Summary
Oral word reading is supported by a neural subnetwork that includes gray matter regions and white matter tracts connected by the regions. Traditional methods typically determine the reading-relevant focal gray matter regions or white matter tracts rather than the reading-relevant global subnetwork. The present study developed a network-based lesion-symptom mapping (NLSM) method to identify the reading-relevant global white matter subnetwork in 84 brain-damaged patients. The global subnetwork was selected among all possible subnetworks because its global efficiency exhibited the best explanatory power for patients' reading scores. This reading subnetwork was left lateralized and included 7 gray matter regions and 15 white matter tracts. Moreover, the reading subnetwork had additional explanatory power for the patients' reading performance after eliminating the effects of reading-related local regions and tracts. These findings refine the reading neuroanatomical architecture and indicate that the NLSM can be a better method for revealing behavior-specific subnetworks.
Subject areas: Biological Science, Neuroscience, Behavioral neuroscience
Graphical Abstract
Highlights
-
•
A network-based lesion-symptom mapping (NLSM) is developed in the current study
-
•
A reading-related global white matter network is identified using the NLSM
-
•
Global efficiency of the network could predict the reading score of patients
-
•
A genetic algorithm is adopted to reduce the computational load
Biological Science, Neuroscience, Behavioral neuroscience
Introduction
Oral word reading refers to the process of generating sounds upon seeing a visual word and is an evolutionarily recent and far-reaching ability of humanity. Modern functional neuroimaging studies using noninvasive techniques (e.g., functional MRI and magnetoencephalography) in healthy subjects and neuropsychological lesion-symptom mapping studies involving brain-damaged patients have generally agreed that this ability is subserved by a left-lateralized brain subnetwork (Bates et al., 2003; Fedorenko and Thompson-Schill, 2014; Hagoort, 2019; Schlaggar and McCandliss, 2007; Seidenberg and McClelland, 1989; Skeide and Friederici, 2016). For instance, these authors found that the “visual word form area” (VWFA) in the left fusiform gyrus is responsible for orthographic processing (Carreiras et al., 2013; Cohen et al., 2000; Dehaene et al., 2010; Li et al., 2020; Schlaggar and McCandliss, 2007; Tan et al., 2005), Wernicke's area in the left superior temporal gyrus (STG) is responsible for phonological processing (Chen et al., 2019; Dewitt and Rauschecker, 2013; Price, 2012), and Broca's area in the left inferior frontal gyrus (IFG) is responsible for articulation and syntactic processing (Price, 2012; Sahin et al., 2009). In addition to these gray matter regions, other scholars have revealed some word reading-relevant white matter tracts, including the left inferior longitudinal fasciculus (ILF), which is responsible for orthographic processing (Epelbaum et al., 2008; Sarubbo et al., 2015; Wang et al., 2020; Yeatman et al., 2013; Zemmoura et al., 2015), the left arcuate fasciculus (AF), which is responsible for phonological processing (Bernal and Ardila, 2009; Catani and Mesulam, 2008; Han et al., 2014; Yeatman et al., 2012), the inferior longitudinal fasciculus (IFOF), which is responsible for semantic processing (Agosta et al., 2010; Han et al., 2013), and the left superior longitudinal fasciculus (SLF), which is responsible for language articulation (Johnson et al., 2015; Kamali et al., 2014; Li et al., 2017).
Although functional neuroimaging studies in healthy individuals could reveal the cortical regions activated during a reading task, whether the activated regions obligatorily participate in word reading remains unknown (Rorden and Karnath, 2004). In contrast, lesion studies in patients should be able to determine the necessary components (e.g., regions and connectivity) by investigating the associations between lesioned components and disrupted behavioral performance (Bates et al., 2003; Friedrich et al., 1998; Han et al., 2013; Rorden and Karnath, 2004; Smith et al., 2006). However, the mass-univariate approach used in previous lesion studies (e.g., voxel-based lesion-symptom mapping [VLSM], Bates et al., 2003 and region-based lesion-symptom mapping [RLSM], Friedrich et al., 1998; Menon, 2011) has been criticized due to the assumption of cortical localizationism. In this case, lesion studies should be inferred based on connectivity data beyond local areas (Catani et al., 2012). Indeed, tract-based lesion-symptom mapping (TLSM) has been used to discover the white matter tracts underlying specific behaviors (Gleichgerrcht et al., 2017; Han et al., 2013; Smith et al., 2006). However, previous studies using lesion methods only identified behavior-related focal gray matter regions or single white matter tracts in the neural network (Friedrich et al., 1998; Han et al., 2013; Menon, 2011).
To overcome the limitations of locality in lesion studies, some researchers began to adopt network-based methods to divide the whole-brain network into different subnetworks (e.g., language subnetwork, attention subnetwork, and default mode subnetwork; Adhikari et al., 2017; Gordon et al., 2016; Griffis et al., 2020; Hagmann et al., 2008; Power et al., 2011). However, whether the detected subnetworks are truly associated with cognitive functions remains unclear. This is because most studies labeled specific cognitive functions to the subnetworks based on the possible roles of individual regions in the subnetworks in the literature, rather than statistical analysis results of the relationship between the detected subnetworks and the behaviors of interest using actual data.
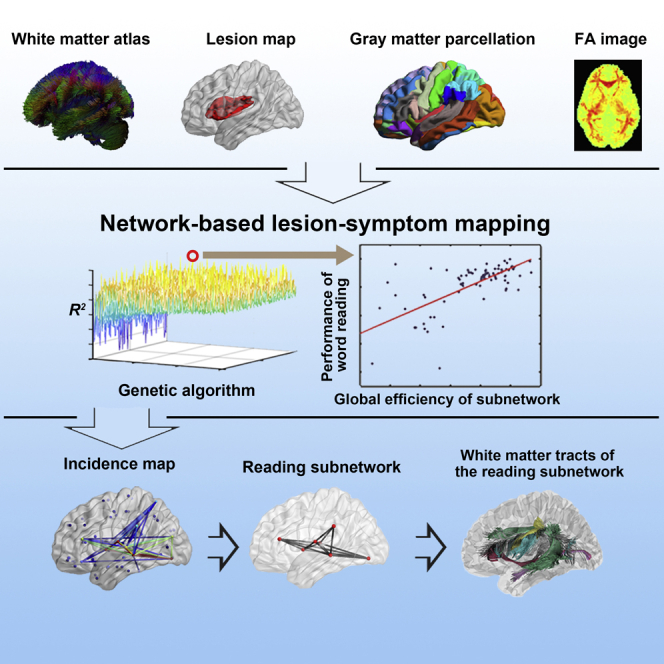
To resolve the above issues, the current study developed a network-based lesion-symptom mapping (NLSM) method to identify the word reading-relevant white matter subnetwork in 84 Chinese speakers with brain damage (Figure 1). The NLSM used a high-dimensional model to obtain the whole reading-related subnetwork instead of the partial components of the subnetwork. Specifically, each patient was instructed to read 140 Chinese words aloud that were used to evaluate their reading performance, and brain neuroimaging data were collected and used to reconstruct their white matter network. Among all white matter subnetworks (i.e., all combinations of the components in the whole brain), we searched for the subnetwork whose global efficiency (Latora and Marchiori, 2001) could optimally account for the variation in oral word-reading performance using unbiased data-driven methods. Specifically, the subnetwork with the highest explanatory power for the variability in reading performance was identified as relevant to reading ability. The number of potential subnetworks was too large (≈1027) to be exhausted under the present computational conditions. A genetic algorithm (GA, Goldberg, 1989) was adopted to reduce the computational load. Moreover, our findings were further validated by performing three additional tests (i.e., permutation test, leave-one-out cross-validation, and repeatability) and controlled for the potential influence of all reading-related focal regions and tracts.
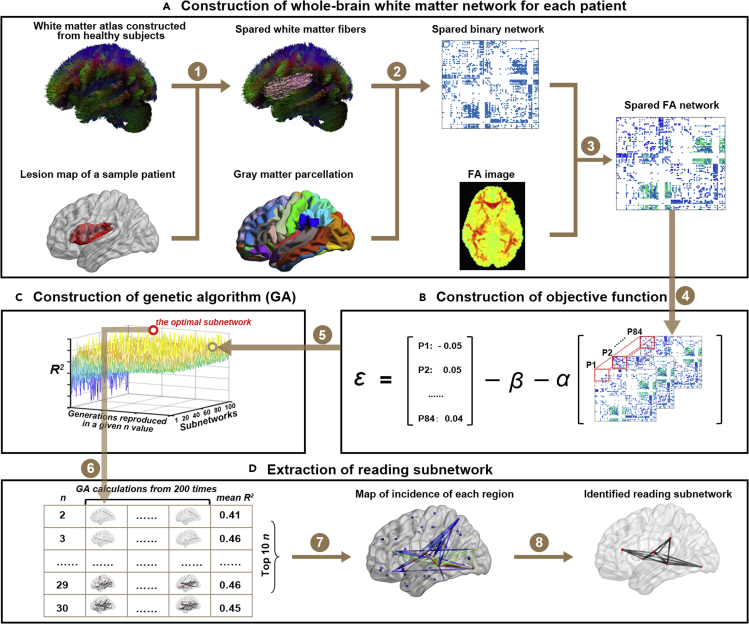
Figure 1.
