Abstract
In clinical and experimental non-alcoholic steatohepatitis (NASH), the origin of the scar-forming myofibroblast is the hepatic stellate cell (HSC). We used foz/foz mice on a Western diet to characterize in detail the phenotypic changes of HSCs in a NASH model. We examined the single cell expression profiles (scRNA-Seq) of HSCs purified from the normal livers of foz/foz mice on a chow diet, in NASH with fibrosis of foz/foz mice on a Western diet, and in livers during regression of NASH after switching back to a chow diet. Selected genes were analyzed using immunohistochemistry, qRT-PCR, and shRNA-knockdown in primary mouse HSCs. Our analysis of the normal liver identified two distinct clusters of quiescent HSCs that correspond to their acinar position of either pericentral vein or periportal vein. The NASH livers had four distinct HSC clusters, including one representing the classic fibrogenic myofibroblast. The three other HSC clusters consisted of a proliferating cluster, an intermediate activated cluster, and an immune and inflammatory cluster. The livers with NASH regression had one cluster of inactivated HSCs, which was similar to, but distinct from, the quiescent HSCs.
Conclusion:
Analysis of scRNA-Seq in combination with an interrogation of previous studies has revealed an unanticipated heterogeneity of HSC phenotypes under normal and injured states.
Keywords: hepatic stellate cells, single-cell RNAseq, fibrosis, non-alcoholic steatohepatitis, regression
Chronic liver injury of any etiology produces inflammation with the subsequent activation of mesenchymal cells to become myofibroblasts. In turn, myofibroblasts secrete the extracellular matrix that produces the fibrous scar of liver fibrosis (1,2). In experimental liver fibrosis including chronic carbon tetrachloride, chronic ethanol intake, and non-alcoholic steatohepatitis (NASH), hepatic stellate cells activate to become myofibroblasts (3-5). Similarly, in human liver diseases, such as Hepatitis B (HBV), Hepatitis C (HCV), and NASH, the origin of myofibroblasts appear to be activated hepatic stellate cells (1,6,7). Other cell types such as mesothelial cells (important in capsular fibrosis) (8), portal fibroblasts (important in cholestatic fibrosis) (9), and fibrocytes (recruited to the inflamed liver) (10) make small contributions to hepatotoxic induced liver fibrosis. Cell fate mapping in the injured and recovered liver has identified three distinct HSC phenotypes, the quiescent HSC in the normal liver, the activated HSC in the fibrogenic liver, and the inactivated HSC in the liver with regression of fibrosis (3,4). Other proposed phenotypes have been less carefully studied, including inflammatory HSCs and senescent HSCs.
Recent reports have used single cell RNA-Seq to develop an atlas of liver cells in humans and mice from normal and diseased livers (5,11-17). These studies have started their analysis with either total liver cells, CD45+ liver cells, or mesenchymal liver cells. Although these atlases have described for the first time the breadth of liver cells, their experimental design did not lead to an in-depth phenotyping of HSCs. While some studies have reported that quiescent HSCs are a homogeneous population (5,14,15), others demonstrate different gene expression patterns along the hepatic acinar gradient (16). Activated HSCs in liver fibrosis models have been reported to show more heterogeneity. A single cell analysis of HSCs during regression of liver fibrosis has not been reported.
Because it closely resembles human NASH (18-20) (and manuscript in preparation), we used foz/foz mice on a Western diet to characterize in detail the phenotypic changes of HSCs in a NASH model. Because HSCs represent only a small percentage of the total hepatic cells in all conditions, we first purified HSCs prior to conducting our analysis (3,4,21). This approach has revealed heterogeneity in the HSC population under different experimental conditions. We examined the single cell expression profiles of HSCs in the normal liver of foz/foz mice on a chow diet, in NASH with fibrosis of foz/foz mice on a Western diet, and in livers during regression of NASH after switching back to a chow diet.
Experimental Procedures and Statistical Analysis
Animals:
Mice with the Alms1 11-base pair truncating mutation (foz/foz mice) on a C57BL6/J genetic background were kindly provided by Dr. Geoffrey C. Farrell, Australian National University Medical School (22). 6 weeks old male foz/foz mice were placed on a Western diet (WD) (AIN-76A; 40% fat, 15% protein, 44% carbohydrates based on caloric content; Test Diet) for 12 weeks to develop NASH-fibrosis (12 weeks WD group). Subsequently, the mice were either euthanized or were switched to a standard chow diet (12% fat, 23% protein, 65% carbohydrates based on caloric content) for an additional 8 weeks for the fibrosis regression study (regression group). An additional group of mice underwent 12 weeks of regression, to demonstrate no further changes were occurring at later time points. 6 weeks old male foz/foz mice (0-week WD group) and 30 weeks old male wild type C57BL6/J mice (24-week WT+Chow group), both on standard chow diet served as the baseline and age-matched control. All animals were maintained under specific pathogen free conditions in filter-topped cages on autoclaved food and water and under 12 light/12 dark cycle, in accordance with NIH guidelines for the use and care of live animals and approved by University of California, San Diego (UCSD) Institutional Animal Care and Use Committee, S07022.
Histology:
The left lateral lobe of mice livers was used for histology. Lobes were fixed in 10% neutral-buffered formalin for 48 hours, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) to evaluate gross morphology, and with Picro Sirius Red to determine fibrosis. For immunohistochemistry, the formalin-fixed paraffin-embedded (FFPE) tissue sections were incubated with primary anti-mouse desmin (Thermo Fisher Scientific, RB-9014-P0, 1:200 dilution), anti-mouse GABRA3 (Sigma-Aldrich, HPA000839; 1:200 dilution) and anti-mouse αSMA (Abcam, ab5694, 1:200 dilution) antibodies. HRP conjugated secondary antibody was obtained from Vector Immpress (anti-rabbit MP-7401). Immunoreactivity was visualized by 3,3'- diaminobenzidine tetrahydrochloride (DAB) staining (Vector Laboratories). Images of seven non-overlapping fields were randomly selected, imaged using Olympus microscope, and quantified using Image J. Quantifications, wherever applicable were normalized to the percentage non-steatotic area of the liver tissue sections.
Quantitative RT-PCR:
Total RNA was isolated using RNeasy columns (Qiagen, MD, USA). Quantitative RT-PCR was performed using a QuantStudio™ 5 Real-Time PCR system (Applied Biosystems). Total RNA was isolated from livers and from mHSCs using Trizol and Pure Link RNA Mini kit (Invitrogen). Expression level of selected genes was calculated and normalized to the housekeeping gene hypoxanthine guanine phosphoribosyl transferase (HPRT) using the ΔΔ CT method (Invitrogen). PCR primers were designed either using an online primer tool, Primer Depot or as previously reported (9). Primer sequences are provided in Table S1. The data are represented as either fold change vs 6 weeks old foz/foz mice on WD for 0-week as controls, or as relative expression to a housekeeping gene (Hprt), mean± SEM, *p < 0.05, ** p < 0.01, *** p < 0.001
HSCs isolation:
Mouse livers were perfused through the inferior vena cava with pronase (Roche 11-459-643-001) 0.33mg/ml 37 degree 10ml/mins for 5 minutes followed by collagenase D (Roche 1108882001) 0.67mg/ml degree 10ml/mins for 5 minutes, and then strained through a 100um mesh. The cell suspension was centrifuged at 50g for 1 minute to remove the hepatocytes, supernatant was collected and centrifuged at 800g for 7 minutes, supernatant was removed, cell pellet washed with GBSS-B buffer at 700g for 7 minutes. HSCs were further isolated by Nycodenz gradient centrifuge 2000g for 20 minutes without break. The HSC layer was then collected and washed with GBSS-b at 800g for 7 minutes.
