Abstract
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) have been responsible for nosocomial outbreaks worldwide and have become endemic in several countries.
Hypothesis/Gap Statement
To better understand the epidemiological trends and characteristics of CRE in the Henan province.
Aim
We assessed the molecular epidemiological characteristics of 305 CRE strains isolated from patients in 19 secondary or tertiary hospitals in ten areas of the Henan province in China.
Methodology
A total of 305 CRE isolates were subjected to multiple tests, including in vitro antimicrobial susceptibility testing, PCR for carbapenemase genes bla KPC, bla NDM, bla IMP, bla VIM, bla OXA-48-like. Tigecycline-resistant genes ramR, oqxR, acrR, tetA, rpsJ, tetX, tetM, tetL were analysed in five tigecycline non-susceptible carbapenem-resistant Klebsiella pneumoniae isolates (TNSCRKP). Additionally, multilocus sequence typing (MLST) was performed for carbapenem-resistant K. pneumoniae (CRKP).
Results
The most common CRE species were K. pneumoniae (234, 77 %), Escherichia coli (36, 12 %) and Enterobacter cloacae (13, 4 %). All strains exhibited multi-drug resistance. Overall, 97 % (295/305) and 97 % (297/305) of the isolates were susceptible to polymyxin B and tigecycline, respectively. A total of 89 % (271/305) of the CRE isolates were carbapenemase gene-positive, including 70 % bla KPC, 13 % bla NDM, 6 % bla IMP, and 1 % combined bla KPC/bla NDM genes. K. pneumoniae carbapenemase (KPC) was the predominant carbapenemase in K. pneumoniae (87 %), whereas NDM and IMP were frequent in E. coli (53 %) and E. cloacae (69 %), respectively. Mutations in the ramR, tetA, and rpsJ genes were detected in five TNSCRKP. Moreover, 15 unique sequence types were detected, with ST11 (74 %), ST15 (9 %) and ST2237 (5 %) being dominant among K. pneumoniae strains.
Conclusion
A high proportion of CRE strains were carbapenemase-positive, and five carbapenem-resistant K. pneumonia isolates were tigecycline non-susceptible, indicating a need for the ongoing surveillance of CRE and effective measures for the prevention of CRE infections.
Keywords: Carbapenem-resistant Enterobacteriaceae , multi-drug resistance, tigecycline-non-susceptible
Introduction
Carbapenems are often used as last-resort drugs to treat multi-drug-resistant (MDR) bacterial infections. In recent years, as carbapenems have become more frequently utilized, CRE strains have emerged worldwide, the management of which has been complicated by the occurrence of antimicrobial resistance [1, 2].
Particularly, many Enterobacteriaceae have acquired carbapenemases, a group of enzymes capable of hydrolysing carbapenems [3, 4]. Despite the global spread of Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), and OXA-48, the prevalence of carbapenemases varies geographically [5]. For instance, carbapenem resistance in Enterobacteriaceae in China has been found to be primarily associated with KPC and metallo-β-lactamases. Specifically, data from 65 hospitals in 25 provinces across China revealed KPC in 77 % of carbapenem-resistant K. pneumoniae isolates, whereas NDM was detected in 75 % of carbapenem-resistant Escherichia coli and 53 % of carbapenem-resistant Enterobacter cloacae [6] Surveillance of antibiotic resistance by the China Antibiotic Resistance Surveillance System (http://www.carss.cn/Report/Details?aId=770) showed that Henan province had the second highest rate of carbapenem-resistant E. coli and highest rate of K. pneumoniae in 2019 (2 % versus 3 % and 11 % versus 33 % for the national versus provincial rates, respectively). Although limited published data from the Henan province on CRE are available, high incidence and endemic spread of NDM-1-producing carbapenem-resistant E. cloacae isolates (73 %, 8/11) have been reported [7]. Indeed, NDM accounts for the most common carbapenemase carried by carbapenem-resistant E. coli (80 %, 4/5) in Henan [8]. Additionally, we previously reported a significant increase in the prevalence of CRE isolates from 13 % in 2014–19 % in 2015 and 23 % in 2016, in a hospital in Henan [9].
