Abstract
Background
The aim of this study is to demonstrate the feasibility of automatic classification of Ki-67 histological immunostainings in patients with squamous cell carcinoma of the vulva using a deep convolutional neural network (dCNN).
Material and methods
For evaluation of the dCNN, we used 55 well characterized squamous cell carcinomas of the vulva in a tissue microarray (TMA) format in this retrospective study. The tumor specimens were classified in 3 different categories C1 (0–2%), C2 (2–20%) and C3 (>20%), representing the relation of the number of KI-67 positive tumor cells to all cancer cells on the TMA spot. Representative areas of the spots were manually labeled by extracting images of 351 × 280 pixels. A dCNN with 13 convolutional layers was used for the evaluation. Two independent pathologists classified 45 labeled images in order to compare the dCNN's results to human readouts.
Results
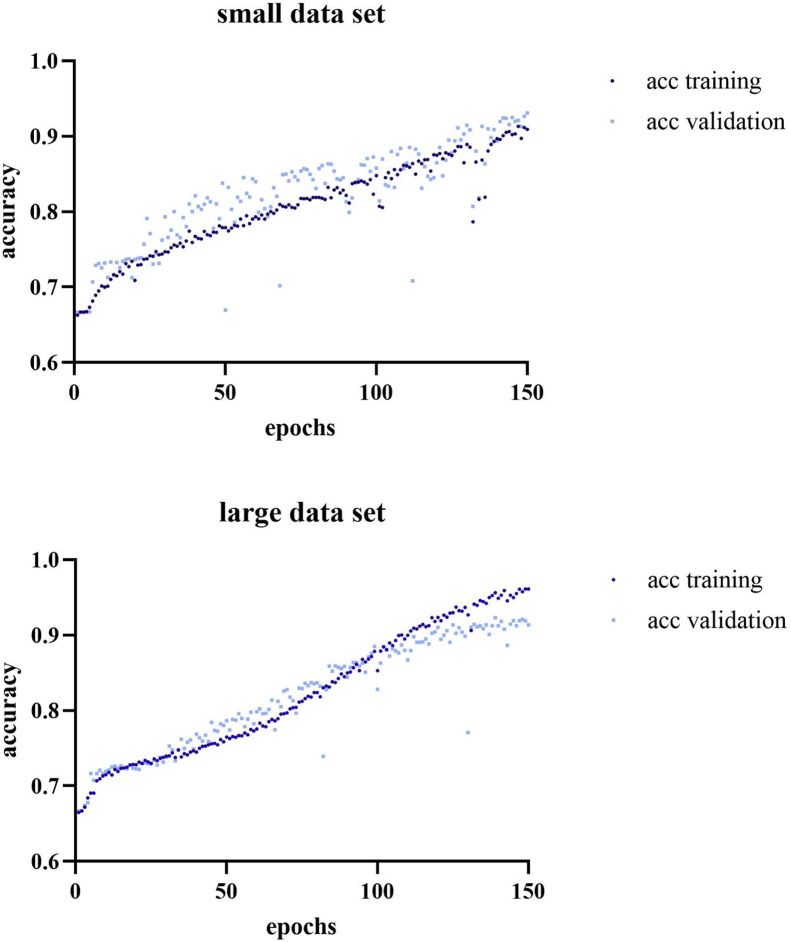
Using a small labeled dataset with 1020 images with equal distribution among classes, the dCNN reached an accuracy of 90.9% (93%) for the training (validation) data. Applying a larger dataset with additional 1017 labeled images resulted in an accuracy of 96.1% (91.4%) for the training (validation) dataset. For the human readout, there were no significant differences between the pathologists and the dCNN in Ki-67 classification results.
Conclusion
The dCNN is capable of a standardized classification of Ki-67 staining in vulva carcinoma; therefore, it may be suitable for quality control and standardization in the assessment of tumor grading.
Keywords: Ki-67, Vulva carcinoma, Deep learning, Convolutional neural network
1. Introduction
The relevance of artificial intelligence in medical image analysis has continuously grown in the past few years. Multiple studies have demonstrated the efficiency of deep convolutional neural networks (dCNNs) regarding its usage for image processing in the medical field [1, 2]. For histopathological image analysis the use of dCNNs is promising, as many studies have shown [3, 4, 5]. Typically, a large amount of high-quality data is required to properly train a dCNN [6]. Quantitative reproducible assessment of biomarkers in histological slides is important for many clinical therapeutic decisions. One of those proteins is Ki-67 as a widely used marker for cell proliferation [7].