Single cell primary analysis:
Gradient layers enriched for hepatic stellate cells were suspended in 0.04% BSA (Sigma-Aldrich: SRE0036-500ml), labeled with 7-AAD (BioLegend), and purified by FACS using a MoFlo Astrios to select “live” singlets with high UV auto-fluorescence (355 nm laser, 558/20 bandpass filter). Hepatic stellate cells were prepared as individual replicates from three mice per group. The preparations were split over two separate dates, 2 mice per group on one date and an additional individual replicate on a separate date. A total of 8,000 cells per biological replicate were targeted per replicate which, as per the manufacturer's protocol, was expected to yield 5,000 single-cell libraries per replicate. After sorting, the resulting ~5,000 cells were used for 10X Genomics 3’ Single Cell RNA-seq (Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1). The resulting scRNA-Seq libraries were balanced and pooled for simultaneous sequencing using a NOVASeq. Resulting FASTQ files were processed with Cell Ranger version 3.0 (23) and each library was mapped to the mm10 genome. The Seurat package (version 3.0) was used for merging read matrices, normalization, and clustering (24). Gene expression of each cell was normalized by total transcript counts and multiplied by 10,000. Expression data were subsequently log transformed and scaled. Cells of suspected low quality were removed, which had too few genes expressed (<500), which had high mitochondrial gene expression (>5%), or which had low Lrat expression, to exclude non-hepatic stellate cells (<0.5 normalized expression units). Following application of all filters 15,423 cells remained in the dataset. Visualization was performed with Uniform Manifold Approximation and Projection (UMAP) (25) dimensionality reduction and the Louvain algorithm was applied with a resolution of 0.5 to cluster the cells (see Table S2 for a description of all identified clusters). We include a link to download the Rdata file containing the raw expression, normalized expression, and cluster identities for the filtered cells: https://drive.google.com/file/d/1u4KX_oPGLkNG1epP0TrSV264sQdEOBzq/view.
Marker gene identification:
Marker genes were identified using the FindMarkers function in Seurat, with min.pct=0.5, using the MAST test option. Genes were classified as marker genes if they were significantly dysregulated (p<0.05) in a focal cluster compared to all other clusters. Top 50 positive marker genes, ranked by log fold change (i.e. those genes which are upregulated in a focal cluster, with Benjamini-Hochberg adjusted p<0.05, and log fold change >0) are included in Tables S3-S12.
Single cell marker gene correspondence with established HSC subtypes:
Cluster characterization was performed by aligning cluster marker genes with a previously published dataset of quiescent (qHSC), activated (aHSC), and inactivated (iHSC) hepatic stellate cells in a carbon tetrachloride (CCl4) mouse model (26). Genes were identified which were both marker genes for a focal cluster and which were differentially expressed in the CCl4 qHSC, aHSC, or iHSC data, with absolute value of the log fold change greater than 0.5. The CCl4 qHSC gene signature was defined by comparing the expression of qHSCs to both aHSCs and iHSCs, while the CCl4 aHSC signature was defined by comparing the expression of aHSCs to both qHSCs and iHSCs, and the CCl4 iHSC gene signature was defined by comparing the expression of iHSCs to both qHSCs and aHSCs. In this way we identified the genes which best define each HSC subtype. The log fold changes for these intersecting genes were then plotted as a scatterplot, with the x-axis representing the log fold change in the CCl4 experiment, and the y-axis representing the log fold change between cells in the focal cluster, and all other cells. We employed a χ2 test to determine if there was consistency between the two datasets (i.e. if there were more genes both upregulated and both downregulated than expected by chance). Clusters were classified by the HSC phenotype with which they had significant alignment.
Interaction networks:
Molecular interactions among marker genes were analyzed using the STRING web interface, with default parameters. Network interconnectivity statistics were calculated using STRING, which measures the number of observed interactions between an input set of genes, compared to the null expectation, using a permutation test.
Heatmap visualization:
Expression of genes of interest were visualized in heatmaps. Expression levels in individual cells were binned first by cluster membership, and then by UMAP dimension 1 within each cluster, to provide a more detailed view of expression within each cluster. The average expression of all cells within each bin is computed, and then the row z-score of these average expression values is displayed in the heatmaps, to visualize the relative expression between and within clusters.
Pseudotime and velocity analysis:
RNA velocity of individual cells was estimated following the default analysis pipeline, from the spliced and unspliced content from the Cell Ranger output (27) The loom files from each sample were merged then smoothed using kNN neighbors pooling (k=500). The gamma fit was calculated using default parameters. Resulting velocities were projected on the UMAP coordinates previously generated using Seurat. Pseudotime trajectories were estimated using Monocle’s DDRTree method, using default parameters (28).
Co-expression and transcription factor analysis:
Regulons were identified using the SCENIC tool (29). The adjacency co-expression matrix was calculated among transcription factors and all genes expressed in at least 100 cells, using the GRNboost2 algorithm included in SCENIC. Co-expression heatmaps displayed in the text were calculated with simple Pearson correlations. Co-expression edges were pruned as recommended in the tool to include only those interactions known to exist between transcription factors and regulated genes. Regulons were identified as transcription factors and the genes they regulate. Regulon activity per cell was assessed by calculating the AUC of expressed genes in each cell, and a binarized version was calculated with the BIC threshold method from SCENIC. In the cases where over half of the cells were found to be active, the median AUC was used as the threshold. Quiescent, activated, and inactivated regulons were defined by the transcription factor analysis from a previous study (30).
shRNA knockdown of TFs in primary mouse HSCs:
shRNA-expressing lentiviral plasmids were co-transfected with plasmids pVSV-G and pCMVd8.2 dvpr into 293T cells. Virus containing media were collected 72 h later, filtered (0.22-μm pore size), concentrated by ultracentrifugation, and purified on a sucrose 20% gradient. Viral titers were measured using immunocapture p24-gag ELISA (Sequencing core facility, LJI). For viral transduction, lentiviral vectors at a multiplicity of infection of 30 + polybrene (10 μg/ml) were added to primary mHSCs (60-70% confluency, 24h after plating) for 36 hours, afterward fresh media containing ± puromycin (5 μg/ml) was added to infected cells for 48 hours. HSCs were harvested, and gene silencing was analyzed by qRT-PCR and RNA-Seq. RNA isolated from HSCs were aligned to the mouse genome (mm10) using STAR, keeping only uniquely alignable reads for subsequent analysis. Differential expression was calculated using HTSeq and DESeq2. Multiple shRNA hairpins were tested: ETS1 (n=5), GATA4 (n=5), GATA6 (n=5), IRF1 (n=7), as described in (30). A minimum of two target sequences with the least off-target knockdowns was selected for each gene (ETS1: TRCN0000042640, TRCN0000042642; IRF1: TRCN0000219079, TRCN0000233968; GATA4: TRCN0000095218, TRCN0000095214; GATA6: TRCN0000085590, TRCN0000085589), compared to non-coding hairpins. Infection with shRNA plasmid DNA control vectors were used to generate infected cells without puromycin treatment as controls (SHC-002 was used as a control for targeting ETS1, GATA4/6 in HSCs; SHC-202 was used as a control for targeting IRF1 in HSCs).
Results
Change of diet causes progression and regression of NASH with fibrosis in foz/foz mice.
A recessive mutation in the Alms1 Gene produces the Alstrom syndrome in patients and the foz/foz strain in mice. Since this mutation produces hyperphagia, obesity, insulin resistance, and NASH in both humans and mice, the foz/foz mice have become a well-characterized model of human NASH with liver fibrosis (18-20).