Current treatment options for infections caused by CRE are severely limited. Tigecycline (TGC), a new class of broad-spectrum glycyl-tetracycline antibiotics, represents the last-line antibiotic to target CRE infections [10], as its clinical use has led to the emergence of resistant strains. Resistance in K. pneumoniae is thought to be primarily mediated by overexpression of genes encoding the AcrAB-TolC efflux pump [11, 12]. The AcrAB efflux pump is regulated by the global transcriptional factor ramA, whereas ramR serves as a local negative regulator of ramA [13]. In addition, an inactivating mutation in oqxR, the local repressor of the OqxAB efflux pump, can cause TGC resistance [14]. Mutations in the rpsJ [15] and tetA [16] genes are also associated with reduced TGC susceptibility. Meanwhile, the plasmid-mediated high-level TGC resistance gene tetX has recently emerged in animals and humans [10, 17]. Furthermore, the CusS-CusR two-component system is reportedly associated with CRKP resistance to TGC [18]. TGC resistance mechanisms have also been found to be caused by overexpression of tetL and tetM in Enterococcus faecium [19]. Although four carbapenem-resistant K. pneumoniae strains have been detected and characterized as non-susceptible to TGC in Henan [20], the underlying resistance mechanism has not been elucidated.
To better understand the epidemiological trends and characteristics of CRE in the Henan province, 305 CRE isolates collected from ten different areas in Henan were analysed. This study provides data that may be clinically useful for controlling and reducing CRE infections within this geographical location.
Methods
Clinical strains
A total of 305 CRE clinical isolates were collected in 2018–2019 from 19 secondary or tertiary hospitals in Henan. These CRE isolates only accounted for a portion of the isolates collected from 19 hospitals. Duplicate isolates were excluded. Samples were isolated from different types of clinical specimens, including blood, sputum, urine, cerebrospinal fluid, etc. All isolates were identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Bruker Daltonics GmbH, Billerica, MA, USA).
Antibiotic susceptibility testing
Antimicrobial susceptibility was evaluated by microdilution methods, and the results were interpreted according to Clinical and Laboratory Standards Institute categories and minimum inhibitory concentration breakpoints [21]. Susceptibility tests were performed using E. coli ATCC 25922 as a quality control strain. The isolates were tested for susceptibility to imipenem (IPM), meropenem (MEM), aztreonam (ATM), ampicillin/sulbactam (SAM), cefazolin (KZ), ertapenem (ETP), cefotaxime (CTX), levofloxacin (LEV), gentamicin (GN), amikacin (AK), trimethoprim-sulfamethoxazole (SXT), TGC, and polymyxin B (PB). CRE were defined as carbapenem-non-susceptible (imipenem, meropenem, or ertapenem) organisms of the Enterobacteriaceae family. The interpretive criteria for TGC (≤2 μg ml−1, susceptible; 4 μg ml−1, intermediate; ≥8 μg ml−1, resistant) and polymyxin (≤2 μg ml−1, susceptible; >2 μg ml−1, resistant) were based on the breakpoints established by the Food and Drug Administration [22] and European Committee on Antimicrobial Susceptibility Testing [23], respectively.
Molecular analysis of carbapenemase genes
All isolates were subjected to PCR to screen for the presence of bla KPC[24], bla IMP[25], bla VIM[25], bla NDM[26] and bla OXA-48[25], as described previously. TGC resistance determinants ramR [11], oqxR [11], acrR [11], tetA [11], tetX [11 ], tetL [11], tetM [11], and rpsJ [11] were amplified in the five TNSCRKP with gene-specific primers, sequenced and compared with the wild-type reference strain K. pneumoniae MGH78578 (CP000647) as described previously [11]. To identify carbapenemase genes, nucleotide sequence analysis was performed using the Basic Local Alignment Search Tool from the National Center for Biotechnology Information.
Multilocus sequence typing
Multilocus sequence typing (MLST) analysis of K. pneumoniae isolates was performed using the Institute Pasteur’s MLST scheme. Seven housekeeping genes, i.e. rpoB, gapA, mdh, pgi, phoE, infB and tonB, were used to characterize carbapenem-resistant K. pneumoniae (CRKP) isolates. CRKP is defined as K. pneumoniae that is non-susceptible to any carbapenems (imipenem, meropenem or ertapenem). Primers and amplification conditions were obtained from https://bigsdb.pasteur.fr/klebsiella/primers_used.html. The allele sequences and sequence types were identified using http://bigsdb.web.pasteur.fr/ klebsiella/ klebsiella.html. Data were analysed using Global Optimal eBURST (goeBURST) version 1.2.1.
