Abstract
Background & Aims
Microfold cells (M cells) are immunosurveillance epithelial cells located in the Peyer’s patches (PPs) in the intestine and are responsible for monitoring and transcytosis of antigens, microorganisms, and pathogens. Mature M cells use the receptor glycoprotein 2 (GP2) to aid in transcytosis. Recent studies have shown transcription factors, Spi-B and SRY-Box Transcription Factor 8 (Sox8). are necessary for M-cell differentiation, but not sufficient. An exhaustive set of factors sufficient for differentiation and development of a mature GP2+ M cell remains elusive. Our aim was to understand the role of polycomb repressive complex 2 (PRC2) as an epigenetic regulator of M-cell development. Estrogen-related–receptor γ (Esrrg), identified as a PRC2-regulated gene, was studied in depth, in addition to its relationship with Spi-B and Sox8.
Methods
Comparative chromatin immunoprecipitation and global run-on sequencing analysis of mouse intestinal organoids were performed in stem condition, enterocyte conditions, and receptor activator of nuclear factor κ B ligand–induced M-cell condition. Esrrg, which was identified as one of the PRC2-regulated transcription factors, was studied in wild-type mice and knocked out in intestinal organoids using guide RNA's. Sox8 null mice were used to study Esrrg and its relation to Sox8.
Results
chromatin immunoprecipitation and global run-on sequencing analysis showed 12 novel PRC2 regulated transcription factors, PRC2-regulated Esrrg is a novel M-cell–specific transcription factor acting on a receptor activator of nuclear factor κB ligand–receptor activator of nuclear factor κB–induced nuclear factor-κB pathway, upstream of Sox8, and necessary but not sufficient for a mature M-cell marker of Gp2 expression.
Conclusions
PRC2 regulates a significant set of genes in M cells including Esrrg, which is critical for M-cell development and differentiation. Loss of Esrrg led to an immature M-cell phenotype lacking in Sox8 and Gp2 expression. Transcript profiling: the data have been deposited in the NCBI Gene Expression Omnibus database (GSE157629).
Keywords: PRC2, Microfold Cells, Esrrg, RankL, Gut Immunity
Abbreviations used in this paper: ChIP-seq, chromatin immunoprecipitation sequencing; FAE, follicle-associated epithelium; ENR500, epidermal growth factor, Noggin, R-spondin 500 ng/mL media; ENRI media, epidermal growth factor, Noggin, R-spondin, Wnt inhibitor IWP2 media; Esrrg, estrogen-related receptor γ; GALT, gut-associated lymphoid tissue; GP2, glycoprotein 2 receptor; Gro-seq, global run-on sequencing; KO, knockout; LTβR, lymphotoxin-β receptor; M cell, Microfold cell; NF-κB, nuclear factor-κB; PBS, phosphate-buffered saline; PP, Peyer’s patch; PRC2, polycomb repressive complex 2; Rank, receptor activator of nuclear factor κB; RankL, receptor activator of nuclear factor κB ligand; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; WENRC media, Wnt, epidermal growth factor, Noggin, R-spondin, Chir media
Graphical abstract
Summary.
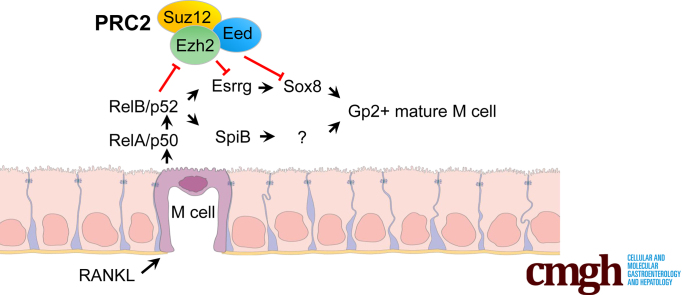
Chromatin immunoprecipitation and global run-on sequencing analysis of gut Microfold cells (M cells) showed 12 previously unknown novel transcription factors and, one of them, estrogen-related–receptor γ, plays a critical role in M-cell differentiation. Lack of estrogen-related–receptor γ showed an immature and nonfunctional M-cell phenotype.
The gut-associated lymphoid tissue (GALT) is involved in immune surveillance of antigens, microorganisms, and foreign pathogens that constantly thrive on the mucosal surface of the intestinal tract. The GALT is the immune initiation site against mucosal antigens and houses specialized gut immune epithelial cells known as microfold cells (M cells). These cells occur on the epithelial surface of lymphoid nodules of the GALT, which includes Peyer's patches (PPs) in the terminal ileum, solitary lymphoid nodules scattered throughout the small intestine, the appendix, and rectal patches in the terminal colon.1, 2, 3 The principal role of M cells is the uptake and transcytosis of luminal antigens into the GALT because they have a high phagocytic and transcytosis capacity, which is responsible for the rapid transport of bacterial antigens to antigen-presenting immature dendritic cells.4,5 Subsequently, these dendritic cells undergo maturation and activate antigen-specific naive T cells, which support B-cell activation, ultimately resulting in the generation of IgA-producing plasma cells.6 It has been shown previously that the absence of M cells or their antigen uptake receptor glycoprotein 2 (GP2) impairs the mucosal immune responses by T cells in mice infected with Salmonella enterica serovar Typhimurium. This is predominantly owing to a lack of bacterial transcytosis by the mature GP2 receptor into the GALT.7,8 Correspondingly, perturbances in transcytosis of Yersinia enterocolitica in PPs were observed in Allograft inflammatory factor 1 (Aif1) mutant mice.9 Recently, it was shown that M cells self-regulate their differentiation by expressing osteoprotegerin, a soluble inhibitor of receptor activator of nuclear factor-κB ligand (RANKL), which suppresses the differentiation of adjacent follicle-associated epithelium (FAE) cells into M cells. This self-regulatory machinery of M-cell density is necessary because Opg−/− mice are highly susceptible to mucosal infection by pathogenic bacteria because of the augmentation of bacterial translocation via M cells.10 Overall, defects in M-cell–dependent antigen uptake led to a decrease in production of antigen-specific secretory IgA in the gut.5,9,11
M-cell differentiation from cycling intestinal crypt cells that express Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 (Lgr5) and receptor activator of nuclear factor κB (RANK) receptors is induced by the RankL. RankL is secreted by stromal cells, also known as M-cell inducer cells or immune cells in the subepithelial dome.1,12 RankL-deficient mice have very few M cells, but exogenous administration of recombinant RankL was able to mitigate that loss.13 RankL binding to Rank receptor leads to the activation of the intracellular adaptor molecule of RANK; TNF Receptor Associated Factor 6 (TRAF6), which in turn leads to activation of both canonical (RelA/p50 heterodimer) and noncanonical nuclear factor-κB (NF-κB) (RelB/p52 heterodimer) activation.14, 15, 16 The canonical RelA/p50 activation led to expression of early M-cell markers such as Marcks like 1 (MarcksL1) and Chemokine (C-C motif) ligand 9 (CCL9), whereas noncanonical RelB/p52 activation led to expression of Spi-B and Sox8 transcription factors, both deemed essential to maturation of M cells.8,16 Along with RankL, expression of Spi-B and Sox8 are essential for the development of GP2-positive M cells. Both Spi-B and Sox8 mutant mice showed the absence of mature M cells with GP2, whereas Marcksl1+AnnexinV+ immature M cells were intact.7,8,17,18 Spi-B-/- still showed Sox8 expression and Sox8-/- mice expressed Spi-B, and even though both mice had activation of both NF-κB transcription pathway, p50/RelA, and p52/RelB, they still showed an immature M-cell phenotype lacking the expression of Gp2.16 Taken together, despite their critical role in the onset of mucosal immune responses, M-cell development and their differentiation into maturity have not yet been fully characterized, partly because of their rarity in the gastrointestinal tract.19 Importantly, the sole overexpression of Spi-B and Sox8 are not sufficient for the induction of GP2 receptor (ie, M-cell maturation), suggesting that additional M-cell–specific transcription factors are needed.8
Intestinal cell differentiation, development, and functionality are regulated by several factors, one of the indispensable ones being polycomb group proteins. Polycomb group proteins are essential for embryonic stem cell self-renewal and pluripotency, but they also are necessary for the maintenance of cell identity and cell differentiation throughout life.20 They broadly form 3 groups of polycomb-repressive complexes (PRCs) known as PRC1, PRC2, and polycomb repressive DeUBiquitinase, each of these complexes reassemble chromatin by explicitly defined mechanisms that involve variable configurations of core and accessory subunits. This configuration is shown by the way PRC2 catalyzes trimethylation of histone H3 lysine 27 (H3K27me3) and presents a binding site for PRC1 in embryonic stem cells.21 Previously, it has been shown that PRC2 played a repressive role of expression of developmental regulators necessary for cell differentiation.22 Interestingly, genes critical for cell identity lose their methylation on H3 lysine K27, whereas genes that regulate alternate cell types keep their methylation and remain repressed.23 In human embryonic stem cells, Wingless/integrated (Wnt)-signaling genes are bound by PRC2, analogously this also is shown in adult tissues (eg, in adipogenesis).24 The integrity and homeostasis of healthy intestine is regulated partly by canonical Wnt signaling and it also has been shown that secretory and absorptive progenitor cells show comparable levels of histone modifications at most of the same cis elements in the genome.25 Our study and others have found out how PRC2 regulates a substantial subset of genes that were involved in canonical Wnt signaling and contributed to the differentiation of Lgr5-expressing stem cells to secretory and absorptive cell types in the intestine.26, 27, 28
To further understand the complexity of M-cell differentiation, we asked if PRC2 regulates M-cell differentiation. We used high-throughput tools such as chromatin immunoprecipitation sequencing (ChIP-seq) and global run-on sequencing (Gro-seq) to identify factors that contribute to the function and development of M cells in the intestine. We identified a total of 12 transcription factors that are regulated by PRC2 exclusively during M-cell differentiation, of which 6 were down-regulated and 6 were up-regulated. One of the M-cell–specific transcription factors, estrogen-related receptor γ (Esrrg), was found to be essential for the differentiation of mature GP2+ M cells in vitro. We found that Esrrg was expressed exclusively in M cells in Peyer's patches and was shown to be critical for the activation of Sox8 transcription factor. Esrrg expression was intact even in Sox8-deficient mice, and was dependent on the activation of noncanonical NF-κB signaling. These observations show that Esrrg is a crucial player in the differentiation and functionality of a mature GP2+ M cell.
