Abstract
Klebsiella pneumoniae strains carrying OXA-48-like carbapenemases are increasingly prevalent across the globe. There is thus an urgent need to better understand the mechanisms that underpin the dissemination of bla OXA-48-like carbapenemases. To this end, four ertapenem-resistant K. pneumoniae isolates producing OXA-48-like carbapenemases were isolated from two patients. Genome sequencing revealed that one sequence type (ST) 17 isolate carried bla OXA-181, whilst three isolates from a single patient, two ST76 and one ST15, carried bla OXA-232. The 50514 bp bla OXA-181-harbouring plasmid, pOXA-181_YML0508, was X3-type with a conjugation frequency to Escherichia coli of 1.94×10−4 transconjugants per donor. The bla OXA-232 gene was located on a 6141 bp ColKP3-type plasmid, pOXA-232_WSD, that was identical in the ST76 and ST15 K. pneumoniae isolates. This plasmid could be transferred from K. pneumoniae to E. coli at low frequency, 8.13×10−6 transconjugants per donor. Comparative analysis revealed that the X3 plasmid acquired the bla OXA-48-like gene via IS3000-mediated co-integration of the ColKP3-type plasmid. Our study highlights how plasmid integration and rearrangements can contribute to the spread of bla OXA-48-like genes, which provides important clues for clinical prevention of the dissemination of K. pneumoniae strains carrying bla OXA-48-like carbapenemases.
Keywords: OXA-181, OXA-232, Klebsiella pneumoniae, carbapenem-resistant Klebsiella pneumoniae , China
The emergence and prevalence of carbapenem-resistant Klebsiella pneumoniae is an increasingly significant threat to public health worldwide and the carbapenemases OXA-48 and its derivatives contribute to the spread of carbapenem resistance in this important opportunistic pathogen [1]. OXA-48 was first found in K. pneumoniae in Turkey [2], and its derivatives such as OXA-181 [3], OXA-232 [4] and OXA-163 [5] were subsequently found in European hospital outbreaks [6]. OXA-48-like carbapenemases appear to have disseminated rapidly via the actions of mobile genetic elements [7]. Broad host range L/M-type and ColE1-like plasmids commonly bear bla OXA-48-like genes in Enterobacteriaceae [8], and the bla OXA-48-like genes are associated with many types of insertion sequences and transposons. For example, the bla OXA-181 gene has been mobilized by ISEcp1 and the resulting structure has been called Tn2013 [9]. However, the mechanisms of bla OXA-48-like gene dissemination and how these important enzymes are spreading remains unclear.
To explore these unclear mechanisms, four K. pneumoniae isolates were collected from rectal swabs or faecal samples taken for surveillance from two patients in Sir Run Run Shaw Hospital (Zhejiang, China) between May and December 2018. YML0508 was isolated from a 60-year-old female while WSD411, WSD2016 and WSD2080 were isolated from an 84-year-old male (Table 1). Antimicrobial susceptibility testing showed that all four isolates were resistant to ertapenem and WSD411 was extensively-drug resistant, only being sensitive to colistin from the panel of antibiotics used in susceptibility testing (Table 2) [10]. Sanger sequencing of PCR products amplified using published primers that detect bla OXA-48-like genes [11] revealed that YML0508 contained bla OXA-181, while the remaining isolates contained bla OXA-232. Strains YML0508, WSD2016 and WSD2080 showed a similar carbapenem susceptibility spectrum, only resistant to ertapenem but sensitive to imipenem and meropenem. As many OXA-48-like producers exhibited only decreased susceptibility to one of the carbapenems, they were called ‘the phantom menace’ in the literature [7]. We emphasize the importance of routinely testing for ertapenem sensitivity as well as imipenem or meropenem in order to avoid missing detection of OXA-48-like carbapenemases in clinical settings.
Table 1.