Results
Strain characteristics
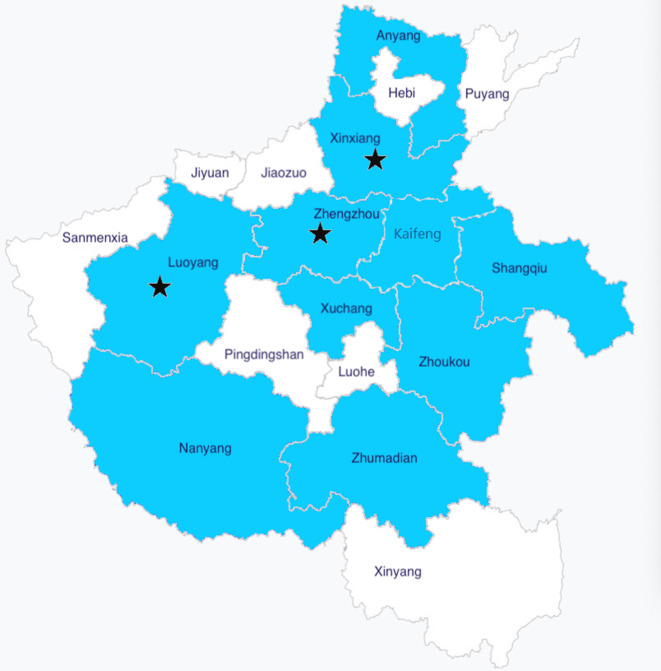
In total, 305 CRE strains from ten different areas in the Henan province were identified in this study (Fig. 1). The most commonly identified CRE species were K. pneumoniae (77 %), followed by E. coli (12 %) and E. cloacae (4 %). Among the CRE, 47 % were isolated from sputum samples, 27 % from blood samples, 7 % from urine samples, and the remaining from other sample types (Table 1).
Fig. 1.
Areas included in the study are shaded. Areas with TNSCRKP isolates are denoted with stars.
Table 1.
Species of 305 carbapenem-resistant Enterobacteriaceae according to the specimen type
|
Species |
Specimen type (n) |
Total |
||||||
|---|---|---|---|---|---|---|---|---|
|
Sputum |
Blood |
Urine |
Ascites |
Wound |
CSF |
Other |
N (%) |
|
|
129 |
58 |
12 |
5 |
5 |
3 |
22 |
234 (77) |
|
|
9 |
12 |
6 |
2 |
2 |
0 |
5 |
36 (12) |
|
|
3 |
6 |
1 |
0 |
2 |
0 |
1 |
13 (4) |
|
|
1 |
2 |
2 |
1 |
0 |
0 |
1 |
7 (2) |
|
|
0 |
3 |
0 |
2 |
0 |
0 |
2 |
7 (2) |
|
|
0 |
0 |
0 |
1 |
0 |
1 |
1 |
3 (1) |
|
|
0 |
1 |
0 |
0 |
0 |
0 |
3 |
4 (1) |
|
|
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 (1) |
|
|
Total |
142 |
82 |
21 |
11 |
9 |
5 |
35 |
305 |
C. freundii, Citrobacter freundii; CSF, cerebrospinal fluid; K. aerogenes, Klebsiella aerogenes; E. cloacae, Enterobacter cloacae; E. coli, Escherichia coli; K. oxytoca, Klebsiella oxytoca; K. pneumoniae, Klebsiella pneumoniae; P. rettgeri, Providencia rettgeri; S. marcescens, Serratia marcescens.
n, number of isolates
Antimicrobial susceptibility of CRE strains
Fig. 2 summarizes the antibiotic susceptibilities of the 305 CRE strains identified in this study. The CRE isolates showed high susceptibility to PB (97 %) and TGC (97 %), however, exhibited lower susceptibility to the other tested antibiotics. The susceptibility rates to SXT, AK, GN and LEV were 34, 29, 11 and 8 %, respectively. The susceptibility rates of IPM, MEM and ATM were all 1 %, while the intermediate rates were 6 % for IPM, 5 % for MEM, and 2 % for ATM, whereas SAM, CTX, KZ and ETP susceptibility rates were zero.
Fig. 2.
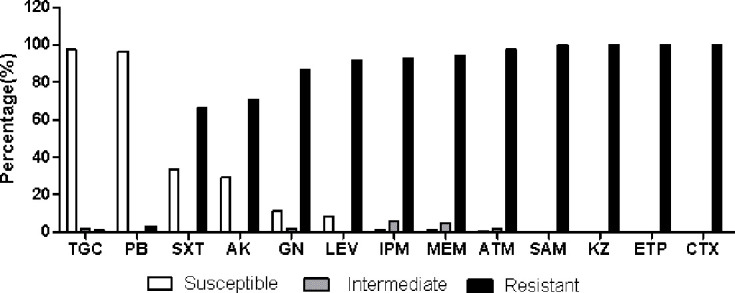
Antimicrobial susceptibility testing results of 305 carbapenem-resistant Enterobacteriaceae isolates.
