 21 | 2,81,65,396 | chr21:28165396:C:A | rs140035545 | A | 0.00619 | 0.86512 | Imputed | 6460 | 3.714 | 2.449 | 5.631 | 6.48E−10 | 9465 | 1.887 | 1.322 | 2.694 | 0.000473 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,65,671 | chr21:28165671:C:G | rs2831578 | G | 0.0066 | 0.86537 | Imputed | 6460 | 3.54 | 2.359 | 5.312 | 1.04E−09 | 9465 | 1.789 | 1.265 | 2.531 | 0.001014 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,65,241 | chr21:28165241:T:A | rs138282731 | A | 0.00622 | 0.86591 | Imputed | 6460 | 3.714 | 2.449 | 5.631 | 6.48E−10 | 9465 | 1.887 | 1.322 | 2.694 | 0.000473 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,67,027 | chr21:28167027:G:A | rs2831584 | A | 0.006 | 0.87016 | Imputed | 6460 | 3.653 | 2.385 | 5.596 | 2.61E−09 | 9465 | 1.878 | 1.306 | 2.699 | 0.000666 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,68,119 | chr21:28168119:C:A | rs2831588 | A | 0.00661 | 0.8744 | Imputed | 6460 | 3.311 | 2.188 | 5.01 | 1.47E−08 | 9465 | 1.746 | 1.235 | 2.468 | 0.001598 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,70,454 | chr21:28170454:C:T | rs2831597 | T | 0.00518 | 0.88171 | Imputed | 6460 | 4.405 | 2.798 | 6.935 | 1.52E−10 | 9465 | 2.218 | 1.501 | 3.277 | 6.37E−05 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,71,717 | chr21:28171717:A:G | rs57177980 | G | 0.00563 | 0.88918 | Imputed | 6460 | 3.752 | 2.382 | 5.908 | 1.15E−08 | 9465 | 1.922 | 1.314 | 2.809 | 0.000749 | ncRNA_intronic | LINC01695 |
| 21 | 2,84,06,331 | chr21:28406331:T:C | rs150168760 | C | 0.0048 | 0.89787 | Imputed | 6460 | 4.157 | 2.613 | 6.614 | 1.80E−09 | 9465 | 1.735 | 1.151 | 2.614 | 0.00844 | Intergenic | LINC01695;LINC00161 |
| 21 | 2,81,87,913 | chr21:28187913:T:C | rs10482989 | C | 0.00602 | 0.93268 | Imputed | 6460 | 3.599 | 2.324 | 5.572 | 9.50E−09 | 9465 | 2.024 | 1.405 | 2.916 | 0.000155 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,89,835 | chr21:28189835:CT:C | C | 0.00604 | 0.93481 | Imputed | 6460 | 3.599 | 2.324 | 5.572 | 9.50E−09 | 9465 | 2.024 | 1.405 | 2.916 | 0.000155 | ncRNA_intronic | LINC01695 | |
| 21 | 2,81,99,184 | chr21:28199184:A:T | rs28883424 | T | 0.00564 | 0.9351 | Imputed | 6460 | 3.761 | 2.405 | 5.881 | 6.35E−09 | 9465 | 2.012 | 1.384 | 2.925 | 0.000252 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,90,781 | chr21:28190781:TAC:T | T | 0.00502 | 0.93737 | Imputed | 6460 | 4 | 2.524 | 6.338 | 3.58E−09 | 9465 | 1.999 | 1.354 | 2.951 | 0.000494 | ncRNA_intronic | LINC01695 | |
| 21 | 2,81,99,308 | chr21:28199308:A:G | rs8130449 | G | 0.00605 | 0.9382 | Imputed | 6460 | 3.599 | 2.324 | 5.572 | 9.50E−09 | 9465 | 2.024 | 1.405 | 2.916 | 0.000155 | ncRNA_intronic | LINC01695 |
| 21 | 2,81,97,976 | chr21:28197976:C:T | rs59067393 | T | 0.00502 | 0.93881 | Imputed | 6460 | 4 | 2.524 | 6.338 | 3.58E−09 | 9465 | 1.999 | 1.354 | 2.951 | 0.000494 | ncRNA_intronic | LINC01695 |