Ki-67 is a nuclear protein which is exclusively expressed in proliferating cells in a cell cycle dependent manner [8]. To date, the exact role of the protein in the cell cycle machinery is unknown. Ki-67 is not expressed equally strong in each stadium of the cell cycle: From G1 to the mitosis the levels intensify; after the mitosis the levels decrease and in the G0 phase Ki-67 cannot be detected [9, 10]. Immunohistochemical examination of Ki-67 in tissue sections is a reliable marker for the growth fraction of a cell population [7].
However, the distribution of Ki-67 positive cells in tumors can vary substantially. Especially tumors with a high growth fraction frequently show a more homogeneous Ki-67 expression. On the other hand, many tumors harbor apparently randomly distributed hot spot regions with clusters of Ki-67 positive cell nuclei, whereas other regions are only minimally active. Another crucial issue is the heterogeneous composition of tumor tissues. In a specific region there are not only proliferating tumor cells but also non-neoplastic cells which can also show proliferation activity and very often inflammatory cells with a high proliferation index. Discrimination of this intimate cell mix can be very challenging for the human eye and electronic systems as well. Nevertheless, Ki-67 is used as a prognostic marker in many types of tumors, including squamous cell carcinomas of various origins [11, 12, 13]. Ki-67 was established as response marker for chemotherapy and prognostic marker in breast cancer [14, 15].
At present, the quantification of Ki-67 is cumbersome and time consuming, e.g. cell counts are carried out manually or estimated semi-quantitatively [16]. The results are examiner-dependent with relatively low reproducibility and high inter- and intraobserver variability [17]. Moreover, the degree of standardization of the reading procedure is low [18]. Thus, a more standardized workflow would be of high interest for clinicopathological purposes.
In the last years, substantial interest and research has been focused on the development of techniques of digital pathology and particularly image analysis using machine learning [19]. Typical tasks for machine learning algorithms in digital pathology comprise detection, classification and segmentation of pathologies. A large variety of different machine learning algorithms is available; and the most suitable Artificial Intelligence technique may be chosen dependent of the individual clinical question and the available data for training [20]. dCNNs are among the most powerful algorithms in machine learning, however requiring a substantial amount of data and computational power [21].
The aim of this study is to test whether a dCNN can be trained to reliably classify Ki-67 immunostains in a standardized manner in vulvar carcinomas by applying a relatively small-sized database of histological immunostainings. In the proposed approach, the Ki-67 immunostains have been subdivided in small images representing homogeneous Ki-67 expression by manual labeling.
2. Material and methods
2.1. Patients
A total of 55 formalin-fixed (buffered neutral aqueous 4% solution), paraffin-embedded squamous cell vulvar carcinomas were available from the Department of Pathology, University Hospital Zurich, Switzerland between 2008 and 2014. Clinicopathological data of the tumor samples are shown in Table 1. Data was retrieved from patients' records, the tumor registry and death certificates. All of them were diagnosed with squamous cell carcinoma of the vulva. There were 2 representative tissue spots per individual sample in a tissue microarray setup assessable for this study. The retrospective study was accepted by the local ethics committee (ZH-Nr 2014-0604).
Table 1.
Clinicopathological patient data is shown including age, tumor type, tumor stage and grading.
| All (n) | |
|---|---|
| Samples | 55 |
| Age (y) | |
| Median |
69 |
| Histologic tumor type | |
| Keratinizing | 26 |
| Nonkeratinizing | 23 |
| Basaloid |
6 |
| Tumor stage | |
| pT1a | 4 |
| pT1b | 31 |
| pT2 | 10 |
| pT3 | 4 |
| pT4 |
3 |
| Nodal stage | |
| pN0 | 17 |
| pN1a,b | 5 |
| pN2a-c |
10 |
| Grading | |
| G1 | 6 |
| G2 | 29 |
| G3 | 16 |
2.2. Immunohistological stainings
Commercially available antibody raised against Ki-67 (clone 30-9, Ventana Medical Systems Inc., Tucson, AZ, USA, 1:200) was used on a Ventana Benchmark automated staining system. For Ki-67, only nuclear staining was regarded to be specific. Immunohistochemistry was evaluated, if possible, in two cores per tumor and the results were combined. Tissue cylinders with a diameter of 0.6 mm and a height of 3–4 mm, corresponding to approximately 2800 pixels, were punched from representative tumor areas of a “donor” tissue block using a custom-built instrument. The Images were digitized using a Hamamatsu NanoZoomer 2.0-HT scanner (Hamamatsu Photonics K.K., Hamamatsu, Japan) and stored as jpegs with 0.235 pixels/micrometer using NDI Viewer (NewTek Inc., San Antonio, TX, USA).