Molecular analysis of carbapenemase genes
A total of five carbapenemases, namely bla KPC, bla IMP, bla VIM, bla NDM and bla OXA-48, were detected in the isolated strains. At least one carbapenemase-encoding gene was detected in 271 (89 %) of the isolates. In 215 (70 %) isolates, bla KPC was detected with PCR; 13 % were bla NDM-positive and 6 % were bla IMP-positive. Two strains contained both bla NDM and bla KPC and 87 % of the CRKP were KPC producers. Meanwhile, 44 % of E. coli and 39 % E. cloacae were NDM positive (Table 2). bla VIM and bla OXA-48 were not detected in any of the isolates.
Table 2.
Distribution of positive carbapenemase gene loci according to bacterial species
|
Species |
KPC |
IMP |
NDM |
Not detected |
No. of isolates |
|---|---|---|---|---|---|
|
K. pneumoniae *, n (%) |
204 (87) |
6 (3) |
9 (4) |
17 (7) |
234 |
|
E. coli , n (%) |
6 (17) |
3 (8) |
16 (44) |
11 (31) |
36 |
|
E. cloacae , n (%) |
2(15) |
4 (31) |
5 (39) |
2 (15) |
13 |
|
C. freundii , n (%) |
2(29) |
2 (29) |
1 (14) |
2 (29) |
7 |
|
K. oxytoca , n (%) |
0 |
2 (29) |
4 (57) |
1 (14) |
7 |
|
P. rettgeri, n (%) |
0 |
0 |
3 (100) |
0 |
3 |
|
K. aerogenes , n (%) |
1 (25) |
0 |
2 (50) |
1 (25) |
4 |
|
S. marcescens , n (%) |
0 |
1 (100) |
0 |
0 |
1 |
|
Total, n (%) |
215 (70) |
18 (6) |
40 (13) |
34 (11) |
305 |
*Two K. pneumoniae coproduce KPC and NDM.
n,number of isolates
TGC resistance determinants
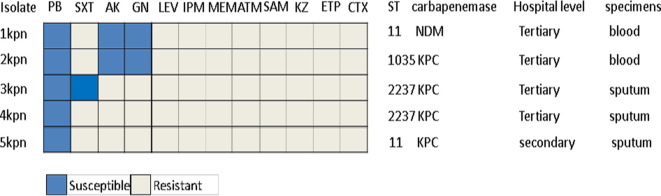
Five TNSCRKP strains with a minimum inhibitory concentration of 4 μg ml−1 were collected from three tertiary hospitals and one secondary hospital (Fig. 1). The five strains were isolated from sputum and blood specimens, of which four were found to carry KPC and one carried NDM. The strains belong to three STs, i.e. ST11, ST2237 and ST1035. Antimicrobial susceptibility testing results showed that two strains were susceptible to amikacin, gentamicin and polymyxin B, one was susceptible to SXT and PB, and two were only susceptible to polymyxin B (Fig. 3).
Fig. 3.
Antimicrobial susceptibility testing results and related characteristics of five TNSCRKP clinical isolates.
Potential TGC resistance determinants ramR, oqxR, tetA, tetX, tetL, tetM, acrR and rpsJ were identified with PCR and sequenced in the five isolates. The genes ramR, tetA, acrR and rpsJ were detected in all isolates; oqxR was found in three (60%) isolates, while tetX, tetL and tetM were not detected in any of the five TNSCRKP isolates. Three isolates showed nucleotide changes or insertion in ramR. Specifically, the A19V and R107C mutations in ramR were identified in two isolates, while one isolate had lysine inserted between 118 glutamate and 119 threonine in ramR. Mutations in acrR, oqxR and rpsJ were also compared to the MGH78578 reference sequence (CP000647). The V57L mutation in rpsJ was detected in three isolates, of which two also combined with the T69S mutation. In addition, all five TNSCRKP isolates had S201A, F202S and V203F mutations in tetA. Meanwhile, mutations in acrR and oqxR were not identified in this study (Table 3).
Table 3.