The clinical characteristics of the clinical strains
|
Patient |
Age (years), sex |
Diagnosis |
Ward |
Antibiotics |
Strain |
Isolation date |
Specimen |
ST |
OXA |
Other resistance genes |
|---|---|---|---|---|---|---|---|---|---|---|
|
Patient 1 |
60, F |
MDS, AML-M2 |
Haematology |
IMP, LZD, VRC, VCV, MXF |
YML0508 |
21 May 2018 |
Rectal swab |
17 |
181 |
bla SHV-94, qnrS1. oqxB. oqxA aac(6')-Ib-cr, fosA |
|
Patient 2 |
84, M |
Fracture, PE, HTN, Af, CRF, ARF |
ICU |
TZP |
WSD411 |
26 August 2018 |
Rectal swab |
15 |
232 |
bla TEM-1B, bla SHV-28, bla CTX-M-15, aph(3')-Ia aac(6')-Ib3 aph(6)-Id rmtF, aph(3'')-Ib rmtB, aadA16 oqxB, oqxA, aac(6')-Ib-cr, qnrB1, fosA, floR, ARR-2, sul2 |
|
WSD2016 |
12 September 2018 |
Faeces |
76 |
232 |
bla SHV-59, bla TEM-1B, oqxB, oqxA, qnrS1, fosA, fosA3, floR, catA2, ARR-3, sul2, tet(A), dfrA27, aph(3'')-Ib, aph(6)-Id, rmtB |
|||||
|
WSD2080 |
22 October 2018 |
Faeces |
76 |
232 |
bla SHV-59, bla TEM-1B, oqxB, oqxA, qnrS1, fosA, fosA3, floR, catA2, ARR-3, sul2, tet(A), dfrA27, aph(3'')-Ib, aph(6)-Id, rmtB |
MDS, myelodysplastic syndrome; AML-M2, acute myelogenous leukaemia; PE, pulmonary embolism; HTN, hypertension; Af, atrial fibrillation; CRF, chronic renal failure; ARF, acute respiratory failure. IMP, imipenem; LZD, linezolid; VRC, voriconazole; VCV, valacyclovir; MXF, moxifloxacin; TZP, piperacillin-tazobactam. All the isolates were identified by the VITEK2 system and matrix assisted laser desorption ionization-time of flight (MALDI-TOF) MS.
Table 2.
The antimicrobial susceptibility results of the clinical strains and transconjugants (mg l−1)
|
Strain |
MIC (mg l−1) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
IMP |
MEM |
ETP |
CTX |
FEP |
LEV |
CIP |
AMK |
GEN |
ATM |
TGC |
CST |
|
|
YML0508 |
0.5 |
0.25 |
4 |
0.25 |
0.25 |
1 |
1 |
1 |
1 |
0.125 |
0.5 |
0.125 |
|
YML0508-C |
0.5 |
0.25 |
1.5 |
0.38 |
0.25 |
4 |
1 |
1 |
0.5 |
0.125 |
0.25 |
0.06 |
|
WSD411 |
4 |
4 |
>64 |
≥32 |
≥256 |
≥32 |
≥512 |
≥256 |
≥512 |
≥256 |
2 |
0.06 |
|
WSD411-C |
0.5 |
0.125 |
1 |
0.125 |
0.25 |
0.19 |
0.25 |
1 |
0.25 |
0.125 |
0.5 |
0.06 |
|
WSD2016 |
0.5 |
0.25 |
8 |
0.75 |
1 |
0.5 |
0.5 |
1 |
1 |
0.125 |
0.25 |
0.125 |
|
WSD2016-C |
0.5 |
0.125 |
0.5 |
0.19 |
0.25 |
0.25 |
0.25 |
1 |
0.5 |
0.25 |
0.125 |
0.125 |
|
WSD2080 |
0.5 |
0.5 |
4 |
0.25 |
0.5 |
0.5 |
0.5 |
1 |
1 |
0.125 |
0.25 |
0.125 |
|
WSD2080-C |
0.5 |
0.125 |
0.38 |
0.064 |
0.25 |
2 |
1 |
1 |
0.5 |
0.125 |
0.25 |
0.06 |
|
EC600 |
0.125 |
0.06 |
0.006 |
0.094 |
0.125 |
0.25 |
0.25 |
1 |
0.5 |
0.125 |
0.125 |
0.06 |
YML0508/WSD411/WSD2016/WSD2080-C: strain EC600 acquired plasmid from strain YML0508/WSD411/WSD2016/WSD2080 by conjugation. IMP: imipenem; MEM: meropenem; ETP: ertapenem; CTX: cefotaxime; FEP: cefepime; CPS: cefperazone/sulbactam; LEV: levofloxacin; CIP: ciprofloxacin; ATM: aztreonam; TGC: tigecycline; CST: colistin. The shaded area represents the antimicrobial susceptibility results of clinical strains. The MICs for IMP, MEM, ETP, TGC and CST were determined using the broth microdilution method following the guidelines from the Clinical and Laboratory Standard Institute (CLSI). MICs were interpreted according to CLSI breakpoints for K. pneumoniae. Susceptibility to other antimicrobial agents was tested via etest (AB bioMérieux) or broth microdilution.
