Abstract
Background & Aims
Gluconeogenesis from amino acids (AAs) maintains glucose homeostasis during fasting. Although glucagon is known to regulate AA catabolism, the contribution of other hormones to it and the scope of transcriptional regulation dictating AA catabolism are unknown. We explored the role of the fasting hormones glucagon and glucocorticoids in transcriptional regulation of AA catabolism genes and AA-dependent gluconeogenesis.
Methods
We tested the RNA expression of AA catabolism genes and glucose production in primary mouse hepatocytes treated with fasting hormones (glucagon, corticosterone) and feeding hormones (insulin, fibroblast growth factor 19). We analyzed genomic data of chromatin accessibility and chromatin immunoprecipitation in mice and primary mouse hepatocytes. We performed chromatin immunoprecipitation in livers of fasted mice to show binding of cAMP responsive element binding protein (CREB) and the glucocorticoid receptor (GR).
Results
Fasting induced the expression of 31 genes with various roles in AA catabolism. Of them, 15 were synergistically induced by co-treatment of glucagon and corticosterone. Synergistic gene expression relied on the activity of both CREB and GR and was abolished by treatment with either insulin or fibroblast growth factor 19. Enhancers adjacent to synergistically induced genes became more accessible and were bound by CREB and GR on fasting. Akin to the gene expression pattern, gluconeogenesis from AAs was synergistically induced by glucagon and corticosterone in a CREB- and GR-dependent manner.
Conclusions
Transcriptional regulation of AA catabolism genes during fasting is widespread and is driven by glucagon (via CREB) and corticosterone (via GR). Glucose production in hepatocytes is also synergistically augmented, showing that glucagon alone is insufficient in fully activating gluconeogenesis.
Keywords: Fasting, Enhancers, Chromatin, Transcription Factors, Endocrinology, Metabolism
Abbreviations used in this paper: AA, amino acid; AOA, aminooxyacetate; cAMP, cyclic adenosine monophosphate; ChIP, chromatin immunoprecipitation; cort, corticosterone; CREB, cAMP responsive element binding protein; CRE, cAMP responsive element; FC, fold change; FGF19, fibroblast growth factor 19; gluc, glucagon; GR, glucocorticoid receptor; GRE, glucocorticoid receptor element; nt, non-treated; PBS, phosphate-buffered saline; PKA, protein kinase A; qPCR, quantitative polymerase chain reaction; SDS, sodium dodecyl sulfate; SI, synergy index; TF, transcriptional factor
Graphical abstract
Summary.
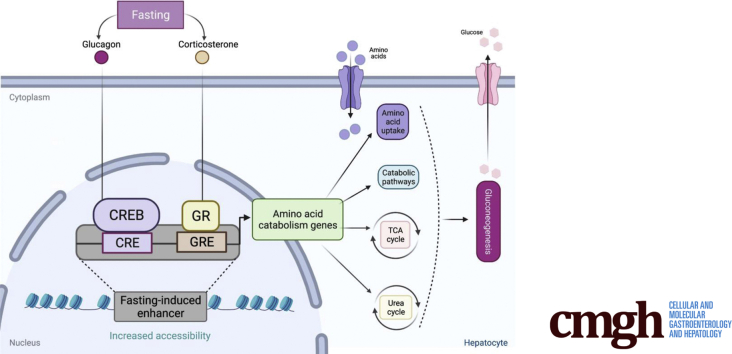
Here we found that all amino acid catabolism routes are transcriptionally regulated during fasting and that fasting hormones glucagon and corticosterone synergistically induce these genes. Accordingly, gluconeogenesis from amino acids only reached its maximal levels in the presence of glucagon and corticosterone.
The liver has a prominent role in maintaining systemic glucose homeostasis. Hepatic glucose production and its secretion are the result of glycogen breakdown (glycogenolysis) and gluconeogenesis, the de novo synthesis of glucose.1 Gluconeogenesis is a multistep enzymatic process occurring mainly in hepatocytes whereby glucose is produced from non-carbohydrate precursors. The 3 major gluconeogenic precursors are amino acids (AAs), lactate, and glycerol. There are 5 catabolic routes through which AAs enter gluconeogenesis. Each route requires certain AA transporters facilitating AA uptake from plasma and several enzymes that convert AAs to TCA cycle intermediates. These intermediates end up as oxaloacetate and enter gluconeogenesis (Figure 1A). Although the contribution of different precursors to gluconeogenesis differs depending on nutritional and endocrine status, it is widely accepted that AAs are major contributors to newly synthesized glucose.1,2
Figure 1.
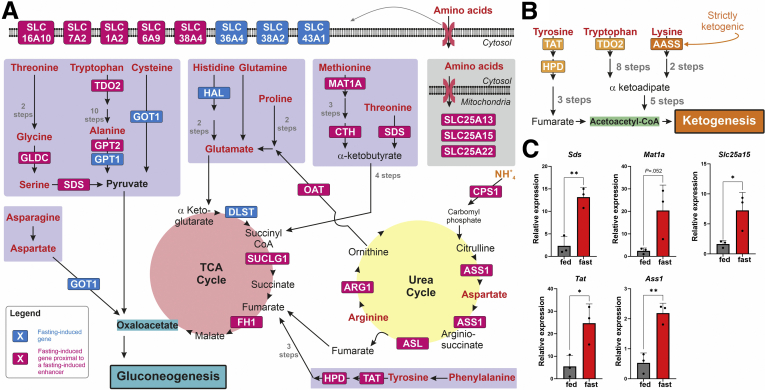
Fasting leads to induction of AA catabolism genes leading to gluconeogenesisand ketogenesis. Scheme showing the fasting-induced genes involved in AA catabolism. AAs (red) are catabolized via 5 catabolic routes (purple rectangles) leading to gluconeogenesis through oxaloacetate. Some of these routes generate TCA cycle intermediates that are channeled to oxaloacetate. AA uptake from the bloodstream and transport across mitochondria are mediated via specific solute carriers (SLCs). AA catabolism relies on the urea cycle that removes the amino group. Genes induced by fasting in mouse liver are depicted with blue background. Genes induced by fasting in mouse liver that are proximal to fasting-induced enhancer are depicted with magenta background. Only enzymes whose gene levels increase during fasting are depicted; other biochemical reactions in the pathways are represented only with black arrows (A). Scheme showing the fasting-induced genes involved in AA catabolism leading to ketogenesis. Genes induced by fasting in mouse liver are depicted with orange background. Only enzymes whose gene levels increase during fasting are depicted; other biochemical reactions in the pathways are represented only with black arrows. The only fasting-induced gene that is strictly ketogenic (Aass) is depicted in dark orange (B). Mice were fasted for 24 hours, and hepatic gene expression was measured (compared with an ad libitum fed control), showing fasting-dependent gene induction of AA catabolism genes (C). Graphs represent data collected from 3 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01.
Hepatic gluconeogenesis occurs both basally and in response to hormonal and nutritional signals. By far the best studied gluconeogenic hormone is glucagon, a peptide hormone secreted from α cells in the pancreas. On its discovery, glucagon was found to increase blood glucose. Further research showed its gluconeogenic capacity and delineated the hepatic cellular components mediating it.3, 4, 5 The gluconeogenic activity of glucagon is mediated in part by its potent effect on hepatic AA uptake and catabolism. Early studies showed that glucagon infusion sharply decreased plasma AA levels, whereas glucagon deficiency increased it.6, 7, 8, 9, 10, 11 More recent studies found that knockout of the glucagon gene or the glucagon receptor as well as glucagon receptor antagonism led to increased plasma and liver AAs.12, 13, 14, 15, 16, 17
The urea cycle is related to AA-based gluconeogenesis as it serves to rid of the amino group in AAs, thus freeing the rest of the molecule for catabolism. It was also suggested that fumarate, released during the urea cycle, feeds gluconeogenesis at significant concentrations.18 Fitting with its role in AA catabolism, glucagon was shown to augment the activity of the urea cycle.19, 20, 21, 22
Glucagon-mediated activation of AA catabolism and the urea cycle are important in 2 nutritional extremes: during fasting and after ingestion of a high-protein meal. Both these conditions lead to increases in glucagon concentration and subsequent AA catabolism.3,23 During fasting, muscle proteins are degraded, and the resulting AAs serve as gluconeogenic precursors in the liver. This process maintains sufficient glucose levels in the lack of glucose entering from meals. After a high-protein meal, the levels of plasma AAs rise because of dietary protein degradation. Glucagon is secreted and activates the hepatic urea cycle to purge excess amino groups. This is coupled with gluconeogenesis that converts the resulting non-amino compounds to glucose.23
The effects of glucagon on fasting hepatocyte metabolism are mediated in part by transcriptional regulation.24 Glucagon signaling increases cyclic adenosine monophosphate (cAMP), activates protein kinase A (PKA), which in turn phosphorylates and activates cAMP responsive element binding protein (CREB).25,26 CREB is long known to induce gluconeogenic genes (such as Pck1, Ppargc1a, and G6pc), thereby contributing to hepatic gluconeogenesis.27, 28, 29
The levels of glucocorticoids (mainly cortisol in humans and corticosterone in rodents) also increase during fasting, leading to activation of the glucocorticoid receptor (GR). Similar to CREB, GR also regulates gluconeogenic genes and increases gluconeogenesis in the liver.29, 30, 31, 32 The prominent role of GR in regulating gluconeogenesis is highlighted by the finding that mice deficient in hepatic GR suffer from hypoglycemia.33 Although glucocorticoids and GR are known as central players in hepatic gluconeogenesis, their contribution to AA catabolism is mostly described in muscle where they facilitate protein degradation and AA release to support hepatic gluconeogenesis.34 In addition to CREB and GR, several other transcriptional factors (TFs) such as FoxO1, FoxA2, C/EBPα, C/EBPβ, and HNF4α were shown to promote gluconeogenic gene expression, largely in a constitutive manner and not in response to hormone stimuli.35,36
Here we explored the transcriptional regulation of genes involved in AA catabolism during fasting. We hypothesized that fasting leads to overt regulation of AA catabolism genes for 2 reasons. (1) Fasting elicits dramatic changes in the expression of thousands of genes.35,37 (2) Glucagon levels increase during fasting, and hepatic glucagon signaling was repeatedly shown to support AA catabolism,6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16 with some reports showing altered gene expression of several genes.12, 13, 14, 15,17,38 We found that the 2 fasting hormones, glucagon and corticosterone, synergistically induce key AA catabolizing enzymes and transporters in primary hepatocytes. These genes are induced during fasting in mice, and their adjacent enhancers become more accessible. This effect is mediated by glucagon-activated CREB and corticosterone-activated GR. The cooperation between glucagon and glucocorticoids was also evident in synergistic hepatic glucose production from various AA precursors. Thus, glucagon alone is unable to fully activate gluconeogenesis, and glucocorticoids serve to maximize its response.