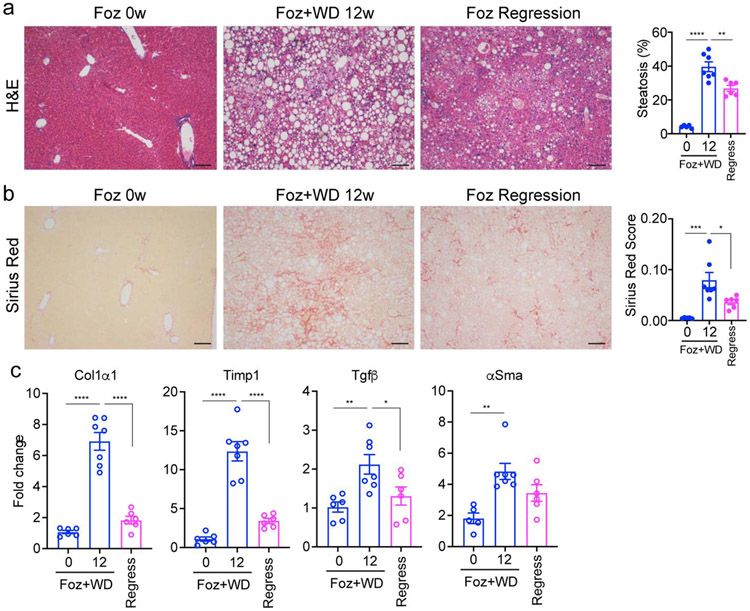
At 5-6 weeks of age, male foz/foz mice were fed a Western diet (AIN-76A) for 12 weeks to establish NASH with fibrosis. To assess the regression of NASH, some mice were returned to a chow diet for an additional 8 weeks. Liver histology revealed that the mice on a chow diet had normal livers without steatosis or fibrosis despite the ALMS1 mutation (Fig.1a, b). After 12 weeks on a Western diet, the foz/foz mice developed steatosis with an inflammatory infiltrate and chicken-wire type fibrosis as demonstrated by sirius red staining (Fig. 1a, b). After returning to a chow diet for 8 weeks following the western diet, the foz/foz mice had greatly diminished steatosis with minimal remaining fibrosis (Fig. 1a, b). Extending the NASH regression timeline from 8 weeks to 12 weeks did not result in an increased reduction of fibrotic gene expression (Fig. S1). Indeed, Chow-fed foz/foz mice, being hyperphagic, develop obesity, diabetes, cholesterol disturbances, and simple steatosis (18,31) which can progress to NASH after 24 weeks (22). We also note that foz/foz mice on western diet develop hepatocellular carcinoma after 6 months (manuscript in preparation).
Figure 1. Progression and regression of fibrosis in foz/foz mice.
Six weeks old foz/foz mice were fed either standard chow (0w) or WD for 12 weeks (12w) or subjected to regression by switching 12w WD fed mice to standard chow for 8 additional weeks. Representative images of liver histology depicting steatosis (a; hematoxylin-eosin (H&E) staining) and fibrosis (b; sirius red staining; quantified as sirius red positive area normalized to non-steatotic area); Scale bar: 100 μm. (c) Total RNA was extracted from the liver lysates and expression of indicated fibrogenic markers were analyzed by qPCR. The data is represented as fold induction versus foz/foz mice on standard chow (0w WD group). All bar graphs represent mean ± SEM, *p <0.05, **p <0.01, ***p <0.001, ****p <0.0001 by ANOVA.
The liver histology was confirmed with gene expression profiling of whole liver in foz/foz mice. This revealed increased expression of fibrotic genes (Col1a1, Timp1, Tgfb, and Acta2) when the mice were fed the Western diet (Fig. 1c). Expression of all fibrogenic genes decreased toward normal levels when the mice were switched back to a chow diet. Thus, this experimental protocol reflects human NASH with diet induced obesity and regression of NASH with dietary restrictions (32).
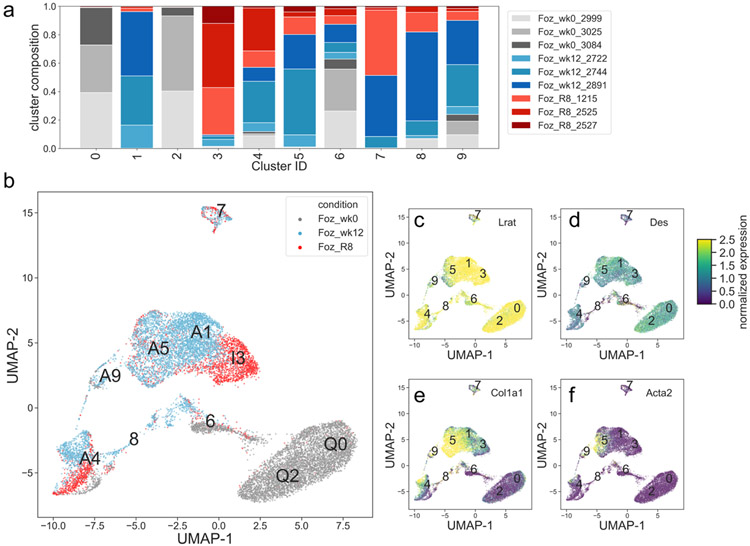
HSCs were purified from three experimental conditions: foz/foz mice fed a chow diet (control), fed a Western diet for 12 weeks, and then returned to a chow diet for an additional 8 weeks. Each experimental condition was performed in triplicate in independent experiments, and then the purified HSCs were subjected to single cell RNA-seq. Cluster analysis revealed nine distinct clusters of single cells with excellent replication within each experimental condition, but with clear separation between the different experimental conditions (Fig. 2a, 2b). As expected, the universal HSC markers Lrat and Desmin was expressed in all clusters (Fig. 2c, 2d), validating the purification methodology. Also, as expected, the activated HSC markers Col1a1 (Fig. 2e) and Acta2 (Fig. 2f) were only expressed in a subset of the HSCs clusters that resulted from the NASH experimental condition, but not in the control normal liver or the NASH regressed liver.
Figure 2: Single cell RNA-seq clusters of HSCs express validated markers.
a) Cell type composition breakdown per cluster (fraction of total cell count per cluster). Three foz/foz week 0 samples shown in gray, three foz/foz week 12 samples shown in blue, and three foz/foz regression samples shown in red. b) Cells plotted in UMAP space, color-coded by condition, cluster identity positioned at the median UMAP-1, UMAP-2 for all cells in the cluster. c-f) Cells plotted in UMAP space, color-coded by Lrat, Des, Acta2, and Col1a1 mRNA expression.
Clustering of single-cell RNA-seq data identifies HSC phenotypes.
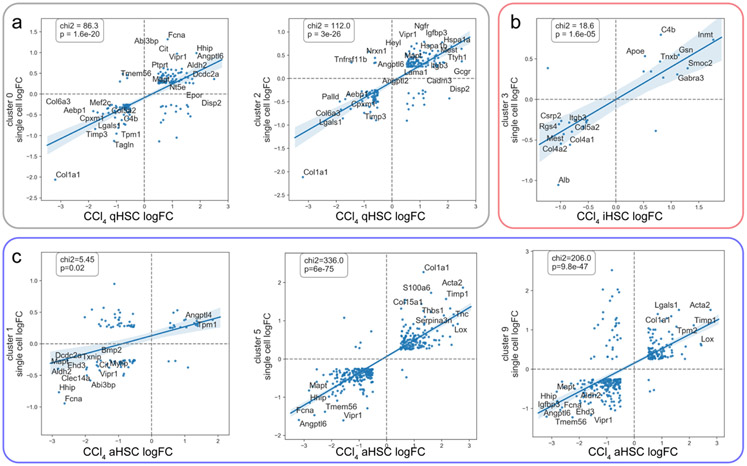
Extensive studies using the CCl4 model of liver fibrosis in mice have identified 3 distinct HSC phenotypes; quiescent (defined by genes such as Ngfr, Hspa1a, and Hspa1b), activated (defined by genes such as Col1a1, Acta2, and Timp1), and inactivated (defined by genes such as Smoc2, Gabra3, and Gsn) (3,4,33). The CCl4 model is an established toxicant induced model of liver fibrosis, but which does not develop steatosis or NASH. We have chosen to extend our analysis into the foz/foz model because it is more pathophysiologically relevant to NASH than CCl4. The gene expression profiles that define these three phenotypes were used to interrogate the nine HSC clusters in the current study. Clusters 0 and 2 tightly correspond to the previously determined quiescent HSC markers (Fig. 3a), and, as expected, contain cells from mainly the control normal liver. Cluster 3 corresponds to inactivated HSC markers (Fig. 3b), and contains cells mainly from the regressed liver. Clusters 1, 5 and 9 correspond most closely to activated HSC markers with varying degrees of correlation (Fig. 3c). Because clusters 6, 7, and 8 were smaller, had lower expression of Desmin and Lrat, and lacked defining characteristics, these clusters were not analyzed further (Fig. 1). In particular, although cluster 7 appears to have a strong correlation with inactivated HSCs, the fact that it expressed Desmin at a much lower level than other clusters, and had fewer expressed genes overall lead us to conclude that these cells may be a different cell type that have some similarities with iHSCs. Table S2 summarizes the relationship between single-cell clusters and previously determined HSC phenotypes. We note that here we are examining cellular subtypes within one larger cell type (HSCs), leading to relatively small overall differences in expression between marker genes, compared to published whole liver analyses, which examine the differences between all cell types within the liver (12). Despite the difference in models and relatively small inter-cluster differences, we are still able to identify convergence between quiescent, activated, and inactivated HSC subtypes across models.