| 21 | 2,82,03,693 | chr21:28203693:G:A | rs78202304 | A | 0.00578 | 0.95837 | Imputed | 6460 | 3.82 | 2.454 | 5.948 | 2.97E−09 | 9465 | 2.141 | 1.478 | 3.101 | 5.69E−05 | ncRNA_intronic | LINC01695 |
| 21 | 2,82,25,654 | chr21:28225654:C:T | rs73897628 | T | 0.00536 | 0.96681 | Imputed | 6460 | 3.635 | 2.309 | 5.721 | 2.45E−08 | 9465 | 1.739 | 1.185 | 2.551 | 0.004699 | ncRNA_intronic | LINC01695 |
| 21 | 2,83,52,351 | chr21:28352351:G:T | rs7278151 | T | 0.00874 | 0.96719 | Imputed | 6460 | 2.909 | 2.029 | 4.172 | 6.37E−09 | 9465 | 1.379 | 1.015 | 1.873 | 0.04014 | Intergenic | LINC01695;LINC00161 |
| 21 | 2,82,06,978 | chr21:28206978:G:A | rs144947925 | A | 0.00515 | 0.97068 | Imputed | 6460 | 3.938 | 2.507 | 6.185 | 2.66E−09 | 9465 | 1.939 | 1.323 | 2.842 | 0.000692 | ncRNA_intronic | LINC01695 |
| 21 | 2,82,06,642 | chr21:28206642:G:C | rs16997642 | C | 0.00558 | 0.97287 | Imputed | 6460 | 3.866 | 2.484 | 6.019 | 2.12E−09 | 9465 | 2.031 | 1.4 | 2.946 | 0.000189 | ncRNA_intronic | LINC01695 |
| 21 | 2,82,22,390 | chr21:28222390:T:C | rs2831662 | C | 0.00587 | 0.97928 | Imputed | 6460 | 3.606 | 2.318 | 5.611 | 1.30E−08 | 9465 | 1.725 | 1.193 | 2.496 | 0.003782 | ncRNA_intronic | LINC01695 |
| 21 | 2,83,36,083 | chr21:28336083:T:C | rs73897689 | C | 0.00493 | 0.98436 | Imputed | 6460 | 3.822 | 2.406 | 6.072 | 1.38E−08 | 9465 | 1.792 | 1.208 | 2.658 | 0.003747 | Intergenic | LINC01695;LINC00161 |
| 21 | 2,83,37,534 | chr21:28337534:T:C | rs73897690 | C | 0.00489 | 0.98684 | Imputed | 6460 | 3.822 | 2.406 | 6.072 | 1.38E−08 | 9465 | 1.807 | 1.217 | 2.683 | 0.003374 | Intergenic | LINC01695;LINC00161 |
| 21 | 3,45,50,989 | chr21:34550989:C:T | rs140276394 | T | 0.0032 | 0.50721 | Imputed | 6460 | 6.028 | 3.245 | 11.2 | 1.31E−08 | 9465 | 1.888 | 1.025 | 3.478 | 0.04147 | Intronic | RCAN1 |
Replication of the PRS model and additional novel loci
Consequently, we switched the two cohorts, i.e. using the second cohort for the statistics of PRS modelling, then we tested the PRS models in the first cohort. The AUCs of different cutoffs of T1D association P values for selection of SNP sets are shown in Table 1b. The best AUC (0.8654) is seen at the cutoff of P value ≤ 1E−05, which repeated the PRS model in the above step. Based on the SNP markers with T1D association P value ≤ 1E−05, a PRS score was acquired for each individual in the independent test cohort. By the maximum MCC (Supplementary Table 3), a PRS cutoff of 7.18E−04 has the maximum MCC (0.6294). A PRS ≤ 7.18E−04 was defined as low risk, and a PRS > 7.18E−04 was defined as high risk. With this threshold, the sensitivity (True positive rate, TPR) for T1D prediction is 66.0%, and the specificity (True negative rate, TFR) for T1D prediction is 93.6%. By PRS ≤ 7.18E−04, 907 (27.5%, including 433 males, 472 females, and 2 cases with undetermined sex) out of 3302 T1D cases had low PRS; and 5567 (90.1%, including 2997 males, 2558 females, and 12 cases with undetermined sex) out of 6181 controls had low PRS.