2.3. Data preparation
The Ki-67-index was determined by visual counting; all stains were made anonymous to the counter. With an internet-based microscope viewer digitalized screenshots from each spot of the vulva cancer array were made. Both, the number of Ki-67 positive and negative cancer cells were semiquantitatively estimated for each spot. The Ki-67 index of each spot was calculated by the ratio of positive cancer nuclei to all tumor nuclei in the individual spot in percent. There is no generally accepted system for classification of Ki-67 in vulva carcinoma. However, it is reasonable to use a cut-off of 20% for high proliferating tumor as widely approved in breast carcinomas and distinguish a group with very low proliferation under 2% [22]. According to the Ki-67 index three groups were formed with low C1 (<2%), medium C2 (2–20%) and high C3 (>20%) Ki-67-values (Figure 1).
Figure 1.
Examples of Ki-67 in vulva carcinoma histological spots for low, intermediate and high distribution (C1–C2) with an enlarged section of C1 visualizing Ki-67 positive and negative nuclei.
2.4. Labeling
A computer routine developed at our institution in the programming language Octave (GNU Octave version 5.1.0, GNU General Public License) was applied for labeling using the images and I/O packages under the Windows 10 operating system (Microsoft, Redmond, WA, USA). Each spot was visualized with a rescale factor ranging from 0.2-0.5 and assigned to the category, from which representative areas were taken for data labeling. To generate sufficient data, for each spot 5 to 20 images of the representative areas were selected for data augmentation. Each labeled image had a matrix size of 351 × 280 pixels. Data augmentation was carried out by the Keras ImageDataGenerator (Keras 2.3.1, Massachusetts Institute of Technology, Cambridge, MA, USA) with the following settings: (rotation_range = 0.1, width_shift_range = 0.05, height_shift_range = 0.05, shear_range = 0.01, zoom_range = 0.01, horizontal_flip = False, fill_mode = 'nearest'). After augmentation, the data was shuffled and divided into two groups, 70% for training and 30% for validating purpose.
2.5. dCNN architecture
Tensorflow 2.0 (Google, Mountain View, CA, USA) was used for the dCNN, similar to a previously described architecture [23]. It is composed of 13 convolutional layers, which were followed by max-pooling, 4 dense layers as demonstrated in Figure 2. Zero-padded convolutional layers were implemented with Nesterov momentum to improve the performance and prevent overfitting. The batch size was set to 40 and 150 training epochs were used.
Figure 2.
Visualization of the deep learning architecture. Using 13 convolutional layers followed by maxpooling and 4 dense layers.
2.6. Generation of probability heatmaps
In order to locally classify the Ki-67 expression in the original immune spots corresponding to one of the classification categories, a sliding window approach was implemented. Thereby, the input images were cropped into an array of 351 × 280 pixel, similar as in the approach used for generating labeling data. The striding for the sliding window amounted 30 px in horizontal and 30 px in vertical dimension (Figure 3). The probability prediction for each sliding window position was performed using the previously trained dCNN model. The determined probability for each class was saved in combination with the center coordinates of the cropped input image. For better visualization of the classification results, probabilities were converted into an RGB image (with channel R representing class 1, channel G class 2, channel B class 3) and overlayed on the input image (Figure 4).
Figure 3.
Explanation of sliding window detection approach. Sections of an image are being cropped and evaluated with deep learning model, probabilities for classes are being predicted and assigned to given image coordinates. In (A) a typical Ki67 stained section is shown, (B) demonstrates a cropped part of the image, in (C) the prediction result of the CNN for the cropped image is presented.
Figure 4.
Overview of image region classification process. Result of classification for all regions with sliding window is displayed with red (C1), green (C2) and blue (C3) channels for each class, with brightness representing probability estimations. The input image shown is overlaid with a color representation according to the result of the classification probabilities. In (A) a typical input image is shown, (B) demonstrates the probability maps per class, and in (C) the final overlay image is presented.