Flow chart of the network-based lesion-symptom mapping (NLSM)
(1) The spared fibers were extracted by removing the tracts that pass through the brain lesion mask of the patient based on the healthy atlas; (2) the spared network matrix was reconstructed by extracting the spared fibers between each pair of the 90 gray matter regions in the AAL90 atlas; (3) the spared network was weighted based on the mean FA value in the tract mask; (4) the objective function of the relationship between the global efficiency of the subnetwork and word reading performance was constructed; (5) the genetic algorithm (GA) procedure was calculated based on the objective function; (6) the optimal subnetwork in a GA procedure for a given n value was selected; (7) an incidence map of the 10 n values with the top mean R2 values in the above subnetwork pool were generated; and (8) the reading subnetwork was identified according to the incidence map (see STAR Methods for details).
Results
Behavioral performance
The mean oral word reading accuracy of the 84 patients was 88% (SD = 19%; range: 19%–100%; see details in Table S3), suggesting that the word reading ability of our patients was impaired.
Whole-brain white matter network of patients
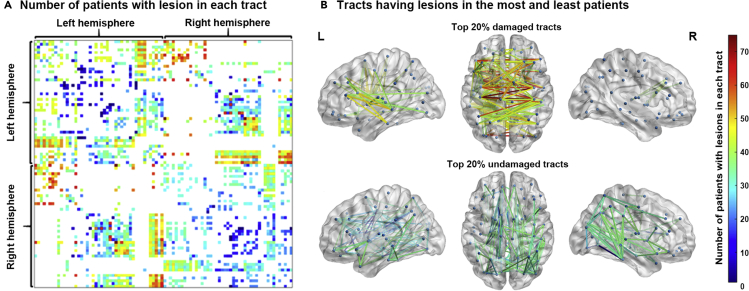
We filtered the white matter atlas of 842 healthy participants (Griffis et al., 2020; Yeh et al., 2018) by 90 preselected regions (automated anatomical labeling [AAL] atlas, Tzouriomazoyer et al., 2002) and obtained 998 white matter tracts. The tracts included 305 left intratracts, 252 right intratracts, and 441 intertracts that connected regions in the left hemisphere, the right hemisphere, and both hemispheres, respectively. Most tracts (990 tracts) were lesioned in at least one patient (left intratracts: 98%; right intratracts: 99%; and intertracts: 100%; Figure 2A). Regarding the most commonly lesioned tracts (i.e., top 20%) based on the number of affected patients, more patients had lesions in the intertracts (158/441, 36%) than in the left intratracts (57/305, 12%; χ2 = 25.82, p < 0.001) and right intratracts (7/252, 3%; χ2 = 96.56, p < 0.001). More patients exhibited damage in the left intratracts than in the right intratracts (χ2 = 34.35, p < 0.001). In contrast, regarding the least commonly lesioned tracts (i.e., bottom 20%) based on the number of affected patients, fewer patients had lesions in the intertracts (48/441, 11%) than in the left intratracts (75/305, 25%; χ2 = 24.60, p < 0.001) and right intratracts (82/252, 32%; χ2 = 49.35, p < 0.001). There was a slight difference in the number of patients with left and right intratract lesions (χ2 = 4.31, p < 0.04; Figure 2B; see Figure S1 for the lesion distribution of the patients).
Figure 2.
Lesion overlap of the white matter tracts in 84 patients
(A) Lesion overlap map of white matter tracts in the 84 patients. The value of each cell denotes the number of patients with lesions on the tract connecting the corresponding two regions. Cells shown in white indicate no white matter connection between two regions. The order of the arrangement of the regions in Figure A is provided in Table S4.
Subnetwork of word reading
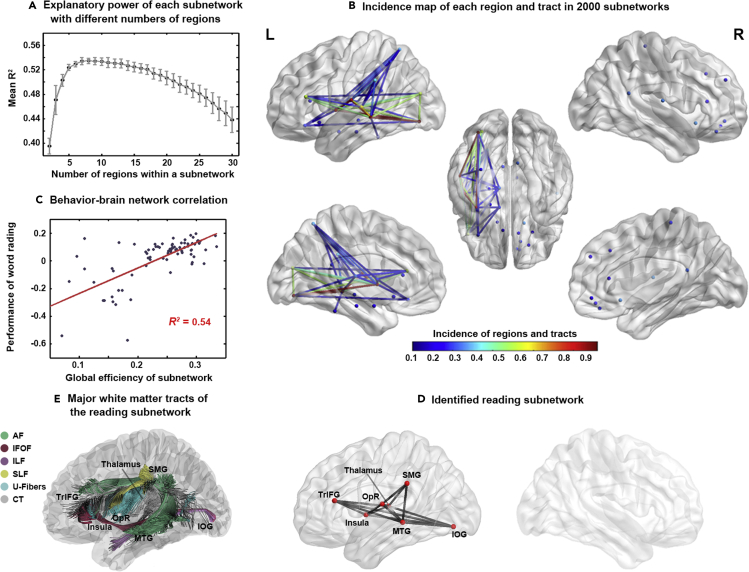
To obtain the optimal reading subnetwork, we investigated the explanatory power (i.e., R2) of the global efficiency of each candidate subnetwork for reading performance using the GA procedure based on the objective function (see STAR Methods for details). For a given n value (the number of nodes included in the subnetwork), we conducted the GA procedure 200 times and obtained the maximum R2 value each time. The n value range was 2–30 (see Figure 1). The R2 values in this range formed an inverted U curve with increasing numbers of regions in the subnetworks (Figure 3A). When the n value was equal to 8, the subnetworks reached the maximum R2 value (mean = 0.53; SD = 0.005). The top 10 mean R2 values corresponded to n values: 6–15. When the 2000 subnetworks with the top 10 R2 values were overlaid, we obtained the incidence of each of the 90 regions (Figure 3B and Table 1). The frequency of the 45 regions in the left hemisphere (mean = 0.19; SD = 0.29) was higher than that of the 45 regions in the right hemisphere (mean = 0.05; SD = 0.07; t = 3.13, p < 0.005). The high-frequency regions were also mainly distributed in the left hemisphere. For instance, the regions with the top 10 highest frequencies, except for the right Heschl gyrus (532/2000 = 27%), were all left lateralized, including the left inferior occipital gyrus (IOG: 99%), middle temporal gyrus (MTG: 97%), insula (92%), Rolandic operculum (92%), STG (66%), triangular inferior frontal gyrus (TrIFG: 55%), middle occipital gyrus (MOG: 55%), supramarginal gyrus (SMG: 34%), and superior partial lobe (SPL: 29%).
Figure 3.
Results of the reading subnetwork identified by network-based lesion-symptom mapping
The error bar in (A) indicates the standard deviation derived by conducting the GA procedure 200 times for a given n value. (B) shows the regions or tracts with an incidence higher than 0.10.
Abbreviations: IOG, inferior occipital gyrus; MTG, middle temporal gyrus; SMG, supramarginal gyrus; TrIFG, triangular inferior frontal gyrus; OpR, Rolandic operculum; AF, arcuate fasciculus; IFOF, inferior longitudinal fasciculus; ILF, inferior longitudinal fasciculus; SLF, superior longitudinal fasciculus; CT, corticothalamic pathway.
Table 1.