Results
Although AA catabolism during fasting is well-documented, whether it is transcriptionally regulated is not clear. To evaluate transcriptional regulation of AA metabolism genes in a genome-wide manner, we analyzed our previously published dataset where the hepatic transcriptome was evaluated via RNA sequencing in mice after 24 hours of fasting.35 Differential expression analysis revealed that 1131 genes were induced, whereas 1742 genes were repressed compared with the ad libitum fed state (fold change [FC] ≥1.5, adjusted P value ≤.05; Supplementary Table 1). To gain insights into the biological pathways enriched during fasting, we analyzed the set of fasting-induced genes via GeneAnalytics (LifeMap Sciences, Walnut, CA).39 We found that AA metabolism was a highly enriched pathway (adjusted P value ≤.0001; ranked the fourth highest enriched pathway out of 47 pathways; Supplementary Table 2). Taking into account the GeneAnalytics output, together with manual curation, we compiled a list of 37 genes that were induced in the liver after fasting and whose encoded proteins play a role in AA metabolism. Strikingly, we found that 31 of 37 genes play a role in AA uptake and catabolism. Within this group, 13 genes participate in AA catabolism into gluconeogenic and/or ketogenic precursors (Cth, Gldc, Got1, Gpt, Gpt2, Hal, Mat1a, Oat, Sds, Tat, Hpd, Tdo2, Aass), 5 genes are part of the urea cycle (Arg1, Asl, Ass1, Cps1, Slc25a15), 3 genes are part of the TCA cycle needed for AA catabolism (Fh1, Dlst, Suclg1), and 10 genes facilitate AA uptake and transport from outside the cell or between organelles (Slc16a10, Slc1a2, Slc25a13, Slc25a22, Slc36a4, Slc38a2, Slc38a4, Slc43a1, Slc6a9, Slc7a2). Because all the above genes eventually promote AA catabolism into gluconeogenic or ketogenic precursors, we collectively term them AA catabolism genes in the rest of the text. Further examination of AA catabolism genes involved in supplying gluconeogenic precursors revealed that all 5 known AA catabolic routes providing gluconeogenic precursors contained between 1 and 6 genes that were induced by fasting (Figure 1A). A similar pattern is observed in AA catabolic routes providing ketogenic precursors. However, except for Aass, which is strictly ketogenic, the other 3 genes can play both a gluconeogenic and a ketogenic role (Figure 1B). To validate gene induction of AA catabolism genes after fasting, we tested gene expression of selected genes after 24 hours of fasting as compared with an ad libitum fed control. In line with the RNA sequencing results, all tested genes were induced after fasting in mouse liver (Figure 1C).
Gene induction during fasting is often accompanied by increased accessibility of enhancers adjacent to the induced gene.35 To examine this in the context of AA catabolism genes, we analyzed our previously published chromatin accessibility data.35 Changes in hepatic chromatin accessibility of mice fasted for 24 hours were evaluated by DNase hypersensitivity followed by sequencing (DNase-seq). Comparing the ad libitum fed state with 24 hours of fasting, we found that 5331 sites in the genome increased their accessibility after fasting, whereas 6472 sites showed decreased accessibility (FC ≥1.5, adjusted P value ≤.05; Supplementary Table 3). Our previous study identified fasting-induced sites as DNA regions with a gene regulatory role and were therefore termed fasting-induced enhancers.35 To find the genes associated with fasting-induced enhancers, we identified the nearest gene to each enhancer. Out of 31 AA catabolism genes induced by fasting, 24 genes were the nearest genes to at least one fasting-induced enhancer (Figure 1A, Supplementary Table 3). Also, GeneAnalytics analysis of genes proximal to fasting-induced enhancers revealed that AA metabolism is ranked twelfth out of 108 enriched pathways (Supplementary Table 3). Taken together, these results show that induction of AA catabolism genes during fasting is widespread and covers all routes of AA catabolism leading to gluconeogenesis. Also, the gene induction pattern of AA catabolism genes during fasting is associated with an increase in the accessibility of adjacent fasting-induced enhancers, suggesting a high degree of transcriptional regulation.
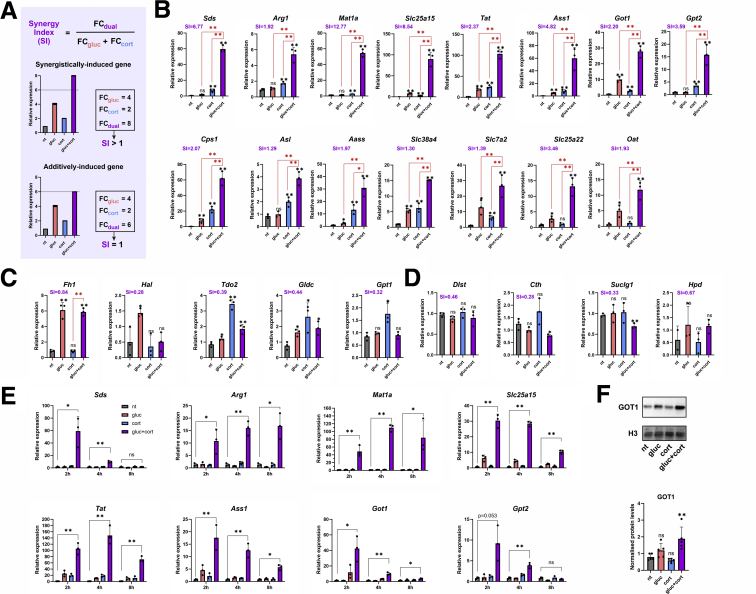
Various studies demonstrated the role of glucagon in promoting both hepatic gluconeogenesis and AA catabolism during fasting. Also, previous studies have shown that glucagon signaling and glucocorticoids together induce genes in a synergistic manner.28,35,40, 41, 42, 43, 44 To explore the possible cooperation between glucagon and glucocorticoids in inducing AA catabolism genes, we treated primary mouse hepatocytes with glucagon, corticosterone (the predominant mouse glucocorticoid increased during fasting), or both in a dual treatment. Cells were treated with hormones for 2 hours to ascertain we are measuring the primary transcriptional effect of these hormones and not secondary effects. Moreover, to measure gene expression changes stemming from transcription (rather than post-transcriptional effects), we measured nascent RNA transcripts throughout the study. We identified a list of 31 fasting-induced AA catabolism genes. From this list we tested all enzyme-encoding genes (n = 20) and a selected group of AA-transporter–encoding genes (n = 4) using quantitative polymerase chain reaction (qPCR). To test for a synergistic effect between the 2 hormones we made these assumptions: In a case where the 2 hormones affect gene expression independently, we could assume that the combined effect of 2 single treatments would be additive, ie, the effect of the dual treatment would amount to the sum of the effects of 2 single treatments. However, in a synergistic relationship where the 2 hormones cooperate to regulate gene expression, the combined effect of the 2 treatments would be above additive, ie, synergistic. On the basis of these assumptions, we distinguished additive from synergistic effects by 2 ways. First, we examined whether the transcript level after a dual treatment is higher than both single treatments in a statistically significant manner. Second, we calculated a synergy index (SI), which measures the FC increase in gene expression after the dual treatment compared with the 2 single treatments. We calculated 3 values: (1) the FC expression of glucagon over non-treated (gluc/nt), (2) the FC expression of corticosterone over non-treated (cort/nt), and (3) the FC expression of dual treatment over non-treated (dual/nt). Then we divided the dual FC value by the sum of the 2 single treatments FC values. Therefore, SI = FCdual / [FCgluc + FCcort]. Thus, if a gene is additively induced, it will have SI = 1, whereas synergistically induced genes will have SI > 1 (Figure 3A). On the basis of these 2 parameters, we found that the majority of tested AA catabolism genes (15/24) showed a synergistic pattern of expression (SI ≥ 1.29) (Figure 3B). Of note, most synergistic genes were induced also by the single treatments. A group of AA catabolism genes were either induced in a non-synergistic manner (n = 5; Figure 3C) or were unresponsive to glucagon or corticosterone treatments (n = 4; Figure 3D). A time course experiment in which cells were treated with hormones for varying periods of time revealed that the synergistic expression pattern is rapid and transient because it subsides by 8 hours of treatment (Figure 3E). The synergistic pattern of expression was evident also at the protein levels for GOT1, translated from the synergistically induced gene Got1 (Figure 3F). Taken together, these findings show that the majority of fasting-induced AA catabolism genes are rapidly induced in a synergistic manner by the 2 major fasting hormones, glucagon and corticosterone.