Results
PRC2 Does Not Restrict M-Cell Differentiation
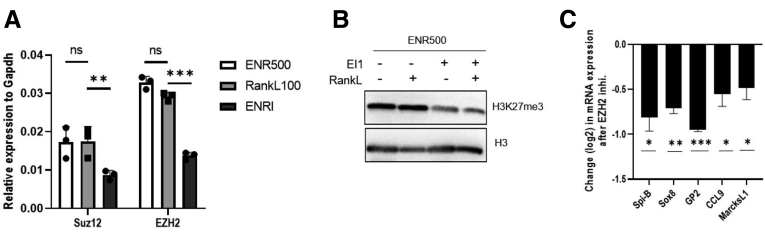
We and others have previously shown that disrupting PRC2 activity leads to a precocious expression of terminal differentiation markers of intestinal epithelium.27, 28, 29 Moreover, PRC2 has been shown to preserve intestinal progenitors and restrict secretory cell differentiation.26 Contrary to absorptive cell differentiation, when organoids were treated with RANKL the level of expression of PRC2 members Enhancer of Zeste homolog 2 (Ezh2) and Supressor of Zeste homolog 12 (Suz12) were comparable with the levels in organoids grown in standard organoid culture media with ENR500 (epidermal growth factor, Noggin, R-spondin 500 ng/mL) (Figure 1A). Next, we asked if PRC2 inhibition (Figure 1B) can augment M-cell differentiation and we saw that, contrary to enterocyte differentiation, expression of all M-cell markers are down-regulated when the activity of PRC2 is inhibited pharmacologically during the RANKL-induced, M-cell differentiation (Figure 1C).27,29
Figure 1.
PRC2 members are expressed in M cells. (A) RT-qPCR analyses of the expression of Suz12 and Ezh2 in mouse intestinal organoids grown in ENR, ENR+RankL, and ENRI conditions. (B) Immunoblot of H3K27me3 and H3 in organoids treated with Ezh2 inhibitor (EI1) inhibitor. Data are representative of 2 independent experiments. (C) RT-qPCR analyses of the expression of M-cell marker genes in RankL-treated organoids with or without EZH2 inhibitor EI1. An unpaired 2-tailed Student t test was performed. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005, N = 3 independent experiments. Values are presented as means ± SD. Gapdh, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.
PRC2 Regulated Genes During M-Cell Differentiation
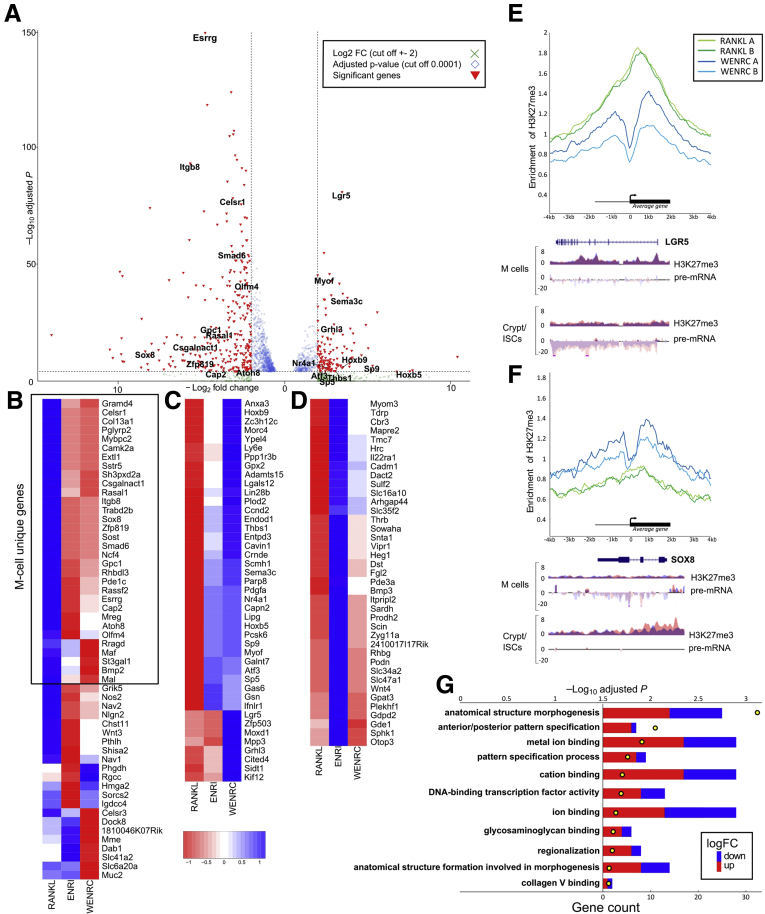
It has been shown previously that PRC2 regulates transcription factors that are necessary to intestinal stem cell maintenance and differentiation (eg, Achaete-Scute Family BHLH Transcription Factor 2 [Ascl2]27,30 and Atonal BHLH Transcription Factor 1 [Atoh1]26). Because PRC2 mainly regulates genes involved in development or signaling,31 we reasoned that identifying genes regulated by PRC2 during M-cell differentiation might show the gene network necessary to this cell type in the intestine. M-cell differentiation was induced in mouse intestinal organoids with recombinant RankL, gene expression was analyzed with Gro-Seq, and PRC2 target genes were identified with ChIP-seq by using H3K27me3 antibody. Genes expressed differentially after RankL treatment are shown in Figure 2A (Supplementary Table 1). ChIP-seq performed with H3K27me3 antibody showed a significant number of genes regulated by PRC2 for M-cell differentiation. When comparing our 3 different Chip-seqs (organoids in Wnt, epidermal growth factor, Noggin, R-spondin, Chir [WENRC] media [stem cell conditions]; epidermal growth factor, Noggin, R-spondin, Wnt inhibitor IWP2 [ENRI] media [enterocyte conditions]; and RankL media [M-cell differentiation]), we observed that, in M cells, 38 (9.2%) and 35 (10.3%) genes were up-regulated but silenced by PRC2 in WENRC and ENRI, respectively. Thirty-two PRC2 target genes were uniquely up-regulated during M-cell differentiation but not in enterocyte differentiation. Forty-six (27.7%) and 52 (11.5%) PRC2 target genes were silenced in organoids treated with RANKL, but expressed in stem cell and differentiation conditions, respectively. Forty-two genes were uniquely silenced in M-cell, but not in enterocyte, differentiation. PRC2 target genes are shown in Figure 2B–D, and the M-cell–specific accumulation and ablation of H3K27me3 signal in the gene promoters are shown in Figure 2E. Gene ontology analyses indicated that PRC2 regulates many DNA-binding transcription factors during M-cell differentiation (Figure 2G). A total of 12 transcription factors were expressed differentially: 6 were silenced by PRC2 in M cells and 6 were expressed specifically in M cells but repressed by PRC2 in organoids grown in both stemness and enterocyte conditions. The 6 PRC2 target genes expressed specifically in M cells were Sox8, Atoh8, Esrrg, Smad6, Maf, and Zfp819. The 6 genes silenced were Hoxb5, Hoxb9, Sp9, Sp5, Nr4a1, and Atf3.
Figure 2.
PRC2-regulated genes during M-cell differentiation. (A) Differentially expressed genes during M-cell differentiation detected with Gro-Seq. Signal is depicted by volcano plot comparing organoids before and after RankL treatment. X-axis and Y-axis indicate the log2 fold change and –log10 adjusted P value. Differentially expressed genes are marked (Gro-seq with log2 fold change cut-off value at ±2, P < .0001). Up-regulated genes from ENRI conditions were removed to show only RankL-specific regulation. (B) Genes up-regulated in M cells compared with stem cells and enterocytes. Heatmap of differentially expressed (Gro-seq with log2 fc cut-off value at ±2 and P value < .0001) and H3K27me3 regulated (log2 fc ±2, P < 10-6) genes in RankL, ENRI, and WENRC-treated organoids showing centered log2 fold change. (C) Genes down-regulated by H3K27me3 in M cells and enterocytes compared with stem cells. (D) Genes down-regulated by H3K27me3 in stem cells and M cells compared with enterocytes. Composite enrichment analysis of H3K27me3 signal density +-4000 bases around transcription start sites in genes (E) specifically silenced by PRC2 in M cells and (F) specifically expressed in M cells. (E) Example genes Lgr5 (below) and (F) Sox8 (below). (G) Gene ontologies enriched (P < .05) in differentially expressed PRC2-target genes between M cells and stem cells (Gro-seq log2 fc ±2, P < .0001, and H3K27me3 log2 fc ±2, P < 10-6) in molecular function and biological process. LogFC shows the direction of gene expression between M cells and stem cells. ISC, intestinal stem cells; mRNA, messenger RNA.