Incidence of 90 anatomical regions in 2000 subnetworks
| Order | Incidence | Anatomical |
Order | Incidence | Anatomical |
Order | Incidence | Anatomical |
|---|---|---|---|---|---|---|---|---|
| Region | Region | Region | ||||||
| 1 | 0.998 | IOG_L | 31 | 0.092 | MidCi_R | 61 | 0.001 | OpR_R |
| 2 | 0.967 | MTG_L | 32 | 0.077 | OrMeFG_L | 62 | 0.001 | Hippocampus_R |
| 3 | 0.92 | STG_L | 33 | 0.07 | Rectus_L | 63 | 0.001 | Angular_L |
| 4 | 0.918 | Insula_L | 34 | 0.07 | Caudate_R | 64 | 0.001 | Amygdala_R |
| 5 | 0.657 | OpR_L | 35 | 0.068 | Pallidum_R | 65 | 0.001 | AntMTG_R |
| 6 | 0.555 | MOG_L | 36 | 0.065 | Hippocampus_L | 66 | 0 | OrSFG_L |
| 7 | 0.555 | TrIFG_L | 37 | 0.065 | SMA_R | 67 | 0 | MFG_L |
| 8 | 0.339 | SMG_L | 38 | 0.062 | MidCi_L | 68 | 0 | SMA_L |
| 9 | 0.287 | SPL_L | 39 | 0.045 | Putamen_R | 69 | 0 | MeSFG_L |
| 10 | 0.266 | Heschl_R | 40 | 0.044 | SOG_L | 70 | 0 | Calcarine _L |
| 11 | 0.262 | SPL_L | 41 | 0.043 | Pallidum_L | 71 | 0 | Lingual_L |
| 12 | 0.227 | Thalamus_L | 42 | 0.038 | Thalamus_R | 72 | 0 | AntSTG_L |
| 13 | 0.215 | OLF_R | 43 | 0.037 | Precuneus _L | 73 | 0 | MFG_R |
| 14 | 0.167 | IPL_L | 44 | 0.034 | OrMeFG_R | 74 | 0 | Insula_R |
| 15 | 0.166 | OrMFG_R | 45 | 0.031 | OrIFG_R | 75 | 0 | Calcarine_R |
| 16 | 0.162 | IPL_L | 46 | 0.029 | PCL_L | 76 | 0 | Lingual_R |
| 17 | 0.162 | Rectus_R | 47 | 0.022 | Heschl_L | 77 | 0 | SOG_R |
| 18 | 0.161 | Caudate_L | 48 | 0.016 | AntMTG_L | 78 | 0 | MOG_R |
| 19 | 0.16 | Caudate_L | 49 | 0.013 | OpIFG_R | 79 | 0 | IOG_R |
| 20 | 0.151 | OrIFG_L | 50 | 0.012 | TrIFG_R | 80 | 0 | Fusiform_R |
| 21 | 0.145 | PosCi_L | 51 | 0.006 | OrMFG_L | 81 | 0 | PoC_R |
| 22 | 0.141 | Fusiform_L | 52 | 0.006 | Cuneus_L | 82 | 0 | SPL_R |
| 23 | 0.14 | PosCi_R | 53 | 0.005 | PrC_L | 83 | 0 | IPL_R |
| 24 | 0.132 | PCL_R | 54 | 0.004 | PHA_R | 84 | 0 | SMG_R |
| 25 | 0.12 | OLF_L | 55 | 0.004 | ITG_L | 85 | 0 | Angular_R |
| 26 | 0.117 | AntCi_L | 56 | 0.004 | Cuneus_R | 86 | 0 | Precuneus _R |
| 27 | 0.116 | OLF_L | 57 | 0.003 | PoC_L | 87 | 0 | STG_R |
| 28 | 0.115 | Fusiform_L | 58 | 0.002 | PrC_R | 88 | 0 | AntSTG_R |
| 29 | 0.111 | Amygdala_L | 59 | 0.001 | SFG_L | 89 | 0 | MTG_R |
| 30 | 0.108 | PHA_L | 60 | 0.001 | Putamen _L | 90 | 0 | ITG_R |
L, left hemisphere; R, right hemisphere. The full name of each region is provided in Table S4.
When we exhausted all possible subnetworks within the top 16 regions in the incidence map, we obtained an optimal subnetwork whose global efficiency could maximally explain the reading performance of the patients (R2 = 0.54; Figure 3C). This subnetwork was considered associated with word reading and included 7 regions and 15 white matter tracts in the left hemisphere (Figure 3D). The seven regions included the left IOG, MTG, operculum rolandic (OpR), insula, TrIFG, SMG, and thalamus. The 15 tracts were distributed in the following 6 major white matter pathways described in the atlas constructed based on healthy subjects (Yeh et al., 2018): the left AF, SLF, ILF, IFOF, U-fibers, and corticothalamic pathway (CT) (Figure 3E). The global efficiency of this subnetwork was significantly positively correlated with the corrected reading scores (i.e., the residual of the accuracy of oral word reading after controlling for demographic variables and disease duration) of the 84 patients (r = 0.74, p < 10−6; Figure 3C).
Validation of the reading subnetwork
To determine whether the reading subnetwork obtained in the above analyses was reliable, we performed the following analyses.
Permutation test
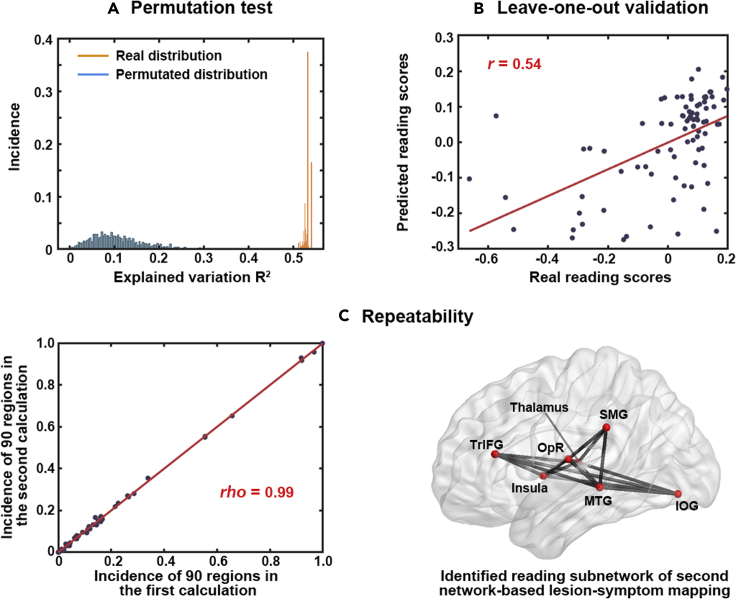
We reconducted the NLSM program using the same analysis described above, except that each patient's reading score was randomly matched with the brain imaging data of another patient. Thus, we obtained a new set of 2000 R2 values from 2000 permuted subnetworks. These permuted R2 values (0.11 ± 0.06) were significantly lower than those of the actual 2000 subnetworks (0.54 ± 0.01; Mann-Whitney U test, U = 54.91, p < 10−6; Figure 4A) and the reading subnetwork that we identified (R2 = 0.54, sign test, z = 44.70, p < 10−6). This finding demonstrates that the obtained reading subnetwork was not a random event.
Figure 4.
Results of the validation analysis of the reading subnetwork
Cross-validation
We used data from 83 patients to build the model, and the remaining patient's data were used to conduct a test at each n value (10 n values in total). For each patient, we obtained a predicted word-reading score per n value. The predicted scores were significantly correlated with the actual scores across the 84 patients per n value (rs = 0.44 to 0.59, median r = 0.54, p < 10−6; Figure 4B). These results suggest that the NLSM procedures used to obtain the reading subnetwork should not have an overfitting problem.
Repeatability
When NLSM was performed again, the newly obtained 2000 subnetworks were highly similar to the original subnetworks. The ranking order of the incidence frequencies of the 90 regions between the original and new subnetworks was significantly correlated (Spearman correlation: rho = 0.99). More importantly, the newly identified reading subnetwork was exactly the same as the original subnetwork, and both subnetworks included the same regions and tracts (Figure 4C). This replication seems to indicate that the random selection and arbitrary parameters of our procedures should not seriously affect our findings.
Controlling for the potential influence of reading-related focal regions and tracts
To further determine whether the optimal reading subnetwork obtained by our NLSM still had additional explanatory power for the patients' reading performance even after eliminating the effects of reading-related local regions or tracts, we correlated the reading scores with the global efficiency of the NLSM-obtained reading subnetwork after regressing out the effects of focal components. Two groups of confounding variables were separately introduced in this correlational analysis. Each group included the lesion volume of each reading-related region and the mean FA value of each reading-related tract. The reading-related regions and tracts in the first group were extracted by RLSM and TLSM methods (see STAR Methods for details), respectively. Those in the second group were derived from the NLSM-obtained reading subnetwork (Figure 3). The first group had 7 reading-related regions (Figure S2A and Table S1) and 11 reading-related tracts (Figure S2B and Table S1) in the left hemisphere, while the second group had 7 regions and 15 tracts. We found that the global efficiency value of the NLSM-obtained reading subnetwork was still significantly correlated with word reading performance of patients even after partialling out the influence of the regions and tracts (the first group: partial r = 0.47, p < 0.0001; the second group: partial r = 0.37, p < 0.005). This demonstrates that the NLSM-obtained reading subnetwork had additional explanatory power for reading performance beyond the simple accumulation of individual reading-relevant regions and tracts.
Discussion
The present study revealed a reading subnetwork containing 7 left hemisphere regions and 15 tracts by applying an NLSM method to 84 brain-damaged patients. In particular, we used global efficiency as a characteristic measure of a subnetwork and identified a subnetwork in the whole brain to optimally account for the variation in reading scores. To save computation time, we applied a heuristic method (i.e., GA) to identify the fittest reading subnetwork. The obtained reading subnetwork was highly reliable, was not accidental, could predict the reading score of new patients, and was replicable. Relative to the measures of focal gray matter regions (lesion percentage) or white matter tracts (mean FA value), the current NLSM could identify a subnetwork whose global efficiency had an additional contribution to reading performance.
Differences between NLSM and traditional lesion-symptom mapping methods
Traditional lesion-symptom mapping methods seek to determine the focal gray matter regions or white matter tracts associated with a given cognitive function (Bates et al., 2003; Friedrich et al., 1998; Medina et al., 2010; Smith et al., 2006). These methods map measures of the local properties of regions or tracts of interest to specific behavioral performances of patients. However, these methods independently consider the effects only in a single region or tract and overlook the interactive function in the whole network.