Figure 3: Single cell clusters match known HSC phenotypes.
a) Clusters 0 and 2 correspond to quiescent HSC markers; x-axis shows log fold change comparing CCl4 quiescent HSCs to activated and inactivated HSCs, y-axis shows log fold change comparing expression in clusters 0 and 2 to all other cells. b) Cluster 3 corresponds to inactivated HSC markers; x-axis shows log fold change comparing CCl4 inactivated HSCs to activated or quiescent HSCs, y-axis shows log fold change comparing expression in cluster 3 to all other cells. c) Clusters 1, 5, and 9 correspond to activated HSC markers; x-axis shows log fold change comparing CCl4 activated HSCs to quiescent and inactivated HSCs, y-axis shows log fold change comparing expression in clusters 1, 5, and 9 to all other cells. For panels (a-c) scatterplot points are shown which are both changed with abs(log fold change)>0.5 in the CCl4 experiment, and which are identified as markers of each cluster in the single cell analysis. The top 15 genes are labeled. A linear regression line was fitted to the data, with 95% confidence intervals shown. The Chi-squared values and corresponding p-values are shown which compare the number of genes observed in each quadrant to what would be expected if there was no correspondence between the two datasets.
Although most studies have reported that the quiescent HSCs in the normal liver are a homogeneous population (3,4,14), our analysis subdivides a large group of related cells into 2 clusters (0 and 2) in the normal liver (Figs. 1 and 2). Of note, a recent study (16) used gene expression and immunohistochemistry to demonstrate that gene expression in the quiescent HSCs changed along the hepatic acinus, such that there are markers that distinguish the central vein associated HSCs from the portal vein associated HSCs. We used these markers (16) to interrogate clusters 0 and 2. This analysis clearly demonstrated that cluster 2 expressed the portal vein associated markers and cluster 0 expressed the central vein associated markers (Fig. 4a). Portal vein associated HSC markers (including Ngfr and Itgb3) (16) were more highly expressed in cluster 2, while central vein associated HSC markers (including Adamtsl2 and RSPO3) (16) were more highly expressed in cluster 0 (Fig. 4b). Marker genes for cluster 2 identified by our analysis highly overlap with the reported portal vein associated HSC markers (33/46 portal vein gene shared), but with no overlap with the central vein associated HSC markers (Fig. 4c). Marker genes for cluster 0 identified by our analysis highly overlap with the reported central vein associated HSC markers (20/26 central vein gene shared), but with no overlap with the portal vein associated HSC markers (Fig. 4d). A co-expression analysis of our data demonstrated that the novel central-vein and portal-vein markers are expressed in the same cells as the known central-vein and portal-vein markers (Fig. S2). This established the acinar location of our novel genes.
Figure 4: Heterogeneity of quiescent HSCs in murine NASH.
a) Previously determined markers of central-vein associated qHSCs (yellow) and portal-vein associated qHSCs (green) correspond to clusters Q0 and Q2 (quiescent clusters) respectively. Heatmap displays relative gene expression in cells binned by UMAP dimension 1, where most of the variance is seen between clusters Q0 and Q2, bins evenly spaced between minimum and maximum UMAP-1 value for cells in clusters Q0 and Q2. Cluster membership annotated by top color bar and dotted line. Inset indicates breakdown of cells in clusters 0 and 2 by sample. b) Individual cell expression plotted in UMAP space for cells in clusters Q0 and Q2, for four selected marker genes of portal-vein HSCs (Ngfr, Itgb3), and central-vein associated HSCs (Adamtsl2, Rspo3). c) Markers of cluster Q2 identified by our analysis overlap with portal-vein markers (33/46 portal-vein genes shared) d) Markers of cluster Q0 identified by our analysis overlap with central vein marker genes (20/26 central-vein genes shared). e) Molecular interactions among cluster Q2 marker genes, from the STRING database. This gene set has more interactions than expected by chance (p<1E-16, permutation test). f) Molecular interactions among cluster Q0 marker genes, from the STRING database. This gene set has more interactions than expected by chance (p<1E-16, permutation test).
Our more clearly defined clusters with deeper sequencing enabled an analysis of molecular interactions that revealed different potential functions of the cluster 0 and 2 quiescent HSCs (Fig. 4e,f). The portal vein associated cluster 2 HSCs genes were more connected than expected by chance (p<1E-16, permutation test), and had a strong interaction at the level of basement membrane type extracellular matrix (ECM) proteins including laminins and type 4 collagens (Fig. 4e). On the other hand, the cluster 0 central vein associated HSCs genes were also highly interconnected (p<1E-16, permutation test), had strong interactions of the AP1 proteins (Jun, JunB, Fos) characteristic of HSCs (30), but not did not include ECM pathway genes (Fig. 4f). Thus, our study combined with a previous study (16) clearly demonstrates 2 subtypes of the quiescent HSC phenotype depending on localization within the hepatic acinus with implications for separate functions.
Fibrogenic myofibroblasts represent only a subset of HSCs in NASH with fibrosis
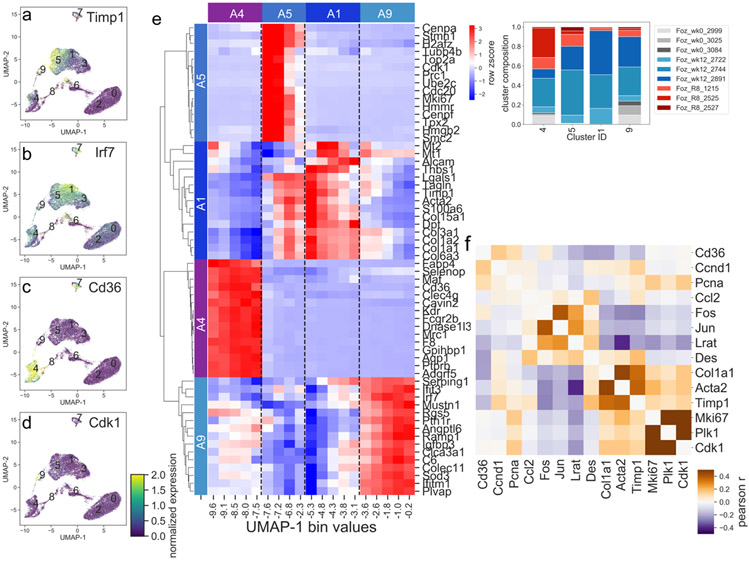
Instead of a single homogenous cluster of activated HSCs as in the CCl4-induced fibrotic liver, as previously described (3,4,13), the NASH-induced activation of HSCs produced four distinct clusters (clusters 1, 4, 5 and 9). Each cluster was defined by distinct expression of gene markers (Fig. 5a-e). Cluster 5 precisely corresponds to the classic CCl4 induced activated HSCs (also called myofibroblasts) (Fig. 5a and Fig. 3c). The fibrogenic genes such as fibrillar collagens, Acta2, and Timp1 (Fig. 5a) define Cluster 5. Cluster 1 has a weaker expression of the fibrogenic gene markers but expresses some other markers at higher levels such as IRF7 (Fig. 5b). Cluster 1 appears to represent an HSC phenotype of intermediate or lower activation. Although cluster 4 represents HSCs from all 3 experimental conditions - normal liver, NASH, and regression, it is predominantly populated by NASH and regression HSCs (Fig. 2a). The presence of both NASH and regression HSCs leads us to believe that cluster 4 represents a cell type which does not regress but persists even after return to a normal diet. While cluster 4 has large populations of both NASH and regression HSCs, subcluster analysis revealed relatively few marker genes distinguishing the cluster 4 NASH cells from cluster 4 regression cells (Fig. S3), suggesting that these two populations are in fact quite similar and should be treated as one cell type. This cluster reflects a novel phenotype that has expression of inflammatory and immune genes such as Cd36, Ly6c, and CLEC proteins, but not fibrogenic genes (Fig. 5c). Finally, cluster 9 expresses genes associated with both fibrosis and proliferation. Many proliferation markers such as Cdk1 are exclusively expressed in this cluster (Fig. 5d). A comparison of co-expression between selected proliferation genes, activation marker genes, and quiescent marker genes demonstrate their correlations in the different clusters (Fig. 5f). Of note, the fibrogenic genes are co-expressed within cells, meaning that these genes tend to be expressed at similar levels within the same cells. For example, a cell that expresses the fibrogenic fibrillar collagens also expresses the activation genes Timp1 and Acta2, consistent with a classic myofibroblast phenotype.