Figure 3.
AA catabolism genes are synergistically induced by glucagon and corticosterone. To separate synergistic from additive effects, we calculated SI for each gene. FC expression of each gene in the dual treatment (FCdual) was divided by the sum of the 2 FC expression values of the single treatments (FCgluc + FCcort). An example for an additive gene expression pattern is shown on the bottom, and a synergistic pattern is shown on the top (A). Primary mouse hepatocytes were treated for 2 hours with glucagon (gluc), corticosterone (cort), or both in a dual treatment (gluc + cort). RNA levels of fasting-induced AA catabolism genes were measured by qPCR. Some genes show synergistic pattern of expression (B), whereas others are induced by only 1 hormone (C) or are unaffected by either hormone (D). Treating cells for varying periods of time (2, 4, and 8 hours) showed peak of induction at 2–4 hours and its decrease at 8 hours (E). Primary mouse hepatocytes were treated for 16 hours with gluc, cort, or both in a dual treatment (gluc + cort). Protein levels of GOT1 were measured via Western blotting, showing a synergistic pattern. GOT1 protein levels were normalized to H3 loading control (F). Black asterisks denote statistical significance compared with non-treated control (nt). Red asterisks denote statistical significance of dual treatment compared with indicated single treatment. Graphs represent data collected from 3 independent replicates except for (F) where 5 replicates were used. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05.
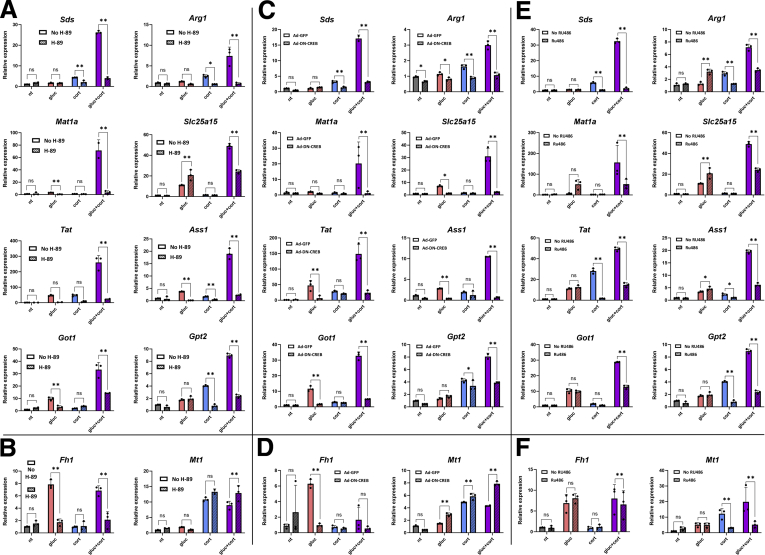
Transcriptional regulation by glucagon is mediated mainly through the cAMP-PKA-CREB pathway,25,26 whereas corticosterone-dependent regulation of genes is brought about by GR.29, 30, 31, 32 To check whether synergistic gene expression of AA catabolism genes is mediated by these 2 pathways, we perturbed them at several points. We treated primary hepatocytes with fasting hormones together with H-89, which inhibits PKA activity. H-89 was able to abolish both glucagon-dependent and synergistic gene induction (Figure 4A). To verify that the responsible TF downstream of PKA is CREB, we perturbed CREB activity via an adenovirally infected dominant-negative peptide (Ad-DN-CREB).45 Indeed, glucagon-dependent as well as synergistic gene induction was abolished (Figure 4C). Of note, both H-89 and Ad-DN-CREB did not affect corticosterone-dependent induction of an unrelated GR target gene (Mt1), suggesting these reagents do not affect GR signaling (Figure 4B and D). Reciprocally, inhibiting GR activity via treatment with RU-486, a GR inhibitor, eliminated corticosterone-dependent as well as synergistic gene induction (Figure 4E). RU-486 treatment did not affect glucagon-dependent induction in a gene induced solely by glucagon (Fh1, Figure 4F). Taken together, these findings show that CREB and GR are both necessary for the synergistic induction of AA catabolism genes by glucagon and corticosterone.
Figure 4.
Synergistic induction of AA catabolism genes is mediated by CREB and GR. Primary mouse hepatocytes were treated for 2 hours with gluc, cort, or both in a dual treatment (gluc + cort), and RNA levels of fasting-induced AA catabolism genes were measured by qPCR. Inactivation of PKA by H-89 abolished synergistic gene expression (A) and non-synergistic, glucagon-mediated gene induction (Fh1) but did not affect corticosterone-mediated gene induction (Mt1) (B). Adenoviral-mediated expression of a dominant negative peptide inactivating CREB (Ad-DN-CREB) abolished synergistic gene expression, whereas adenoviral-mediated expression of green fluorescent protein did not (C). Ad-DN-CREB also affected non-synergistic, glucagon-mediated gene induction (Fh1) but did not affect corticosterone-mediated gene induction (Mt1) (D). Inactivation of GR by RU-486 abolished synergistic gene expression (E) and non-synergistic, corticosterone-mediated gene induction (Mt1) but did not affect glucagon-mediated gene induction (Fh1) (F). Graphs represent data collected from 3 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05.
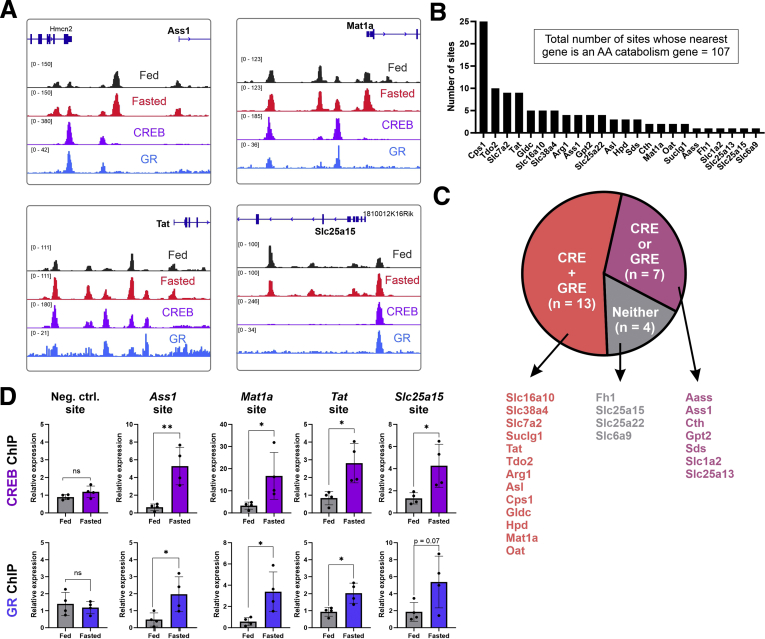
Twenty-four AA catabolism genes were the nearest genes to at least one fasting-induced enhancer (Figure 1A, Supplementary Table 3). As an example, the loci of 4 genes are depicted in Figure 2A, and an obvious increase in hepatic chromatin accessibility is observed on fasting. Further analysis revealed that most AA catabolism genes had several fasting-induced enhancers adjacent to them, reaching a total of 107 fasting-induced enhancers whose nearest gene is an AA catabolism gene (Figure 2B). This observation further emphasizes the high degree of transcriptional regulation imposed on these genes. Considering the involvement of CREB and GR in regulating AA catabolism gene expression (Figure 4), we examined the occurrence of their recognition motif sequences (cAMP response element [CRE] and glucocorticoid response element [GRE]) in fasting-induced enhancers. Out of 24 genes with fasting-induced enhancers, 20 genes were adjacent to at least one fasting-induced enhancer harboring a CRE and/or a GRE. Of these 20 genes, 13 had a fasting-induced enhancer harboring both a CRE and a GRE (Figure 2C). To examine whether CREB and GR bind these motifs after glucagon and corticosterone treatment, we analyzed our previously published CREB and GR chromatin immunoprecipitation assay coupled with sequencing (ChIP-seq) data in primary mouse hepatocytes co-treated with these 2 hormones.35 We found evident binding of both CREB and GR after hormone treatment at fasting-induced enhancers (Figure 2A). To show that these binding events occur also in liver after fasting, we performed ChIP for CREB and GR in livers of mice fasted for 24 hours (compared with the ad libitum fed control). We found that after fasting, both CREB and GR showed increased binding at fasting-induced enhancers adjacent to synergistically induced AA catabolism genes (Figure 2D). Together, these results show that fasting-induced enhancers accommodate CREB and GR binding next to AA catabolism genes during fasting and after hormone treatment. This binding is associated with increased expression of these genes.
Figure 2.