However, the present NLSM method assumes that reading ability relies on distributed brain areas and white matter tracts among these areas (Fedorenko and Thompson-Schill, 2014; Hagoort, 2019; Skeide and Friederici, 2016). We adopted global efficiency to measure the information flow in the subnetwork and found a subnetwork that could maximally explain the variation in reading deficits across patients. Furthermore, the significant correlation between the global efficiency of the identified reading subnetwork and reading performance persisted even when ruling out the effects of all areas and tracts in the subnetwork.
Reading subnetwork identified by the NLSM method
The 7 regions in the reading subnetwork identified by the NLSM method have been repeatedly reported to be involved in language information processing. Remarkably, although our sample included few patients with lesions in the posterior occipital area, we still observed robust inclusion of the left IOG in the reading subnetwork. The left IOG is adjacent to an important reading area, i.e., the VWFA (Cohen et al., 2000; Dehaene and Cohen, 2011). The left MTG has been reported to engage in reading processing (Price, 2012; Wu et al., 2012). The locations of the left insula and TrIFG are close to the classic Broca's area (Dronkers, 1996; Sahin et al., 2009), the left SMG and OpR in the dorsal pathway are critical for speech processing (Dewitt and Rauschecker, 2013; Price, 2012), and the left thalamus might be involved in semantic, syntactic, and articulation processing (Johnson and Ojemann, 2000; Price, 2012; Wahl et al., 2008). Our results suggest that the interactive function (evaluated by global efficiency) among these areas is highly related to word-reading abilities in literate individuals.
The 15 tracts in our optimal reading subnetwork were distributed in six major white matter pathways described in the atlas constructed from healthy subjects (Yeh et al., 2018). Four of these pathways (the left AF, SLF, ILF, and IFOF) were discovered to participate in word reading-relevant processing (Fang et al., 2018; Hagoort, 2019; Han et al, 2013, 2014; Wang et al., 2020; Yeatman et al., 2013). The corticothalamic pathways mainly connected the thalamus and cortical regions, and the U-fibers primarily connected two adjacent region areas, such as the left insula-TrIFG, in our data.
We identified a functionally specialized subnetwork supporting oral word reading. However, word reading involves multiple processes, such as orthographic recognition, orthographic to phonology mapping, and oral production (Price, 2012). In general, each process is not subserved by a single region or tract. Instead, these processes are supported by the complex coupling mechanism of many regions and tracts. For example, visual word form recognition relies on an interaction between the ventral occipital-temporal cortex and other higher-level language regions (Carreiras et al., 2013; Li et al., 2020; Price and Devlin, 2011; see the opposite view in Dehaene and Cohen, 2011). Therefore, the connectivity pattern among different cortical areas plays a more critical role in supporting cognitive functions (Fedorenko and Thompson-Schill, 2014).
Notably, our reading task was performed in the Chinese language. Prior studies found that alphabetic and morpho-syllabic writing systems share common regions (e.g., the left ventral visual cortex, superior posterior temporal area, and IFG). However, Chinese word reading might include additional involvement of the left middle frontal area (MFG) and the right ventral occipital-temporal cortex (Bolger et al., 2005; Tan et al., 2001; Wu et al., 2012; Zhu et al., 2014). Our optimal reading subnetwork mainly contained the regions shared by two writing systems (e.g., the left IOG, MTG, and TrIFG) but not Chinese-specific regions (e.g., the left MFG and right ventral occipito-temporal cortex). Therefore, the reading subnetwork identified in the present study should be language-general.
In addition, our materials in the reading task contained various semantic categories. Prior studies have uncovered that these categories might rely on dissociated brain tissues. We also observed that our patients presented categorical differences in reading performance and reading subnetworks (see details in Figure S3 and Table S2). Briefly, a higher reading accuracy was observed in response to common objects (0.85–0.91) and faces (0.89) than actions (0.85). Similarly, living categories (0.89–0.91) had higher accuracy than nonliving categories (0.85–0.88). The reading subnetwork identified by collapsing all categories (i.e., the mean reading network) had 7 regions. Four (the left IOG, MTG, OpR, and insula) of the 7 regions were shared by the reading networks of each category, and each of the other three regions appeared in the networks of at least 5 categories (the left SMG and thalamus: 5 categories; TrIFG: 6 categories). However, we observed the following five category-specific regions outside the mean reading network: the left caudate, left STG, left MOG, left IPL, and left SPL. Note that the limited number of items in each of the categories (20 items) might not be robust enough to reflect the validated behavioral performance for these categories. In addition, the NLSM approach might also lead to some noise in the results due to the random sample features (such as the VLSM approach, Lorca-Puls et al., 2018). Therefore, the detailed components for category-specific anatomical structure should be further investigated with more reliable behavioral measures and larger sample sizes of the patients.
Further methodological considerations regarding the NLSM
The purpose of the NLSM is to select the subnetwork most relevant to behavior from all possible subnetworks. Specifically, in relation to the other subnetworks, the selected subnetwork has the maximum power to explain the variation in the behavior (i.e., the optimal subnetwork). Ideally, we should first calculate the explanatory power of each subnetwork and then select the optimal subnetwork. However, the computation amount was too large (approximately 1027) to be completed using the current computer ability. To solve this problem, we adopted an approximate calculation procedure (i.e., GA) to reduce the computational load. This procedure included some arbitrary steps. In the final arbitrary step, we selected and included 16 regions from the 90 regions in the subsequent analyses. Indeed, the selection of the 16 regions was not determined by complete subjectivity. Instead, we mainly considered the following two factors: (1) the number of regions in the optimal subnetwork among the 2000 subnetworks and (2) the computational capability of modern computers. We found that the optimal subnetwork included 8 regions (Figure 3A). This finding suggests that the optimal subnetwork probably contained approximately 8 regions. The top 16 regions (twice as many as 8 regions) in the incidence map should contain the optimal combination of regions in the optimal subnetwork. Moreover, modern computers could exhaustively calculate all possible subnetworks of the 16 nodes (n = 65,535). Therefore, we included the 16 regions with the highest incidence in the subsequent analyses. Notably, we should select as many regions in this step under the condition of an acceptable computation time. The bottom line shows that the number of selected regions was not smaller than that in the optimal subnetwork.
The above description of NLSM shows that NLSM differs from typical permutation-based inference. First, the number of built models differs between these approaches (NLSM: multiple models; permutation-based inference: one model). Second, the approach of selecting the behavior-related components in the whole network differs between these approaches. The NLSM selects the behavior-related subnetwork by comparing the explanatory effects of all possible subnetworks. Therefore, NLSM selects the best subnetwork among all subnetworks and does not need to set the significance threshold. In contrast, permutation-based inference selects behavior-related regions or connections by setting a certain significant threshold in a distribution of a model obtained in a permutation (e.g., the 95th percentile).
Caution should be taken when making inferences regarding the results of NLSM analyses. After all, the selected subnetwork was only the most relevant to the behavioral performance under a given patient sample and specific measures. The reliability and meaningfulness of the identified subnetwork should be further estimated in depth. In the present study, multiple analyses (e.g., permutation test, leave-one-out cross-validation, and repeatability) were performed to confirm the psychological reality of the results. In addition, we observed that not all seven regions in the optimal reading subnetwork had the highest incidence in the incidence map (Figure 3B and Table 1). Therefore, some regions with a high incidence (e.g., the left STG with the fourth highest incidence) did not appear in the optimal subnetwork, which may suggest that reading processing is supported by multiple separate subnetworks and that the missing regions in the optimal subnetwork were pivotal nodes in other subnetworks. In brief, the interpretation and inference of the results of NLSM should consider various factors (e.g., the validation results, incidence map, and explanatory power of each subnetwork).
Conclusion
We propose a method (NLSM) for identifying the reading subnetwork in a group of patients with brain damage. The method considered the integrated effect rather than the local effect of the network and identified an elaborate reading subnetwork with high ecological validity and high explanatory power, thereby providing direct evidence of the systematic function of the subnetwork in reading processing. This new method could be widely used to reveal potential subnetworks responsible for specific behaviors in lesion studies.
Limitations of the study
This study has at least the following caveats. (1) The global efficiency of the identified reading subnetwork accounted for only a part (54%) but not all of the reading performance of the patients likely. This may be because the global efficiency may not be the most sensitive to measuring the integrity of a neural network; the 90 AAL regions were very large in size (e.g., STG) such that some reading-unrelated cortical issues or white matter fibers were involved in our analyses; or the effects of the reading-relevant regions or tracts without lesions were not considered. (2) When the whole-brain white matter network of a patient was reconstructed, a tract was defined to exist if it included at least one intact fiber (Figure 1A). This cut-off might not be optimal. (3) Our present analyses were based on the assumption that the global efficiency of the reading network should linearly correlate with the reading performance of the patients. If the correlation is actually nonlinear, our findings require modification. (4) The distribution of the reading scores was highly skewed with a ceiling effect, and this lack of behavioral variation likely impacted the power of the results of the current NLSM.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Brain data and code | This paper | Mendeley Data (https://data.mendeley.com/datasets/mycvb6dy8r/draft?a=7c3885ea-a3c0-4c51-b1e4-2771fd432a78) |
| Software and algorithms | ||
| MATLAB | Mathworks | https://www.mathworks.com/ |
| SPM | FIL | https://www.fil.ion.ucl.ac.uk/spm/ |
| BrainVoyager | Goebel, 2012 | http://www.brainvoyager.com/ |
| PNADA(MATLAB) | Cui et al., 2013 | www.nitrc.org/projects/panda/ |
| DSI studio | Fang-Cheng Yeh | http://dsi-studio.labsolver.org/publications |
| BCT(MATLAB) | Rubinov and Sporns, 2010 | https://www.nitrc.org/projects/bct/ |
| BrianNet Viewer(MATLAB) | Xia et al., 2013 | http://www.nitrc.org/projects/bnv/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, ZaiZhu Han (zzhhan@bnu.edu.cn).
