Abstract
Sampling campaigns in Greenland and Svalbard were executed to explore fungal diversity in cold habitats. Three very abundant groups of strains were discovered, consisting either of recently described or of yet-undescribed psychrophilic and oligotrophic yeasts and dimorphic fungi, accounting for around 50 % of the total cultivable diversity of basidiomycetes in our studies. The occurrence of these taxa has also been demonstrated by culture-independent methods. Based on phylogenetic analyses of ribosomal gene cluster sequences (D1/D2 domains of 28S (LSU), 18S (SSU), ITS with 5.8S rDNA) and sequences of protein-coding genes for elongation factor one alpha (TEF), cytochrome b (CYTB) and two subunits of the RNA polymerase II (RPB1 and RPB2) obtained from pure cultures, the isolated taxa presented in this study belong to Basidiomycota, subphylum Pucciniomycotina, class Microbotryomycetes, family Camptobasidiaceae. The dataset of the sequences supported the recognition of three species: Camptobasidium gelus, Camptobasidium arcticum sp. nov. (ex-type strain EXF-12713) and Psychromyces glacialis gen. and sp. nov. (ex-type strain EXF-13111). Camptobasidium gelus was found in the Svalbard and Greenland samples, while representatives of the here proposed new species, C. arcticum, were found only in the Greenland Ice Sheet. Psychromyces gen. nov. was erected for the dimorphic/filamentous isolates found in Svalbard and Greenland glacial environments. The taxon, for which the invalid name ‘Rhodotorula svalbardensis’ has been used, belongs to this genus. Based on ribosomal genes, Camptobasidium arcticum and Psychromyces glacialis are related, phylogenetically most closely related to the genera Glaciozyma and Cryolevonia. Seven genes phylogeny restricted to taxa with available sequences, supported the placement of Psychromyces to Camptobasidiaceae.
Keywords: psychrophiles, yeast, filamentous, Svalbard, Greenland Ice Sheet, glacial ice, Microbotryomycetes
Basidiomycetous yeasts have a widespread distribution in cold environments, particularly in polar areas [1]. Psychrotolerant and psychrophilic species can be endemic or cosmopolitan [2]. Due to their ability to survive in extreme conditions they have become model organisms in searches for potentially novel secondary metabolites and in studies addressing the evolution of life strategies at low temperatures [3–8]. In recent years, the diversity of yeast species in glacial habitats of polar (Antarctic and Arctic) and non-polar (e.g. Alpine) areas has been investigated with culture-dependent and independent methods [9–18]. These studies reported considerable numbers of new taxa, for instance, in Bannozyma—B. arctica [19]; Mrakia – M. robertii, M. blollopsis, M. psychrophila [20, 21]; Rhodotorula—R. arctica [22]; Phenoliferia – P. psychrophenolica, P. psychrophila, P. glacialis [23]; Glaciozyma—G. watsonii, G.martinii, G. antarctica, G. litorale [24, 25]; Camptobasidium – C. gelus [18]; and Cryolevonia—C. schafbergensis, C. giraudoae [17, 18]. These studies exemplify the rich and still largely undiscovered diversity of glacial environments, especially among Microbotryomycetes (Pucciniomycotina). This class contains ‘anther smuts’, formerly classified in Ustilaginomycotina, and numerous anamorphic yeasts (reviewed in [26]). It contains the orders Microbotryales (predominantly teliospore-forming plant parasites), Kriegeriales (plant parasites, aquatic fungi or saprobes from cold, temperate and tropical environments), Sporidiobolales, Leucosporidiales (anamorphic or teliospore-forming yeasts isolated from various habitats and surfaces), Heterogastridiales (filamentous fungi isolated from decaying plant material and mushrooms), Rosettozymales (yeasts from phyllosphere) and Heitmaniales (yeasts from phyllosphere) [19]. The description of the ballistospore-forming yeast from neotropical forests, Meredithblackwellia eburnea, and its relatedness to Kriegeria eriophori and Camptobasidium hydrophilum, led to the erection of the order Kriegeriales for the families Kriegeriaceae and Camptobasidiaceae. The taxonomic position of numerous species of the Kriegeriales could not be classified on the family level and above because their phylogenetic relationships were studied on the level of ribosomal genes only (e.g. [17, 18]), which is problematic due to low variability in LSU and high variability in ITS sequences [27]. Although the recent approaches employed multi-gene analyses [19, 27, 28], the taxonomic position of many taxa in the Microbotryomycetes remains unresolved.
Sampling campaigns in Greenland and Svalbard in 2017, investigating subglacial and glacial environments [15, 16], led to the isolation of 168 yeasts and yeast-like isolates, 150 of them belonging to the phylum Basidiomycota. Approximately half of these isolates could not be reliably identified at the species level. About half of the cultivable taxa were also detected through amplicon sequencing of the fungal internal transcribed spacer 2 (ITS2) [15, 16]. In an effort to continue the characterization of yeasts from Greenland and Svalbard collections, a new species of Camptobasidium, C. arcticum, is described as the phylogenetic sister of the recently described Camptobasidium gelus [18]. A new genus and species, Psychromyces glacialis, is also proposed to accommodate isolates identical in ribosomal sequences to ‘Rhodotorula svalbardensis’. Its taxonomy and status were discussed in several studies following the application of the ‘one fungus=one name’ system [27].