Esrrg Is Expressed in M Cells and Induced by Rank–RankL Signaling
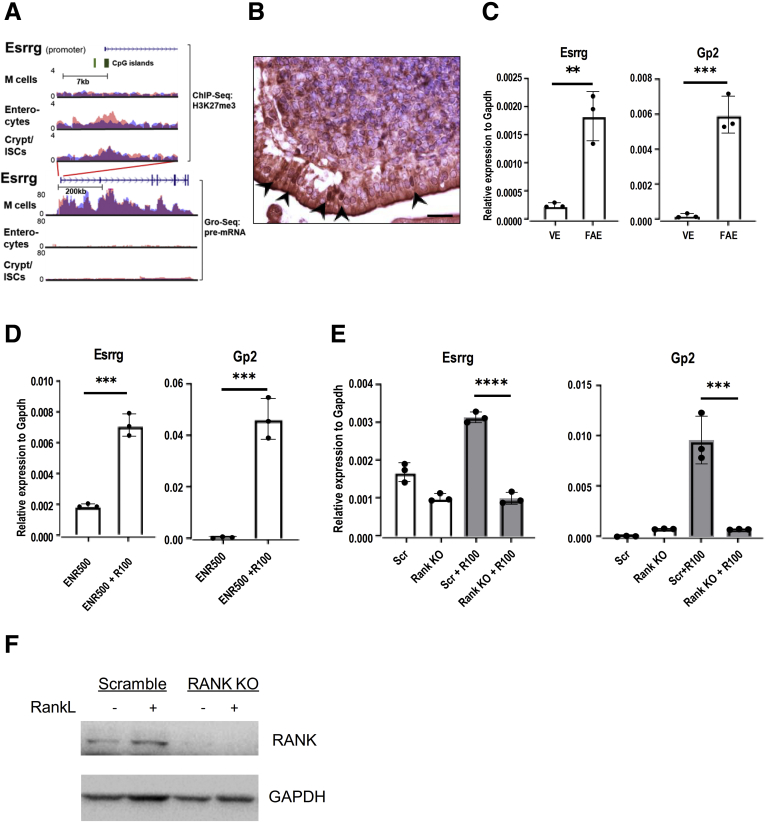
Of the 6 transcription factors that were expressed specifically in M cells in a PRC2-dependent manner (Figure 2A and B), Esrrg turned up as one of the most highly expressed PRC2-regulated transcription factors during M-cell differentiation (log2 fold changes, –6.64 RankL vs ENRI and –4.76 in RankL vs WENRC comparisons) (Figure 3A). Immunohistochemistry analysis for Esrrg in PP showed that Esrrg was localized in the FAE cells (Figure 3B). RNA was isolated from FAE isolated from PP and villus epithelium and the reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis confirmed that the Esrrg was enriched significantly in FAE (Gp2 as a marker) when compared with villus epithelium (Figure 3C). To ascertain that Esrrg is a novel RankL-induced M-cell gene, RT-qPCR analyses were performed for organoids before and after 4 days of RankL treatment. The expression of Esrrg, together with M-cell marker Gp2, was induced significantly with RankL treatment (Figure 3D). To confirm that Esrrg expression was regulated specifically by Rank-Rankl signaling, we generated Rank-deficient mouse intestinal organoids using the Lenti-V2 CRISPR/Cas9 plasmid and found that in Rank-deficient organoids, RankL treatment did not induce the expression of Esrrg (Figure 3E). The Rank knockout (KO) organoids were validated by immunoblot analysis (Figure 3F).
Figure 3.
Esrrg is expressed in FAE in PPs and is dependent on Rank–RankL signaling. (A) H3K27me3 occupancy at CpG islands spanning the promoter and first exon of the Esrrg gene in organoids treated with RankL (M cells) or inhibited with IWP2 (enterocytes) or treated with Wnt3a and Chir99021 (crypt/intestinal stem cells). Below, pre–messenger RNA (mRNA) expression of Esrrg in organoids treated as described earlier (y-axis: normalized tag count, ENR500 = R-spondin 500 ng/mL, R100 = Rankl 100 ng/mL). (B) Section of PP from wild-type mice stained with Esrrg antibody. Arrowheads indicating Esrrg expression in the nuclei of M cells in FAE. (C) RT-qPCR analysis of Esrrg and Gp2 in the FAE and villous epithelium (VE) from C57BL/6JRj mice (N = 3 from wild-type mice). (D) Organoids generated from wild-type mice were stimulated with 100 ng RankL for 4 days. Esrrg and Gp2 expression was examined by qPCR analysis. (E) Rank KO organoids and Scrambled organoids generated by lentiCRISPR v2 were incubated with RankL for 4 days, Esrrg and Gp2 expression was analyzed by quantitative RT-qPCR. (C–E) An unpaired 2-tailed Student t test was performed for 3 independent experiments. ∗∗∗∗P < .0005, ∗∗∗P < .005, and ∗∗P < .01. Values are presented as means ± SD. (F) RANK protein expression in RANK KO and Scrambled cells generated by lentiCRISPR v2 genome editing in C57BL/6JRj intestinal organoids. Organoid lysates were analyzed by Western blot. Gapdh, glyceraldehyde-3-phosphate dehydrogenase.
Noncanonical NF-κB Activation Is Necessary and Sufficient for Esrrg Expression
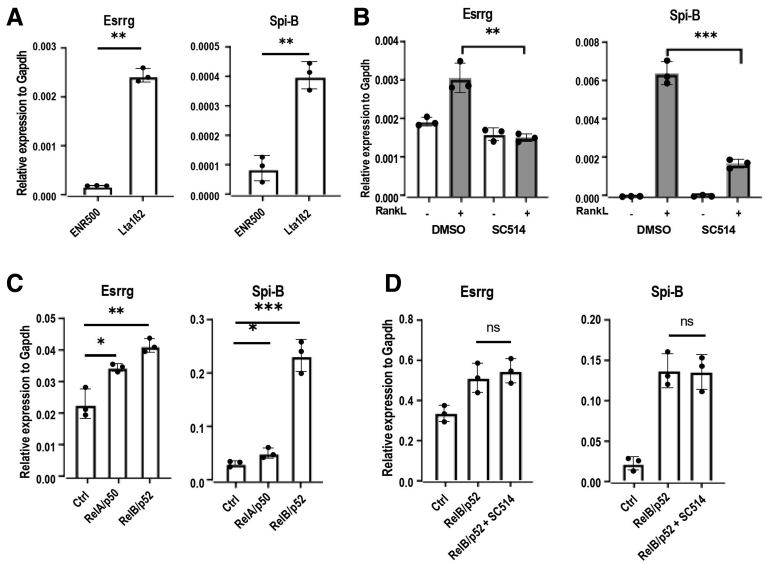
Rank–RankL signaling was shown previously to activate canonical as well as noncanonical NF-κB pathways. Lymphotoxin-β receptor (LTβR) signaling was implicated in inducing both classic p50–RelA and noncanonical p52–RelB heterodimers in PP.32 Mouse intestinal organoids were grown for 3 days in the presence of LTα1β2, the ligand of LTβR, and we observed that, similar to Spi-B, Esrrg expression also was increased significantly (Figure 4A). To identify if canonical NF-κB had a role in Esrrg expression specifically, mouse organoids were grown in the presence and absence of RankL and SC-514, a specific inhibitor of IκB kinase-β (inhibitor of nuclear factor kappa B).33 We found that inhibiting canonical NF-κB with SC-514 completely abrogated the expression of Esrrg (Figure 4B), as was reported similarly for Spi-B.16 Both p50-RelA and p52-RelB overexpression led to increased expression of Esrrg (Figure 4C). It has been shown previously that p50/RelA (canonical NF-κB) directly targets the transcription of RelB.34 The treatment of p52/RelB overexpression organoids with SC-514 could not suppress the activation of Esrrg (Figure 4D). To conclude, these data indicate that noncanonical NF-κB is necessary and sufficient to induce Esrrg.
Figure 4.
Esrrg expression is induced by RelB/p52 activation. (A) Ltα1β2 prominently up-regulated the expression of Esrrg in organoids. Organoids from C57BL/6JRj mice were stimulated with LTα1β2 for 3 days, and the gene expression was analyzed by qPCR. (B) Organoids from wild-type mice were stimulated with RankL for 3 days in the absence or presence of 125 μmol/L SC-514. Gene expression was analyzed by qPCR. (C) Organoids were transduced to express classic and noncanonical NF-κB and the expression of Esrrg and Spi-B (control) are represented relative to glyceraldehyde-3-phosphate dehydrogenase (Gapdh). (D) Organoids expressing p52 and RelB in the presence of SC-514 for 3 days, and the expression of Esrrg and Spi-B was analyzed by qPCR. Values in all are presented as the means ± SD and an unpaired 2-tailed Student t test was performed, N = 3 independent experiments. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005. Ctrl, control; DMSO, dimethyl sulfoxide.
Esrrg Expression Is Required for Sox8 Activation and Maturation of M Cells
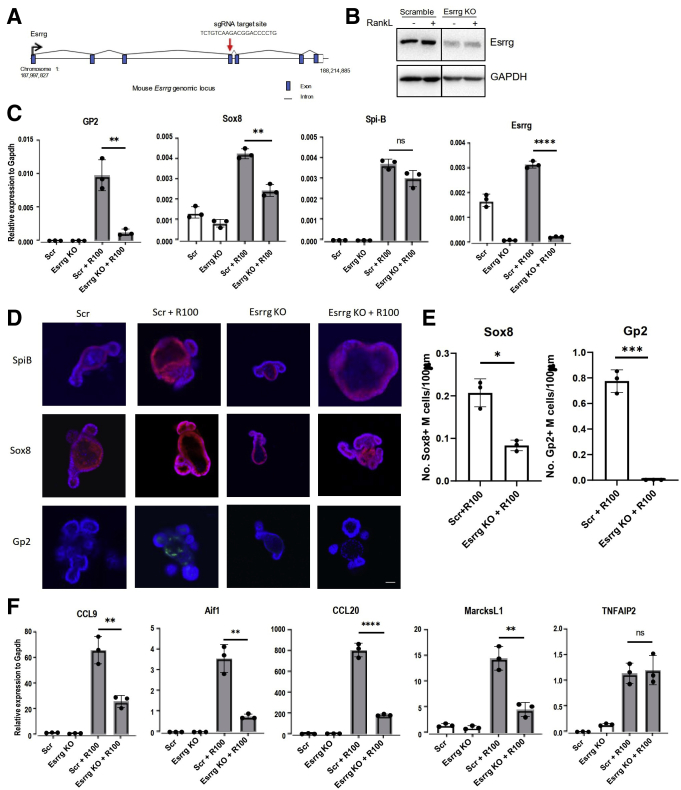
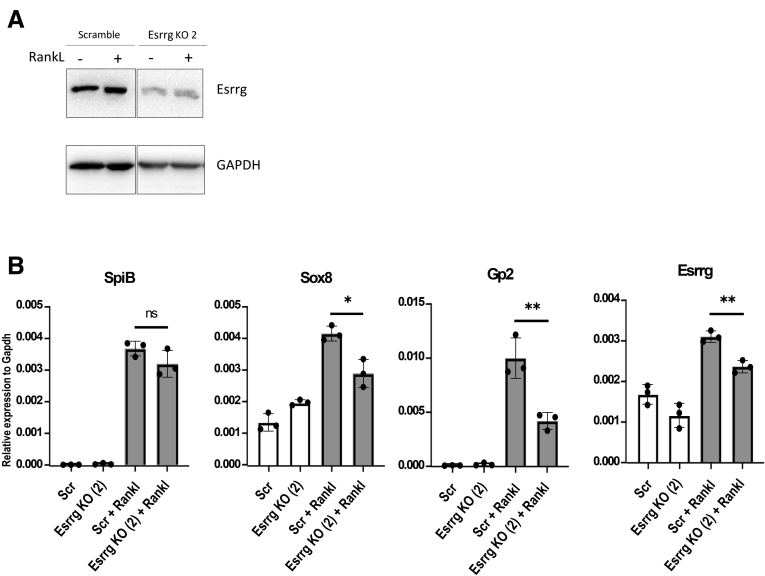
Given that Esrrg was expressed prominently in PP and regulated by noncanonical NF-κb expression, we sought to see if abolition of Esrrg had any effect on M-cell differentiation and development. To investigate this, mouse intestinal organoids deficient in Esrrg were generated by LentiCRISPR V2 CRISPR-Cas9 genome editing. Targeting of Esrrg by the guide RNA selected (Figure 5A) resulted in a significant reduction in expression of Esrrg protein (Figure 5B). Esrrg-deficient organoids were grown in the presence and absence of RankL for 3 days. RT-qPCR analysis showed that Gp2 was nearly absent and Sox8 expression was reduced significantly in Esrrg-targeted organoids (Figure 5C). Gp2 and Sox8 immunostaining in Esrrg-targeted organoids showed an absent and reduced expression, respectively (Figure 5D and E). However, expression of Spi-B and TNF Alpha Induced Protein 2 (Tnfaip2) remained unaffected in the Esrrg-deficient organoids (Figure 5C and E). Early M-cell differentiation markers such as CCL20, CCL9, and MarcksL1 were affected significantly by the lack of Esrrg as well (Figure 5F). Aif1, which is a regulatory gene for transcytosis in M cells, also was found to be affected severely by lack of Esrrg expression (Figure 5F). Our observations showed that Esrrg is required for the expression of Sox8 and for the early markers as well as late maturation steps in M-cell differentiation. To validate for off-target effects, we knocked out Esrrg with a second guide RNA; comparable results were observed (Figure 6A and B).