Related resistance determinants of tigecycline non-susceptible clinical isolates examined in the present study
|
Isolate |
TGC* MIC (μg ml−1) |
Presence of TGC resistance determinants (mutations occurring in the protein sequence) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
ramR |
acrR |
oqxR |
rpsJ |
tetA |
tetX |
tetL |
tetM |
||
|
1kpn |
4 |
+ |
+ |
− |
+(V57L) |
+(S201A, F202S, V203F) |
− |
− |
− |
|
2kpn |
4 |
+(118_119ins†) |
+ |
+ |
+ |
+(S201A, F202S, V203F) |
− |
− |
− |
|
3kpn |
4 |
+(A19V, R107C) |
+ |
+ |
+(V57L, T69S) |
+(S201A, F202S, V203F) |
− |
− |
− |
|
4kpn |
4 |
+(A19V, R107C) |
+ |
+ |
+(V57L, T69S) |
+(S201A, F202S, V203F) |
− |
− |
− |
|
5kpn |
4 |
+ |
+ |
− |
+ |
+(S201A, F202S, V203F) |
− |
− |
− |
*TGC, tigecycline; †ins, insertion; +, presence of PCR product and no change in the nucleotide/amino acid sequences; −, absence of PCR product;
Multilocus sequence typing
A total of 234 CRKP were analysed by MLST, and 15 STs were detected. ST11 (74 %) was the most frequent type, followed by ST15 (9 %). ST2237 (5 %), ST48 (3 %), ST438 (2 %), ST147 (1 %), ST530 (1 %) and ST36 (1 %). ST23, ST37, ST17, ST290, ST1035, ST1 and ST18 were detected in only one strain each. Moreover, 88 % (153/174) of ST11 isolates, 86 % (18/21) of ST15 isolates, and 67 % (8/12) of ST2237 isolates produced KPC carbapenemase; whereas 4 % (7/174) of ST11 isolates and 17 % (2/12) of ST2237 isolates produced NDM carbapenemase. Additionally, 2 % (4/174) of ST11 and 67 % (2/3) of ST147 isolates were IMP producers (Table 4). The goeBURST analysis further demonstrated that ST290, ST15, ST11, ST36 and ST1035 are triple-locus variants (TLVs) of ST18, while ST17 is a double-locus variant (DLV) of ST18 and ST530 is a singleton (Fig. 4).
Table 4.
Different MLSTs with carbapenemase gene distribution of CRKP isolates
|
ST |
Total |
KPC |
NDM |
IMP |
KPC+NDM |
Negative |
|---|---|---|---|---|---|---|
|
ST11, no.(%) |
174 |
151 (87) |
5 (3) |
4 (2) |
2 (1) |
12 (7) |
|
ST15, no.(%) |
21 |
18 (86) |
0 |
0 |
3 (14) |
|
|
ST2237, no.(%) |
12 |
8 (67) |
2 (17) |
0 |
2 (17) |
|
|
ST48, no.(%) |
6 |
6 (100) |
||||
|
ST438, no.(%) |
5 |
5 (100) |
||||
|
ST147, no.(%) |
3 |
1 (33) |
2 (67) |
|||
|
ST530, no.(%) |
3 |
3 (100) |
||||
|
ST36, no.(%) |
3 |
3 (100) |
||||
|
ST23, no.(%) |
1 |
1 (100) |
||||
|
ST37, no.(%) |
1 |
1 (100) |
||||
|
ST17, no.(%) |
1 |
1 (100) |
||||
|
ST290, no.(%) |
1 |
1 (100) |
||||
|
ST1035, no.(%) |
1 |
1 (100) |
||||
|
ST18, no.(%) |
1 |
1 (100) |
||||
|
ST1, no.(%) |
1 |
1 (100) |
||||
|
Total |
234 |
202 |
7 |
6 |
2 |
17 |
Fig. 4.
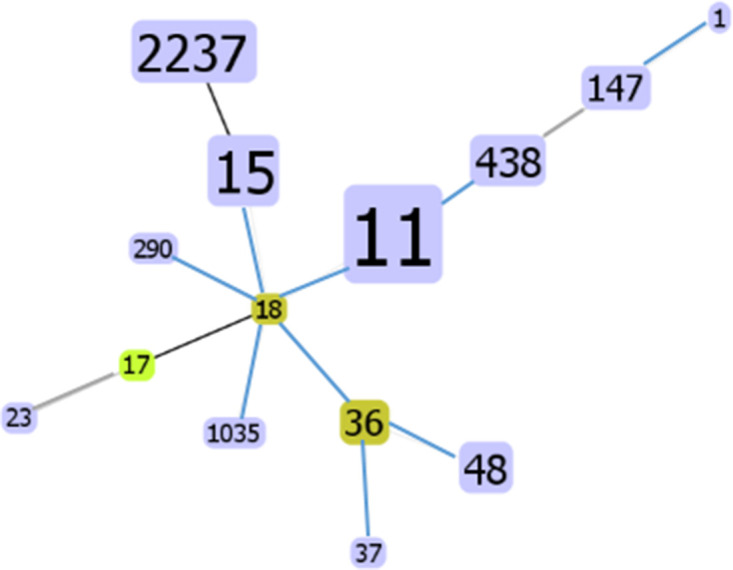
MLSTs of CRKP isolates at the TLV (triple-locus variants, TLVs) level using goeBURST. The yellow circle denotes the subgroup founder. Number size is proportional to the ST abundance.