CREB and GR bind fasting-induced enhancers adjacent to fasting-induced AA catabolism genes. Loci of 4 fasting-induced genes are presented. Black and red tracks represent normalized DNase accessibility of hepatic chromatin in the fed vs 24-hour fasted states, respectively. Violet and blue tracks represent normalized ChIP-seq of CREB and GR, respectively, after treatment of glucagon and corticosterone in primary mouse hepatocytes. Increased chromatin accessibility as well as evident CREB and GR binding at enhancers is observed adjacent to fasting-induced AA catabolism genes (A). Number of fasting-induced enhancers adjacent to each AA catabolism gene is depicted (7 genes with no fasting-induced enhancers are not shown) (B). Genes with adjacent fasting-induced enhancers were divided into 3 groups: genes whose proximal enhancer contain both a CRE and a GRE (n = 13), either a CRE or a GRE (n = 7), or neither (n = 4) (C). Chromatin immunoprecipitation of CREB and GR was performed on mice fasted for 24 hours and on ad libitum fed mice. CREB and GR showed increased binding at sites near fasting-induced, synergistic AA catabolism genes. Graphs represent data collected from 4 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05 (D).
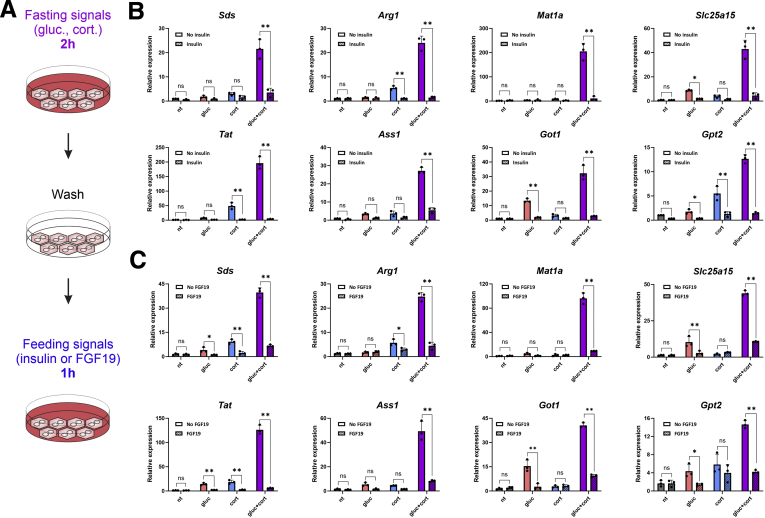
Glucagon and glucocorticoids are the major fasting hormones encountered by the liver. Conversely, on refeeding with a carbohydrate meal, insulin is secreted and quickly halts gluconeogenesis and other fasting-related pathways.46 Fibroblast growth factor 19 (FGF19) is an intestinal endocrine factor secreted after a meal that negatively regulates gluconeogenesis, similar to insulin.47 To examine whether these 2 feeding signals can repress the induction of AA catabolism genes, we treated primary hepatocytes with glucagon and corticosterone for 2 hours, washed the cells with phosphate-buffered saline (PBS), and treated them with either insulin or FGF19 for 1 hour (Figure 5A). The synergistic induction of all AA catabolism genes was potently repressed by either insulin or FGF19, bringing their expression levels to basal or near-basal levels. Insulin and FGF19 also dampened the induction seen in the single treatments by either glucagon or corticosterone (Figure 5B and C). Thus, although fasting hormones induce AA catabolism genes, feeding hormones rapidly and potently de-induce them back to basal levels.
Figure 5.
Feeding hormones de-induce AA catabolism gene expression back to basal levels. Primary hepatocytes were treated for 2 hours with gluc, cort, or both in a dual treatment (gluc + cort). Then hepatocytes were washed with PBS and treated with insulin or FGF19 for 1 hour. RNA was isolated, and levels of fasting-induced AA catabolism genes were measured by qPCR (A). Synergistic gene induction, as well as single-treatment gene induction, was rapidly and potently repressed by insulin (B) and FGF19 (C). Graphs represent data collected from 3 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05.
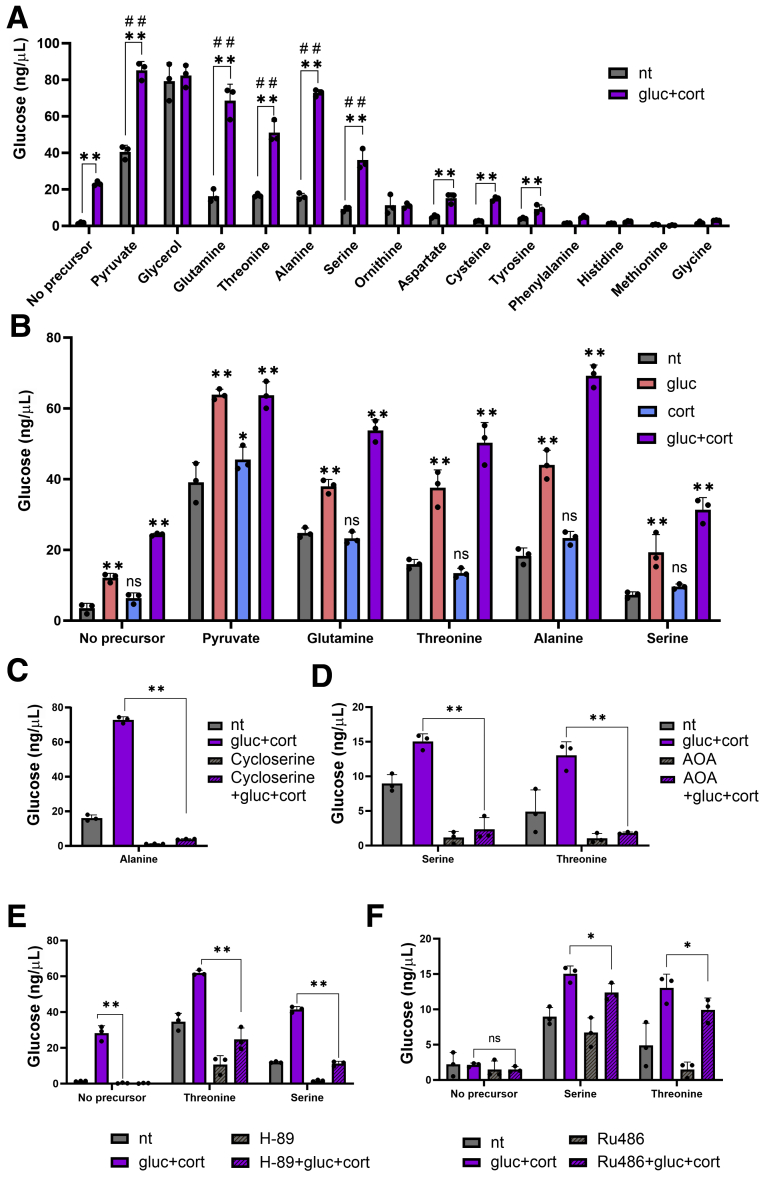
The widespread cooperation between glucagon and glucocorticoid in regulating AA catabolism genes is expected to augment the gluconeogenic capacity of the liver from AA precursors. To examine this hypothesis, we measured glucose production from primary mouse hepatocytes after a dual hormone treatment. Hepatocytes were incubated with glucagon and corticosterone together with a gluconeogenic precursor. As a control, we used 2 non-AA precursors, pyruvate and glycerol. Glucose production from pyruvate after a dual hormone treatment showed a substantial increase compared with pyruvate alone, as previously reported.35 This is assumed to occur partly because of induction of gluconeogenic genes such as Pck1. Glucose production from glycerol was high regardless of hormone treatment, presumably because gluconeogenesis from glycerol requires fewer steps and does not necessitate the synergistically induced gene Pck1. To examine gluconeogenesis from AA precursors, we incubated hepatocytes with all AAs whose catabolism is suspected to require at least one fasting-induced gene (n = 12; Figure 1A). Notably, these AAs are elevated in the plasma of mice with perturbed glucagon signaling.12,14, 15, 16 We found that glucose production from 4 AA precursors (serine, threonine, glutamine, and alanine) was increased after treatment with glucagon and corticosterone (Figure 6A). Glucose production from other AA precursors increased after a dual hormone treatment but not to a higher extent compared with the control (to which no precursor was added; Figure 6A). Next, we tested whether glucagon and corticosterone synergize in glucose production from the 4 AAs mentioned above (serine, threonine, glutamine, and alanine). Glucagon alone led to increased glucose production from all 4 AAs, whereas corticosterone did not. However, a dual treatment led to a synergistic increase in glucose production from all tested AAs (Figure 6B), in line with the synergistic gene induction pattern. To determine whether this synergistic effect is dependent on AA catabolism routes, we examined glucose production after inhibition of AA catabolic enzymes. L-cycloserine is an inhibitor of aminotransferases with selectivity toward alanine aminotransferase48 (encoded by the synergistically induced gene Gpt2). Glucose production from alanine was abolished by L-cycloserine at both the unstimulated and hormone-stimulated conditions (Figure 6C). Aminooxyacetate (AOA) is an inhibitor of pyridoxal 5-phosphate–dependent reactions.49 Many AA catabolic reactions are pyridoxal 5-phosphate–dependent, including the reaction catalyzed by serine dehydratase (encoded by the synergistically induced gene Sds). Glucose production from serine or threonine (whose catabolism relies on Sds) was eliminated by AOA (Figure 6D).
Figure 6.