2.7. Validation by human read-out
With the purpose of comparing the dCNN's results to human readouts, two experienced gynecopathologists (MC, AG) independently classified 15 labelled images per category (n = 45), which were presented in a randomized order. The interrater agreement was calculated with Microsoft Excel (version 1902, Microsoft Corporation, Redmond, WA, USA) using Cohen's Kappa:
With p0 meaning the relative observed agreement among readers, and pc the hypothetical probability of chance agreement. To test for significant differences, a paired student's t-test was applied.
3. Results
3.1. Data generation
To Category 1 (n = 9), Category 2 (n = 53) and Category 3 (n = 44) histological spots were manually assigned, as described in the methods section. Four spots couldn't be utilized, due to empty images. From originally 106 spots, we generated 2037 images through labelling. The 2037 images were assigned to the following groups: C1 (n = 175), C2 (n = 940) and C3 (n = 922). After data augmentation, we reached a total of 10′279 images, depicted as “large data” in Table 2. The labelling was performed in two steps, whereby the number of the spots were divided by two. In the first stage C1 (n = 89), C2 (n = 511), C3 (n = 420) images were used, in the second C1 (n = 86), C2 (n = 429), C3 (n = 502) images. The first augmentation process generated 1′638 images in class C1, 1′846 images in C2, and 1′892 images in C3. Through the second augmentation we were able to maintain 2′844 images in C1, 3′745 images in C2, 3′690 images in class C3 (Table 2).
Table 2.
Results from the two-stage data augmentation comparing labeled and augmented data in C1, C2 and C3.
| Labeled | Augmented | Labeled | Augmented | ||||
|---|---|---|---|---|---|---|---|
| Small data | C1 | 89 | 1638 | Large data | C1 | 175 | 2844 |
| C2 | 511 | 1846 | C2 | 940 | 3745 | ||
| C3 | 420 | 1892 | C3 | 922 | 3690 | ||
| Total | 1′020 | 5′376 | Total | 2′037 | 10′279 |
3.2. Training of the dCNN
The dCNN was trained in two steps. Starting with a small labeled dataset with 1′020 (class 1: 89, class 2: 511, class 3: 420) augmented to 5′376 images, with equal distribution among classes. After 150 epochs the final accuracy reached 90.9% (93%) for training (validation) data. Adding a second data set with additional 1′017 labeled images (class 1: 86, class 2: 429, class 3: 502), a total of 10′279 images was generated through augmentation. 96.1% (91.4%) was reached for the training (validation) dataset after 150 epochs. The learning curves are visualized in Figure 5. Typical probability analyses for the dCNN to classify an unknown image section for each category are demonstrated in Figure 6.
Figure 5.
Learning curves for the small and large data set. Depicting the association between numbers of epochs and accuracy.
Figure 6.
Typical examples of image analyses of sections with low, intermediate and high Ki-67 expression are shown. The first image section was allocated with a 94.31% probability to C1, 5.29% to C2, and 0.39% to C3. The second and third sections were assigned to C2 with a 95.73% probability and to C3 with a probability of 99.99% (Figure 4), respectively.
3.3. Generation of probability heatmaps
Heatmaps of Ki-67 distribution could be generated in excellent image quality. Typical examples are displayed in Figure 7 for two spots which received an overall classification from Ki-67 counting of C1 and C3. It can be seen that the inhomogeneous distribution of the Ki-67 positive cells is well detected by the sliding-window approach of the dCNN with different color encoding for areas with low, intermediate and high Ki-67 content.
Figure 7.
Visualization of Ki-67 distribution using sliding window and heatmap method in C1 and C3. Examples showing categorization result of different regions of distributions based on probabilities evaluated from cropped subsamples for given coordinates. Red showing low, green intermediate and blue high Ki-67 distribution.
3.4. Validation by human read-out
For further validation of the trained dCNN we decided to use 45 randomly selected images from our analyses as mentioned in the method section. Two independent pathologists categorized these images according to the three Ki-67 proliferation classes used to train the dCNN in this study (Table 3). Compared to the human reader labeling the data, the pathologists classified 62.2%, and 68.9% of the images identically, the dCNN classified 51.1% of the images in the same category. The interobserver agreement between the two pathologists was substantial with a kappa value of 0.83. The agreement in the image assignment between the pathologists and the dCNN was only 0.33 and 0.38, respectively. However, there were no significant differences between the pathologists and the dCNN in Ki-67 classification results (p > 0.05). Confusion matrices of the human readers and the dCNN are shown in Tables 4a, b, c.