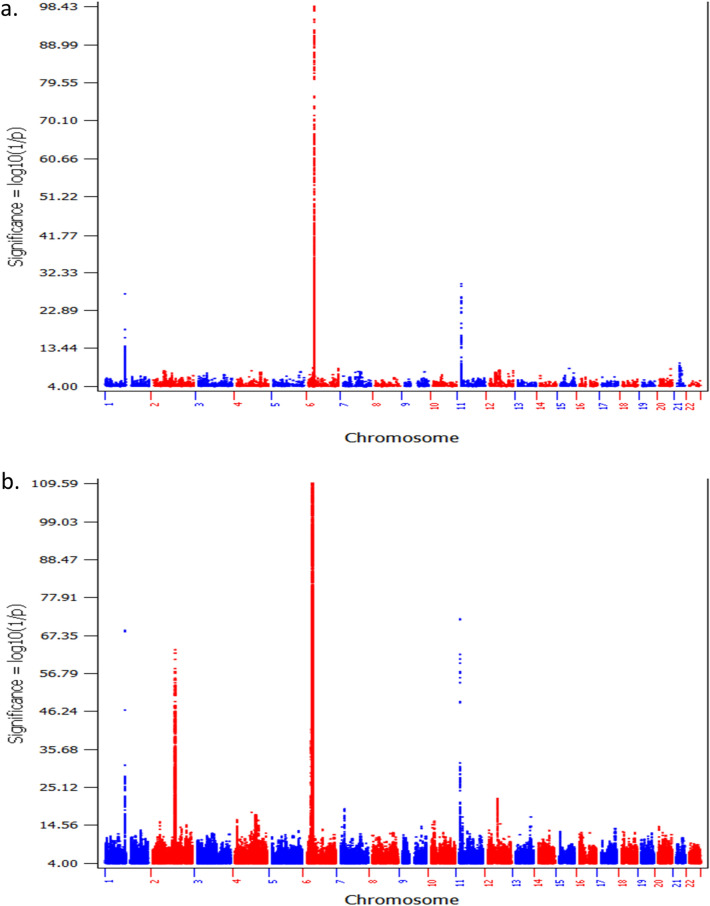
As expected from the above results, in the switched cohort, the GWAS of T1D patients with low T1D PRS compared to controls with low T1D PRS identified a large number of SNPs associated with T1D with genome-wide significance (P ≤ 5.0 × E−08) as well (Supplementary Table 4, Fig. 3). Among these loci, 4 loci have been established of T1D association by previous studies, including HLA, INS, PTPN22, IKZF4/RPS26/ERBB3, and the locus (Table 2b). Consistent to the first GWAS results listed above, by looking at the established leading T1D signal of each locus, the frequencies of the predisposing alleles of HLA, PTPN22 and IKZF4 were lower in the low T1D PRS cohort, while the protective allele of INS were higher in the low T1D PRS cohort. The effect size of the leading HLA SNP was significantly smaller in the low PRS cases (P = 5.49E−11). Besides these established T1D loci, 18 loci associated with low PRS T1D were identified in this cohort (Table 3b). LocusZoom plots for genetic association signals within each locus are shown in Supplementary Figures 8–25. Among the 18 loci, 16 loci are novel, while the Notch ligand Delta-like 1 (DLL1) locus, with the gene function essential for pancreatic islet homeostasis23, have been identified by our gene-based association study on low PRS T1D24. The other locus, containing the UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 gene (B3GNT2) and transmembrane protein 17 gene (TMEM17), is ~ 200 kb from the Eps15 homology domain binding protein 1 locus (EHBP1) that has been identified of genome-wide significance in our study on low T1D GRS patients21.