Genomic DNA from each of the K. pneumoniae isolates was sequenced on the Illumina HiSeq2000 platform. The mean coverage and N50 of contigs of assembly of whole-genome sequencing data was >90× and >48 kb, respectively (Table S1, available in the online version of the paper). According to multi-locus sequence typing (MLST) using the PubMLST database, YML0508 was sequence type (ST)17, WSD411 was ST15, and WSD2016 and WSD2080 were ST76. WSD2016 and WSD2080 have identical core genome (cg)MLST profiles while the other isolate from patient 2, WSD411, differs by 1990 alleles (Fig. 1). Draft genomes were screened for antibiotic resistance genes using ResFinder on the CGE server. This confirmed the presence of bla OXA-181 in YML0508, and bla OXA-232 in WSD411, WSD2016 and WSD2080. Additional antibiotic resistance genes consistent with phenotypic data were detected in each of the draft genomes (Table 1).
Fig. 1.
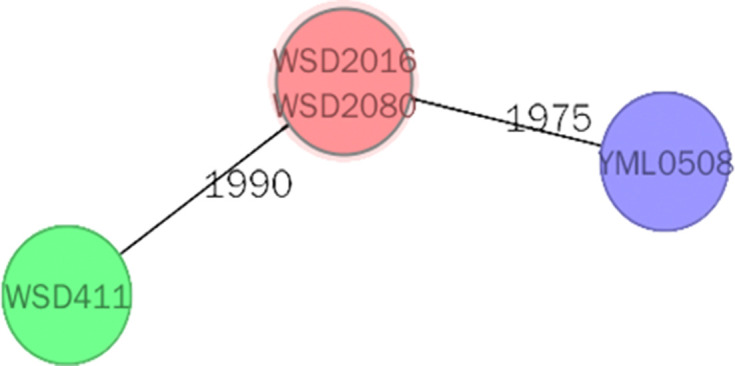
Minimum spanning tree based on cgMLST allelic profiles of four K. pneumoniae isolates. Each circle represents an allelic profile, and the numbers on the connecting lines illustrate the numbers of target genes with different alleles. The cgMLST scheme was executed as described in a previous study [17].
In the draft genomes of WSD411, WSD2016 and WSD2080, the bla OXA-232 gene was found in identical contigs with overlapping sequences at their ends. As such configurations represent circular plasmid sequences, one copy of the overlapping sequence was removed in each case. This revealed that the same 6141 bp plasmid, which was named pOXA-232_WSD, was present in all three isolates. According to PlasmidFinder, the replicon type of pOXA-232_WSD is ColKP3. pOXA-232_WSD does not contain any antibiotic resistance genes apart from bla OXA-232. The presence of an identical plasmid in two, unrelated K. pneumoniae STs within a single patient is clearly indicative of horizontal transfer. However, the transmission pathway could not be precisely determined with the data generated here.