Discussion
Carbapenems are antibiotics used to treat severe infections caused by MDR Enterobacteriaceae . However, CRE has been increasingly reported worldwide over the past ten years, posing a serious threat to public health. Particularly, the increasing prevalence of carbapenem-resistant K. pneumoniae is a major source of concern [27]. Globally, the most common carbapenemases in CRE are KPC, NDM and OXA-48; however, only scattered reports of epidemiological data on CRE in the Henan province in China is available. For instance, five carbapenemase-producing E. coli strains were screened from 1014 isolates, and the positive rate of bla NDM-1 was 80 % in Xinxiang, Henan [8]. Moreover, the prevalence of CRE in a hospital in Henan rapidly increased from 2014 to 2016 according to our previous study [9]. Meanwhile, the results of the current study, which represents the first multicentre study in the Henan province, agreed with other reports in China [6, 28, 29], showing that 89 % of CRE were carbapenemase-producers, of which 70, 13 and 6 % were KPC-, NDM- and IMP-producers, respectively. Moreover, 11 % common carbapenemase-negative strains were identified, which may carry unknown types of metallo-lactamase or overexpress extended-spectrum β-lactamases and/or AmpC enzymes combined with outer-membrane porin loss [30–32].
CRE strains observed in this study were highly resistant to the most common antimicrobial agents, except TGC and PB, which are considered as last-line treatments for CRE infections [33]. Two TNSCRKP strains harboured the A19V mutation combined with R107C in ramR, and one strain had 118_119ins in ramR. In fact, the A19V mutation is reportedly the most common ramR mutation in TGC-resistant K. pneumoniae [11, 34]. Additionally, the R107C mutation and 118_119ins were detected in ramR in the current study; however, the R107H mutation was previously reported in one laboratory-evolved TGC-resistant K. pneumoniae strain [35]. In addition, a V57L mutation in rpsJ was detected in three TNSCRKP, of which two were combined with T69S mutation; meanwhile, the V57L mutation in rpsJ, which is related to TGC resistant, was also reported in a laboratory-evolved TGC-resistant K. pneumoniae strain in China [35] and Korea [36], as well as in a 59-year-old male patient infected with KPC-producing K. pneumonia following TGC therapy [15]. However, the T69S mutation was newly identified in the current study. All five TNSCRKP strains harboured S201A, F202S and V203F mutations in the tetA, which was reported previously, and designated as a novel Type 2 TetA variant, that commonly combined with other mutations, including I5R, A93V, G151S and G268A [11]. However, it was reported that the 201nd to the 203rd amino acids in the sequence were substituted from serine, phenylalanine and valine (SFV) to alanine, serine and proline (ASP) in tigecycline-susceptible CRKP isolates [37]. Considering that no mutations were detected in oqxR or acrR, in this study, the ramR and rpsJ mutations may represent the primary contributor to the TGC non-susceptible phenotype observed in this study. However, one TNSCRKP strain did not harbour ramR or rpsJ mutations, indicating that other regulatory mechanisms may also exist in this strain. Moreover, only five TNSCRKP were detected in this study; hence, further research is needed to determine if the new mutations identified in this study are frequent and contribute to TGC non-susceptibility among TNSCRKP strains in Henan.
The prevalent CP-Kps STs also vary geographically. In Asia, particularly in China and Taiwan, the spread of KPC-producing K. pneumoniae is associated with ST11 [29, 38]. With the exception of the most common ST11, 14 different STs were detected in CRKP. ST15 and ST2237 were the second and third most common sequence types in CRKP in Henan, detected in this study. Other STs, such as ST23, ST490, ST15, ST1, ST37, ST36 and ST147, were also detected in 70 K . pneumoniae strains and associated with ventilator-associated pneumonia in Henan [39]. Meanwhile, 88 % of ST11 strains were KPC producers, whereas 4 % of ST11 and 17 % of ST2237 were NDM-positive in this study; however, ST17 isolates were more likely to produce NDM carbapenemase (91 %) in CRKP strains in a national CRE Study in China[6]. IMP was also detected in six CRKP strains in this study, indicating its low prevalence in CRKP in Henan.