Figure 5: Heterogeneity of activated HSCs in murine NASH.
a-d) Individual cell expression of selected marker genes (Timp1, Irf7, Cd36, and Cdk1) for each of the 4 activated clusters (clusters A1, A4, A5, and A9). e) Heatmap displays relative gene expression in cells binned by UMAP dimension 1, where most of the variance is seen among clusters A4, A9, A5, and A1, bins evenly spaced between minimum and maximum UMAP-1 value for cells in clusters A4, A9, A5, and A1. Relative expression for top 15 marker genes for each cluster is shown. Cluster membership annotated by top color bar and dotted line. Inset indicates breakdown of cells in clusters A4, A9, A5, and A1 by sample. f) Heatmap showing the co-expression between selected proliferation genes and activated and quiescent marker genes. Co-expression was calculated between all cells from the foz/foz week 12 condition.
Does the acinar position of the quiescent HSCs (clusters 0 and 2) determine their activated phenotype in NASH? To address this question, the gene expression in clusters 4 (inflammatory and immune HSCs) and cluster 5 (classic activated HSCs) were interrogated with respect to the expression of central vein associated HSCs (cluster 0), and portal vein associated HSCs (cluster 2). Surprisingly, we found that the two quiescent HSC clusters contributed equally to the fibrogenic activated HSC cluster (Fig. S4), as demonstrated in this heat map. Although clusters A4 and A5 separate clearly, the representation of the quiescent HSC genes from clusters 0 and 2 are distributed uniformly throughout the activated phenotypes. Perhaps this reflects that the fibrogenic activated HSCs in our NASH model are located diffusely throughout the hepatic acinus, as identified by aSMA immunohistochemistry (see below). This contrasts with the previous scRNA-Seq study with CCl4-induced fibrosis, where the activated HSCs were primarily located in the pericentral vein region (acinus zone 3). This is more easily demonstrated by examining the portal vein associated HSC markers Ngf2 and Itgb3 (Fig. S4b,d) and central vein associated markers Rspo3 and Adamtsl2 (Fig. S4c,e), which do not demonstrate an association with any particular cluster generated by the activated HSC in NASH.
Inactivated HSCs have a gene expression pattern that is similar to, but distinct from, quiescent HSCs.
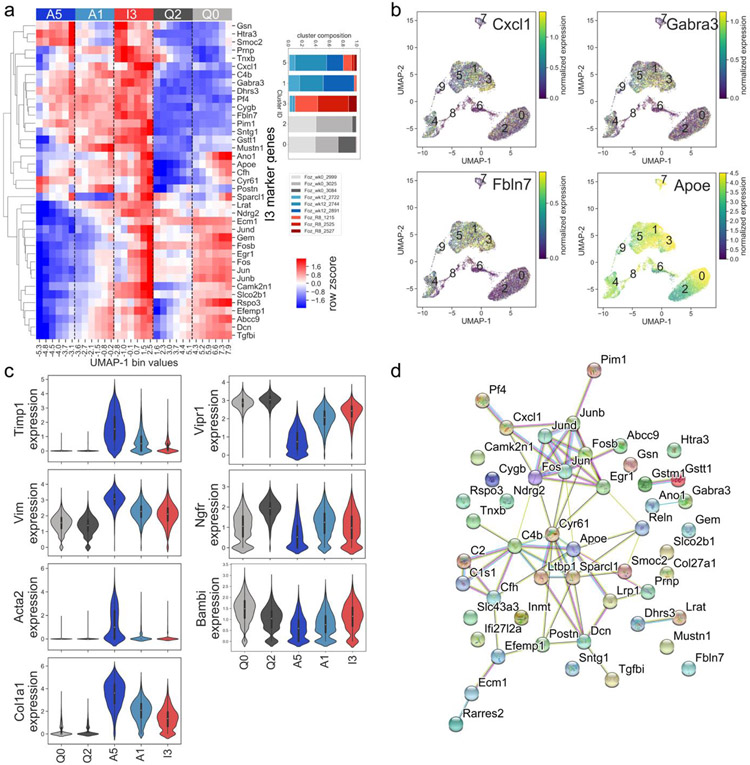
The previously established inactivated HSC phenotype (3,4) correlates closely with cluster 3 (Fig. 2 and Fig. 6). This is demonstrated by the relative expression of the top 40 cluster I3 marker genes compared to clusters A1 and A5 (Fig. 6a). Some genes are uniquely expressed in the inactivated HSCs, but not in quiescent HSCs, such as Cxcl1, Gabra3, and Fbln7 (Fig. 6b). On the other hand, other genes are upregulated in quiescent HSCs, decreased in activated HSCs and then reappear in inactivated HSCs, such as Bambi, Vipr1 (Fig. 6c) and ApoE (Figs. 6a,b). Established markers of activated HSCs, such as Acta2, Timp1, and Col1a1 are highly induced in the myofibroblast HSCs (cluster A5) compared to the quiescent HSCs (clusters Q0 and Q2), and then decrease back towards baseline in the inactivated HSCs (cluster I3). The intermediate activated HSCs (cluster A1) has an expression lever intermediate between the myofibroblast HSCs and the inactivated HSCs (Fig 6c). Molecular interactions (Fig. 6d) demonstrate that cluster I3 marker genes are more interconnected than expected by chance (p<1E-16, permutation test), and express of pathways related to mesenchymal cell phenotype and AP1 expression similar to quiescent cells.
Figure 6: Characterization of inactivated HSCs in murine NASH.
a) Heatmap displays relative gene expression in cells binned by UMAP dimension 1, for cells in clusters A5, A1 (activated), I3 (inactivated), Q0, and Q2 (quiescent), bins evenly spaced between minimum and maximum UMAP-1 value for cells in clusters A5, A1, I3, Q0, and Q2. Relative expression for top 40 I3 marker genes for each cluster is shown. Expression levels of cells in the A1 cluster are intermediate between A5 and I3. Relative expression for top 15 marker genes for each cluster is shown. Cluster membership annotated by top color bar and dotted line. Inset indicates breakdown of cells in A5, A1, I3, Q0, and Q2 by sample. b) Individual cell expression of selected I3 marker genes. c) Violin plots demonstrating cluster-specific expression of selected marker genes in quiescent, activated, and inactivated clusters. d) Molecular interactions among cluster I3 marker genes, from the STRING database. This gene set has more interactions than expected by chance (p<1E-16, permutation test).