Table 3.
Classification results of 45 randomly selected images representing the three Ki-67 proliferation classes in our analysis in comparison between two experienced gynecopathologists and the trained dCNN used in this study.
| Ki-67 category | Total of images | Correctly classified images |
p-value | ||
|---|---|---|---|---|---|
| MC | AG | dCNN | |||
| <2% | 15 | 11 | 10 | 5 | p > 0.05 |
| 2–20% | 15 | 7 | 10 | 6 | |
| >20% | 15 | 10 | 11 | 12 | |
Table 4a.
Confusion matrix: reader 1 versus dCNN.
| dCNN | ||||
|---|---|---|---|---|
| Reader 1 | category | 1 | 2 | 3 |
| 1 | 4 | 8 | 2 | |
| 2 | 2 | 8 | 6 | |
| 3 | 0 | 2 | 13 | |
Table 4b.
Confusion matrix: reader 2 versus CNN.
| dCNN | ||||
|---|---|---|---|---|
| Reader 2 | category | 1 | 2 | 3 |
| 1 | 3 | 7 | 1 | |
| 2 | 3 | 10 | 6 | |
| 3 | 0 | 1 | 14 | |
Table 4c.
Confusion matrix: reader 1 versus reader 2.
| Reader 2 | ||||
|---|---|---|---|---|
| Reader 1 | category | 1 | 2 | 3 |
| 1 | 11 | 3 | 0 | |
| 2 | 0 | 15 | 1 | |
| 3 | 0 | 1 | 14 | |
4. Discussion
In this study we proved the feasibility to classify Ki-67 immunostains automatically from patients with squamous cell carcinoma of the vulva with the help of a dCNN. Using 106 immunohistologically stained tumor specimens of a vulvar cancer TMA, we generated approximately 2′000 small sections by manual labeling subdivided in three categories with low, intermediate and high Ki-67 density. Using a dCNN, an accuracy of 96% on the training data and 91% on the validation data could be achieved.
This result is comparable to the results of a recently published study, which showed the feasibility of automatically quantified Ki-67 in cervical intraepithelial lesions by a fully convolutional network [4]. Moreover, Saha et al recently described a deep learning technique, which detects single Ki-67 positive cell using a relatively small matrix of 71 × 71 pixels [24]. Typically, the amount of high-quality data correlates with the precision of the algorithm [6] and reduces the risk of overfitting [2]. Therefore, increasing the amount of data may be applied as a method to improve the performance of the dCNN. On the other hand, our study indicates that a relatively small dataset is already suitable to train an artificial network to recognize and quantify Ki-67 positive nuclei in immunohistochemically stained slides. To increase the amount of data, unsupervised learning might be used in the future to obtain data directly without being labeled beforehand, making a fully automatic approach possible [25].
The current evaluation method of Ki-67 in routine pathology setting is problematic as results suffer from low reproducibility, caused by inter- and intraobserver variability [17]. In our validation cohort with 45 randomly selected images representing three different Ki-67 proliferation classes, the dCNN used in this study showed a relatively low interreader agreement to the trained gynecopathologists of 0.3–0.4 whereas the two experienced gynecopathologists exhibited an agreement of 0.83. The main reason for the low recognition rate could be the usage of a relatively small test section of the whole tumor spot in our cancer TMA. Well trained pathologists are able to classify small sections of images reliably. When it comes to larger images (i.e. more than fractions of an image), classification becomes a more complex procedure, as there is much more room for interpretation, due to heterogeneity of the distribution and color intensity of Ki-67 [24]. Remarkably, the classification results were much better in the third category of tumors with a proliferation rate >20%. In rapidly proliferating tumors the distribution of Ki-67 positive cell nuclei is more homogeneous. This could be the explanation for the improved results in this group and the more ambiguous estimations of Ki-67 in lower proliferating tumors with more heterogeneous distribution of Ki-67 positive tumor cell nuclei [24]. The high reproducibility of Ki-67 counts in fast proliferating tumors could be of clinical relevance as these tumors have a poor prognosis and a high proliferation serves as a valuable marker for chemotherapy response [11]. This underpins the usability of a dCNN in the diagnostic process for proliferation measurements of tumors.