Materials availability
The study did not generate new unique reagents.
Data and code availability
-
•
The data (e.g., 3D lesion images, FA images, and intermediate NLSM results) used in this study area available at the Mendeley Data (https://data.mendeley.com/datasets/mycvb6dy8r/draft?a=7c3885ea-a3c0-4c51-b1e4-2771fd432a78).
-
•
The code (e.g., source codes and guidance) used in this study are also available at the Mendeley Data (see above link)
-
•
Any additional information is available from the lead contact upon request
Experimental model and subject details
Participants
Eighty-four patients with brain damage (65 males) from the China Rehabilitation Research Center were recruited using the following criteria. The patients suffered from first-time brain injury at least one month postonset (mean = 6.25 months, SD = 12.36 months; range: 1–86 months), had no other neurological or psychiatric diseases (e.g., schizophrenia or depression), and were able to follow the task instructions. Most of patients (n = 69) suffered from stroke, and the other patients suffered from traumatic brain injury. The patients’ mean age and education were 44.65 years (SD = 13.42 years) and 12.79 years (SD = 3.32 years), respectively. All patients were native Chinese speakers and right handed (Oldfield, 1971). The detailed demographic information of each patient is shown in Table S3. The lesions of the patients were widely distributed, with most patients having lesions in the insula and surrounding white matter tissues (see details in Figure 2 and Figure S1). Sex and/or gender might have potential influence on our results. For example, language-related deficits exhibit clear sex differences (Wallentin, 2009). Hence, the gender in our NLSM analyses was treated as a covariate to eliminate its confounding effects.
The patients provided informed written consent. The study was approved by the institutional review board of the National Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University. Most patients have been reported in previous focal lesion studies.
Method details
Behavioral data collection and preprocessing
An oral word-reading task was adopted. We selected 140 objects, namely, 20 objects from each of seven object categories (i.e., animals, tools, common artifacts, fruits and vegetables, large nonmanipulable objects, actions and famous people). The Chinese written names were presented on the screen, and the patients were instructed to read the words aloud.
The task was operated by DMDX (Forster and Forster, 2003). Each patient was individually tested in a quiet room. Pauses for rest were allowed upon request. The word presentation order was pseudorandomized but identical across the patients.
The first complete response to each word was scored. To exclude the influence of demographic factors and disease duration on reading performance, the raw accuracy of each patient was corrected by regressing out four variables (age, gender, education level and disease duration). The corrected scores were treated as the patients’ reading ability measures in the following analyses.
MRI data collection
The patients were scanned at the China Rehabilitation Research Center with an 8-channel split head coil in a 1.5 T GE SIGNA EXCITE scanner. The following three types of images were collected: (1) T1-weighted 3D magnetization-prepared rapid acquisition with gradient echo (MPRAGE) images on the sagittal plane with the following parameters: matrix size = 512 × 512, voxel size = 0.49 × 0.49 × 0.70 mm3, repetition time (TR) = 12.26 ms, echo time (TE) = 4.2 ms, inversion time = 400 ms, field of view (FOV) = 250 × 250 mm2, flip angle = 15°, and slice number = 248; (2) fluid-attenuated inversion recovery (FLAIR) T2-weighted images on the axial plane with the following parameters: matrix size = 512 × 512, voxel size = 0.49 × 0.49 × 5 mm3, TR = 8002 ms, TE = 127.57 ms, inversion time = 2 s, FOV = 250 × 250 mm2, flip angle = 90°, and slice number = 28; and (3) diffusion-weighted images (DWI), which had two separate sequences with different diffusion weighting direction sets such that 32 directions were covered in total. The first sequence had the following parameters: 15 diffusion weighting directions, matrix size = 128 × 128, voxel size = 1.95 × 1.95 × 2.6 mm3, TR = 13000 ms, TE = 69.3 ms, inversion time = 0 s, FOV = 250 × 250 mm2, flip angle = 90°, and slice number = 53. The other sequence had the same parameters, except that the sequence included 17 different directions. The first two volumes were b0 volumes, and the b-value of the other volumes was 1000 s/mm2 in each sequence. All sequences except for the FLAIR T2 images were scanned twice to improve the quality of the images.
Quantification and statistical analysis
Imaging data preprocessing
Structural magnetic resonance imaging data
We first coregistered the two T1-weighted structural imaging data on the same native space using the trilinear interpolation method applied in SPM5 to obtain the averaged structural image (http://www.fil.ion.ucl.ac.uk/spm/software/spm5) and then coregistered and resliced the FLAIR T2 images to the averaged structural image using the trilinear interpolation method in SPM5. The lesion contours of each patient were drawn on the T1 structural image by two trained persons slice-by-slice by visually referring to the FLAIR T2 images. This procedure was supervised by a superior radiologist. The patients were excluded if they had very diffuse damage, and we could not precisely demark the boundary of the lesion. Then, the structural images were resliced into 1 × 1 × 1 mm voxel sizes. For normalization, some studies adopt an automatic normalization method through which the local detailed information of brain images is automatically evaluated and matched (e.g., Price et al., 1998), but this method cannot exclude the effect of lesions in the brain, which might cause extra distortions in the images. To resolve this problem, one method is to mask the lesions and use the remaining intact issues for normalization processing (Brett et al., 2001). In contrast, manual registration methods might overcome such limitations. Thus, a manual method was adopted in the present study. Specifically, each patient’s structural images were registered into the Talairach space via the ‘3D Volume Tools’ in BrainVoyager QX v2.0 (www.brainvoyager.com; Goebel, 2012), and we manually marked the anterior commissure to posterior commissure plane and the borders of the cerebrum. The affine transformation matrix between the native and Talairach spaces was extracted with the ANTs software package (Advanced Normalization Tools, www.picsl.upenn.edu/ANTS/). The lesion images were transformed into the Talairach space using this matrix with the ‘WarpImageMultiTransform’ program. Given that the registration procedures were based on anatomical landmarks without evaluating the local detailed information of the brain, this process was not affected by the lesions. The lesion image was finally transformed into the MNI space using the affine transformation matrix between the MNI and Talairach spaces using a similar method.
Diffusion magnetic resonance imaging data
We first merged the two paired sequences into a single 4D nifty-1 format file and their diffusion-weighted gradient tables. Then, we executed BET: skull removal; Eddycorrect (correction of eddy current distortion) and DTIFIT (build diffusion tensor models) with a pipeline tool, PANDA (Cui et al., 2013) (www.nitrc.org/projects/panda/). Fractional anisotropy (FA) maps in the individual space were generated after performing the above pipeline. Then, we registered these fractional anisotropy maps with the FMRIB fractional anisotropy template in the MNI space with ANTs (version 1.9). The normalization included the following two parts: 1) linear rigid affine transform, which first obtained an affine transform.txt file for each participant and then produced the fractional anisotropy map in the MNI space with the ‘WarpImageMultiTransform’ program, and 2) nonlinear transform registration, which obtained a more fine-grained normalized fractional anisotropy map of each patient in the MNI space with the shell script ‘buildtemplate’.
Construction of the whole-brain white matter network of each patient
To identify the white matter subnetwork of word reading, we first reconstructed the anatomical whole-brain lesion network of each patient (Figure 1A). This process was performed using a white matter connectome atlas based on healthy people (Griffis et al., 2020; Yeh et al., 2018; http://brain.labsolver.org/diffusion-mri-templates/tractography). This atlas was created on the basis of high angular DWI data based on 842 healthy subjects in the Human Connectome Project and contained expert-vetted streamline trajectories in the MNI space. Specifically, for each patient, we first extracted the spared fibers in the healthy atlas, which bypassed the brain lesion mask of the patient using DSI studio software (http://dsi-studio.labsolver.org/). Then, we reconstructed the spared network by extracting the spared fibers between each pair of 90 gray matter regions in the Automated Anatomical Labeling atlas (AAL90 without the cerebellum, Tzouriomazoyer et al., 2002; see details in Table S4). Finally, we built the FA network by masking all fibers in each pair of regions that had spared fibers and calculating the mean FA value in the tract mask (i.e., averaged FA values based on the DWI data of each voxel of the mask per patient). The FA value of a weighting tract in our patient sample could introduce more individual differences in the connectivity matrix. Therefore, the FA value might increase the variation in the integrity value of the network across patients and render the network lesion-deficit correlation reliable. The FA value of the tract connecting the two regions reflected the white matter connectivity strength of the tract. As a result, a whole-brain FA network containing 90 regions (i.e., a 90 ∗ 90 matrix) was obtained for each patient.