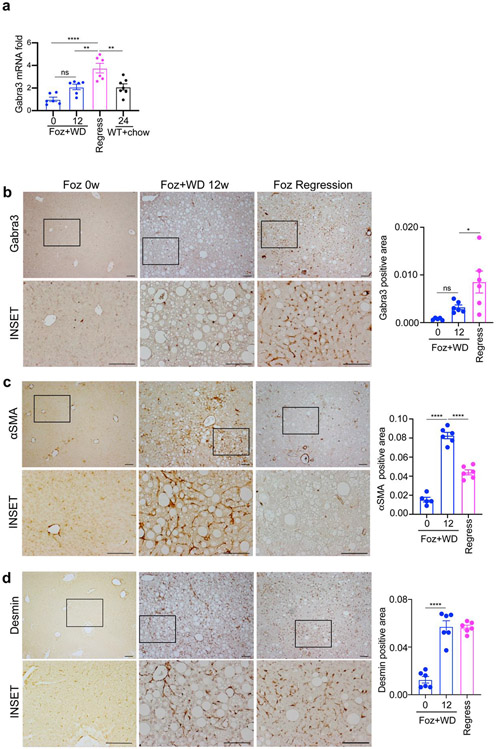
The gene Gabra3 was selected for further analysis, as it had a high correspondence with the established iHSC CCl4 gene signature (Figure 3b), was only expressed in HSCs in the liver, and had an immunohistochemistry suitable antibody. Gabra3 is highly expressed in inactivated cluster 3 but less so in activated HSCs and even further down regulated in quiescent HSCs (Fig. 6a,b). Consistently, there was a significant increase in Gabra3 gene expression in the liver tissues of mice undergoing regression compared to both normal and NASH livers (Fig. 7a). To characterize for the first time the distribution of the Gabra3 protein in HSCs, immunohistochemistry was performed in mouse livers from normal controls, NASH, and after regression of NASH. Consistent with the gene expression, immunohistochemistry of Gabra3 revealed minimal detection in the normal livers and NASH livers, with uniform detection throughout the hepatic acinus in HSCs in the livers with regression (Fig. 7b). In contrast the levels of aSMA protein, a classical marker of activated HSC, was much higher in the NASH liver (Fig. 7c), (which were Gabra3 negative). As expected, Desmin, a universal HSC marker, was detected in normal, NASH, and regression livers (Fig. 7d). The contrasting expression of aSMA and Gabra3, in NASH and regression livers respectively, further establishes Gabra3 as a specific marker of inactivated HSC populating the livers undergoing NASH regression.
Figure 7. Inactivated hepatic stellate cells are characterized by high GABRA3, low aSMA expression.
a) GABRA3 gene expression in the indicated groups was analyzed by qPCR. Liver sections from the indicated groups were stained with b) anti-GABRA3, c) anti-aSMA and d) anti-DESMIN, antibody and quantified as normalized to the non-steatotic area. Scale bar: 100 μm. Data are represented as fold induction versus foz/foz mice on standard chow (0w WD group). Bar graphs represent mean ± SEM, *p <0.05, **p <0.01, ***p <0.001, ****p <0.0001 by ANOVA.
To assess the temporal relationship between the activated and inactivated HSC phenotypes, we performed RNA velocity vector analysis and pseudotime analysis (Fig. S5). RNA velocity (34) is a tool which infers the stage of a gene’s transcriptional activity from the ratio of spliced to unspliced reads. Genes which are just beginning to be transcribed have a lower ratio of spliced to unspliced reads, whereas genes which have stopped being transcribed have a higher ratio of spliced to unspliced reads. This information can be used to infer the predicted temporal progression of cells through clusters (Fig. S5a, arrows). Pseudotime analysis (28) is a complementary approach to measure the progress of cells through a biological process (here the process from the activated to inactivated state). From both approaches it appears that cluster A5 (classic activated) progresses to cluster A1 (intermediate activated), and then to cluster I3 (inactivated) (Fig. S5).
Expression of specific transcription factors correlate with HSC phenotypes in NASH.
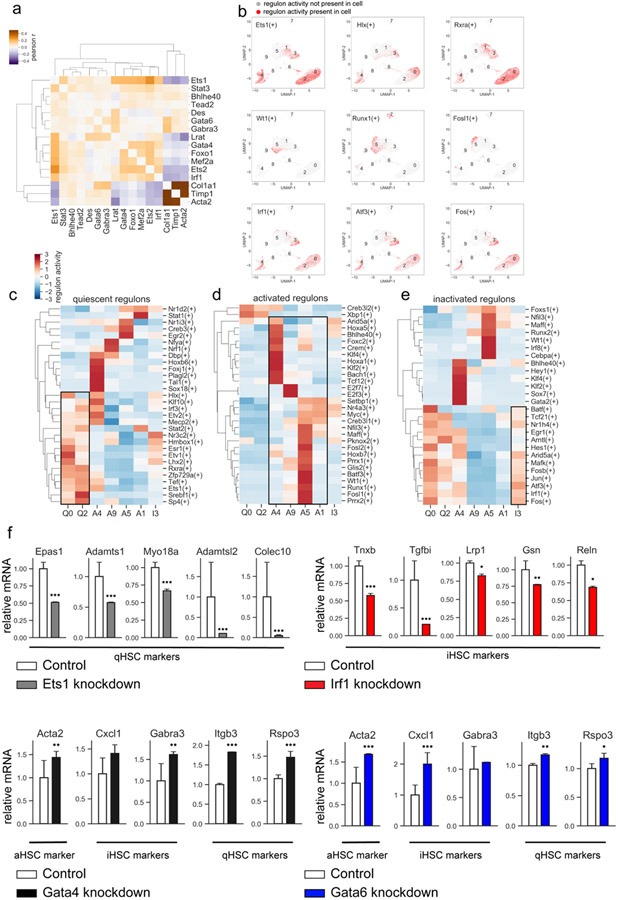
Finally, we used previously published CCl4 induced liver fibrosis and regression epigenetic and gene expression studies(30) to analyze the regulon activity (which predicts the transcriptional activity of transcription factors that control expression of specific genes) in our foz/foz scRNA-Seq data. Transcription factors (TFs) that correlate with HSC signature genes and activation status of qHSCs, aHSCs, and iHSCs were revealed (Fig. 8 a-b). As expected, high activity of quiescence-associated TFs (Ets1, Srebf1, Sp4, Irf3, Stat2, and Esr1) was observed in Clusters 0 and 2 (Fig. 8c). The aHSC phenotype (Cluster 5) was linked to activation of Fosl1/2, Foxs1, Runx, Glis2, Hoxb7, and WT1 TFs, while inflammatory Cluster 4 was linked to Foxc2, Sox7, Crem, Klf4, Tcf12, and Bach1 TFs. The regulon profile of iHSCs (cluster I3) was associated with increased transcriptional activity of Irf1, Atf3, Hes1, Egr1, and Tcf21 (Fig. 8d). Although iHSC and qHSC regulons share some similarities, iHSCs failed to activate Ets1 and other TFs previously implicated in the maintenance of quiescent-like phenotype. (Fig. 8e).
Figure 8: Analysis of transcription factors.
a) Heatmap displaying the correlation in expression (Pearson r) between selected transcription factors and selected markers of stellate cell activation. b) Top row: regulon activity of selected quiescent transcription factors, middle row: regulon activity of selected activated transcription factors, bottom row: regulon activity of selected inactivated transcription factors. c-e) Relative regulon activity per cluster for quiescent (c), activated (d), or inactivated (e) regulons. f) Primary HSCs (1 x 106 cells) were infected with TF-specific shRNA- or non-targeting lentiviruses (>2 targeted and control vectors were tested), followed by ± puromycin (5μg/ml) and analyzed by RNA-Seq. Selected quiescent HSC marker genes observed to be downregulated upon knockdown of Ets1 (top left), selected inactivated HSC marker genes observed to be downregulated upon Irf1 knockdown (top right), selected markers of activated, inactivated, and quiescent HSCs observed to be significantly upregulated upon knockdown of Gata4 (bottom left) or Gata6 (bottom right). Relative expression normalized to mean of control mice. Error bars indicate 1 standard deviation. *p <0.05, **p <0.01, ***p <0.001, adjusted p-value from DESeq2 analysis of RNAseq data.