Glucose production from AA precursors is synergistically augmented by glucagon and corticosterone. Primary hepatocytes were treated with gluconeogenic precursors and with gluc, cort, or both in a dual treatment (gluc + cort) for 20 hours. Dual treatment increased hepatic glucose production from several AA precursors (A). Glucose production was unaffected by corticosterone alone. Glucagon treatment increased glucose production, and this effect was synergistically augmented by corticosterone (B). Effect of glucagon and corticosterone on glucose production from alanine was abolished when cells were treated with L-cycloserine, an inhibitor of alanine aminotransferase (C). Effect of glucagon and corticosterone on glucose production from serine and threonine was abolished when cells were treated with AOA, an inhibitor of pyridoxal 5-phosphate–dependent reactions (D). Synergistic glucose production was impaired on inhibition of glucagon signaling by H-89 (E) or corticosterone signaling by RU-486 (F). Graphs represent data collected from 3 independent replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. Single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. Double pound signs denote statistical significance of adjusted P value ≤.01 compared with gluc + cort condition with no precursor added. In (B) asterisks denote statistical significance compared with non-treated (nt control) under the same AA precursor.
To show that the increase in gluconeogenesis after glucagon and corticosterone is dependent on CREB and GR, we measured glucose production in cells treated with either H-89 or RU-486. These 2 inhibitors inhibited hormone-dependent glucose production from both serine and threonine (Figure 6E and F), highlighting the importance of gene regulation in gluconeogenesis from AA precursors.
Discussion
Hepatic catabolism of AAs during fasting is long known. However, the gene expression program that supports it and the crosstalk between fasting hormones mediating it are not well-elucidated. In this study we explored the fasting-dependent transcriptional regulation of genes involved in AA catabolism. We show that 31 genes directly related to AA catabolism are transcriptionally regulated. Moreover, we describe a crosstalk between glucagon and glucocorticoids driving a synergistic gene expression pattern of AA catabolism genes as well as augmented gluconeogenesis from AA precursors.
The synergistic cooperation between glucagon and glucocorticoids was described in early studies showing that co-infusion of glucagon together with cortisol to dogs led to synergistic glucose production,50 a finding that was soon replicated in humans (in this case, the co-infusion also included epinephrine).51 Years later, the synergistic cooperation between glucagon-cAMP-PKA signaling and glucocorticoids was reported to take part in the transcriptional regulation of several genes. First, Pck1 was observed to be synergistically induced by the 2 signals in H4IIE cells, the responsible CRE and GRE motifs were identified, and a physical interaction between CREB and GR was shown.40 The synergistic induction of Pck1 was later replicated in HepG2 cells28 and in primary mouse hepatocytes.35 The urea cycle gene Cps1 was also shown to be synergistically induced by the 2 signals,41 as was G6pc.42 The synergistic relationship between glucagon signaling and glucocorticoids is apparent also outside the liver because studies have shown synergistic induction of Sst43 and Cyp1944 in non-liver cells.
Although synergy between glucagon and glucocorticoids was reported in several genes, this study identifies a set of synergistically induced genes all serving to bring about a specific metabolic outcome—AA catabolism toward gluconeogenesis. Moreover, we show here that this synergistic pattern of expression leads to synergistic glucose production from AA precursors. Together with the previously reported synergistic induction of gluconeogenic genes, these data suggest a transcriptionally based mechanism for the reported synergistic effect of the 2 hormones on glucose production in vivo.50
The models suggested to promote the synergy between glucagon and glucocorticoids were (1) physical interaction between GR and CREB40. (2) An indirect effect mediated via glucagon-induced coactivators of GR. The glucagon-CREB induced coactivator peroxisome proliferator-activated receptor gamma coactivator 1α (Ppargc1a) was shown to promote GR-dependent induction of Pck1.28 Similarly, CREB regulated transcription coactivator 2 (Crtc2), also induced by the glucagon-CREB axis, was found to promote the synergistic induction of G6pc.42 (3) Assisted loading, a model of cooperative binding,52 was previously suggested by us whereby GR increases chromatin accessibility at specific enhancers and thus potentiates CREB binding.35
We show here that the synergy between glucagon and glucocorticoids is apparent in most, but not all, AA catabolism genes we examined. This gene-selective type of cooperation suggests that the crosstalk between the 2 hormones is brought about by TF cooperation at specific enhancers, affecting only a subset of AA catabolism genes. In support of this, we show that CREB and GR co-bind specific enhancers adjacent to synergistically induced genes. Importantly, we show that perturbing GR abolishes synergy but does not affect unrelated glucagon-dependent gene induction. Reciprocally, inactivation of CREB abolishes synergy as well but leaves non-synergistic, corticosterone-induced genes unaffected. Thus, our data strongly support an enhancer-specific model of cooperation (such as the assisted loading model) whereby CREB and GR cooperate on some enhancers to synergistically induce one set of genes but also regulate a different set of genes independently.
We show here that CREB and GR binding is found also at sites lacking their binding motif, a finding that was described elsewhere for both TFs.53,54 Tools for motif searches rely on similarity to the consensus motif, and thus motifs that are less similar to the consensus motif (but may nonetheless confer TF binding) might be missed by these tools. We therefore surmise that the lack of detectable consensus motifs at sites bound by CREB and GR does not rule out direct binding of these TFs to DNA. In addition, the assisted loading model reconciles the lack of detectable motifs in enhancers bound by GR and CREB. We have previously shown that assisted loading of a different TF (signal transducer and activator of transcription 3 [Stat3]) is more prevalent at enhancers where its motif is weaker (ie, less similar to the consensus motif).55 A plausible explanation for this phenomenon is that sites where a TF is weakly bound are those that benefit most from assistance by another TF. Therefore, sites with weaker motifs (that may not be detected in motif searches) may rely on assisted loading for efficient binding. This may be the case in CREB-GR assisted loading because CREB binding sites are associated with GR binding genome-wide, and more than one-third of CREB binding sites lack a detectable CRE.53
Importantly, although GR is able to assist CREB binding by increasing chromatin accessibility,35 it is able to do so only on predetermined enhancers that were made accessible by lineage-determining factors.56 This is exemplified for both CREB and GR, whose binding is compromised in the lack of FoxA2, a liver lineage-determining factor.57 Further research showed that CREB binding to enhancers and promoter regions is only marginally increased after fasting53 or activation by cAMP.58 We show here (as also shown by others57) that certain CREB binding sites do show increased binding on fasting. This supports a scenario in which most CREB binding is constitutive, reaching maximal binding capacity regardless of cAMP, whereas some sites only reach maximal binding on signal, possibly because of cooperative binding with GR. It is also important to note that glucagon activates additional transcription factors other than CREB,24 and that there is redundancy in the role of CREB in regulating gluconeogenesis.59 However, in our experimental settings, perturbing CREB resulted in very similar results to inhibiting PKA, suggesting it plays a role in glucagon-mediated AA catabolism gene induction.
Insulin is well-recognized to repress gluconeogenesis and other hepatic fasting-related pathways such as fatty acid oxidation and ketogenesis. However, there is an ongoing debate of whether the effects of insulin are brought about by direct hepatic insulin signaling or by effects of insulin in non-hepatic tissues. Whereas many studies clearly point to an indirect role of insulin,46,60 other evidence shows a role for direct hepatic signaling.61 Specifically, hepatic insulin signaling was shown to repress CREB.62,63 The role of insulin in GR signaling is not straightforward, with both antagonistic and cooperative mechanisms described.64,65 Similar to insulin, FGF19 is a feeding signal whose inhibitory activities include suppressing CREB-mediated gluconeogenesis.66 Our results show that direct hepatic signaling of insulin and FGF19 strongly repress the transcriptional effects of the glucagon-CREB axis and the corticosterone-GR axis.
When examining hepatic glucose production from specific AA precursors, we found that serine, threonine, alanine, and glutamine led to the highest glucose production after glucagon and corticosterone treatments. These results are in line with previous in vivo reports and human studies. First, glutamine and alanine are considered major contributors to gluconeogenesis because of the conversion of branched-chain AAs in muscle to glutamine and alanine, followed by their release to the blood.67 Second, glutamine was recently shown to heavily contribute to glucagon-mediated gluconeogenesis.68 Third, threonine and serine plasma levels (together with glycine) show the largest decrease on glucagon infusion.6,8 Finally, the uptake of glutamine, alanine, serine, and threonine is robustly mediated by glucagon as evidenced by their increased plasma levels on perturbation of glucagon signaling.12,14,15
Interestingly, one of the most synergistically induced genes is Tat, which is involved in tyrosine catabolism. However, gluconeogenesis from tyrosine was similar to the no-precursor control. A possible explanation for this observation is that tyrosine catabolism is brought about to reduce the signaling effect of tyrosine rather than to direct it to gluconeogenesis. Pancreatic α cells secrete glucagon in response to high plasma AA levels,69 and tyrosine is suggested to play an instrumental role in stimulating glucagon secretion.70 Thus, hepatic tyrosine catabolism via increased Tat expression may reduce plasma tyrosine levels, serving to relieve glucagon secretion. This potential negative feedback loop warrants further investigation.
In addition to enzymes directly involved in AA catabolic pathways, our analysis found a major transcriptional induction of urea cycle enzymes and AA transporters during fasting. This is in accordance with the central role of AA uptake and the urea cycle in AA catabolism. Therefore, fasting hormones cooperate in transcriptional induction of core AA catabolic enzymes as well as supporting processes such as AAs uptake, transport, and amino group removal. Three TCA cycle enzymes needed for AA catabolism were also induced by fasting. However, 2 of them (Dlst, Suclg1) were not induced by either glucagon or corticosterone, whereas Fh1 was induced only by glucagon. This suggests that the induction of TCA cycle enzymes during fasting is not mediated by corticosterone and that glucagon plays a minor role in it.