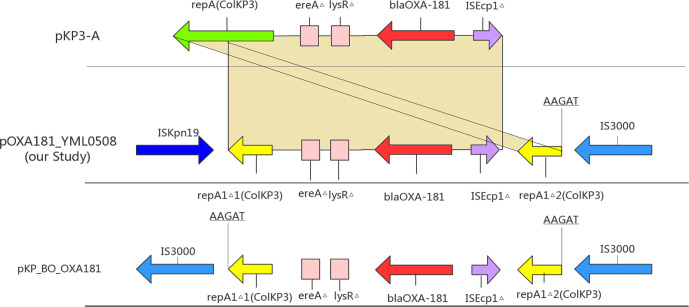
The bla OXA-181 gene in the YML0508 draft genome was found in a short contig that did not represent a complete plasmid or chromosomal sequence. To determine the location of this gene, genomic DNA from YML0508 was sequenced using Oxford Nanopore MinION long-read sequencing technology. In the hybrid-assembled genome of YML0508, the bla OXA-181 gene was found in a 50 514 bp X3-type plasmid, designated pOXA-181_YML0508. pOXA-181_YML0508 also contained a partial ColKP3-type replicon, and the qnrS1 gene associated with low-level resistance to fluoroquinolones. The sequence of pOXA-181_YML0508 was identical to many plasmids in the GenBank non-redundant nucleotide database, such as pOXA181_EC14828 (accession number KP400525), EC-p266917_2_04 (CP026727) and pOXA-181_29144 (KX523903). In pOXA-181_YML0508, the bla OXA-181 gene is part of a 3173 bp segment flanked on the left by a copy of ISKpn19 and on the right by a 2553 bp fragment of IS3000 that has been truncated by a copy of IS26 (Fig. 2). At either end of the segment that includes bla OXA-181 are fragments of the ColKP3 replication initiation gene, repA1Δ1(ColKP3) and repA1Δ2(ColKP3). The 3173 bp segment also includes a 264 bp fragment of ISEcp1, and the entire region is identical to a region in plasmid pKP3-A (Fig. 2). Part of pOXA-232_WSD is also identical to this segment, apart from the one nucleotide substitution that distinguishes bla OXA-181 and bla OXA-232.
Fig. 2.
Scheme of the genetic environment of bla OXA-181 gene. △: incomplete. The yellow shadow represents the homology sequence between pKP3-A and pOXA-181.
This configuration surrounding bla OXA-181 in X3 plasmids has been described before [12]. In that study the role of IS3000 in the co-integration of a ColKP3 plasmid into the X3 plasmid backbone was suspected, but could not be confirmed with the sequence data available at the time. However, using the sequence of this region from pOXA-181_YML0508 to query the GenBank nucleotide database revealed that a likely ancestral structure is found in E. coli plasmid pKP_BO_OXA181 (GenBank accession MG228426). In pKP_BO_OXA181, the ColKP3-derived sequence is flanked by two copies of IS3000 (Fig. 2). In pKP_BO_OXA181, a 5 bp duplication (AAGAT) in the ColKP3-derived sequence shows that IS3000 transposed into the ColKP3 backbone, indicating that IS3000 is probably responsible for the integration of the ColKP3 plasmid into the X3 plasmid. The insertion sequence ISKpn19 in pOXA-181_YML0508 and other plasmids has probably inserted after the integration event, and subsequently deleted one copy of IS3000.
To evaluate the transferability and dissemination risk of the bla OXA-181- and bla OXA-232-containing plasmids from the clinical K. pneumoniae described here, conjugation experiments were performed using a filter mating method with rifampin-resistant E. coli EC600 as the recipient strain. Transconjugants were selected on Mueller-Hinton (MH) agar containing ampicillin (100 mg l−1) and rifampicin (1024 mg l−1). In all cases, the carbapenemase genes transferred from clinical K. pneumoniae to EC600. The mean conjugation frequencies of each plasmid, calculated from five independent experiments, were 8.13×10−6 transconjugants per donor for pOXA-232_WSD and 1.94×10−4 transconjugants per donor for pOXA-181_YML0508. The ertapenem MICs of transconjugants increased at least 60-fold relative to those of EC600, whereas the increase of the MICs of imipenem and meropenem were less pronounced, only about 2- to 8-fold (Table 2).