In vitro shRNA-knockdown of GATA4/6, ETS1, and IRF1 results in activation of targeted HSCs.
ETS1, GATA4, GATA6, and IRF1 were originally identified as the putative lineage-determining factors regulating HSC biology (30), and implicated in the maintenance of quiescent HSC phenotype, as well driving inactivation of CCl4-injured aHSC. Here we confirm that the same TFs play a critical role in NASH-induced HSC activation in foz/foz mice. To determine the specific functions of these TFs, GATA4, GATA6, ETS1, and IRF1 were shRNA knocked down in primary mouse HSCs (30). At least two shRNA sequences with the highest knockdown efficiency and lowest off target effects (30) were selected to knockdown each TF in primary HSCs. The gene expression profiles of targeted HSCs were compared to control HSCs (infected with non-targeting shRNA-expressing lentiviruses) by RNA-Seq. The effect of each individual TF knockdown on quiescence, activation, and inactivation-associated gene expression in HSCs was determined. Upon knockdown of ETS1, a quiescent-associated TF, a number of known and newly identified markers of quiescent HSCs were significantly downregulated, including Epas1, Adamts1, Myo18a, Adamtsl2, and Colec10 (Fig. 8f), confirming that ETS1 plays a role in regulation of quiescent HSC markers. Similarly, upon knockdown of IRF1, an inactivation-associated TF, a number of newly identified markers of inactivated HSCs were significantly downregulated, including Tnxb, Tgfbi, Lrp1, Gsn, and Reln (Fig. 8f). Moreover, shRNA-knockdown of GATA4 and GATA6, two TFs known to regulate HSCs from the literature, but not identified as associated with a specific HSC subtype here, critically induced expression of marker genes Acta2, Gabra3, Itgb3, and Rspo3 (Fig. 8f). These data are in concordance with previous findings (30,35), and suggest that ETS1, IRF1, GATA4/6 are key regulators of HSC phenotype in NASH-injured foz/foz mice.
Discussion
Mesenchymal cells such as HSCs and fibroblasts are generally characterized into 2 phenotypes; quiescent, found in normal tissues, and activated, found in injured tissues. However, recent studies primarily using single cell RNA-Seq in a variety of tissues and disease states have demonstrated that these cells are more heterogeneous. Our study in a mouse model of NASH and regression clearly demonstrates heterogeneity in both the quiescent HSC population and in the activated HSC population. Furthermore, an inactivated HSC population emerges during regression of NASH. iHSCs originate from activated HSCs after the fibrogenic stimulus is removed (CCl4, ethanol, or Western diet) (3,4). iHSCs occupy the same location in the Space of Disse as qHSCS and fulfill the same functions; to act as pericytes and store vitamin A. By interrogating our new data with genetic cell fate mapping from previous CCl4 induce liver fibrosis, we were able to clearly identify different HSC phenotypes. This approach can now be extended to study HSC phenotypes in other experimental models and in patients with NASH. Furthermore, we demonstrated that shRNA-knockdown of selected transcription factors in targeted HSCs resulted in a more activated, less quiescent phenotype.
Quiescent HSCs have retinol containing lipid droplets, express characteristic marker genes L-rat and desmin, and are localized in the Space of Disse. They function as the vitamin A storage site for the body and as pericytes for the hepatic sinusoids. Although originally believed to be a homogenous cell population distributed across the hepatic acinus, a recent study (16) demonstrated distinct gene expression profiles for central vein associated HSCs (acinar zone 3) and portal vein associated HSCs (acinar zone 1). Our analysis revealed two distinct quiescent HSC clusters in which cluster 0 corresponds to central vein associated HSC and cluster 2 corresponds to portal vein associated HSCs. In CCl4 induced fibrosis it was reported that only central vein associated HSCs are activated into fibrogenic myofibroblasts (16). However, in our study the quiescent HSCs from both clusters are identified within the activated myofibroblast cluster 5. Perhaps this reflects that CCl4 acts primarily on acinar zone 3, while the metabolic insult in NASH is across the acinus resulting in diffuse fibrosis.
Activated mesenchymal cells, such as HSCs and fibroblasts, have been traditionally labeled as myofibroblasts that express Acta2 and extracellular matrix proteins. However, studies in cancer-associated fibroblasts (CAF) have demonstrated that these ACTA2+ myofibroblasts represent only a subset of all CAFs (36-38). This has led to the identification of three distinct CAFs based on the gene expression profiling and functional characteristics, which are defined as myofibroblastic CAFs, inflammatory CAFs, and antigen presenting CAFs (36-38). Our analysis of scRNA-Seq data identifies four distinct clusters of activated HSCs that are present in the fibrotic NASH liver, but not in the normal liver and highly depleted after regression. In our study, cluster 5 contains classic myofibroblasts expressing the established myofibroblast markers such as Acta2, Timp1, fibrillar collagens and S100A6. There is remarkable direct association between the CCl4 activated HSCs and cluster 5 in the current NASH with fibrosis study. Furthermore, there is a tight direct correlation between the gene expression of the established myofibroblast markers Col1a1, Timp1, and Acta2. In other words, the classic activation markers are co-expressed together at high levels in the same cells. Although, cluster 1 is similar to cluster 5, cluster 1 does not express classic myofibroblasts markers such as Acta2 and some of the fibrillary collagens at a much lower level. RNA velocity analysis demonstrates that cluster 1 is intermediate between activated myofibroblast cluster 5 and inactivated cluster 3. Such an intermediate activated phenotype was not detected in the CCl4 induced fibrosis model. Perhaps the stronger injury induced by CCl4 with massive hepatocyte death leads to a stronger activation of HSCs compared to the milder gradual injury induced by the metabolic syndrome in NASH.
Although cluster 4 is mainly populated by HSCs from NASH with fibrosis, it differs markedly from the classic activated myofibroblast cluster 1 and the intermediate cluster 1, as it is dominated by genes involved in signaling and secretory pathways. While cells in cluster 4 express some genes characteristic of sinusoidal endothelial cells (e.g. Kdr, Cavin2, and Aqp1), they also have high expression levels of canonical HSC marker genes (including Lrat) and purify on a gradient consistent with containing retinoid lipid droplets, suggesting these cells are a novel type of inflammatory HSC. We note that endothelial cells were identified as a separate cluster and eliminated in the upstream analysis. In fact, cluster 4 resembles the inflammatory cluster recently described in cancer associated fibroblasts (CAFs). The markers expressed in both iCAFs and cluster 4 include Lif, multiple CLEC proteins, (C-type lectin domain containing proteins) and LY6G (LY6G6C, LY6G6D, and LY6G6F). The CLEC proteins have a role in inflammation and immunity. The function of this novel HSC phenotype is unknown, but it may be relevant to the eventual HCC known to develop in foz/foz mice after 6 months on western diet.
Finally, a discrete cluster 9 closely resembles the classic activated myofibroblast cluster 5. However, the two clusters differ in that only cluster 9 expresses the markers of cellular proliferation such as CDK1. Thus, only activated HSCs appear to be undergoing proliferation in the livers.