An important finding in our study is the insufficiency of glucagon to fully activate gluconeogenesis. Our gene expression analyses and glucose production assays clearly show a synergistic effect between glucagon and corticosterone in regulating gluconeogenesis. Gluconeogenesis from AA precursors becomes substantial only in prolonged fasting (18 hours) after glycogen stores are depleted.71 This is simultaneous with the increase of corticosterone during fasting35 and aligns with our results showing that corticosterone is needed to reach maximal glucose production from AA precursors, whereas glucagon alone leads to submaximal glucose levels.
In conclusion, we found that 31 AA catabolism genes are induced during fasting, with their enhancers activated. This is mediated by the fasting hormones glucagon and corticosterone that synergistically induce AA catabolism genes via CREB and GR, leading to increased gluconeogenesis. Although glucagon undoubtedly plays a central role in gluconeogenesis from AA precursors, it is only together with corticosterone that maximal gluconeogenic capacity is achieved.
Materials and Methods
Reagents and Materials
Cells were treated with the following reagents at the specified concentrations: glucagon 100 nmol/L (RayBiotech, Peachtree Corners, GA; cat# 228-10549-1); corticosterone 1 μmol/L (Sigma-Aldrich, St Louis, MO; 50-22-6); insulin 1 μmol/L (Biological Industries, Beit HaEmek, Israel; 41-975-100); FGF19 80 ng/mL (R & D, Minneapolis, MN; 969-FG); H-89 20 μmol/L (Sigma-Adlrich; B1427); RU-486 2 μmol/L (Sigma-Aldrich; 84371-65-3); L-cycloserine 300 μmol/L (Sigma-Aldrich; 339-72-0); and AOA 1 mmol/L (Cayman Chemical Company, Ann Arbor, MI; 28298).
Primary Mouse Hepatocyte Isolation
Isolation of primary mouse hepatocytes was performed as detailed in our published protocol with no modifications.72 Male 8-week-old mice (C57BL/6JOlaHsd) were used for isolations. All animal procedures were compatible with the standards for care and use of laboratory animals. The research has been approved by the Hebrew University of Jerusalem Institutional Animal Care and Use Committee. The Hebrew University of Jerusalem is accredited by the National Institutes of Health and by Association for Assessment and Accreditation of Laboratory Animal Care International to perform experiments on laboratory animals (NIH approval number: OPRR-A01-5011).
RNA Preparation, Reverse Transcription, and qPCR
Total RNA was isolated from primary mouse hepatocytes using NucleoSpin kit (Macherey-Nagel, Duren, Germany; cat# 740955.25) according to the manufacturer’s protocol. For qPCR, 1 μg total RNA was reverse transcribed to generate cDNA (Quantabio, Beverly, MA; cat# 76047-074). Quantitative PCR was performed using a C1000 Touch thermal cycler CFX96 instrument (Bio-Rad, Hercules, CA) using SYBR Green (Quantabio; 101414-276). Gene values were normalized with a housekeeping gene (Tbp or Rpl13). The primers used in this study (except for Tbp and Rpl13) were designed to amplify nascent transcripts (ie, the amplified region span exon-intron junctions) as a proxy for transcription and to avoid confounding post-transcriptional events. The primers used in this study are detailed in Supplementary Table 4.
Adenoviral Infections
Four hours after plating primary mouse hepatocytes, adenoviral particles containing a dominant-negative peptide against CREB45 or green fluorescent protein as a control (Vector Biolabs, Burlingame, CA; cat# 1060) were added to the cultures (cells plated in 6-well plates, 4 × 105 cells per well, 1 × 106 pfu per well). Twenty-four hours after infection, cells were treated for 2 hours with combinations of glucagon and corticosterone, followed by collection and RNA isolation.
ChIP
Liver samples (200 mg) from mice (male, 8-week-old, C57BL/6JOlaHsd) either fed ad libitum or fasted for 24 hours were homogenized and cross-linked with 1% formaldehyde for 10 minutes at room temperature and quenched with 0.125 mol/L glycine. Crosslinked samples were washed in PBS, resuspended in ChIP lysis buffer (0.5% sodium dodecyl sulfate [SDS], 10 mmol/L EDTA, 50 mmol/L Tris-HCl pH 8), and sonicated (Bioruptor; Diagenode, Denville, NJ) to release ~300 base pair fragments. Antibodies (4 μg per sample) against CREB (Cell Signaling Technology, Danvers, MA; #9197S) and GR (Cell Signaling Technology; #12041S) were conjugated to magnetic beads (Sera-Mag; Merck, Kenilworth, NJ; #GE17152104010150) for 2 hours at 4°C. Chromatin was immunoprecipitated with antibody-bead conjugates over night at 4°C. Immunocomplexes were washed sequentially with the following buffers: low-salt buffer (0.01% SDS, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris-HCl pH 8, 150 mmol/L NaCl), high-salt buffer (0.01% SDS, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris-HCl pH 8, 500 mmol/L NaCl), low-salt buffer, and TE buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA pH 8). Chromatin was de-proteinized with proteinase K (Hy Labs, Israel; EPR9016) for 2 hours at 55°C and de-crosslinked overnight at 65°C. DNA was subsequently phenol-chloroform purified and ethanol precipitated. Primers used for quantification are detailed in Supplementary Table 4.
Western Blotting
Cells were lyzed with RIPA buffer, and protein samples were loaded on 12% polyacrylamide SDS gels. Proteins were transferred (Trans Blot Turbo, Bio-Rad; cat# 1704158) to a nitrocellulose membrane (Trans-Blot Turbo Transfer Pack, Bio-Rad; cat# 1704158), blocked for 1 hour with 5% low-fat milk, and incubated with primary antibody (1:1000 anti-GOT1 Cell Signaling Technologies, cat# 34423S; 1:2000 anti-histone H3, Active Motif, cat# 39763) solution (Tris-buffered saline 0.5% Tween, 5% bovine serum albumin) for 16 hours. Membranes were incubated with secondary Peroxidase AffiniPure goat anti-rabbit immunoglobulin G (1:2000, Jackson Laboratory, Bar Harbor, ME; cat# 111-035-144) or anti-mouse (1:10000, Jackson Laboratory; cat# 115-035-146) for 1 hour, followed by washes and 1-minute incubation with Western blotting luminol reagent (Santa Cruz Biotechnologies, Santa Cruz, CA; cat# sc-2048). Imaging and quantification were done with ChemiDoc (Bio-Rad).
Hepatic Glucose Production
Twenty-four hours after isolation, primary hepatocytes (plated in 12-well plates; 2.5 × 105 cells per well) were incubated for 9 hours with Dulbecco modified Eagle medium lacking glucose, pyruvate, glutamine, and phenol red (Gibco, Waltham, MA; A14430-01). Then cells were washed with PBS and treated for 20 hours with medium containing combinations of glucagon (100 nmol/L) and corticosterone (1 μmol/L) together with one gluconeogenic precursor (20 mmol/L): pyruvate (Biological Industries; cat# 03-042-1B), glycerol (Biolab; cat# 07120501), L-ornithine (Sigma-Aldrich; cat# O2375-5G), L-asparagine (Formedium, Norfolk, UK; DOC0114), L-tyrosine (Formedium; DOC0190), L-methionine (Formedium; DOC0166), L-aspartic-acid (Formedium; DOC0166), L-serine (Formedium; DOC0178), L-alanine (Formedium; DOC0102), L-histidine (Formedium; DOC0142), L-phenylalanine (Formedium; DOC0170). After 20 hours, 40 μL of medium was sampled, and glucose was measured using the glucose oxidase colorimetric method according to the manufacturer’s instructions (Sigma-Aldrich; GAGO20).
Bioinformatic Analyses
RNA-seq, ChIP-seq, and DNase-seq analyses were performed on our previously published data35 (deposited in GEO; series# GSE72087). Fastq files were mapped to the mm10 genome assembly using Bowtie2. Differential gene expression was performed using DEseq2 via the HOMER package with default parameters. DNase hypersensitive sites were called using MACS2, and differential hypersensitivity was determined using DEseq2 via the HOMER package with default parameters. The HOMER package was used to detect the nearest gene to each hypersensitive site. Genes whose transcription start site was more the 100 kb away from the hypersensitive site were omitted. The HOMER package was used to identify CREs and GREs. Selected gene loci were visualized by the integrated genome browser (IGV).
Statistics
All conditions in all of the described experiments were performed in 3 replicates or more. Error bars represent standard deviation of biological replicates. Statistical significance was determined by t test; P value was adjusted for multiple comparisons using the Holm-Sidak method. A single asterisk denotes statistical significance of adjusted P value ≤.05. Double asterisks denote statistical significance of adjusted P value ≤.01. ns denotes P value >.05.
All authors had access to the study data and had reviewed and approved the final manuscript.
Acknowledgments
The authors thank Dana Goldberg for help in bioinformatics analyses.
CRediT Authorship Contributions
Noga Korenfeld (Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Equal; Writing – review & editing: Supporting)
Maya Finkel (Formal analysis: Equal; Investigation: Equal; Methodology: Equal)
Nufar Buchshtab (Investigation: Supporting; Methodology: Supporting)
Meirav Bar-Shimon, PhD (Project administration: Equal)
Meital Charni-Natan, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Lead; Supervision: Equal; Writing – review & editing: Equal)
Ido Goldstein, PhD (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Project administration: Supporting; Supervision: Lead; Validation: Supporting; Visualization: Supporting; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the Israel Science Foundation (ISF, grants no. 1469/19 and 3533/19), the Canadian Institutes of Health Research (CIHR), the International Development Research Centre (IDRC), the Zuckerman STEM Leadership Program, the Azrieli Foundation, and the Abisch-Frenkel Foundation. I-G is a Zuckerman Faculty Scholar. M-CN was supported by the Golda Meir Fellowship and the Leo & Julia Forcheimer Endowment. None of the funding agencies were involved in study design, collection, analysis, or interpretation of data.