Figure 5.
Abolition of Esrrg impairs Sox8 activation and the functional maturation of M cells. (A) Schematic representation of Esrrg KO design by CRISPR-Cas9 genome editing in mouse intestinal organoids. Exon, intron, and genomic position are indicated. (B) Esrrg protein expression in Esrrg KO and Scrambled cells generated by lentiCRISPR v2 genome editing in C57BL/6JRj intestinal organoids. Organoid lysates were analyzed by Western blot. (C) qPCR analysis of M-cell–associated genes expressed in Scrambled and Esrrg KO stimulated by RankL for 4 days. (D) Immunostaining images for Spi-B (red), Sox8 (red), and GP2 (green) in Scrambled organoids with and without 100 ng RankL for 4 days and Esrrg KO with and without 100 ng RankL. Scale bars: 100 μm. (E) The number of GP2+ M cells and Sox8+ M cells per length of epithelium of organoids was compared between Scr+R100– and Esrrg KO+R100–treated organoids (n = 3). Images are representative of 3 independent experiments. (F) qPCR analysis of early markers of M-cell–associated genes expressed in vitro in the presence of RankL for 4 days. Values in all are presented as the means ± SD. Unpaired 2-tailed Student t test, N = 3 independent experiments. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ∗∗∗∗P < .0005. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; sgRNA, single guide RNA.
Figure 6.
Esrrg abolition by genomic RNA (gRNA) 2 impairs Sox8 activation and the functional maturation of M cells. (A) Esrrg protein expression in Esrrg KO2 with gRNA 2 and Scrambled cells generated by lentiCRISPR v2 genome editing in C57BL/6JRj intestinal organoids. Organoid lysates were analyzed by Western blot. (B) qPCR analysis of M-cell–associated genes expressed in Esrrg KO2 and Scrambled intestinal organoids stimulated by RankL for 4 days. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Values in all are presented as the means ± SD. Unpaired 2-tailed Student t test, N = 3 independent experiments ∗P < .05, ∗∗P < .01.
Esrrg Acts Upstream of Sox8 Expression
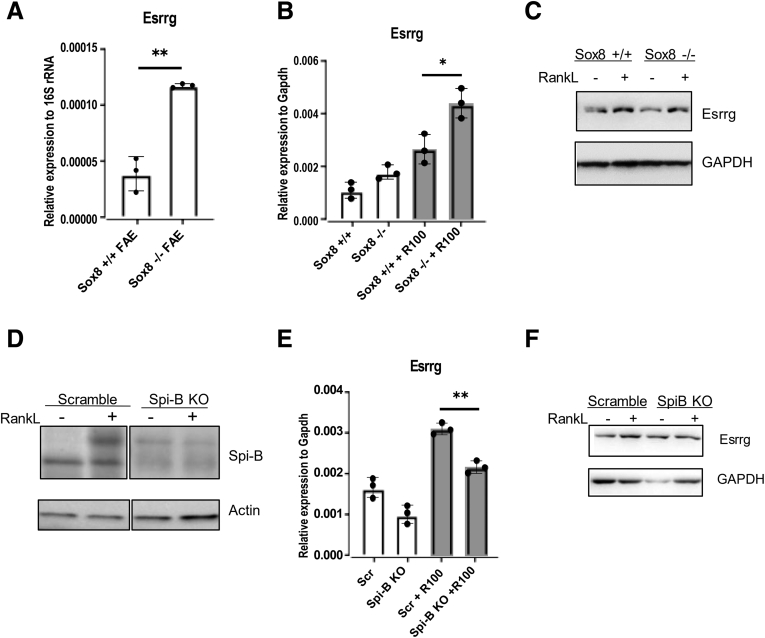
Spi-B and Sox8 were found to be 2 key transcription factors involved and essential for M-cell differentiation and regulation of expression of other M-cell–associated genes.7,8,17,18 Spi-B and Sox8 mutant mice lacked GP2+ mature M cells and were unable to transcytose antigens. However, it was observed that Spi-B was dispensable to the expression of Sox8 even though Spi-B expression was reduced moderately in the Sox8 mutant mice.8 To investigate if Esrrg is affected by knockout of Sox8, RNA was isolated from FAE of PPs from Sox8+/+ and Sox8-/- mice. RT-qPCR analysis of FAEs from PPs of Sox8+/+ and Sox8-/- showed that the expression of Esrrg was higher in Sox8-/- mice (Figure 7A). Organoids isolated from Sox8+/+ and Sox8-/- mice were treated with RankL and expression of Esrrg expression was analyzed by RT-qPCR and Western blot. The analysis showed Esrrg expression was intact and similar to the in vivo data (Figure 7B and C). This suggests that Esrrg acts upstream of Sox8 and could play a role in the activation of Sox8. Next, we explored how Esrrg was affected by Spi-B. Spi-B–deficient organoids were generated by LentiCRISPR V2 genome editing and grown in the presence and absence of RankL. qPCR analysis showed that Esrrg was affected moderately by the lack of Spi-B (Figure 7E). Western blot analysis also indicated that Esrrg was expressed less in Spi-B–deficient organoids (Figure 7F). Spi-B KO organoids were validated by immunoblot analysis (Figure 7D).
Figure 7.
Esrrg expression and its relation to other M-cell developmental markers Spi-B and Sox8. (A) Esrrg expression was unaffected by lack of Sox8 expression. qPCR analysis of Esrrg and Gp2 in the FAE and villous epithelium from Sox8+/+ and Sox8-/- mice. (B) Organoids generated from Sox8+/+ and Sox8-/- mice were stimulated with and without RankL for 4 days. Esrrg expression was examined by qPCR analysis. (C) Organoids isolated from Sox8 wild-type and Sox8 KO mice were lysed and analyzed by Western blot for Esrrg expression. (D) Spi-B protein expression in Spi-B KO and Scrambled cells generated by lentiCRISPR v2 genome editing in C57BL/6JRj intestinal organoids. Organoid lysates were analyzed by Western blot. (E) qPCR analysis of Esrrg in a Spi-B KO intestinal organoid by lentiCRISPR v2. (F) Organoid lysates for Esrrg in Scrambled and SpiB KO organoids were analyzed by Western blot. Values in all are presented as the means ± SD. Unpaired 2-tailed Student t test, N = 3. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; rRNA, ribosomal RNA.
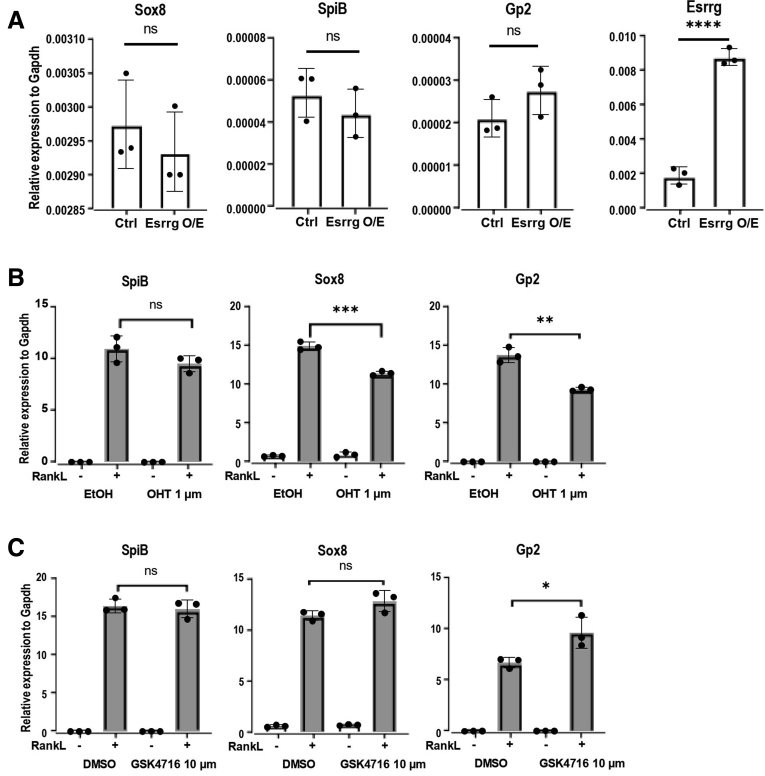
Overexpression of Esrrg Is Not Sufficient for Gp2+ M Cells but Esrrg Agonist Augmented Gp2 Expression
Spi-B expression in Sox8 KO mice and Sox8 expression in Spi-B KO mice did not lead to a mature GP2+ M-cell phenotype in either of these mice.7,8 Because we found that the expression of Gp2 was dependent on Esrrg (Figure 5C), we sought to investigate if the overexpression of Esrrg alone could lead to up-regulation of Sox8 or Gp2 expression. Esrrg cloned into CSII-CMV-MCS-IRES2-Bsd overexpression vector was transduced into mouse intestinal organoids. RT-qPCR analysis showed that Esrrg alone was not adequate enough to induce Gp2 or other M-cell–specific transcription factor such as Spi-B or Sox8 (Figure 8A). Esrrg is an orphan nuclear receptor without known natural ligands. However, 4-hydroxytamoxifen has been shown to bind and inhibit Esrrg activity and phenolic acyl hydrazones; GSk4716 was identified as an agonist that enables the activation of other co-activators and other downstream targets.35,36 The antagonist 4-hydroxytamoxifen along with RankL induced a similar result to the lack of Esrrg protein, Spi-B remained unaffected, although Sox8 and Gp2 were affected significantly (Figure 8B). Organoids treated with agonist GSK4716 and RankL did not show a significant increase in Sox8 or Spi-B expression, however, Gp2 expression was augmented slightly with GSK4716 (Figure 8C).