In conclusion, K. pneumoniae and E. coli were the most common species of CRE in the Henan province of China. KPC was primarily detected in K. pneumoniae , whereas NDM and IMP were the most common carbapenemases among other CRE species. In addition, ST11, ST15 and ST2237 were the first, second and third most common STs in CRKP isolates, and 88 % of ST11 strains were KPC producers in Henan. The ramR and rpsJ mutations may contribute to the TGC-non-susceptible phenotype in K. pneumoniae strains, which should be considered further in a surveillance study. Taken together, the results of this study indicate that surveillance of CRE and carbapenemases should be improved to monitor CRE on an ongoing basis, and prevention and control measures must be implemented to reduce the spread of CRE strains.
Funding information
This work was supported by the Henan Provincial Key Programmes in Science and Technology [grant number 182102311241]; and Science and Technology Research Project of Henan Province [grant number SBGJ2018084].
Acknowledgements
We would like to thank the following hospitals for their participation in CRE strains collection: Xuchang people’s Hospital, Xiangcheng county people’s Hospital, Anyang sixth people’s Hospital, Kaifeng central Hospital, Kaifeng people’s Hospital, Luoyang central Hospital, Ruyang county people’s Hospital, Nanyang nanshi Hospital, Nanyang central Hospital, the 83 Army Hospitals of PLA, Xinxiang central Hospital, Nanyang chest Hospital, Henan hongli Hospital, Yongcheng people’s Hospital, Zhengzhou central Hospital, Gongyi people’s Hospital, Zhoukou central Hospital, Zhumadian first people’s Hospital. We would like to thank Editage (www.editage.cn) for English language editing.
Author contributions
Conceptualization: Y.L.,W.J.Y., S.M.W. Data curation: W.J.Y., N.J., Q.M. Formal analysis: W.J.Y., N.J., Y.H.Y. Funding acquisition: Y.L., W.J.Y. Investigation: A.L.L., L.H.C, B.M. Writing - original draft: W.J.Y., J.H.X. Writing - review and editing: Q.Z., J.F.Z., Y.L.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This work has been approved by the Ethics Committee of Henan Provincial People’s Hospital, Henan, China (20190050); No samples were collected specifically for this research. As only anonymized clinical isolates collected during routine hospital procedures prior to this research were used for this study.
Footnotes
Abbreviations: IMP, Imipenemase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA48-like, oxacillinases 48-like; VIM, verona integron-encoded metallo-β-lactamase.
References
- 1.Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae . Drug Resist Updat. 2016;29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivalingam P, Poté J, Prabakar K. Environmental prevalence of carbapenem resistance Enterobacteriaceae (CRE) in a tropical ecosystem in India: human health perspectives and future directives. Pathogens. 2019;8:174. doi: 10.3390/pathogens8040174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durante-Mangoni E, Andini R, Zampino R. Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect. 2019;25:943–950. doi: 10.1016/j.cmi.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: detection and antimicrobial therapy. Front Microbiol. 1823;2019:10. doi: 10.3389/fmicb.2019.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase‐producing Enterobacteriaceae : a review. Ann N Y Acad Sci. 2019;1457:61–91. doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Wang X, Wang J, Ouyang P, Jin C, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016) Clin Infect Dis. 2018;67:S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, Qin S, Xu H, Xu L, Zhao D, et al. New Delhi Metallo-β-Lactamase 1(NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China. PLoS One. 2015;10:e0135044. doi: 10.1371/journal.pone.0135044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang W-juan, Liu H-ying, Duan G-C, Zhao Y-xin, Chen S-yin, et al. Emergence and mechanism of carbapenem-resistant Escherichia coli in Henan, China, 2014. J Infect Public Health. 2018;11:347–351. doi: 10.1016/j.jiph.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Sun Q-ling, Shen Y, Zhang Y, Yang J-wen, et al. Rapid increase in prevalence of carbapenem-resistant Enterobacteriaceae (CRE) and emergence of colistin resistance Gene mcr-1 in CRE in a hospital in Henan, China. J Clin Microbiol. 2018;56:e01932–17. doi: 10.1128/JCM.01932-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He T, Wang R, Liu D, Walsh TR, Zhang R, et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol. 2019;4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 11.Chiu S-K, Huang L-Y, Chen H, Tsai Y-K, Liou C-H, et al. Roles of ramR and tet(A) mutations in conferring Tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2017;61:e00391–17. doi: 10.1128/AAC.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park Y, Choi Q, Kwon GC, Koo SH. Molecular epidemiology and mechanisms of tigecycline resistance in carbapenem‐resistant Klebsiella pneumoniae isolates. J Clin Lab Anal. 2020;34:e23506. doi: 10.1002/jcla.23506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elgendy SG, Abdel Hameed MR, El-Mokhtar MA. Tigecycline resistance among Klebsiella pneumoniae isolated from febrile neutropenic patients. J Med Microbiol. 