Construction of the objective function of the relationship between the global efficiency of the subnetwork and the word reading performance
To calculate the global efficiency of a subnetwork, we first calculated the weighted shortest path length between any pairs in the networks as follows (BCT; Rubinov and Sporns, 2010):
where is the shortest path (geodesic) between regions i and j, is the intermediate path of the whole shortest path, and is the weighted value of path . is a map (e.g., an inverse) from the .
The global efficiency of a subnetwork can be calculated using the following formula (Latora and Marchiori, 2001; Rubinov and Sporns, 2010):
where N is the set of regions in the subnetwork, n is the number of regions, and is the weighted shortest path length between regions i and j. Efficiency provides a precise quantitative measure of the information flow in the network (Latora and Marchiori, 2001). This approach enabled us to estimate the influence of brain damage on the information exchange ability of the whole subnetwork rather than individual gray matter regions or white matter tracts. Therefore, the relationship between the oral word reading (Sbhv) behavioral performance and the global efficiency values of the network (Ew) could be expressed in the following regression equation:
where α and β are the regression coefficients, and ε is the error term. By combining the above two equations, we obtain the following formula:
This formula represents the objective function that needed to be optimized (Figure 1B). In the function, Sbhv is a known value (i.e., word-reading scores of the patients). We needed to identify the subnetwork (i.e., the combinations of regions and tracts in the whole network) that could best explain the variation in the word-reading scores of the patients, i.e., identify the subnetwork that had the minimum error term ε in the function. The structure of a given subnetwork in the function was determined by the regions and tracts. We attempted to determine the subnetwork whose global efficiency would result in the lowest ε value. Because the regions and tracts were restricted by the above white matter connectome atlas based on healthy people in the current study, our aim was to determine the set of regions N to minimize the ε value in the above objective function.
To quantitatively depict the degree to which the global efficiency of a reading subnetwork could account for the variation in reading scores among the patients, we further calculated the explanatory power of the global efficiency of each reading subnetwork with regard to the variation in reading scores as the coefficient of determination R2 as follows:
where was an observed value (word-reading scores), was the mean value of all , was the fitted value by the above objective function corresponding to the minimum ε value, and was the number of all patients. Thus, the higher the R2 value of the model, the higher the explanatory power of the subnetwork for reading scores. Therefore, we attempted to obtain the subnetwork with the maximum R2 value (see Figure 1C).
Construction of the procedures used to search for the reading subnetwork: genetic algorithm (GA)
To identify the subnetwork that best explained the variation in the word-reading performance of the patients, an ideal way was to calculate the R2 values in the objective function of each potential subnetwork (i.e., all combinations of the 90 regions and their tracts in the whole network) and select the best subnetwork with the maximum R2 value. However, the number of potential subnetworks was too large (≈ 1027) to be exhausted under the present conditions. To reduce the computation time and obtain the best subnetwork, we adopted a GA (Goldberg, 1989) (Figure 1C). A GA is a heuristic often used to resolve optimization or search problems based on the principles of natural evolution and survival of the fittest and commonly includes biologically inspired procedures, such as crossover, mutation and selection. A GA is able to produce approximate results in less computation time. A GA has been used in different fields, such as complex networks (Pizzuti, 2012), experimental design optimization (Wager and Nichols, 2003) and engineering problems (Ahmadi and Dincer, 2010).
We employed the GA function in MATLAB software to determine the fittest word-reading subnetwork from the whole-brain network. This process underwent the following steps. Notably, most parameters in the steps were derived from the default values in the MATLAB procedure.
Step 1. Creating the subnetworks of the initial generation through random selection
For a given n value in the objective function (e.g., n = 10 for a subnetwork with 10 regions), we did not exhaust all sets of subnetworks in the whole network (i.e., all combinations of 10 regions among the 90 regions, N = 5.72 ∗ 1012). Instead, using the GA, we first created 100 initial subnetworks (i.e., the first generation; each subnetwork included 10 regions randomly selected among the 90 regions). Then, the R2 value of each subnetwork was calculated using the above objective function. Finally, the 5 subnetworks with the maximum R2 values were selected as better subnetworks because they were more likely to contain the critical regions that could explain the behavioral variation. We also randomly selected 80 subnetworks from the remaining subnetworks. The 85 selected subnetworks, including the 5 better subnetworks, were treated as parents to reproduce the new generation in the following step.
Step 2. Creating the subnetworks of the second generation through a crossover between parent subnetworks and mutation of a small proportion of subnetworks
We performed the following steps. 1) The 85 parent subnetworks were randomly paired, and 100 pairs of subnetworks were selected. 2) Each pair of subnetworks produced a new subnetwork by combining half of the regions randomly extracted from each parent subnetwork. If the new subnetwork included the same regions, the combining procedure was repeatedly conducted until the extracted regions differed. Thus, we obtained 100 new subnetworks. 3) To increase the search space, one of the 100 new subnetworks was mutated such that 25% of the regions were replaced with other regions randomly selected from among the 90 regions. 4) The 100 new subnetworks, including the mutated subnetwork, consisted of the second generation. 5) The 85 new parent subnetworks from this generation were extracted using the method described in Step 1 and used to reproduce the next generation.
Step 3. Selecting the best reading subnetwork of the family through the reproduction of many generations in the family
The above second step was repeated until the coefficients of determination, i.e., R2 values, of 50 continuous newly reproduced generations did not further increase. All subnetworks generated in the above steps formed a family for a given n value (i.e., the set of subnetworks with n regions). The subnetwork with the maximum R2 value was a potentially optimal reading-related subnetwork for that particular n value.
Extraction of the reading subnetwork
To avoid sampling bias in a subnetwork from a single family for a given n value and obtain a reliable reading subnetwork, we subsequently carried out the following steps (Figure 1D).
Step 1. Obtaining the subnetwork pool by conducting the GA procedure multiple times for each n value
The above GA procedure was conducted 200 times per n value (number of regions) of the subnetworks, and a mean R2 value was obtained for each n value. Theoretically, we should have conducted the procedures for all n values (i.e., 2 - 90). Practically, to reduce computations, we conducted the procedure with only values from 2-30 because the subnetworks in this range should include those with the maximum R2 values according to the results of the above analyses.
Step 2. Extracting the incidence map of each region
The 10 n values with the top mean R2 values from the above subnetwork pool were selected as these subnetworks most likely include the word reading-related regions and tracts. Thus, we obtained 2000 subnetworks (200 subnetworks/n value ∗ 10 n values) in total and a map of the incidence of each of the 90 regions from the 2000 subnetworks. The higher the incidence of a region, the higher the possibility that this region participates in the word-reading subnetwork.
Step 3. Identifying the reading subnetwork
We extracted a new network whose regions were selected at a given threshold. (i.e., the top 16 regions in incidence) and tracts with connections between the selected regions. The perfect method to identifying the optimal subnetwork could follow the logic of leave-last-nodes out iteration as follows: each time we obtained the results of the above incidence of nodes, we could remove some nodes with the lowest incidence because these nodes likely had no significant contribution to the reading task. After many iterations, we obtained a subnetwork that had the maximal explanatory power for the reading task (the optimal reading subnetwork). However, to save computation time, we directly proceeded with 16 nodes because the above results showed that a subnetwork with 8 nodes had the maximum mean R2 value, indicating that the optimal subnetwork probably contained approximately 8 nodes. The top 16 nodes (twice as many as 8 nodes) in the incidence map should contain the optimal combination of regions in the optimal subnetwork. More importantly, all possible subnetworks structured by the 16 nodes (n = 65,535) could be exhausted under the present commutating capacity of modern computers. Therefore, the 16 nodes were introduced to the NLSM program to extract the optimal reading subnetwork.
To further visualize the shape of the identified reading subnetwork, we outlined the trajectories of the reading tracts. For each reading tract, we extracted all fibers between the two AAL regions connecting the tract in the healthy white matter atlas (Griffis et al., 2020; Yeh et al., 2018). To understand how the reading tracts were related to the classic major white matter pathways, we clustered and labeled these tracts in accordance with the 80 main tracts in the atlas constructed based on healthy subjects (Yeh et al., 2018).
The BrainNet Viewer package (Xia et al., 2013) was used to display the brain network and DIS studio (http://dsi-studio.labsolver.org/) was used to display the major white matter tracts.
Validation of the reading subnetwork
To determine whether the reading subnetwork obtained from the above analyses was reliable, we performed the following validation analyses.
Permutation test
To examine whether the reading subnetwork was an accidental event, we conducted the GA again as described above, except for the corrected reading scores of the 84 patients were randomly paired with another patient’s brain images. Furthermore, to save computing time, the GA only processed the top 10 n values that were selected in the above analyses. Similarly, each n value was processed by the GA 200 times, and in total, 2000 subnetworks and 2000 R2 values were obtained. These 2000 R2 values from the permuted data were compared with the 2000 R2 values from the above actual data (using the Mann-Whitney U test) and the R2 value from the subnetwork obtained in the above analyses (using the sign test).