Figure 8.
Esrrg alone is not sufficient for maturation of Gp2+ M cells. (A) Intestinal organoids were dissociated and transduced by lentivirus encoding Esrrg. qPCR analysis of Spi-B, Sox8, and GP2 showed no significant changes. (B) Organoids were grown in the presence and absence of 100 ng/mL Rankl and 1 um tamoxifen-antagonist of Esrrg. Spi-B, Gp2, and Sox8 expression were analyzed with RT-qPCR. (C) Intestinal organoids from mice were grown in the presence and absence of 100 ng/mL Rankl and 10 um GSK 4718-agonist of Esrrg for 3 days. Spi-B, Gp2, and Sox8 were analyzed with RT-qPCR. Values are presented as the means ± SD. Unpaired 2-tailed Student t test, N = 3. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ∗∗∗∗P < .0005. Ctrl, control; DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; O/E, overexpression.
Discussion
Our data show the PRC2-regulated genes during the differentiation of intestinal microfold cells. Because PRC2 is the master regulator of development, it is very likely that many of the identified PRC2 targets that are induced specifically in M cells contribute to the maturation of this cell type. Transcription factors usually are expedient to development, and we identified 6 PRC2-regulated transcription factors up-regulated specifically after the RankL-induced M-cell differentiation. Among those was previously identified transcription factor Sox8, which we also here showed to be necessary for M-cell differentiation.8 Because Esrrg was clearly the most highly induced PRC2 target gene during M-cell differentiation, we studied it in more detail in intestinal organoids and showed that it is indispensable to the maturation of intestinal stem cells into GP2+ M cells. Esrrg is a member of the ESRR nuclear-receptor family, which also includes ESRRA and ESRRB.37 This subfamily of orphan nuclear receptors has been shown to share target genes, coregulatory ligands, and sites of action with ERs. Esrrg was implicated to control macrophage function indirectly through regulation of intracellular iron. In response to Salmonella typhimurium infection, hepatic expression of the hormone hepcidin is up-regulated by ERRγ downstream on interleukin 6 signaling.34,38 However, no prior data about the expression and function of Esrrg in M cells and M-cell induced transcytosis of antigens exist. We defined the specific expression of Esrrg by M cells in mouse FAEs and how its expression was up-regulated by induction of RankL and was under the influence of the RankL–Rank pathway. The loss of Esrrg led to a lack of expression of the GP2 receptor in Esrrg KO organoids, which is characteristic of a mature M cell, and a lack of GP2 in M cells has been shown to result in attenuation of antigen sampling and transcytosis. This distribution of Esrrg along with the phenotype of Esrrg KO organoid highlights a critical role for Esrrg in the maturation of functional M cells.
RelB/p52 activation was shown to up-regulate Spi-B.16 Similarly, here we show that Esrrg also is regulated by the activation of the noncanonical NF-κB pathway. We believe that the expression of Esrrg and Spi-B is regulated downstream in parallel by RankL-Rank-RelB/p52 signaling (Figure 9). Esrrg has been shown to behave as a constitutive activator of transcription.39 Here, we show that Esrrg is needed for the activation of Sox8. Sox8 was discovered to be indispensable for the expression of GP2, and Sox8 KO mice showed a decrease in uptake of antigens and a significant decrease of IgA+ immunoglobulins. Sox8 also was shown to bind directly to the Gp2 promoter along with SpiB.8 The significant decrease in Sox8 expression could explain why Esrrg KO organoids were not able to activate Gp2. Sox8 expression alone cannot lead to an increase in Gp2 expression because enhancer activation by SOX proteins require DNA binding partners specific for each member of the SOX family. These DNA binding partners aid in stabilizing the SOX family to their target regions.40 This suggests that Esrrg-activated Sox8 requires another molecule downstream of Spi-B or through another pathway to bind to Gp2 to induce its expression, however, further exploration is required to confirm this. Esrrg overexpression alone did not lead to expression of Sox8, confirming the need of a ligand and/or other factors to activate downstream targets.
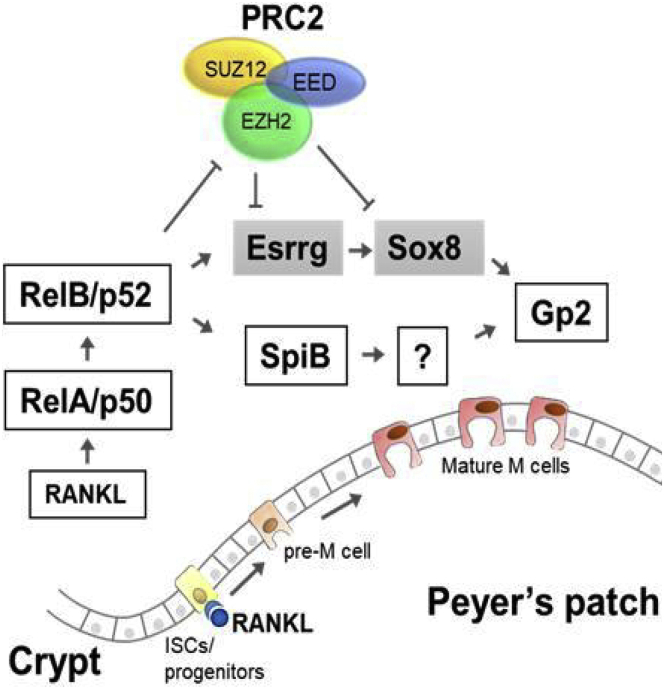
Figure 9.
PRC2 regulates the differentiation of M cells. In the absence of Rank–RankL signaling, Esrrg and Sox8 genes are repressed by PRC2 in the intestinal epithelium. When migrating progenitors with Rank receptors bind to RankL in PPs, this induces NF-κB signaling, leading to loss of H3K27me3 from the gene promoters and activation of Esrrg and Sox8. Sustained expression of Sox8 and GP2 and differentiation of M cells is dependent on Esrrg. ISC, intestinal stem cells.
A major portion of our investigation of Esrrg in M cells was conducted in vitro and Esrrg ablation in mouse models is required to further ascertain its role. Interestingly, Sox8 deficiency did not affect early M-cell markers, however, Esrrg KO organoids showed a drastic decrease in expression of early M-cell markers such as CCL9 and MarcksL1. Mature marker Gp2-receptor expression was decreased significantly as well. The loss of Aif1 expression in Esrrg KO means that transcytotic capacity of the M cells would be affected as well. Esrrg KO organoids also impaired the expression of Sox8, and in the Sox8 KO mice we observed that Esrrg expression still was intact and even observed to have a higher expression in vivo and in vitro, suggesting that Esrrg was not affected by the absence of Sox8 and possibly could be acting upstream of it. The significance of Esrrg was confirmed further with our antagonist and agonist experimental study. Although tamoxifen is known to trigger multiple signaling pathways in the cell, it has been identified as an antagonist for Esrrg receptor; tamoxifen was able to significantly decrease the expression of Gp2 and Sox8 similar to the Esrrg KO organoids. The treatment of intestinal organoids with RankL and Esrrg agonist GSK4716 augmented the expression of Gp2 when compared with organoids with just RankL treatment. This could be because GSK4716 binds to Esrrg and activates several other unknown downstream targets that combine to attenuate Gp2 expression. Esrr family members are known to be orphan receptors, meaning they might not need a ligand for its function, or the ligand remains unknown. Overexpression of Esrrg did not lead to an increase in Sox8 expression or other transcription factors necessary for mature M-cell differentiation, presumably because a specific ligand might be necessary. However, further studies are needed to prove this.
In conclusion, we identified several previously unknown PRC2-regulated genes implicated in M-cell differentiation. One of the genes we identified, Esrrg, is a key transcription factor, and therefore is required for the functional development and M-cell differentiation that is pertinent for constant surveillance of the mucosal lining of the gastrointestinal tract. We believe that the further exploration of other activators of Gp2 will lead to better elucidation of M-cell maturation and antigen transcytosis. This will create the potential to provide strategic innovation in support of mucosal/oral vaccine advancement.
Materials and Methods
Animals
C57BL/6JRj mice were purchased from Janvier labs (Le Genest-Saint-Isle, France) and were maintained with constant breeding. The Bac-Cre-ERT2;Sox9f/f;Sox8-/-;Rosa26Eyfp mice were a gift from Raphael Jimenez (University of Granada, Granada, Spain). These mice were backcrossed with C57BL/6JRj to isolate the Sox8-/- allele. Sox8+/- were bred to obtain Sox8-/- and littermate controls: Sox8+/- and Sox8+/+. F1–4 mice were used for gene or protein expressions. Genotyping of the wild-type, heterozygous, and deleted alleles was performed by PCR with the following primers: F1, 5’-GTCCTGCGTGGCAACCTTGG-3’; R1, 5’-GCCCACACCATGAAGGCATTC-3’; and F3, 5’-TAAAAATGCGCTCAGGTCAA-3’. Conventional conditions were observed for the maintenance of these mice at the pathogen-free animal facility of the faculty of Medicine and Health Technology. All animal experiments were approved by the Finnish National Animal Experiment Board (permit ESAVI/5824/2018).