The automatic generation of heatmaps is an advantage of using a neural network for measurement of tumor proliferation activity. This feature opens the possibility to visualize the distribution of Ki-67 positive cells in a tumor. Furthermore, heatmaps can help to better localize proliferation hotspots in whole tissue slides as recently described [26]. This data could potentially better forecast the behavior of a specific tumor than simple estimations of the proliferation activity as done so far. In vulvar cancer, there are some hints that the distribution of proliferation hotspots may be of clinical relevance [27]. Consequently, deep learning could be used to automate a prognosis and therapy recommendation. By implementing an adequate therapy at the right time, good prognosis might be expected [28]. The use of Ki-67 as a prognostic parameter in vulva carcinoma could be interesting, since advanced tumors have a poor prognosis and treatment decisions are complex [29, 30]. In this way, the cases that can benefit from a chemotherapy could be detected at early stage.
Some limitations of this study should be mentioned. (1) The available data was limited; statements regarding the accuracy in larger datasets cannot be made. However, it is remarkable that we could achieve such a high accuracy even though usually a lot more data is required. (2) No cross-validation was done, which would have improved the validation. Also, data augmentation might have introduced a sampling bias. (3) Furthermore, there were not the same number of spots per category. (4) Since the manual evaluation was made by one person, the classification was not performed objectively nor standardized. Despite the examiner-dependence a high accuracy was achieved in distinguishing the different classes in the training of the dCNN. (5) A variable rescaling factor was used in the labeling, and potentially different results could have been generated by using a constant rescale factor. (6) Since no classification for Ki-67 in vulva carcinoma has been established, the results are not directly applicable to the clinical routine. (7) No other machine learning technique was examined and used; therefore, no statement can be made if other algorithms would have brought up more precise results. (8) In the human readout, no gold standard could be applied for the classification of the images; therefore, interreader variability was applied for validation. (9) Finally, no independent dataset was available for validation and data from the same spot might have been used for training and validation due to the limited amount of data; therefore, the results may be overoptimistic regarding the generalization.
In conclusion, we were able to demonstrate the feasibility of a standardized classification of Ki-67 through a dCNN with an accuracy of 96.1% after 150 epochs. The implementation of the dCNN in the pathologist's daily routine could be used to simplify the pathologist's workflow by producing examiner-independent and standardized results. It cannot replace pathologists but could be a support by accelerating the workflow and help decrease the assigned workload. It can be used for quality control, or to train less experienced pathologists.
Declarations
Author contribution statement
Matthias Choschzick: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mariam Alyahiaoui: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Alexander Ciritsis, Cristina Rossi: Contributed reagents, materials, analysis tools or data; Wrote the paper.
André Gut: Analyzed and interpreted the data.
Patryk Hejduk: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Andreas Boss: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Clinical Research Priority Program Artificial Intelligence in oncological Imaging of the University of Zurich.
Data availability statement
The authors do not have permission to share data.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Litjens G. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Yasaka K. Deep learning with convolutional neural network in radiology. Jpn. J. Radiol. 2018;36(4):257–272. doi: 10.1007/s11604-018-0726-3. [DOI] [PubMed] [Google Scholar]
- 3.Araujo T. Classification of breast cancer histology images using Convolutional Neural Networks. PloS One. 2017;12(6) doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheikhzadeh F. Automatic labeling of molecular biomarkers of immunohistochemistry images using fully convolutional networks. PloS One. 2018;13(1) doi: 10.1371/journal.pone.0190783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehteshami Bejnordi B. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 2018;31(10):1502–1512. doi: 10.1038/s41379-018-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueda D., Shimazaki A., Miki Y. Technical and clinical overview of deep learning in radiology. Jpn. J. Radiol. 2019;37(1):15–33. doi: 10.1007/s11604-018-0795-3. [DOI] [PubMed] [Google Scholar]