Note: to access the supplementary materials accompanying this article, visit the online version of Cellular and Molecular Gastroenterology and Hepatology at www.cmghjournal.org.
Supplementary Material
References
- 1.Petersen M.C., Vatner D.F., Shulman G.I. Regulation of hepatic glucose metabolism in health and disease. Nature Reviews Endocrinology. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tetrick M.A., Odle J. What constitutes a gluconeogenic precursor? J Nutr. 2020;150:2239–2241. doi: 10.1093/jn/nxaa166. [DOI] [PubMed] [Google Scholar]
- 3.Campbell J.E., Drucker D.J. Islet α cells and glucagon: critical regulators of energy homeostasis. Nature Reviews Endocrinology. 2015;11:329–338. doi: 10.1038/nrendo.2015.51. [DOI] [PubMed] [Google Scholar]
- 4.Habegger K.M., Heppner K.M., Geary N., Bartness T.J., DiMarchi R., Tschöp M.H. The metabolic actions of glucagon revisited. Nature Reviews Endocrinology. 2010;6:689–697. doi: 10.1038/nrendo.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahrén B. Glucagon: early breakthroughs and recent discoveries. Peptides. 2015;67:74–81. doi: 10.1016/j.peptides.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Boden G., Rezvani I., Owen O.E. Effects of glucagon on plasma amino acids. J Clin Invest. 1984;73:785–793. doi: 10.1172/JCI111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liljenquist J.E., Lewis S.B., Cherrington A.D., Sinclair-Smith B.C., Lacy W.W. Effects of pharmacologic hyperglucagonemia on plasma amino acid concentrations in normal and diabetic man. Metabolism Clinical Experimental. 1981;30:1195–1199. doi: 10.1016/0026-0495(81)90041-x. [DOI] [PubMed] [Google Scholar]
- 8.Marliss E.B., Aoki T.T., Unger R.H., Soeldner J.S., Cahill G.F., Jr. Glucagon levels and metabolic effects in fasting man. J Clin Invest. 1970;49:2256–2270. doi: 10.1172/JCI106445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landau R.L., Lugibihl K. Effect of glucagon on concentration of several free amino acids in plasma. Metabolism Clinical Experimental. 1969;18:265–276. doi: 10.1016/0026-0495(69)90047-x. [DOI] [PubMed] [Google Scholar]
- 10.Couet C., Fukagawa N.K., Matthews D.E., Bier D.M., Young V.R. Plasma amino acid kinetics during acute states of glucagon deficiency and excess in healthy adults. Am J Physiol. 1990;258(Pt 1):E78–E85. doi: 10.1152/ajpendo.1990.258.1.E78. [DOI] [PubMed] [Google Scholar]
- 11.Wolfe B.M., Culebras J.M., Aoki T.T., O’Connor N.E., Finley R.J., Kaczowka A., Moore F.D. The effects of glucagon on protein metabolism in normal man. Surgery. 1979;86:248–257. [PubMed] [Google Scholar]
- 12.Watanabe C., Seino Y., Miyahira H., Yamamoto M., Fukami A., Ozaki N., Takagishi Y., Sato J., Fukuwatari T., Shibata K., Oiso Y., Murata Y., Hayashi Y. Remodeling of hepatic metabolism and hyperaminoacidemia in mice deficient in proglucagon-derived peptides. Diabetes. 2012;61:74–84. doi: 10.2337/db11-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J., Okamoto H., Huang Z., Anguiano G., Chen S., Liu Q., Cavino K., Xin Y., Na E., Hamid R., Lee J., Zambrowicz B., Unger R., Murphy A.J., Xu Y., Yancopoulos G.D., Li W.H., Gromada J. Amino acid transporter Slc38a5 controls glucagon receptor inhibition-induced pancreatic α cell hyperplasia in mice. Cell Metabolism. 2017;25:1348–1361.e8. doi: 10.1016/j.cmet.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean E.D., Li M., Prasad N., Wisniewski S.N., Von Deylen A., Spaeth J., Maddison L., Botros A., Sedgeman L.R., Bozadjieva N., Ilkayeva O., Coldren A., Poffenberger G., Shostak A., Semich M.C., Aamodt K.I., Phillips N., Yan H., Bernal-Mizrachi E., Corbin J.D., Vickers K.C., Levy S.E., Dai C., Newgard C., Gu W., Stein R., Chen W., Powers A.C. Interrupted glucagon signaling reveals hepatic α cell axis and role for L-glutamine in α cell proliferation. Cell Metabolism. 2017;25:1362–1373.e5. doi: 10.1016/j.cmet.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solloway M.J., Madjidi A., Gu C., Eastham-Anderson J., Clarke H.J., Kljavin N., Zavala-Solorio J., Kates L., Friedman B., Brauer M., Wang J., Fiehn O., Kolumam G., Stern H., Lowe J.B., Peterson A.S., Allan B.B. Glucagon couples hepatic amino acid catabolism to mTOR-dependent regulation of α-cell mass. Cell Rep. 2015;12:495–510. doi: 10.1016/j.celrep.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y., Wang M.Y., Du X.Q., Charron M.J., Unger R.H. Glucagon receptor knockout prevents insulin-deficient type 1 diabetes in mice. Diabetes. 2011;60:391–397. doi: 10.2337/db10-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tellez K., Hang Y., Gu X., Chang C.A., Stein R.W., Kim S.K. In vivo studies of glucagon secretion by human islets transplanted in mice. Nat Metab. 2020;2:547–557. doi: 10.1038/s42255-020-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ipata P.L., Pesi R. Understanding the interrelationship between the synthesis of urea and gluconeogenesis by formulating an overall balanced equation. Adv Physiol Educ. 2017;41:286–290. doi: 10.1152/advan.00180.2016. [DOI] [PubMed] [Google Scholar]
- 19.Winther-Sørensen M., Galsgaard K.D., Santos A., Trammell S.A.J., Sulek K., Kuhre R.E., Pedersen J., Andersen D.B., Hassing A.S., Dall M., Treebak J.T., Gillum M.P., Torekov S.S., Windeløv J.A., Hunt J.E., Kjeldsen S.A.S., Jepsen S.L., Vasilopoulou C.G., Knop F.K., Ørskov C., Werge M.P., Bisgaard H.C., Eriksen P.L., Vilstrup H., Gluud L.L., Holst J.J., Wewer Albrechtsen N.J. Glucagon acutely regulates hepatic amino acid catabolism and the effect may be disturbed by steatosis. Mol Metab. 2020;42:101080. doi: 10.1016/j.molmet.2020.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husson A., Buquet C., Vaillant R. Induction of the five urea-cycle enzymes by glucagon in cultured foetal rat hepatocytes. Differentiation. 1987;35:212–218. doi: 10.1111/j.1432-0436.1987.tb00171.x. [DOI] [PubMed] [Google Scholar]
- 21.Snodgrass P.J., Lin R.C., Müller W.A., Aoki T.T. Induction of urea cycle enzymes of rat liver by glucagon. J Biol Chem. 1978;253:2748–2753. [PubMed] [Google Scholar]
- 22.Vilstrup H., Hansen B.A., Almdal T.P. Glucagon increases hepatic efficacy for urea synthesis. J Hepatol. 1990;10:46–50. doi: 10.1016/0168-8278(90)90072-y. [DOI] [PubMed] [Google Scholar]
- 23.Bankir L., Bouby N., Speth R.C., Velho G., Crambert G. Glucagon revisited: coordinated actions on the liver and kidney. Diabetes Res Clin Pract. 2018;146:119–129. doi: 10.1016/j.diabres.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein I., Hager G.L. The three Ds of transcription activation by glucagon: direct, delayed, and dynamic. Endocrinology. 2018;159:206–216. doi: 10.1210/en.2017-00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh K.J., Han H.S., Kim M.J., Koo S.H. CREB and FoxO1: two transcription factors for the regulation of hepatic gluconeogenesis. BMB Reports. 2013;46:567–574. doi: 10.5483/BMBRep.2013.46.12.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakravarty K., Cassuto H., Reshef L., Hanson R.W. Factors that control the tissue-specific transcription of the gene for phosphoenolpyruvate carboxykinase-C. Crit Rev Biochem Mol Biol. 2005;40:129–154. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 28.Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A., Rudolph D., Schutz G., Yoon C., Puigserver P., Spiegelman B., Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein I., Hager G.L. Transcriptional and chromatin regulation during fasting: the genomic era. Trends Endocrinol Metab. 2015;26:699–710. doi: 10.1016/j.tem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Præstholm S.M., Correia C.M., Grøntved L. Multifaceted control of GR signaling and its impact on hepatic transcriptional networks and metabolism. Frontiers in Endocrinology. 2020;11:572981. doi: 10.3389/fendo.2020.572981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greulich F., Hemmer M.C., Rollins D.A., Rogatsky I., Uhlenhaut N.H. There goes the neighborhood: assembly of transcriptional complexes during the regulation of metabolism and inflammation by the glucocorticoid receptor. Steroids. 2016;114:7–15. doi: 10.1016/j.steroids.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratman D., Vanden Berghe W., Dejager L., Libert C., Tavernier J., Beck I.M., De Bosscher K. How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Mol Cell Endocrinol. 2013;380:41–54. doi: 10.1016/j.mce.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Opherk C., Tronche F., Kellendonk C., Kohlmuller D., Schulze A., Schmid W., Schutz G. Inactivation of the glucocorticoid receptor in hepatocytes leads to fasting hypoglycemia and ameliorates hyperglycemia in streptozotocin-induced diabetes mellitus. Mol Endocrinol. 2004;18:1346–1353. doi: 10.1210/me.2003-0283. [DOI] [PubMed] [Google Scholar]