Intestinal Organoid Culture
Intestinal crypt isolation and culture techniques were observed as previously established by the protocols of Sato and Clevers41 and de Lau et al.17 Mouse duodenum were cut longitudinally, and the villi were gently scraped off with 2 glass slides. After a couple of phosphate-buffered saline (PBS) washes, they were cut into 2-mm pieces and pipetted up and down 5 times in 15 mL PBS with a 10-mL pipette, this step was repeated 3 times with fresh PBS. The pieces were incubated in 10 mmol/L EDTA in PBS for 20 minutes, rocking at room temperature. The pieces were vigorously suspended in cold PBS and the mixture was strained through a 70-μm cell strainer (cat no: 22363548; Fisher Scientific, Waltham, MA). This mixture was enriched to crypt fraction through centrifugation at 150 × g for 5 minutes. The enriched crypts were embedded in Matrigel (cat no: 356255, lot 9119006; Corning, Corning, NY), and 30 uL were plated on a 24-well plate. Crypts were cultured in an optimal medium consisting of advanced Dulbecco’s modified Eagle medium/F12 (cat no: 12634010; Thermo Fisher Scientific, Waltham, MA) that contained HEPES (10 mmol/L, cat no: 15630-080; Sigma-Aldrich, St. Louis, MO), Glutamax (2 mmol/L, cat no: 35050-038; Thermo Fisher Scientific), penicillin-streptomycin (100 U/mL, cat no: 11659990; Sigma-Aldrich), B-27 supplement minus vitamin A (cat no: 17504-044; Thermo Fisher Scientific), N-2 supplement (cat no: 17502-001; Thermo Fisher Scientific), N-acetylcysteine (1 mmol/L, cat no: A9165, lot SLCB3719; Sigma-Aldrich), recombinant murine epidermal growth factor (50 ng/mL, cat no: PMG8043, lot 2135273; Gibco, Waltham, MA), recombinant murine Noggin (100 ng/mL, cat no: 250-38; PeproTech), and recombinant mouse R-spondin 1 (1 μg/mL, cat no: 120-38; R&D Systems, Minneapolis, MN). Media were changed every 2 days. For M-cell differentiation, recombinant mouse RankL (100 ng/mL, cat no: 315-11; PeproTech, Cranbury, NJ) was added to the media and incubated for 4 days. PRC2 was inhibited by addition of 5 μm of Ezh2 inhibitor (cat no: CAS 1418308-27-6 C; Calbiochem Chemicals, San Diego, CA). For activation of NF-κB, human LTα1β2 (1 μg/mL, cat no: 8884-LY; R&D Systems) was added into organoid cultures. Restriction of IκB kinase-β activity was achieved by adding SC-514 (125 μmol/L, cat no: 354812-17-2; Selleckchem, Houston, TX) for 3 days.42 (Z)-4-Hydroxytamoxifen (1 μm, cat no: 68392-35-8; Sigma-Adrich) was used as an antagonist of Esrrg, and GSK 4716 (10 μm, cat no: 101574-65-6; Sigma-Aldrich) was used as an agonist of Esrrg.
ChIP-Seq Analysis
Intestinal organoids (in ENR500, ENRI, and RankL culturing conditions) were isolated from Matrigel with Cell Recovery Solution (Corning). This was followed by washes with cold PBS and dissociation into single-cell suspension using TrypLE Express (cat no: 12604013; Thermo Fisher Scientific) and counted. Cells (10 × 106) of each condition were cross-linked with formaldehyde, after which nuclei were isolated with lysis buffers 1, 2, and 3 as described,43 and sonicated with a Covaris S220 ultrasonicator (Woburn, MA). The resulting nuclear extract was incubated with Dynal protein G beads, which were preincubated with 5 μg H3K27me3 (cat no: ab6002; Abcam, Cambridge, UK) or H3 antibody (cat no: ab1791; Abcam), respectively, at 4°C overnight. After washing and elution of bound complexes from the beads, cross-links were reversed by heating to 65°C. Immunoprecipitation and input DNA then were purified by treatment with RNase A, proteinase K, and phenol:chloroform extraction. The NEBnext UltraDNA-library preparation kit for Illumina (cat no: E7805; NEB, Ipswich, MA) was used to construct libraries from immunoprecipitation and input DNA and subjected to 50-bp, single-end, read sequencing with an Illumina (Ipswich, MA) Hiseq 2000 at EMBL Genecore (Heidelberg, Germany).
Gro-Seq Analysis
ENR500 and RankL-treated organoids were harvested (as in ChIP-Seq) and GRO-Seq was performed for an equal number of isolated nuclei. Nuclei extraction and a run-on reaction were performed as previously established.44 For each replicate, 3 million cells were suspended to a final volume of 80–200 μL of freezing buffer. TRIzol LS (cat no: 10296028; Life Technologies, Carlsbad, CA) was used to extract RNA and fragmented for 13 minutes in 70°C using RNA fragmentation reagents (cat no: AM8740; Life Technologies), and later purified by running through a RNase-free P-30 column (cat no: 7326250; Bio-Rad, Hercules, CA). T4 Polynucleotide Kinase was used to dephosphorylate RNA for 2 hours (cat no: M02014; New England Biolabs, Ipswich, MA), followed by heat-inactivation. A total of 65 μL of blocking solution was added (5× volume of 0.25 × SSPE, 1 mmol/L EDTA, 37.5 mmol/L NaCl, 0.05% Tween-20, 0.1% Polyvinylpyrrolidone, and 0.1% ultrapure bovine serum albumin for 1 hour in room temperature), the anti-bromodeoxyuridine bead slurry (cat no: sc-500780; Santa Cruz Biotech, Dallas, TX) suspended in 500 μL binding buffer (0.25 × Sodium Chloride-Sodium Phosphate-EDTA [SSPE], 1 mmol/L EDTA, 37.5 mmol/L NaCl, 0.05% Tween-20) was used to purify the dephosphorylated reaction. After binding for an hour in room temperature, the beads were washed 2× with binding buffer, 2× with low-salt buffer (0.2 × SSPE, 1 mmol/L EDTA, 0.05% Tween-20), 1× with high-salt buffer (0.2 × SSPE, 1 mmol/L EDTA, 135 mmol/L NaCl, 0.05% Tween-20), and, lastly, 2× with Tris elution buffer (1 × TE, 0.05% Tween-20). The elution of the RNA was completed with 130 μL elution buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 0.1% sodium dodecyl sulfate, 1 mmol/L EDTA, and 20 mmol/L dithiothreitol, followed by ethanol precipitation overnight. All buffers were supplemented with SUPERase ln (2 μL/10 mL, cat no: AM2694; Life Technologies). Library preparations were performed the next day as previously described.45 The library was amplified with 14 cycles and the final product of 190–135 bp was extracted from a 10% Tris/Borate/EDTA gel. The DNA was purified from the gel using the Gel Extraction Kit (cat no: KO961, ThermoFisher) and eluted in TE buffer (TE 0.1% Tween + 150 mmol/L NaCl). ChIP DNA Clean and Concentrator Kit (cat no: D5205; Zymo Research Corporation, Irvine, CA) was used to purify the library, the DNA was quantified with the Qubit fluorometer (Waltham, MA), and sequenced with an Illumina HiSeq 2000 at EMBL Genecore.
ChIP- and Gro-Seq Data Analyses
Analyses were performed as described previously.27,44,45 The data have been deposited in the NCBI Gene Expression Omnibus database (GSE157629). Gro-seq and ChIP-seq data for the individual genes are shown in Supplementary Tables 1 and 2, respectively.
Immunohistochemistry and Immunofluorescence
PPs from the ileum were isolated and washed with cold PBS and embedded into paraffin blocks. Sections from the blocks were rehydrated and washed with PBS. After incubation with 1% PBS/bovine serum albumin supplemented with 5% normal donkey serum for blocking, antigen retrieval was processed with citrate buffer, pH 6.0 (121°C for 5 min), and stained overnight at 4°C for Esrrg (cat no: ab49129; Abcam) and GP2 (cat no: D278-3; MBL, Woburn, MA) antibodies. This was followed by anti-rabbit secondary for Esrrg (cat no: A32731; ThermoFisher) and anti-rat secondary for GP2 (cat no: A48261; ThermoFisher). The sections were examined with a light microscope. For whole-mount immunostaining, crypt organoids were plated in an 8-well chamber and cultured for 4 days, after which they were fixed with 4% paraformaldehyde, followed by permeabilization with 0.1% Triton (Gibco) X-100. The organoids were stained with the following primary antibodies overnight at 4°C: rabbit anti–Spi-B (Spi-B [D3C5E], cat no: 14223; CST, Danvers, MA), rabbit anti-Sox8 (cat no: ab221053; Abcam), and rat anti-GP2 (cat no: D278-3; MBL). This was followed by incubation with secondary antibody anti-rabbit Alexa Fluor 568 (cat no: A-11011; ThermoFisher) for Spi-B and Sox8 and anti-rat Alexa Fluor 488 (cat no: A-11006; ThermoFisher) for GP2. Cells were analyzed with a Nikon (Melville, NY) A1R+ Laser Scanning Confocal Microscope after mounting with ProLong Diamond with 4′,6-diamidino-2-phenylindole mounting solution (cat no: 15810083; ThermoFisher).
Isolation of Villous Epithelium and FAE Cells
Villous epithelium and FAE were prepared by isolating ileal PPs and small pieces of ileum from the intestine. These pieces were washed in cold PBS and later incubated in 30 mmol/L EDTA (cat no: 1557-038; Gibco), 5 mmol/L dithiothreitol (cat number: R0861; ThermoFisher) in PBS, and gently shaken in ice on a rocker for 20 minutes. After which, surrounding epithelial cells were peeled off from lamina propria and PPs. FAE was carefully cleaned off from surrounding villous epithelium tissues with a 26-gauge needle under a stereo microscope.