Cluster 3 represents the inactivated HSCs. The cluster only appears during regression of NASH when the mice are switched from a Western diet back to a chow diet. This represents the first documentation of inactivated HSCs in NASH. Thus, in CCl4, ethanol and diet induced fibrosis, removing the etiological agent leads to inactivated of HSCs. Of note, our experimental protocol in which the foz/foz mice are switched back to the chow diet have a stable obese weight, yet the liver regresses back towards normal with respect to steatosis, inflammation and fibrosis. The quiescent phenotype is similar to but distinct from the quiescent HSC phenotype. Some genes (such as Bambi) are expressed high in quiescence, decrease in activation, and return to high levels during inactivation. Other genes have low expression in quiescence, rise with activation, and then return toward baseline with inactivation (Col1a1, Timp1, Acta2). However, there are some genes that are low in quiescence remain low in activation but are uniquely induced in activation. A newly described gene that follows the latter sequence is Gabra3, which is expressed in inactivated HSCs distributed throughout the hepatic acinus. Interestingly, markers of senescence (such as p53, p21, p16ink4a, and Bmi-1) were never expressed at detectable levels. Therefore, this study has not demonstrated any evidence for a role for senescent HSCs in NASH development and regression.
HSCs are critical for normal homeostasis, functioning as pericytes and as storage for vitamin A, so that indiscriminate ablation of HSCs might be deleterious (unpublished observations). However, HSCs are also the primary cell driving pathological liver fibrosis. Our study and others have revealed an unanticipated heterogeneity of HSC phenotypes under normal and injured states. Further studies may reveal that a specific cluster drives fibrosis, while other clusters are required for homeostasis and repair. Such observations would provide new insights into the targeted treatment of liver fibrosis.
Supplementary Material
Acknowledgment.
We thank Sara Morimoto for help preparing the manuscript.
Financial support:
This work is supported by the NIDDK-funded San Diego Digestive Diseases Research Center (P30 DK120515), the NIAAA-funded Southern California Research Center for ALPD and Cirrhosis (P50 AA011999), the ACTRI and NIH (KL2TR001444), and the NIEHS-funded Superfund Center (P42 ES010337). Additional support from the NIH and NIDDK funded grants R01 DK091183, P30 DK063491 and T32 DK007044.
Abbreviations:
- NASH
non-alcoholic steatohepatitis
- scRNA-Seq
single-cell RNA sequencing
- HSC
hepatic stellate cell
- WD
western diet
- H&E
hematoxylin and eosin
- FFPE
formalin-fixed paraffin-embedded
- DAB
diaminobenzidine tetrahydrochloride
- HPRT
hypoxanthine guanine phosphoribosyn transferase
- UMAP
uniform manifold approximation and projection
- CCl4
carbon tetrachloride
- qHSC
quiescent hepatic stellate cell
- aHSC
activated hepatic stellate cell
- iHSC
inactivated hepatic stellate cell
- ECM
extracellular matrix
- TF
transcription factor
- CAF
cancer-associated fibroblast
- CLEC
C-type lectin domain containing
Footnotes
COI: UC San Diego receives grant support from Janssen
References
- 1.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol 2017;14:397–411. [DOI] [PubMed] [Google Scholar]
- 2.Koyama Y, Brenner DA. Liver inflammation and fibrosis. J. Clin. Invest 2017;127:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc. Natl. Acad. Sci. U. S. A 2014;111:E3297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mederacke I, Hsu CC, Troeger JS, Huebener P, Mu X, Dapito DH, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat. Commun 2013;4:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 2019;75:644–660.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatun M, Ray RB. Mechanisms underlying hepatitis C virus-associated hepatic fibrosis. Cells. 2019;8:1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Su Y, Hua X, Xie C, Liu J, Huang Y, et al. Levels of hepatic Th17 cells and regulatory T cells upregulated by hepatic stellate cells in advanced HBV-related liver fibrosis. J. Transl. Med 2017;15:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balog S, Li Y, Ogawa T, Miki T, Saito T, French SW, et al. Development of capsular fibrosis beneath the liver surface in humans and mice. Hepatology. 2020;71:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishio T, Hu R, Koyama Y, Liang S, Rosenthal SB, Yamamoto G, et al. Activated hepatic stellate cells and portal fibroblasts contribute to cholestatic liver fibrosis in MDR2 knockout mice. J. Hepatol 2019;71:573–585. [DOI] [PubMed] [Google Scholar]
- 10.Scholten D, Reichart D, Paik YH, Lindert J, Bhattacharya J, Glass CK, et al. Migration of fibrocytes in fibrogenic liver injury. Am. J. Pathol 2011;179:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacParland SA, Liu JC, Ma X-Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong X, Kuang H, Liu T, Lin JD. A single-cell perspective of the mammalian liver in health and disease. Hepatology. 2020;71:1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krenkel O, Hundertmark J, Ritz TP, Weiskirchen R, Tacke F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, et al. Single-cell transcriptomics uncovers zonation of function in the mesenchyme during liver fibrosis. Cell Rep. 2019;29:1832–1847.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran P, Matchett KP, Dobie R, Wilson-Kanamori JR, Henderson NC. Single-cell technologies in hepatology: new insights into liver biology and disease pathogenesis. Nat. Rev. Gastroenterol. Hepatol 2020;17:457–472. [DOI] [PubMed] [Google Scholar]
- 18.Arsov T, Silva DG, O’Bryan MK, Sainsbury A, Lee NJ, Kennedy C, et al. Fat Aussie—A new Alström syndrome mouse showing a critical role for ALMS1 in obesity, diabetes, and spermatogenesis. Mol. Endocrinol 2006;20:1610–1622. [DOI] [PubMed] [Google Scholar]
- 19.Van Rooyen DM, Larter CZ, Haigh WG, Yeh MM, Ioannou G, Kuver R, et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology. 2011;141:1393–403, 1403.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mridha AR, Wree A, Robertson AAB, Yeh MM, Johnson CD, Van Rooyen DM, et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J. Hepatol 2017;66:1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jophlin LL, Koutalos Y, Chen C, Shah V, Rockey DC. Hepatic stellate cells retain retinoid-laden lipid droplets after cellular transdifferentiation into activated myofibroblasts. Am. J. Physiol. Gastrointest. Liver Physiol 2018;315:G713–G721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell GC, Mridha AR, Yeh MM, Arsov T, Van Rooyen DM, Brooling J, et al. Strain dependence of diet-induced NASH and liver fibrosis in obese mice is linked to diabetes and inflammatory phenotype. Liver Int. 2014;34:1084–1093. [DOI] [PubMed] [Google Scholar]
- 23.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 26.Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl. Acad. Sci. U. S. A 2012;109:9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensson V, Pachter L. RNA velocity: Molecular kinetics from single-cell RNA-seq. Mol. Cell 2018;72:7–9. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 2017;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Xu J, Rosenthal S, Zhang L-J, McCubbin R, Meshgin N, et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158:1728–1744.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arsov T, Larter CZ, Nolan CJ, Petrovsky N, Goodnow CC, Teoh NC, et al. Adaptive failure to high-fat diet characterizes steatohepatitis in Alms1 mutant mice. Biochem. Biophys. Res. Commun 2006;342:1152–1159. [DOI] [PubMed] [Google Scholar]
- 32.Vilar-Gomez E, Martinez-Perez Y, Calzadilla-Bertot L, Torres-Gonzalez A, Gra-Oramas B, Gonzalez-Fabian L, et al. Weight loss through lifestyle modification significantly reduces features of nonalcoholic steatohepatitis. Gastroenterology. 2015;149:367–78.e5; quiz e14–5. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Xu J, Brenner DA, Kisseleva T. Reversibility of liver fibrosis and inactivation of fibrogenic myofibroblasts. Curr. Pathobiol. Rep 2013;1:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560:494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knittel T, Kobold D, Dudas J, Saile B, Ramadori G. Role of the Ets-1 transcription factor during activation of rat hepatic stellate cells in culture. Am. J. Pathol 1999;155:1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett R, Puré E. Cancer-associated fibroblasts: key determinants of tumor immunity and immunotherapy. Curr. Opin. Immunol 2020;64:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol 2020;17:487–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.