- 34.Kuo T., Harris C.A., Wang J.C. Metabolic functions of glucocorticoid receptor in skeletal muscle. Mol Cell Endocrinol. 2013;380:79–88. doi: 10.1016/j.mce.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein I., Baek S., Presman D.M., Paakinaho V., Swinstead E.E., Hager G.L. Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 2017;27:427–439. doi: 10.1101/gr.212175.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int J Biochem Cell Biol. 2012;44:33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Kinouchi K., Magnan C., Ceglia N., Liu Y., Cervantes M., Pastore N., Huynh T., Ballabio A., Baldi P., Masri S., Sassone-Corsi P. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 2018;25:3299–3314.e6. doi: 10.1016/j.celrep.2018.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erion D.M., Kotas M.E., McGlashon J., Yonemitsu S., Hsiao J.J., Nagai Y., Iwasaki T., Murray S.F., Bhanot S., Cline G.W., Samuel V.T., Shulman G.I., Gillum M.P. cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 2 (CRTC2) promotes glucagon clearance and hepatic amino acid catabolism to regulate glucose homeostasis. J Biol Chem. 2013;288:16167–16176. doi: 10.1074/jbc.M113.460246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Ari Fuchs S., Lieder I., Stelzer G., Mazor Y., Buzhor E., Kaplan S., Bogoch Y., Plaschkes I., Shitrit A., Rappaport N., Kohn A., Edgar R., Shenhav L., Safran M., Lancet D., Guan-Golan Y., Warshawsky D., Shtrichman R. GeneAnalytics: an integrative gene set analysis tool for next generation sequencing, RNAseq and microarray data. Omics. 2016;20:139–151. doi: 10.1089/omi.2015.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imai E., Miner J.N., Mitchell J.A., Yamamoto K.R., Granner D.K. Glucocorticoid receptor-cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J Biol Chem. 1993;268:5353–5356. [PubMed] [Google Scholar]
- 41.Christoffels V.M., Grange T., Kaestner K.H., Cole T.J., Darlington G.J., Croniger C.M., Lamers W.H. Glucocorticoid receptor, C/EBP, HNF3, and protein kinase A coordinately activate the glucocorticoid response unit of the carbamoylphosphate synthetase I gene. Mol Cell Biol. 1998;18:6305–6315. doi: 10.1128/mcb.18.11.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill M.J., Suzuki S., Segars J.H., Kino T. CRTC2 is a coactivator of GR and couples GR and CREB in the regulation of hepatic gluconeogenesis. Mol Endocrinol. 2016;30:104–117. doi: 10.1210/me.2015-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J.L., Papachristou D.N., Patel Y.C. Glucocorticoids activate somatostatin gene transcription through co-operative interaction with the cyclic AMP signalling pathway. Biochem J. 1994;301(Pt 3):863–869. doi: 10.1042/bj3010863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe M., Ohno S., Nakajin S. Forskolin and dexamethasone synergistically induce aromatase (CYP19) expression in the human osteoblastic cell line SV-HFO. Eur J Endocrinol. 2005;152:619–624. doi: 10.1530/eje.1.01882. [DOI] [PubMed] [Google Scholar]
- 45.Ahn S., Olive M., Aggarwal S., Krylov D., Ginty D.D., Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Titchenell P.M., Lazar M.A., Birnbaum M.J. Unraveling the regulation of hepatic metabolism by insulin. Trends in Endocrinology and Metabolism. 2017;28:497–505. doi: 10.1016/j.tem.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadaleta R.M., Moschetta A. Metabolic messengers: fibroblast growth factor 15/19. Nat Metab. 2019;1:588–594. doi: 10.1038/s42255-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 48.Cornell N.W., Zuurendonk P.F., Kerich M.J., Straight C.B. Selective inhibition of alanine aminotransferase and aspartate aminotransferase in rat hepatocytes. Biochem J. 1984;220:707–716. doi: 10.1042/bj2200707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.John R.A., Charteris A. The reaction of amino-oxyacetate with pyridoxal phosphate-dependent enzymes. Biochem J. 1978;171:771–779. doi: 10.1042/bj1710771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eigler N., Saccà L., Sherwin R.S. Synergistic interactions of physiologic increments of glucagon, epinephrine, and cortisol in the dog: a model for stress-induced hyperglycemia. J Clin Invest. 1979;63:114–123. doi: 10.1172/JCI109264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shamoon H., Hendler R., Sherwin R.S. Synergistic interactions among antiinsulin hormones in the pathogenesis of stress hyperglycemia in humans. J Clin Endocrinol Metab. 1981;52:1235–1241. doi: 10.1210/jcem-52-6-1235. [DOI] [PubMed] [Google Scholar]
- 52.Voss T.C., Schiltz R.L., Sung M.H., Yen P.M., Stamatoyannopoulos J.A., Biddie S.C., Johnson T.A., Miranda T.B., John S., Hager G.L. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everett L.J., Le Lay J., Lukovac S., Bernstein D., Steger D.J., Lazar M.A., Kaestner K.H. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC Genomics. 2013;14:337. doi: 10.1186/1471-2164-14-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phuc Le P., Friedman J.R., Schug J., Brestelli J.E., Parker J.B., Bochkis I.M., Kaestner K.H. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genetics. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein I., Paakinaho V., Baek S., Sung M.H., Hager G.L. Synergistic gene expression during the acute phase response is characterized by transcription factor assisted loading. Nature Communications. 2017;8:1849. doi: 10.1038/s41467-017-02055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L., Rubins N.E., Ahima R.S., Greenbaum L.E., Kaestner K.H. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metabolism. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X., Odom D.T., Koo S.H., Conkright M.D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J.R., Emerson B., Hogenesch J.B., Unterman T., Young R.A., Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee D., Le Lay J., Kaestner K.H. The transcription factor CREB has no non-redundant functions in hepatic glucose metabolism in mice. Diabetologia. 2014;57:1242–1248. doi: 10.1007/s00125-014-3203-2. [DOI] [PubMed] [Google Scholar]
- 60.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576:51–60. doi: 10.1038/s41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 61.Hatting M., Tavares C.D.J., Sharabi K., Rines A.K., Puigserver P. Insulin regulation of gluconeogenesis. Ann N Y Acad Sci. 2018;1411:21–35. doi: 10.1111/nyas.13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He L., Sabet A., Djedjos S., Miller R., Sun X., Hussain M.A., Radovick S., Wondisford F.E. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell. 2009;137:635–646. doi: 10.1016/j.cell.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J.M., Han H.S., Jung Y.S., Harris R.A., Koo S.H., Choi H.S. The SMILE transcriptional corepressor inhibits cAMP response element-binding protein (CREB)-mediated transactivation of gluconeogenic genes. J Biol Chem. 2018;293:13125–13133. doi: 10.1074/jbc.RA118.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pierreux C.E., Rousseau G.G., Lemaigre F.P. Insulin inhibition of glucocorticoid-stimulated gene transcription: requirement for an insulin response element? Mol Cell Endocrinol. 1999;147:1–5. doi: 10.1016/s0303-7207(98)00238-x. [DOI] [PubMed] [Google Scholar]
- 65.Kalvisa A., Siersbaek M.S., Praestholm S.M., Christensen L.J.L., Nielsen R., Stohr O., Vettorazzi S., Tuckermann J., White M., Mandrup S., Grontved L. Insulin signaling and reduced glucocorticoid receptor activity attenuate postprandial gene expression in liver. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Potthoff M.J., Boney-Montoya J., Choi M., He T., Sunny N.E., Satapati S., Suino-Powell K., Xu H.E., Gerard R.D., Finck B.N., Burgess S.C., Mangelsdorf D.J., Kliewer S.A. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metabolism. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cahill G.F., Jr. Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 68.Miller R.A., Shi Y., Lu W., Pirman D.A., Jatkar A., Blatnik M., Wu H., Cárdenas C., Wan M., Foskett J.K., Park J.O., Zhang Y., Holland W.L., Rabinowitz J.D., Birnbaum M.J. Targeting hepatic glutaminase activity to ameliorate hyperglycemia. Nat Med. 2018;24:518–524. doi: 10.1038/nm.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noguchi G.M., Huising M.O. Integrating the inputs that shape pancreatic islet hormone release. Nat Metab. 2019;1:1189–1201. doi: 10.1038/s42255-019-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suppli M.P., Bagger J.I., Lund A., Demant M., van Hall G., Strandberg C., Kønig M.J., Rigbolt K., Langhoff J.L., Wewer Albrechtsen N.J., Holst J.J., Vilsbøll T., Knop F.K. Glucagon resistance at the level of amino acid turnover in obese subjects with hepatic steatosis. Diabetes. 2020;69:1090–1099. doi: 10.2337/db19-0715. [DOI] [PubMed] [Google Scholar]
- 71.Wang Y., Kwon H., Su X., Wondisford F.E. Glycerol not lactate is the major net carbon source for gluconeogenesis in mice during both short and prolonged fasting. Mol Metab. 2020;31:36–44. doi: 10.1016/j.molmet.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charni-Natan M., Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR Protoc. 2020;1:100086. doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.