CRISPR–Cas9 Gene Editing of Intestinal Organoids
Guide RNAs for Rank, Spi-B, and Esrrg were designed with the CRISPR design tool (http://crispr.mit.edu).46 The guides were cloned into lentiCRISPR v2 vector (52961; Addgene, Watertown, MA). The cloned vector was transfected into 293FT cells (cat no: R7007; ThermoFisher) and the supernatant was collected at 48 hours and concentrated with Lenti-X concentrator (cat number: 631231; Clontech, Mountain View, CA). The 293FT cell line was found to be negative for mycoplasma. Cultured intestinal organoids were grown in EGF, Noggin, Chir, Y-27632; epidermal growth factor, Noggin, Chir-99021 (cat no: S1263; Selleckchem) and Y-27632 (cat no: 72304; Selleckchem) 2 days before transduction. Organoids were dissociated into single cells mechanically along with TrypLE Express (Thermo Fisher Scientific) supplemented with 1000 U/mL DNase I for 5 minutes at 32°C. The single-cell suspension was washed once with Advanced Dulbecco’s modified Eagle medium and resuspended in transduction medium (ENR media supplemented with 1 mmol/L nicotinamide, Y-27632, Chir99021, 8 μg/mL polybrene, cat no: 28728-55-4; Sigma-Aldrich) and mixed with concentrated virus. The cell–virus mixture was spinoculated for 1 hour at 600 × g at 32°C followed by a 2- to 4-hour incubation at 37°C, after which they were collected and plated on 60% Matrigel overlaid with transduction medium without polybrene. Transduced organoids were selected with 2 rounds of 2 μg/mL of puromycin (cat no: P8833, 10 mg; Sigma-Aldrich) on day 2 and day 4, after which clones were expanded in maintenance ENR medium. KO was confirmed by Western blot to check for the expression of deleted gene.
Oligonucleotides used for generation of genomic RNAs were as follows: Esrrg (1) CACCGTCTGTCAAGACGGACCCCTG, AACCAGGGGTCCGTCTTGACAGAC, Esrrg (2) CACCGTGGCGTCGGAAGACCCACCA; AAACCAGGGGTCCGTCTTGACAGAC. Spi-B (1) CACCGAGACTCCTTCTGGGTACTGG, AAACCCAGTACCCAGAAGGAGTCTC; and Rank (1) CACCGAAAGCTAGAAGCACACCAG, AAACCTGGTGTGCTTCTAGCTTTC.
Lentivirus Infection for Overexpression
RelB, p52, RelA, and p50 plasmids were a gift from Hiroshi Ohno’s laboratory (RCIMS, Kangawa, Japan) and Esrrg complementary DNA was cloned by Twist Bioscience (San Francisco, CA). These were cloned into CSII-CMV-MCS-IRES2-Bsd vector, which was kindly provided by the RIKEN Bioresource Center (Ibaraki, Japan) and Hiroyuki Miyoshi. The same protocol for Crispr-Cas9 lentiviral generation and transduction was followed and cells were embedded into Matrigel and incubated for 2–3 days.
Immunoblotting
Organoids were recovered from Matrigel with Cell Recovery media (cat no: 354253; Corning). Organoids were washed with PBS and the cells were lysed with 2× Laemmli solution and boiled at 98ºC. Protein concentrations were measured by a Pierce (Waltham, MA) 660 nm Protein Assay Reagent and IDCR (cat no: 22660; ThermoFisher Scientific). Samples were loaded equally in terms of protein concentration into 10% Bis-Tris protein gels (cat no: 4561033; Bio-Rad) and blotted on nitrocellulose membranes. Membranes were incubated with primary antibodies: anti-Esrrg (cat no: ab49129; Abcam); anti–Spi-B (Spi-B D4V9S, cat no: 14337; CST); anti-H3K27me3 (cat no: ab192985; Abcam); anti-H3 (cat no: ab1791; Abcam), anti-Rank (cat no: MBS9133424; MyBioSource, San Diego, CA), and anti-glyceraldehyde-3-phosphate dehydrogenase (cat no: ab8245; Abcam) at 4°C overnight, and horseradish-peroxidase–conjugated anti-rabbit (1:5000, cat no: RABHRP1-10UL; Sigma-Aldrich) or anti-mouse (1:1000, cat no: 7076; CST) for 1 hour at room temperature. Signal was detected using ECL reagent (cat no: 2232; Amersham, Amersham, UK).
Real-Time RT-qPCR
Total RNA was prepared using TRIzol (cat no: 15596018; Life Technologies) from intestinal organoids and epithelium isolated from mice. Isolated RNA was transcribed to first-strand complementary DNA using the iScript complementary DNA synthesis kit (1708891; Bio-Rad). qPCR amplification was detected using Ssofast evergreen supermixes (172-5203; Bio-Rad). The specific primers used are listed in Table 1.
Table 1.
List of Oligonucleotides Primers Used for RT-qPCR
| Oligonucleotide | Sequence, 5’ to 3’ |
|---|---|
| Gapdh_fwd | TGTGTCCGTCGTGGATCTG |
| Gapdh_rev | CCTGCTTCACCACCTTCTTGA |
| Suz 12_fwd | GATGAGAAAGATCCAGAATGGC |
| Suz12_rev | ATAATTTTCTACAAACAGCATACAGGC |
| Ezh2_fwd | GTCTGATGTGGCAGGCTGG |
| Ezh2_rev | GCCCTTTCGGGTTGCATC |
| Spi-B_fwd | GGAGTCTTCTACGACCTGGACAG |
| Spi-B_rev | GCAGGATCGAAGGCTTCATAGG |
| Sox8_fwd | GGACCAGTACCCGCATCTCC |
| Sox8_rev | TTCTTGTGCTGCACACGGAGC |
| GP2_fwd | GTGTACAAGTTACAGGGTACCCC |
| GP2_rev | GACAAGTAATCTCACAATTCTTGG |
| CCL9_fwd | GCCCAGATCACACATGCAAC |
| CCL9_rev | AGGACAGGCAGCAATCTGA |
| MarcksL1_fwd | CCCGTGAACGGAACAGATGA |
| MarcksL1_rev | CCCACCCTCCTTCCGATTTC |
| Esrrg_fwd | GTGTCTCAAAGTGGGCATGC |
| Esrrg_rev | GCTGTTCTCAGCATCTATTCTGC |
| Aif1_fwd | GGATTTGCAGGGAGGAAAA |
| Aif1_rev | TGGGATCATCGAGGAATTG |
| CCL20_fwd | TGTACGAGAGGCAACAGTCG |
| CCL20_rev | TCTGCTCTTCCTTGCTTTGG |
| TNFAIP2_fwd | GTGCAGAACCTCTACCCCAATG |
| TNFAIP2_rev | TGGAGAATGTCGATGGCCA |
| 18s rRNA_fwd | GTAACCCGTTGAACCCCATT |
| 18s rRNA_rev | CCATCCAATCGGTAGTAGCG |
fwd, forward; rev, reverse.
Data Availability
The ChIP-seq and Gro-seq data have been deposited in the NCBI Gene Expression Omnibus database (GSE157629).
CRediT Authorship Contributions
Joel Johnson (Conceptualization: Lead; Data curation: Lead; Investigation: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Mikko Oittinen (Data curation: Lead; Formal analysis: Equal; Investigation: Supporting; Validation: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Laura Martin-Diaz (Formal analysis: Equal; Investigation: Supporting; Visualization: Supporting; Writing – review & editing: Equal)
Veronika Zapilko (Investigation: Supporting; Writing – review & editing: Supporting)
Sharif Iqbal (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Terhi Rintakangas (Investigation: Supporting; Writing – review & editing: Supporting)
Fabio Tadeu Arrojo Martins (Investigation: Supporting; Writing – review & editing: Supporting)
Henri Niskanen (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Pekka Katajisto (Methodology: Supporting; Writing – review & editing: Supporting)
Minna Kaikkonen (Methodology: Supporting; Writing – review & editing: Supporting)
Keijo Viiri, Ph.D. (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Acknowledgments
The authors thank Raphael Jimenez from the Departamento de Genética e Instituto de Biotecnología, Universidad de Granada, Granada, Spain, for providing the Bac-Cre-ERT2;Sox9f/f;Sox8-/-;Rosa26Eyfp mice. The authors also thank Takshi Kanaya and Hiroshi Ohno from the Laboratory for Intestinal Ecosystem, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan, for graciously providing the RelB, p52, RelA, and p50 plasmids for the lentiviral transfection experiments.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Academy of Finland (310011), Tekes (Business Finland) (658/31/2015), Paediatric Research Foundation, Sigrid Jusélius Foundation, Mary och Georg C. Ehrnrooths Stiftelse, and Laboratoriolääketieteen Edistämissäätiö sr. The funding sources played no role in the design or execution of this study or in the analysis and interpretation of the data.