In this study, we screened four K. pneumoniae isolates harbouring bla OXA-48-like genes. One isolate, YML0508, contained the bla OXA-181 gene. The bla OXA-181 gene of YML0508 is in an X3 plasmid, and previous studies have described isolates of K. pneumoniae , E. coli and K. variicola [13, 14] that carry bla OXA-181-containing plasmids of the same type. We have provided strong evidence that the bla OXA-181 gene was acquired by an ancestral X3 plasmid along with a small ColKP3-type in an event facilitated by IS3000, and that ISKpn19 inserted later, before causing a deletion that complicated previous analyses of this region [12]. We also found an identical 6 141 bp ColKP3-type plasmid harbouring bla OXA-232 in ST15 and ST76 K. pneumoniae isolates from the same patient, suggesting horizontal transfer of this small plasmid. Consistent with this plasmid being transferrable, we showed that pOXA-232_WSD transferred from K. pneumoniae to E. coli in the laboratory at low frequency. As pOXA-232_WSD does not contain genes sufficient for self-transfer, it must utilize mobilization [15] for horizontal dissemination, and it will be interesting to determine which plasmid (or other element) might be mobilizing it. This is an important observation, as mobilization of relatively small plasmids is an underappreciated mechanism for carbapenemase gene dissemination. Furthermore, the presence of bla OXA-232, which produces a single amino acid variant of OXA-181, in a small plasmid might suggest that this mutation occurred in this context, as it is believed that the high copy number of small plasmids facilitates the emergence of antibiotic resistance gene variants [16]. The integration and transmission of plasmids with bla OXA-48-like genes contribute to the increasing prevalence of K. pneumoniae strains carrying bla OXA-48-like carbapenemases in China.
Supplementary Data
Funding information
This work was supported by grants from the National Natural Science Foundation of China (grant numbers: 81830069 and 81802043), the Key Research and Development Programme of Zhejiang (grant number: 2015C03046), the National Science and Technology Major Project (grant number: 2018ZX10714002) and the Medical Research Council (grant number: MR/S013660/1).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by the Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (20180802-5). Consent to publish has been obtained.
Footnotes
Abbreviations: Af, atrial fibrillation; AML-M2, acute myelogenous leukemia; ARF, acute respiratory failure; ATM, aztreonam; CFR, chronic renal failure; cgMLST, core genome multi-locus sequence typing; CIP, ciprofloxacin; CLSI, Clinical and Laboratory Standard Institute; CPS, cefperazone/sulbactam; CST, colistin; CTX, cefotaxime; ETP, ertapenem; FEP, cefepime; HTN, hypertension; IMP, imipenem; LEV, levofloxacin; LZD, linezolid; MDS, myelodysplastic syndrome; MEM, meropenem; MXF, moxifloxacin; PE, pulmonary embolism; ST, sequence type; TGC, tigecycline; TZP, piperacillin-tazobactam; VCV, valacyclovir; VRC, voriconazole.
One supplementary table is available with the online version of this article.
References
- 1.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18:413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae . Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, et al. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY antimicrobial surveillance program, 2006-2007. Antimicrob Agents Chemother. 2011;55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae . Int J Antimicrob Agents. 2013;41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Castanheira M, Carrër A, Rodriguez CP, Jones RN, et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55:2546–2551. doi: 10.1128/AAC.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo L, de Been M, Rogers MRC, Schürch AC, Scharringa J, et al. Sequence-based epidemiology of an OXA-48 plasmid during a hospital outbreak. Antimicrob Agents Chemother. 2019;63:e01204–01219. doi: 10.1128/AAC.01204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, et al. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae . Antimicrob Agents Chemother. 2011;55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing; Twenty-eighth Informational Supplement. Document M100-S28. Wayne, PA: CLSI; 2018. [Google Scholar]
- 11.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 12.McGann P, Snesrud E, Ong AC, Appalla L, Koren M, et al. War wound treatment complications due to transfer of an IncN plasmid harboring bla(OXA-181) from Morganella morganii to CTX-M-27-producing sequence type 131 Escherichia coli . Antimicrob Agents Chemother. 2015;59:3556–3562. doi: 10.1128/AAC.04442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greig DR, Dallman TJ, Hopkins KL, Jenkins C. MinION nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli . Microb Genom. 2018;4:e000213. doi: 10.1099/mgen.0.000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zurfluh K, Poirel L, Nordmann P, Klumpp J, Stephan R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob Resist Infect Control. 2015;4:38. doi: 10.1186/s13756-015-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsay JP, Firth N. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr Opin Microbiol. 2017;38:1–9. doi: 10.1016/j.mib.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 16.San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol. 2016;1:10. doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Liu W, Qin T, Liu C, Ren H. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae . Front Microbiol. 2017;8:371. doi: 10.3389/fmicb.2017.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.