Figure 3.
The Manhattan plots of cohort A. (a) The plot of the GWAS of T1D patients with low T1D PRS compared to controls with low T1D PRS (907 cases vs. 5567 controls); (b) the plot of the GWAS of all T1D patients compared to all controls (3302 cases vs. 6181 controls).
Discussion
Altogether, rare variants (MAF < 5%) from 22 novel loci were identified in the low PRS T1D cases with genome-wide significance (P < 5.00E−08), in addition to the 4 established T1D loci, 2 reported loci in the low GRS patients, and 1 locus by our gene-based study. The genome-wide significant association signals of these loci are only seen in low PRS T1D cases, but not in the T1D cases overall, thus were missed previously due to rare allele frequencies and diluted genetic effects in the general T1D cohort. A number of genetic associations with body mass index (BMI), obesity, and autoimmunity, have been reported in the flanking regions of 300 kb on each side of these new loci according to the GWAS Catalog (https://www.ebi.ac.uk/gwas/, Supplementary materials for review). Further discussion on these novel loci is focused on 13 loci with high imputation quality (i.e. Quality Score r2 > 0.9). Among these loci, 9 loci are related to obesity traits (T1bD mechanism), 2 loci are related to glucose homeostasis (T1bD mechanism), and 2 loci are related to autoimmunity (T1aD mechanism).
Obesity-related/ T1bD-related loci
FAM49A/RAD51AP2 tagged by rs56806432
Two coding genes in this locus are the CYFIP related Rac1 interactor A gene (CYRIA) and the RAD51 associated protein 2 gene (RAD51AP2). CYRIA is highly expressed in brain and thyroid, while RAD51AP2 has restricted expression toward testis25. Previous GWAS has identified association of this locus with subcutaneous adipose tissue26.
NFIB tagged by rs10961435
The nuclear factor I B gene (NFIB) encodes a transcription factor in the FOXA1 transcription factor network. NFIB has been shown to play critical roles in lung and brain development. A previous study has shown that NFIB can bind with FoxA1 and modulate the transcriptional activity of FoxA127, while the later has been suggested to play a role in pancreatic and ß-cell function and non-autoimmune diabetes as discussed above. The nuclear factor I B gene (NFIB) has ubiquitous expression in fat, brain, and other tissues25. This locus has been reported of association with BMI by several GWA studies28–30.
LINC00841/C10orf142 tagged by rs746298
The two genes at this locus, LINC00841/C10orf142, encode two long intergenic non-protein coding RNAs (lincRNA). While the function of these two genes remain unknown, this locus has been reported of association with obesity-related traits31.
FAM136A/TGFA tagged by rs77418738
The family with sequence similarity 136 member A gene (FAM136A) encodes a mitochondrially localized protein32. The transforming growth factor alpha gene (TGFA) mediates cell–cell adhesion and activates cell proliferation, differentiation and development33. This region has been reported of association with obesity-related traits31.
CALN1 tagged by rs118182411
The calneuron 1 gene (CALN1), encoding a protein with high similarity to the calcium-binding proteins of calmodulin, is highly expressed in brain and adrenal25. This genetic region has established association with BMI by previous studies29,30.
EPHB4 tagged by rs3890144
The EPH receptor B4 gene (EPHB4) has ubiquitous expression in multiple tissues, and is involved in numerous developmental processes34. EPHB4 plays critical roles in vascular development35 and lymphatic valve development36. Previous GWAS has identified association of this locus with BMI37 and waist circumference adjusted for BMI30.
TXN/TXNDC8 tagged by rs10816957
The thioredoxin gene (TXN) has ubiquitous expression in multiple tissues, while the thioredoxin domain containing 8 gene (TXNDC8) has restricted expression toward testis25. Thioredoxin plays a protective role against oxidative stresses38. Thioredoxin interacting protein (TXNIP) has been implicated in β cells death in diabetes and is a novel potential therapeutic target of diabetes39. Previous GWAS has identified association of this locus with BMI40 and waist-to-hip ratio adjusted for BMI30.