Supplementary Material
References
- 1.Mabbott N.A., Donaldson D.S., Ohno H., Williams I.R., Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neutra M.R., Frey A., Kraehenbuhl J.P. Epithelial M cells: gateways for mucosal infection and immunization. Cell. 1996;86:345–348. doi: 10.1016/s0092-8674(00)80106-3. [DOI] [PubMed] [Google Scholar]
- 3.Owen R.L. Uptake and transport of intestinal macromolecules and microorganisms by M cells in Peyer’s patches: a personal and historical perspective. Semin Immunol. 1999;11:157–163. doi: 10.1006/smim.1999.0171. [DOI] [PubMed] [Google Scholar]
- 4.Neutra M.R., Mantis N.J., Kraehenbuhl J.P. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat Immunol. 2001;2:1004–1009. doi: 10.1038/ni1101-1004. [DOI] [PubMed] [Google Scholar]
- 5.Rios D., Wood M.B., Li J., Chassaing B., Gewirtz A.T., Williams I.R. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 2016;9:907–916. doi: 10.1038/mi.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraehenbuhl J.P., Neutra M.R. Epithelial M cells: differentiation and function. Annu Rev Cell Dev Biol. 2000;16:301–332. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 7.Kanaya T., Hase K., Takahashi D., Fakuda S., Katsuaki H., Sasaki I., Hemmi H., Knoop K.A., Kumar N., Sato M., Katsuno T., Yokosuka O., Toyooka K., Nakai K., Sakamoto A., Kitahara Y., Jinnohara T., McSorley S.J., Kaisho T., Williams I.R., Ohno H. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat Immunol. 2012;13:729–736. doi: 10.1038/ni.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura S., Kobayashi N., Nakamura Y., Kanaya T., Takahashi D., Fujiki R., Mutoh M., Obata Y., Iwanaga T., Nakagawa T., Kato N., Sato S., Kaisho T., Ohno H., Hase K. Sox8 is essential for M cell maturation to accelerate IgA response at the early stage after weaning in mice. J Exp Med. 2019;216:831–846. doi: 10.1084/jem.20181604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishikawa S., Sato S., Kaneto S., Uchino S., Kohsaka S., Nakamura S., Kiyono H. Allograft inflammatory factor 1 is a regulator of transcytosis in M cells. Nat Commun. 2017;8:14509. doi: 10.1038/ncomms14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura S., Nakamura Y., Kobayashi N., Shiroguchi K., Kawakami E., Mutoh M., Takahashi-Iwanaga H., Yamada T., Hisamoto M., Nakamura M., Udagawa N., Sato S., Kaisho T., Iwanaga T., Hase K. Osteoprotegerin-dependent M cell self-regulation balances gut infection and immunity. Nat Commun. 2020;11:234. doi: 10.1038/s41467-019-13883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hase K., Kawano K., Nochi T., Pontes G.S., Fukuda S., Ebisawa M., Kadokura K., Tobe T., Fujimura Y., Kawano S., Yabashi A., Waguri S., Nakato G., Kimura S., Murakami T., Iimura M., Hamura K., Fukuoka S.I., Lowe A.W., Itoh K., Kiyono H., Ohno H. Uptake through glycoprotein 2 of FimH + bacteria by M cells initiates mucosal immune response. Nature. 2009;462:226–230. doi: 10.1038/nature08529. [DOI] [PubMed] [Google Scholar]
- 12.Nagashima K., Sawa S., Nitta T., Prados A., Koliaraki V., Kollias G., Nakashima T., Takayanagi H. Targeted deletion of RANKL in M cell inducer cells by the Col6a1-Cre driver. Biochem Biophys Res Commun. 2017;493:437–443. doi: 10.1016/j.bbrc.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Knoop K.A., Kumar N., Butler B.R., Senthilkumar K.S., Rebekah T.T., Tomonori N., Hisaya A., Hideo Y., Hiroshi K., Ifor R.W. RANKL is necessary and sufficient to initiate development of antigen-sampling M cells in the intestinal epithelium. J Immunol. 2009;183:5738–5747. doi: 10.4049/jimmunol.0901563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lernbecher T., Kistler B., Wirth T. Two distinct mechanisms contribute to the constitutive activation of RelB in lymphoid cells. EMBO J. 1994;13:4060–4069. doi: 10.1002/j.1460-2075.1994.tb06723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh M.C., Choi Y., Hong J., Teitelbaum S.L. Biology of the RANKL-RANK-OPG system in immunity, bone, and beyond. Front Immunol. 2014;5:511. doi: 10.3389/fimmu.2014.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanaya T., Sakakibara S., Jinnohara T., Masami H., Naoko T., Shinya H., Takashi K., Shunsuke K., Toshihiko I., Tomoo N., Tatsuro K., Naoya K., Taishin A., Toshiro S., Ifor R.W., Hiroshi O. Development of intestinal M cells and follicle-associated epithelium is regulated by TRAF6-mediated NF-κB signaling. J Exp Med. 2018;215:501–519. doi: 10.1084/jem.20160659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lau W., Kujala P., Schneeberger K., Sabine M., Vivian S.W.L., Nick B., Anton M., Frans H., Rodney P.D., Peter J.P., Edward N., Hans C. Peyer’s patch M cells derived from Lgr5+ stem cells require SpiB and are induced by RankL in cultured “miniguts. Mol Cell Biol. 2012;32:3639–3647. doi: 10.1128/MCB.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato S., Kaneto S., Shibata N., Takahashi Y., Okura H., Yuki Y., Kunisawa J., Kiyono H. Transcription factor Spi-B dependent and independent pathways for the development of Peyer's patch M cells. Mucosal Immunol. 2013;6:838–846. doi: 10.1038/mi.2012.122. [DOI] [PubMed] [Google Scholar]
- 19.Kanaya T., Ohno H. The mechanisms of M-cell differentiation. Biosci Microbiota Food Health. 2014;33:91–97. doi: 10.12938/bmfh.33.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuettengruber B., Cavalli G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 21.Cao R., Wang L., Wang H., Hengbin W., Li X., Hediye E.B., Paul T., Richard S.J., Yi Z. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 22.Lee T.I., Jenner R.G., Boyer L.A., Matthew G.G., Stuart S.L., Roshan M.K., Brett C., Sarah E.J., Megan F.C., Isono K., Koseki H., Fuchikami T., Abe K., Murray H., Zucker J.P., Yuan B., Bell G.W., Herbolsheimer E., Hannett N.M., Sun K., Odom D.T., Otte A.T., Volkert T.L., Bartel D.P., Melton D.A., Gifford D.K., Jaenisch R., Young R.A. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein E., Duncan E.M., Masui O., Gil J., Heard E., Allis C.D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Jin Q., Lee J.E., Su I.H., Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci U S A. 2010;107:7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T.H., Li F., Ferreiro-Neira I., Ho L.L., Luyten A., Nalapareddy K., Long H., Verzi M., Shivdasani R.A. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiacchiera F., Rossi A., Jammula S., Zanotti M., Pasini D. PRC 2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016;35:2301–2314. doi: 10.15252/embj.201694550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oittinen M., Popp A., Kurppa K., Lindfors K., Mäki M., Kaikkonen M.U., Viiri K. Polycomb repressive complex 2 enacts Wnt signaling in intestinal homeostasis and contributes to the instigation of stemness in diseases entailing epithelial hyperplasia or neoplasia. Stem Cells. 2017;35:445–457. doi: 10.1002/stem.2479. [DOI] [PubMed] [Google Scholar]
- 28.Vizán P., Beringer M., Di Croce L. Polycomb-dependent control of cell fate in adult tissue. EMBO J. 2016;35:2268–2269. doi: 10.15252/embj.201695694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benoit Y.D., Laursen K.B., Witherspoon M.S., Lipkin S.M., Gudas L.J. Inhibition of PRC2 histone methyltransferase activity increases TRAIL-mediated apoptosis sensitivity in human colon cancer cells. J Cell Physiol. 2013;228:764–772. doi: 10.1002/jcp.24224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., van der Flier L.G., Cuppen E., Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Ram O., Goren A., Amit I., Shoresh N., Yosef N., Ernst J., Kellis M., Gymrek M., Issner R., Coyne M., Durham T., Zhang X., Donaghey J., Epstein C.B., Regev A., Bernstein B.E. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yilmaz Z.B., Weih D.S., Sivakumar V., Weih F. RelB is required for Peyer’s patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 2003;22:121–130. doi: 10.1093/emboj/cdg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishore N., Sommers C., Mathialagan S., Guzova J., Yao M., Hauser S., Huynh K., Bonar S., Mielke C., Albee L., Weier R., Graneto M., Hanau C., Perry T., Tripp C.S. A selective IKK-2 inhibitor blocks NF-B-dependent gene expression in interleukin-1-stimulated synovial fibroblasts. J Biol Chem. 2003;278:32861–32871. doi: 10.1074/jbc.M211439200. [DOI] [PubMed] [Google Scholar]
- 34.Bren G.D., Solan N.J., Miyoshi H., Pennington K.N., Pobst L.J., Paya C.V. Transcription of the RelB gene is regulated by NF-κB. Oncogene. 2001;20:7722–7733. doi: 10.1038/sj.onc.1204868. [DOI] [PubMed] [Google Scholar]
- 35.Coward P., Lee D., Hull M.V., Rgen J., Lehmann M. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor. Proc Natl Acad Sci U S A. 2001;98:8880–8884. doi: 10.1073/pnas.151244398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuercher W.J., Gaillard S., Orband-Miller L.A., Chao E.Y.H., Shearer B.G., Jones D.G., Miller A.B., Collins J.L., McDonnell D.P., Willson T.M. Identification and structure-activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERRβ and ERRγ. J Med Chem. 2005;48:3107–3109. doi: 10.1021/jm050161j. [DOI] [PubMed] [Google Scholar]
- 37.Giguère V. To ERR in the estrogen pathway. Trends Endocrinol Metab. 2002;13:220–225. doi: 10.1016/s1043-2760(02)00592-1. [DOI] [PubMed] [Google Scholar]
- 38.Kim D.K., Jeong J.H., Lee J.M., Kwang S.K., Park S.H., Kim Y.D., Koh M., Shin M., Jung Y.S., Kim H.S., Lee T.H., Oh B.C., Kim J.I., Park H.T., Jeong W.I., Lee C.H., Park S.B., Min J.J., Jung S.I., Choi S.Y., Choy H.E., Choi H.S. Inverse agonist of estrogen-related receptor γ controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat Med. 2014;20:419–424. doi: 10.1038/nm.3483. [DOI] [PubMed] [Google Scholar]
- 39.Matsushima A., Kakuta Y., Teramoto T., Koshiba T., Liu X., Okada H., Tokunaga T., Kawabata S., Kimura M., Shimohigashi Y. Structural evidence for endocrine disruptor bisphenol A binding to human nuclear receptor ERRγ. J Biochem. 2007;142:517–524. doi: 10.1093/jb/mvm158. [DOI] [PubMed] [Google Scholar]
- 40.Kamachi Y., Cheah K.S.E., Kondoh H. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol Cell Biol. 1999;19:107–120. doi: 10.1128/mcb.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato T., Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 42.Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., Matsumoto M., Nitta T., Takahama Y., Inoue J.I. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:427–437. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 43.Lee T.I., Johnstone S.E., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaikkonen M.U., Niskanen H., Romanoski C.E., Kansanen E., Kivelä A.M., Laitalainen J., Heinz S., Benner C., Glass C.K., Ylä-Herttuala S. Control of VEGF-A transcriptional programs by pausing and genomic compartmentalization. Nucleic Acids Res. 2014;42:12570–12584. doi: 10.1093/nar/gku1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and Gro-seq data have been deposited in the NCBI Gene Expression Omnibus database (GSE157629).