Cross validation
This procedure was employed to estimate whether the procedure used to obtain the above reading subnetwork had an overfitting problem. We adopted a leave-one-out cross-validation method to evaluate whether the above objective function, which was established on the basis of 83 patients, could predict the reading performance of the remaining patient when his or her imaging data were introduced into the function. Specifically, the above GA used to process the word reading and brain imaging data of the 83 patients was conducted again while the data from one patient were omitted. To reduce time consumption, the GA only processed the top 10 n values selected in the above analyses. Each n value was processed once. The objective function with the maximum R2 value was selected as the testing function. We input the imaging data from the remaining patient into the testing function and obtained the predicted reading score of the patient. Thus, each patient obtained a predicted reading score per n value. We computed the correlation between the actual and predicted reading scores per n value across the 84 patients.
Repeatability
Some random selections and arbitrary parameters were included in the procedures used identify the above reading subnetwork [e.g., initial number of subnetworks (100); the number of GA procedures we performed (200); the number of n values with the top mean R2 values (10); and the threshold of the nodes used in the final GA procedures (16)]. To verify whether these random and arbitrary factors influenced our findings, we conducted the above procedures of extracting the reading subnetwork once again. We examined whether the newly obtained reading subnetwork was the same as the original subnetwork in structure, and the correlation between the incidences of the regions in the incidence maps was high.
Controlling for the potential influence of reading-related focal regions and tracts
To test whether the reading subnetwork obtained by the present NLSM method had an additional contribution to the patients’ reading performance even after ruling out the effects of reading-related local regions or tracts, we correlated the reading scores with the global efficiency of the NLSM-obtained reading subnetwork after controlling for potential confounding variables across the 84 patients. Two groups of confounding variables were separately introduced in the correlational analyses. Each group included the lesion volumes of each reading-related region (i.e., the number of voxels with lesions in the region) and the mean FA volumes of each reading-related tract (i.e., average FA values of all voxels in the tract). The regions and tracts in the two groups were acquired by traditional focal lesion-symptom mapping methods (i.e., RLSM and TLSM) and the NLSM-obtained reading subnetwork. Specifically, for the traditional methods, we first extracted the reading-relevant regions and tracts using RLSM and TLSM (i.e., regionsRLSM and tractsTLSM), respectively. In RLSM, we correlated the corrected reading scores with the lesion volumes of each gray matter region in the AAL90 atlas across the 84 patients. The regions reaching a significant level (Bonferroni-corrected p < 0.01) were treated as regionsRLSM. TractsTLSM were identified using identical procedures to the above region-based analysis, except for the 90 regions were replaced with the 998 tracts from the white matter atlas and the lesion volumes were replaced with the mean FA values. For the NLSM-obtained reading subnetwork, the lesion volumes of each region and mean FA values of each white matter tract in the NLSM-obtained reading subnetwork were used as control variables.
Acknowledgments
We would like to thank the BNU-CN Lab members for the data collection. We are also grateful to all research participants. This work was supported by the National Key Research and Development Program of China (2018YFC1315200) and the National Natural Science Foundation of China (31872785; 81972144).
Author contributions
M.L. and Z.H. conceived the study. L.S., Y.Z., and Z.H. implemented the experiment and collected the data. M.L. performed the analyses. Z.H. and M.L. discussed the results and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102862.
Supplemental information
References
- Adhikari M.H., Hacker C.D., Siegel J.S., Griffa A., Hagmann P., Deco G., Corbetta M. Decreased integration and information capacity in stroke measured by whole brain models of resting state activity. Brain. 2017;140:1068–1085. doi: 10.1093/brain/awx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta F., Henry R.G., Migliaccio R., Neuhaus J., Miller B.L., Dronkers N.F., Brambati S.M., Filippi M., Ogar J.M., Wilson S.M., Gorno-Tempini M.L. Language networks in semantic dementia. Brain. 2010;133:286–299. doi: 10.1093/brain/awp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi P., Dincer I. Exergoenvironmental analysis and optimization of a cogeneration plant system using Multimodal Genetic Algorithm (MGA) Energy. 2010;35:5161–5172. doi: 10.1016/j.energy.2010.07.050. [DOI] [Google Scholar]
- Bates E., Wilson S.M., Saygin A.P., Dick F., Sereno M.I., Knight R.T., Dronkers N.F. Voxel-based lesion-symptom mapping. Nat. Neurosci. 2003;6:448–450. doi: 10.1038/nn1050. [DOI] [PubMed] [Google Scholar]
- Bernal B., Ardila A. The role of the arcuate fasciculus in conduction aphasia. Brain. 2009;132:2309–2316. doi: 10.1093/brain/awp206. [DOI] [PubMed] [Google Scholar]
- Bolger D.J., Perfetti C.A., Schneider W. Cross-cultural effect on the brain revisited : Universal structures plus writing system variation. Hum. Brain Mapp. 2005;104:92–104. doi: 10.1002/hbm.20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M., Leff A.P., Rorden C., Ashburner J. Spatial normalization of brain images with focal lesions using cost. Funct. Masking. 2001;500:486–500. doi: 10.1006/nimg.2001.0845. [DOI] [PubMed] [Google Scholar]
- Carreiras M., Armstrong B.C., Perea M., Frost R. The what, when, where, and how of visual word recognition. Trends Cogn. Sci. 2013;18:90–98. doi: 10.1016/j.tics.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Catani M., Dell’Acqua F., Bizzi A., Forkel S.J., Williams S.C., Simmons A., Murphy D.G., Thiebaut de Schotten M. Beyond cortical localization in clinico-anatomical correlation. Cortex. 2012;48:1262–1287. doi: 10.1016/j.cortex.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Catani M., Mesulam M. The arcuate fasciculus and the disconnection theme in language and aphasia: History and current state. Cortex. 2008;44:953–961. doi: 10.1016/j.cortex.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wassermann D., Abrams D.A., Kochalka J., Gallardo-Diez G., Menon V. The visual word form area (VWFA) is part of both language and attention circuitry. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-13634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen L., Dehaene S., Naccache L., Lehéricy S., Dehaene-Lambertz G., Hénaff M.A., Michel F. The visual word form area. Spatial and temporal characterization of an initial stage of reading in normal subjects and posterior split-brain patients. Brain. 2000;123:291–307. doi: 10.1093/brain/123.2.291. [DOI] [PubMed] [Google Scholar]
- Cui Z., Zhong S., Xu P., He Y., Gong G. PANDA: a pipeline toolbox for analyzing brain diffusion images. Front. Hum. Neuroence. 2013;7:42. doi: 10.3389/fnhum.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaene S., Cohen L. The unique role of the visual word form area in reading. Trends Cogn. Sci. 2011;15:254–262. doi: 10.1016/j.tics.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Dehaene S., Pegado F., Braga L.W., Ventura P., Nunes Filho G., Jobert A., Dehaene-Lambertz G., Kolinsky R., Morais J., Cohen L. How learning to read changes the cortical networks for vision and language. Science. 2010;330:1359–1364. doi: 10.1126/science.1194140. [DOI] [PubMed] [Google Scholar]
- Dewitt I., Rauschecker J.P. Wernicke’s area revisited: Parallel streams and word processing. Brain Lang. 2013;127:181–191. doi: 10.1016/j.bandl.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkers N.F. A new brain region for coordinating speech production. Nature. 1996;384:14. doi: 10.1038/384159a0. [DOI] [PubMed] [Google Scholar]
- Epelbaum S., Pinel P., Gaillard R., Delmaire C., Perrin M., Dupont S., Dehaene S., Cohen L. Pure alexia as a disconnection syndrome: new diffusion imaging evidence for an old concept. Cortex. 2008;44:962–974. doi: 10.1016/j.cortex.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Fang Y., Wang X., Zhong S., Song L., Han Z., Gong G., Bi Y. Semantic representation in the white matter pathway. PLoS Biol. 2018;16:1–21. doi: 10.1371/journal.pbio.2003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko E., Thompson-Schill S.L. Reworking the language network. Trends Cogn. Sci. 2014;18:120–126. doi: 10.1016/j.tics.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster K.I., Forster J.C. DMDX: a Windows display program with millisecond accuracy. Behav. Res. Methods Instrum. Comput. 2003 doi: 10.3758/BF03195503. [DOI] [PubMed] [Google Scholar]
- Friedrich F.J., Egly R., Rafal R.D., Beck D. Spatial attention deficits in humans: a comparison of superior parietal and temporal-parietal junction lesions. Neuropsychology. 1998;12:193–207. doi: 10.1037/0894-4105.12.2.193. [DOI] [PubMed] [Google Scholar]
- Gleichgerrcht E., Fridriksson J., Rorden C., Bonilha L. Connectome-based lesion-symptom mapping (CLSM): a novel approach to map neurological function. Neuroimage Clin. 2017;16:461–467. doi: 10.1016/j.nicl.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel R. BrainVoyager - past, present, future. Neuroimage. 2012;62:748–756. doi: 10.1016/j.neuroimage.2012.01.083. [DOI] [PubMed] [Google Scholar]