2018;67:972–975. doi: 10.1099/jmm.0.000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. Characterization of RaRA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012;56:4450–4458. doi: 10.1128/AAC.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He F, Shi Q, Fu Y, Xu J, Yu Y, et al. Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect Genet Evol. 2018;66:188–191. doi: 10.1016/j.meegid.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Du X, He F, Shi Q, Zhao F, Xu J, et al. The rapid emergence of tigecycline resistance in blaKPC–2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front Microbiol. 2018;9:648. doi: 10.3389/fmicb.2018.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Chen C, Cui C-Y, Zhang Y, Liu X, et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol. 2019;4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Zhao Y, Qiu Y, Xiao L, He H, et al. CusS-CusR two-component system mediates tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae. Front Microbiol. 2019;10:3159. doi: 10.3389/fmicb.2019.03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, et al. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet (L) and tet (M) J Antimicrob Chemother. 2016;71:871–881. doi: 10.1093/jac/dkv420. [DOI] [PubMed] [Google Scholar]
- 20.Qin S, Fu Y, Zhang Q, Qi H, Wen JG, et al. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother. 2014;58:4275–4282. doi: 10.1128/AAC.02813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute M100‐S30. Performance Standards for Antimicrobial Susceptibility Testing: Thirty Edition [S] Wayne, PA: CLSI; 2020. [Google Scholar]
- 22.U.S. Food and Drug Administration Antibacterial susceptibility test interpretive criteria. 2020-10-13.
- 23.European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0.2020-01‐01. n.d. [Google Scholar]
- 24.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae . J Antimicrob Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 25.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman M, Shukla SK, Prasad KN, Ovejero CM, Pati BK, et al. Prevalence and molecular characterisation of New Delhi metallo-β-lactamases NDM-1, NDM-5, NDM-6 and NDM-7 in multidrug-resistant Enterobacteriaceae from India. Int J Antimicrob Agents. 2014;44:30–37. doi: 10.1016/j.ijantimicag.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang L, Lu X, Xu H, Ma X, Chen Y, et al. Epidemiology and risk factors for carbapenem-resistant Enterobacteriaceae colonisation and infections: case-controlled study from an academic medical center in a southern area of China. Pathog Dis. 2019;77:FTZ034. doi: 10.1093/femspd/ftz034. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R, Liu L, Zhou H, Chan EW, Li J, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36:182–186. doi: 10.1053/j.semdp.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Nordmann P, Poirel L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin Infect Dis. 2019;69:S521–S528. doi: 10.1093/cid/ciz824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3:15–21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betts JW, Phee LM, Hornsey M, Woodford N, Wareham DW. In vitro and in vivo activities of tigecycline-colistin combination therapies against carbapenem-resistant Enterobacteriaceae . Antimicrob Agents Chemother. 2014;58:3541–3546. doi: 10.1128/AAC.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Z-K, Hu F, Wang W, Guo Q, Chen Z, et al. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2014;58:6982–6985. doi: 10.1128/AAC.03808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang L, Chen Q, Shi K, Li X, Shi Q, et al. Step-Wise increase in tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR, Lon and rpsJ. PLoS One. 2016;11:e0165019. doi: 10.1371/journal.pone.0165019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, Lee H, Shin D, Ko KS. Change of Hypermucoviscosity in the development of tigecycline resistance in hypervirulent Klebsiella pneumoniae sequence type 23 strains. Microorganisms. 2020;8:1562. doi: 10.3390/microorganisms8101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y, Choi Q, Kwon GC, Koo SH. Molecular epidemiology and mechanisms of tigecycline resistance in carbapenem‐resistant Klebsiella pneumoniae isolates. J Clin Lab Anal. 2020;34:e23506. doi: 10.1002/jcla.23506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y-H, Chou S-H, Liang S-W, Ni C-E, Lin Y-T, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. Antimicrob Chemother. 2018;73:2039–2046. doi: 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 39.Guo S, Xu J, Wei Y, Xu J, Li Y, et al. Clinical and molecular characteristics of Klebsiella pneumoniae ventilator-associated pneumonia in mainland China. BMC Infect Dis. 2016;16:608. doi: 10.1186/s12879-016-1942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]