SYT10/ALG10 tagged by rs4142676
The synaptotagmin 10 gene (SYT10) encodes a membrane protein of secretory vesicles expressed in pancreas, lung and kidney41. The ALG10 alpha-1,2-glucosyltransferase gene (ALG10) encodes a membrane-associated protein that adds the third glucose residue to the lipid-linked oligosaccharide precursor for N-glycosylation in endoplasmic reticulum (ER)42. As discussed above in the ZNF804B locus, N-glycosylation of IgG, cytokines and proteases is also a regulatory mechanism in inflammation and autoimmunity43,44 associated with different autoimmune diseases. Several previous GWASs have identified association of this locus with waist-to-hip ratio and waist-to-hip ratio adjusted for BMI30,45.
CHFR/LOC101928530/ZNF605 tagged by rs12230138
The checkpoint with forkhead and ring finger domains gene (CHFR) encodes an E3 ubiquitin-protein ligase and is involved in the DNA damage response and checkpoint regulation46. The structure and function of the gene LOC101928530 is still uncharacterized. The function of the zinc finger protein 605 gene (ZNF605) may be related to Herpes Simplex Virus 1 infection (https://pathcards.genecards.org/card/herpes_simplex_virus_1_infection). This region has been reported of association with BMI by previous study28.
Genetic loci related to glucose homeostasis (T1bD-related)
LOC730100 tagged by rs28957087
LOC730100 encodes a long non-coding RNA (ncRNA), a competing endogenous RNA for human microRNA 760 (miR-760)47. The latter inhibits the expression of the Forkhead Box A1 gene (FOXA1). As a hepatocyte nuclear factor, FOXA1, also known as HNF3A or TCF3A, regulates tissue-specific gene expression in liver and many other tissues48. FoxA1 is essential for normal pancreatic and ß-cell function and a negative regulator of the hepatocyte nuclear factor-1 (HNF1) homeobox A gene (HNF1A) and the hepatocyte nuclear factor 4, alpha gene (HNF4A)49,50. HNF1A and HNF4A are established genes causing maturity-onset diabetes of the young (MODY). The FOXA1 mutation Ser448Asn has been suggested of association with impaired glucose homeostasis50.
LINC01695/LINC00161 tagged by rs2831597
Function of the long intergenic non-protein coding RNA 1695 gene (LINC01695) is still uncharacterized. The long intergenic non-protein coding RNA 161 gene (LINC00161) encodes a functional RNA that regulates Mitogen-activated protein kinase 1 (MAPK1) expression51. The MAPK1/STAT3 pathway has been proposed as a novel diabetes target for its critical role in glucose homeostasis52.
Autoimmune-related loci
In addition to the above ALG10 locus associated with both autoimmune diseases and obesity-related traits, two other loci were identified in the low PRS T1D cases. The rare variants in these loci may represent rare forms of autoimmune diabetes with low T1D PRS53.
LINC02432/IL15 tagged by rs9790756
The long intergenic non-protein coding RNA 2432 gene (LINC02432) has higher expression in kidney and pancreas25. Interleukin 15 (IL-15) encoded by the gene IL15 is essential for regulating activation and proliferation of T and natural killer cells, and supporting lymphoid homeostasis54. IL-15 and interleukin 2 (IL-2) share many biological activities and receptor components with IL-255. IL-2 is a powerful growth factor for both T and B lymphocytes56. Both IL2 and the α chain of the IL2 receptor complex gene (IL2RA) has been established of genetic association with T1D by previous studies57–59.
ZNF804B tagged by rs76060515
The zinc finger protein 804B gene (ZNF804B) has been reported of association with N-linked glycosylation of human immunoglobulin G (IgG), which modulates its binding to Fc receptors43. N-glycosylation of cytokines and proteases is also a regulatory mechanism in inflammation and autoimmunity44. Changes in N-glycosylation have been associated with different autoimmune diseases, including rheumatoid arthritis60, type 1 diabetes61, Crohn's disease62.