- Goldberg D.E. Addison-Wesley Pub. Co; 1989. Genetic Algorithm in Search, Optimization, and Machine Learning. [Google Scholar]
- Gordon E.M., Laumann T.O., Adeyemo B., Huckins J.F., Kelley W.M., Petersen S.E. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis J.C., Metcalf N.V., Corbetta M., Shulman G.L. Damage to the shortest structural paths between brain regions is associated with disruptions of resting-state functional connectivity after stroke. Neuroimage. 2020;210 doi: 10.1016/j.neuroimage.2020.116589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Van Wedeen J., Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:1479–1493. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagoort P. The neurobiology of language beyond single-word processing. Science. 2019;366:55–58. doi: 10.1126/science.aax0289. [DOI] [PubMed] [Google Scholar]
- Han Z., Ma Y., Gong G., He Y., Caramazza A., Bi Y. White matter structural connectivity underlying semantic processing: evidence from brain damaged patients. Brain. 2013;136:2952–2965. doi: 10.1093/brain/awt205. [DOI] [PubMed] [Google Scholar]
- Han Z., Ma Y., Gong G., Huang R., Song L., Bi Y. White matter pathway supporting phonological encoding in speech production: a multi-modal imaging study of brain damage patients. Brain Struct. Funct. 2014:577–589. doi: 10.1007/s00429-014-0926-2. [DOI] [PubMed] [Google Scholar]
- Johnson C.P., Juranek J., Swank P.R., Kramer L., Cox C.S., Ewing-Cobbs L. White matter and reading deficits after pediatric traumatic brain injury: a diffusion tensor imaging study. Neuroimage Clin. 2015;9:668–677. doi: 10.1016/j.nicl.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.D., Ojemann G.A. The role of the human thalamus in language and memory : evidence from electrophysiological studies. Brain Cogn. 2000;230:218–230. doi: 10.1006/brcg.1999.1101. [DOI] [PubMed] [Google Scholar]
- Kamali A., Flanders A.E., Brody J., Hunter J.V., Hasan K.M. Tracing superior longitudinal fasciculus connectivity in the human brain using high resolution diffusion tensor tractography. Brain Struct. Funct. 2014;219:269–281. doi: 10.1007/s00429-012-0498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V., Marchiori M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001;87:198701–198704. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Li M., Xu Y., Luo X., Zeng J., Han Z. Linguistic experience acquisition for novel stimuli selectively activates the neural network of the visual word form area. Neuroimage. 2020;215:116838. doi: 10.1016/j.neuroimage.2020.116838. [DOI] [PubMed] [Google Scholar]
- Li M., Zhang Y., Song L., Huang R., Ding J., Fang Y., Xu Y., Han Z. Structural connectivity subserving verbal fluency revealed by lesion-behavior mapping in stroke patients. Neuropsychologia. 2017;101:85–96. doi: 10.1016/j.neuropsychologia.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Lorca-Puls D.L., Gajardo-Vidal A., White J., Seghier M.L., Leff A.P., Green D.W., Crinion J.T., Ludersdorfer P., Hope T.M.H., Bowman H., Price C.J. The impact of sample size on the reproducibility of voxel-based lesion-deficit mappings. Neuropsychologia. 2018;115:101–111. doi: 10.1016/j.neuropsychologia.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Kimberg D.Y., Chatterjee A., Coslett H.B. Inappropriate usage of the Brunner-Munzel test in recent voxel-based lesion-symptom mapping studies. Neuropsychologia. 2010;48:341–343. doi: 10.1016/j.neuropsychologia.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn. Sci. 2011;15:483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Oldfield R.C. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971 doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Pizzuti C. A multiobjective genetic algorithm to find communities in complex networks. IEEE Trans. Evol. Comput. 2012;16:418–430. doi: 10.1109/TEVC.2011.2161090. [DOI] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.J. A review and synthesis of the first 20years of PET and fMRI studies of heard speech, spoken language and reading. Neuroimage. 2012;62:816–847. doi: 10.1016/j.neuroimage.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.J., Devlin J.T. The Interactive Account of ventral occipitotemporal contributions to reading. Trends Cogn. Sci. 2011 doi: 10.1016/j.tics.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.J., Howard D., Patterson K., Warburton E.A., Friston K.J., Frackowiak S.J., Frackowiak R.S.J. A functional neuroimaging description of two deep dyslexic patients. J. Cogn. Neurosci. 1998;10:303. doi: 10.1162/089892998562753. [DOI] [PubMed] [Google Scholar]
- Rorden C., Karnath H.O. Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat. Rev. Neurosci. 2004;5:812–819. doi: 10.1038/nrn1521. [DOI] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sahin N.T., Pinker S., Cash S.S., Schomer D., Halgren E. Sequential processing of lexical, grammatical, and phonological information within broca’s area. Science. 2009;326:445–449. doi: 10.1126/science.1174481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarubbo S., De Benedictis A., Merler S., Mandonnet E., Balbi S., Granieri E., Duffau H. Towards a functional atlas of human white matter. Hum. Brain Mapp. 2015;36:3117–3136. doi: 10.1002/hbm.22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaggar B.L., McCandliss B.D. Development of neural systems for reading. Annu. Rev. Neurosci. 2007;30:475–503. doi: 10.1146/annurev.neuro.28.061604.135645. [DOI] [PubMed] [Google Scholar]
- Seidenberg M.S., McClelland J.L. A distributed, developmental model of word recognition and naming. Psychol. Rev. 1989;96:523–568. doi: 10.1037/0033-295X.96.4.523. [DOI] [PubMed] [Google Scholar]
- Skeide M.A., Friederici A.D. The ontogeny of the cortical language network. Nat. Rev. Neurosci. 2016;17:323–332. doi: 10.1038/nrn.2016.23. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Johansen-Berg H., Rueckert D., Nichols T.E., Mackay C.E., Watkins K.E., Ciccarelli O., Cader M.Z., Matthews P.M., Behrens T.E.J. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Tan L.H., Laird A.R., Li K., Fox P.T. Neuroanatomical correlates of phonological processing of Chinese characters and alphabetic words : a meta-analysis. Hum. Brain Mapp. 2005;91:83–91. doi: 10.1002/hbm.20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L.H., Liu H.L., Perfetti C.A., Spinks J.A., Fox P.T., Gao J.H. The neural system underlying Chinese logograph reading. Neuroimage. 2001;13:836–846. doi: 10.1006/nimg.2001.0749. [DOI] [PubMed] [Google Scholar]
- Tzouriomazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wager T.D., Nichols T.E. Optimization of experimental design in fMRI: a general framework using a genetic algorithm. Neuroimage. 2003;18:293–309. doi: 10.1016/S1053-8119(02)00046-0. [DOI] [PubMed] [Google Scholar]
- Wahl M., Marzinzik F., Friederici A.D., Hahne A., Kupsch A., Schneider G.H., Saddy D., Curio G., Klostermann F. The human thalamus processes syntactic and semantic language violations. Neuron. 2008;59:695–707. doi: 10.1016/j.neuron.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Wallentin M. Putative sex differences in verbal abilities and language cortex: a critical review. Brain Lang. 2009;108:175–183. doi: 10.1016/j.bandl.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Wang K., Li X., Huang R., Ding J., Song L., Han Z. The left inferior longitudinal fasciculus supports orthographic processing: evidence from a lesion-behavior mapping analysis. Brain Lang. 2020;201:104721. doi: 10.1016/j.bandl.2019.104721. [DOI] [PubMed] [Google Scholar]
- Wu C., Ho M.R., Chen S.A. NeuroImage A meta-analysis of fMRI studies on Chinese orthographic , phonological , and semantic processing. Neuroimage. 2012;63:381–391. doi: 10.1016/j.neuroimage.2012.06.047. [DOI] [PubMed] [Google Scholar]
- Xia M., Wang J., He Y. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman J.D., Dougherty R.F., Ben-shachar M., Wandell B.A. Development of white matter and reading skills. Proc. Natl. Acad. Sci. U S A. 2012;109:17756–17757. doi: 10.1073/pnas.1206792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman J.D., Rauschecker A.M., Wandell B.A. Anatomy of the visual word form area: adjacent cortical circuits and long-range white matter connections. Brain Lang. 2013;125:146–155. doi: 10.1016/j.bandl.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.C., Panesar S., Fernandes D., Meola A., Yoshino M., Fernandez-Miranda J.C., Vettel J.M., Verstynen T. Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage. 2018;178:57–68. doi: 10.1016/j.neuroimage.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmoura I., Herbet G., Moritz-Gasser S., Duffau H. New insights into the neural network mediating reading processes provided by cortico-subcortical electrical mapping. Hum. Brain Mapp. 2015;36:2215–2230. doi: 10.1002/hbm.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Nie Y., Chang C., Gao J.H., Niu Z. Different patterns and development characteristics of processing written logographic characters and alphabetic words: an ALE meta-analysis. Hum. Brain Mapp. 2014;35:2607–2618. doi: 10.1002/hbm.22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data (e.g., 3D lesion images, FA images, and intermediate NLSM results) used in this study area available at the Mendeley Data (https://data.mendeley.com/datasets/mycvb6dy8r/draft?a=7c3885ea-a3c0-4c51-b1e4-2771fd432a78).
-
•
The code (e.g., source codes and guidance) used in this study are also available at the Mendeley Data (see above link)
-
•
Any additional information is available from the lead contact upon request