- 7.Scholzen T., Gerdes J. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 2000;182(3):311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Endl E., Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp. Cell Res. 2000;257(2):231–237. doi: 10.1006/excr.2000.4888. [DOI] [PubMed] [Google Scholar]
- 9.Gerdes J. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 1984;133(4):1710–1715. [PubMed] [Google Scholar]
- 10.Yang C. Ki67 targeted strategies for cancer therapy. Clin. Transl. Oncol. 2018;20(5):570–575. doi: 10.1007/s12094-017-1774-3. [DOI] [PubMed] [Google Scholar]
- 11.Li L.T. Ki67 is a promising molecular target in the diagnosis of cancer (review) Mol. Med. Rep. 2015;11(3):1566–1572. doi: 10.3892/mmr.2014.2914. [DOI] [PubMed] [Google Scholar]
- 12.Takkem A. Ki-67 prognostic value in different histological grades of oral epithelial dysplasia and oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2018;19(11):3279–3286. doi: 10.31557/APJCP.2018.19.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gioacchini F.M. The clinical relevance of Ki-67 expression in laryngeal squamous cell carcinoma. Eur. Arch. Oto-Rhino-Laryngol. 2015;272(7):1569–1576. doi: 10.1007/s00405-014-3117-0. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura R. Clinical significance of Ki-67 in neoadjuvant chemotherapy for primary breast cancer as a predictor for chemosensitivity and for prognosis. Breast Cancer. 2010;17(4):269–275. doi: 10.1007/s12282-009-0161-5. [DOI] [PubMed] [Google Scholar]
- 15.Urruticoechea A., Smith I.E., Dowsett M. Proliferation marker Ki-67 in early breast cancer. J. Clin. Oncol. 2005;23(28):7212–7220. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 16.Forster S., Tannapfel A. [Use of monoclonal antibodies in pathological diagnostics] Internist (Berl) 2019;60(10):1021–1031. doi: 10.1007/s00108-019-00667-1. [DOI] [PubMed] [Google Scholar]
- 17.Shui R. An interobserver reproducibility analysis of Ki67 visual assessment in breast cancer. PloS One. 2015;10(5) doi: 10.1371/journal.pone.0125131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung Y.R. Interobserver variability of Ki-67 measurement in breast cancer. J. Pathol. Transl. Med. 2016;50(2):129–137. doi: 10.4132/jptm.2015.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madabhushi A. Digital pathology image analysis: opportunities and challenges. Imag. Med. 2009;1(1):7–10. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barragan-Montero A. Artificial intelligence and machine learning for medical imaging: a technology review. Phys. Med. 2021;83:242–256. doi: 10.1016/j.ejmp.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y., Zhao H., Wong S.T.C. Deep learning powers cancer diagnosis in digital pathology. Comput. Med. Imag. Graph. 2021;88:101820. doi: 10.1016/j.compmedimag.2020.101820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustreo S. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: a large case series study with a long-term follow-up. Breast Canc. Res. Treat. 2016;157(2):363–371. doi: 10.1007/s10549-016-3817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciritsis A. Determination of mammographic breast density using a deep convolutional neural network. Br. J. Radiol. 2019;92(1093):20180691. doi: 10.1259/bjr.20180691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha M. An advanced deep learning approach for Ki-67 stained hotspot detection and proliferation rate scoring for prognostic evaluation of breast cancer. Sci. Rep. 2017;7(1):3213. doi: 10.1038/s41598-017-03405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raina R. Self-taught learning: transfer learning from unlabeled data. Proc. Twenty-fouth Int. Conf. Mach. Learn. 2007;227 [Google Scholar]
- 26.Wessel Lindberg A.S. Quantitative tumor heterogeneity assessment on a nuclear population basis. Cytometry. 2017;91(6):574–584. doi: 10.1002/cyto.a.23047. [DOI] [PubMed] [Google Scholar]
- 27.Hantschmann P. Tumor proliferation in squamous cell carcinoma of the vulva. Int. J. Gynecol. Pathol. 2000;19(4):361–368. doi: 10.1097/00004347-200010000-00011. [DOI] [PubMed] [Google Scholar]
- 28.Canavan T.P., Cohen D. Vulvar cancer. Am. Fam. Physician. 2002;66(7):1269–1274. [PubMed] [Google Scholar]
- 29.Woelber L. Clinical management of primary vulvar cancer. Eur. J. Canc. 2011;47(15):2315–2321. doi: 10.1016/j.ejca.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Rogers L.J., Cuello M.A. Cancer of the vulva. Int. J. Gynaecol. Obstet. 2018;143(Suppl 2):4–13. doi: 10.1002/ijgo.12609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors do not have permission to share data.