In summary, in the genetic regions containing the 13 novel loci with high imputation quality disclosed by this study, 9 of these regions have been reported of association with obesity-related traits, BMI, or waist circumference. The correlation with obesity related traits or impaired glucose homeostasis is in keeping with non-autoimmune roles in the diabetes patients with low T1D PRS. Interestingly, the genes ZNF804B and ALG10 related to N-linked glycosylation are highlighted in this study, which may suggest the role of N-glycosylation in impaired glucose homeostasis and pediatric diabetes, while N-glycosylation is commonly altered in diabetes63. In addition, 3 loci encoding long intergenic non-protein coding RNAs (lncRNA) identified in this study emphasize the importance of lncRNAs in these diabetes patients. However, we admit that this study has limitations related to the bottleneck of sample size and data resources. The novel loci reported in this study still need replication in independent samples. In addition, the functional mechanisms of these genetic loci in diabetes warrant experimental investigation. Due to the lack of data of T1D autoantibodies in the subjects, the mixture of rare forms of autoimmune diabetes (e.g. monogenic autoimmunity53) in addition to non-autoimmune diabetes may exist as suggested by the identification of rare variants in autoimmune-related genes.
Supplementary Information
Acknowledgements
Dr. Hakon Hakonarson is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Author contributions
Conceptualization, H.H., and H.Q.Q.; literature search, J.Q. and H.Q.Q.; Figures and Tables, J.Q., and H.Q.Q.; data analysis, J.Q., H.Q.Q., and J.P.B.; data interpretation, J.Q., H.Q.Q., J.P.B., J.T.G., X.C., L.T., M.M., J.J.C., J.D.R., P.M.A.S., and H.H.; original draft writing, J.Q., H.Q.Q., and H.H.; review and revision, J.Q., H.Q.Q. and H.H.; supervision, H.H. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by Institutional Development Funds from the Children’s Hospital of Philadelphia to the Center for Applied Genomics and The Children’s Hospital of Philadelphia Endowed Chair in Genomic Research to HH.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94994-9.
References
- 1.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd JA, Bell JI, McDevitt HO. HLA-DQ[beta] gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329:599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 3.Baisch JM, et al. Analysis of HLA-DQ genotypes and susceptibility in insulin-dependent diabetes mellitus. N. Engl. J. Med. 1990;322:1836–1841. doi: 10.1056/NEJM199006283222602. [DOI] [PubMed] [Google Scholar]
- 4.Todd JA. Genetic analysis of type 1 diabetes using whole genome approaches. PNAS. 1995;92:8560–8565. doi: 10.1073/pnas.92.19.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble JA, et al. The role of HLA class II genes in insulin-dependent diabetes mellitus: Molecular analysis of 180 Caucasian, multiplex families. Am. J. Hum. Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 6.She J-X. Susceptibility to type I diabetes: HLA-DQ and DR revisited. Immunol. Today. 1996;17:323. doi: 10.1016/0167-5699(96)10014-1. [DOI] [PubMed] [Google Scholar]
- 7.Bell GI, Horita S, Karam JH. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33:176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 8.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: Advances and prospects. Nat. Rev. Genet. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 10.Rich SS, et al. The type 1 diabetes genetics consortium. Ann. N. Y. Acad. Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 11.Leslie RD, Atkinson MA, Notkins AL. Autoantigens IA-2 and GAD in type I (insulin-dependent) diabetes. Diabetologia. 1999;42:3–14. doi: 10.1007/s001250051105. [DOI] [PubMed] [Google Scholar]
- 12.Ehtisham S, Hattersley A, Dunger D, Barrett T. First UK survey of paediatric type 2 diabetes and MODY. Arch. Dis. Child. 2004;89:526–529. doi: 10.1136/adc.2003.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019;28:R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- 14.Hakonarson H, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 15.Bradfield JP, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated loci. PLoS Genet. 2011;7:e1002293. doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:s13742-015-0047-8. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi SW, O'Reilly PF. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience. 2019 doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Dvorkin D, Da Y. SNPEVG: A graphical tool for GWAS graphing with mouse clicks. BMC Bioinform. 2012;13:319. doi: 10.1186/1471-2105-13-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinform. (Oxf. Engl.) 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Diaz R, Caicedo A. Neural control of the endocrine pancreas. Best Pract. Res. Clin. Endocrinol. Metab. 2014;28:745–756. doi: 10.1016/j.beem.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Qu, H.-Q. et al. Genetic architecture of type 1 diabetes with low genetic risk score informed by 41 unreported loci. Commun Biol4, 908 (2021). [DOI] [PMC free article] [PubMed]
- 22.Sharp SA, et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care. 2019;42:200–207. doi: 10.2337/dc18-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubey M, et al. DLL1- and DLL4-mediated notch signaling is essential for adult pancreatic islet homeostasis. Diabetes. 2020;69:915–926. doi: 10.2337/db19-0795. [DOI] [PubMed] [Google Scholar]
- 24.Qu J, et al. Association of DLL1 with type 1 diabetes in patients characterized by low polygenic risk score. Metab. Clin. Exp. 2020 doi: 10.1016/j.metabol.2020.154418. [DOI] [PubMed] [Google Scholar]
- 25.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. MCP. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox CS, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boachie AM, et al. Abstract 1231: Nuclear Factor I family members interact with FoxA1 to regulate androgen responsive promoters. Can. Res. 2010;70:1231–1231. doi: 10.1158/1538-7445.am10-1231. [DOI] [Google Scholar]
- 28.Kichaev G, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulit SL, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2020;145:537–549. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comuzzie AG, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS ONE. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Requena T, et al. Identification of two novel mutations in FAM136A and DTNA genes in autosomal-dominant familial Meniere's disease. Hum. Mol. Genet. 2015;24:1119–1126. doi: 10.1093/hmg/ddu524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anklesaria P, et al. Cell-cell adhesion mediated by binding of membrane-anchored transforming growth factor alpha to epidermal growth factor receptors promotes cell proliferation. Proc. Natl. Acad. Sci. 1990;87:3289–3293. doi: 10.1073/pnas.87.9.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arvanitis D, Davy A. Eph/ephrin signaling: Networks. Genes Dev. 2008;22:416–429. doi: 10.1101/gad.1630408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erber R, et al. EphB4 controls blood vascular morphogenesis during postnatal angiogenesis. EMBO J. 2006;25:628–641. doi: 10.1038/sj.emboj.7600949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, et al. EphB4 forward signalling regulates lymphatic valve development. Nat. Commun. 2015;6:1–10. doi: 10.1038/ncomms7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turcot V, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: Current research with special reference to human disease. Biochem. Biophys. Res. Commun. 2010;396:120–124. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 39.Wondafrash DZ, et al. Thioredoxin-interacting protein as a novel potential therapeutic target in diabetes mellitus and its underlying complications. Diabetes Metab. Syndr. Obes. Targets Ther. 2020;13:43. doi: 10.2147/DMSO.S232221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boardman JD, et al. Is the gene-environment interaction paradigm relevant to genome-wide studies? The case of education and body mass index. Demography. 2014;51:119–139. doi: 10.1007/s13524-013-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao E, et al. Cloning and characterization of human synaptotagmin 10 gene. DNA Seq. J. DNA Seq. Mapp. 2003;14:393–398. doi: 10.1080/10425170310001608407. [DOI] [PubMed] [Google Scholar]
- 42.Burda P, Aebi M. The ALG10 locus of Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- 43.Lauc G, et al. Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 2013;9:e1003225. doi: 10.1371/journal.pgen.1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van den Steen P., Rudd P.M., Dwek R.A., Van Damme J., Opdenakker G. (1998) Cytokine and Protease Glycosylation as a Regulatory Mechanism in Inflammation and Autoimmunity. In: Axford J.S. (eds) Glycoimmunology 2. Advances in Experimental Medicine and Biology, vol 435. Springer, Boston, MA. [DOI] [PubMed]
- 45.Lotta LA, et al. Association of genetic variants related to gluteofemoral vs abdominal fat distribution with type 2 diabetes, coronary disease, and cardiovascular risk factors. JAMA. 2018;320:2553–2563. doi: 10.1001/jama.2018.19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Lu J, Xia J, Wen M, Wang C. Long non-coding RNA LOC730100 enhances proliferation and invasion of glioma cells through competitively sponging miR-760 from FOXA1 mRNA. Biochem. Biophys. Res. Commun. 2019;512:558–563. doi: 10.1016/j.bbrc.2019.03.124. [DOI] [PubMed] [Google Scholar]
- 48.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 49.Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science (N. Y.) 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- 50.Navas MA, et al. The human HNF-3 genes: Cloning, partial sequence and mutation screening in patients with impaired glucose homeostasis. Hum. Hered. 2000;50:370–381. doi: 10.1159/000022943. [DOI] [PubMed] [Google Scholar]
- 51.Xu M, Zhou K, Wu Y, Wang L, Lu S. Linc00161 regulated the drug resistance of ovarian cancer by sponging microRNA-128 and modulating MAPK1. Mol. Carcinog. 2019;58:577–587. doi: 10.1002/mc.22952. [DOI] [PubMed] [Google Scholar]
- 52.Kinoshita T, et al. Knowledge-based identification of the ERK2/STAT3 signal pathway as a therapeutic target for type 2 diabetes and drug discovery. Chem. Biol. Drug Des. 2011;78:471–476. doi: 10.1111/j.1747-0285.2011.01151.x. [DOI] [PubMed] [Google Scholar]
- 53.Johnson MB, et al. A type 1 diabetes genetic risk score can discriminate monogenic autoimmunity with diabetes from early-onset clustering of polygenic autoimmunity with diabetes. Diabetologia. 2018;61:862–869. doi: 10.1007/s00125-018-4551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burkett PR, et al. Coordinate expression and trans presentation of interleukin (IL)-15Rα and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J. Exp. Med. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giri JG, et al. IL-15, a novel T cell growth factor that shares activities and receptor components with IL-2. J. Leukoc. Biol. 1995;57:763–766. doi: 10.1002/jlb.57.5.763. [DOI] [PubMed] [Google Scholar]
- 56.Lowenthal JW, Zubler RH, Nabholz M, MacDonald HR. Similarities between interleukin-2 receptor number and affinity on activated B and T lymphocytes. Nature. 1985;315:669–672. doi: 10.1038/315669a0. [DOI] [PubMed] [Google Scholar]
- 57.Vella A, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu H-Q, Montpetit A, Ge B, Hudson TJ, Polychronakos C. Toward further mapping of the association between the IL2RA locus and type 1 diabetes. Diabetes. 2007;56:1174–1176. doi: 10.2337/db06-1555. [DOI] [PubMed] [Google Scholar]
- 59.Plagnol V, et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 2011;7:e1002216. doi: 10.1371/journal.pgen.1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa H, et al. Detection of altered N-glycan profiles in whole serum from rheumatoid arthritis patients. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007;853:133–137. doi: 10.1016/j.jchromb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Bermingham ML, et al. N-glycan profile and kidney disease in type 1 diabetes. Diabetes Care. 2018;41:79–87. doi: 10.2337/dc17-1042. [DOI] [PubMed] [Google Scholar]
- 62.Trbojević Akmačić I, et al. Inflammatory bowel disease associates with proinflammatory potential of the immunoglobulin G glycome. Inflamm. Bowel Dis. 2015;21:1237–1247. doi: 10.1097/mib.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudman N, Gornik O, Lauc G. Altered N-glycosylation profiles as potential biomarkers and drug targets in diabetes. FEBS Lett. 2019;593:1598–1615. doi: 10.1002/1873-3468.13495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.