Sampling sites and isolation methods
Cryoconite, snow, supraglacial ice (clear ice), supraglacial ice dominated by dark pigmented glacier algae (dark ice) and supraglacial water samples were collected from the southwest margin of the Greenland Ice Sheet during a fieldwork campaign in July 2017 [15] (Table 1). The fieldwork site was located at around 60 km east of Kangerlussuaq and was within the so-called ‘dark zone’, characterized by extensive algal blooms [29]. Subglacial ice and glacial meltwater were collected from three polythermal glaciers (Midtre Lovénbreen, Pedersenbreen and Vestre Brøggerbreen), situated in the Kongsfjorden area, Ny-Ålesund (Svalbard, Norway) in August 2017 [16] (Table 1).
Table 1.
Data of collected samples
|
Sampling location |
Sample type |
Sampling time |
|---|---|---|
|
Greenland, Kangerlussuaq, Ice Sheet (67° 04′ 43″ N 49° 20′ 29″ W) |
Snow |
July 2017 |
|
Dark ice | ||
|
Clear ice | ||
|
Supraglacial water | ||
|
Norway, Svalbard, Ny-Ålesund: |
August 2017 |
|
|
Midtre Lovénbreen (78° 53′ 37″ N 12° 04′ 13″ E) |
Subglacial ice |
|
|
Midtre Lovénbreen (78° 53′ 25″ N 12° 03′ 15″ E) |
Glacial meltwater |
|
|
Vestre Brøggerbreen (78° 54′ 55″ N 11° 45′ 48″ E) |
Subglacial ice |
|
|
Pedersenbreen (78° 52′ 46″ N 12° 17′ 57″ E) |
Subglacial ice |
All samples were collected with sterilized tools and handled with sterile nitrile gloves to avoid contamination. Samples were collected in sterile Whirl-Pak plastic bags or sterile plastic bottles and processed within a few hours of their collection at the primary ice camp on the Greenland Ice Sheet or the NERC Arctic Research Station (Ny-Ålesund). The samples were filtered and incubated at 15 °C on six different media, listed in Table S1 (available in the online version of this article), as described by Perini et al. [15, 16]. A total of 150 basidiomycetous yeasts and yeast-like fungi were isolated from all the above listed samples (Svalbard and Greenland [15, 16]). The majority of the strains reported in this study (53 of 74) was isolated from media with high water activity (aw=1) and poor in nutrients, such as minimal medium (MM) [30] and synthetic nutrient agar (SNA) [31], both supplemented with chloramphenicol (50 mg l−1), and from Reasoner’s 2A (R2A) agar [32]. The remaining strains (21 of 74) were isolated from media richer in nutrients, such as dichloran rose bengal chloramphenicol agar (DRBC) [33] and media with lower aw (MY10–12, DG18) [34].
All strains were deposited in the Ex Culture Collection of the Infrastructural Centre Mycosmo (MRIC UL), Slovenia (www.ex-genebank.com) at the Department of Biology, Biotechnical Faculty, University of Ljubljana, Slovenia (Table S1). Around 15 % of the strains (21 of 74) could not be revived after 3 years of preservation at −80 °C using commercial preservation kit.
The holotype of the new species from Svalbard, Psychromyces glacialis, was deposited in the CBS culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, as CBS 16467.
Phenotypic characterization
Morphological characters of pure cultures were observed on potato dextrose agar (PDA) [35], oatmeal agar (OA) [36] and corn meal agar (CMA) [37] (Dalmau plating) incubated at 15 °C. For the codes used to describe colony colour we refer to [38]. Microscopic characters were observed with Nomarski interference contrast optics on Olympus BX-51 microscope, micrographs were recorded with DP72 camera (Olympus). To test for the occurrence of teleomorphic structures in the yeast group of isolates described in this manuscript as Camptobasidium arcticum and identified as C. gelus, 14-day-old yeast cultures of selected representatives were pairwise-crossed on malt extract agar (MEA), yeast extract malt extract agar (YMA), soytone–glucose agar (SG) and water agar [39] and incubated at 15 °C for 6 months. The production of hyphae and teliospores was checked regularly.
Fermentation of d-glucose was tested in liquid medium with a 2 % (w/v) solution of sugar [39]. Assimilation of carbon and nitrogen sources were evaluated in Camptobasidium gelus (group 1 isolates: EXF-12745, EXF-12576, EXF-12594, EXF-13102, EXF-13103), Camptobasidium arcticum (group 2 isolates: EXF-12713T, EXF-12522, EXF-12524, EXF-12711) and Psychromyces glacialis (group 3 isolates: EXF-13111T, EXF-12886, EXF-12419, EXF-12991). The assimilation of selected C and N sources was tested in liquid media according to Kurtzman et al. [39]; C sources: d-glucose, d-galactose, l-sorbose, d-glucosamine, d-ribose, d-xylose, l-arabinose, d-arabinose, sucrose, maltose, α-trehalose, cellobiose, salicin, arbutin, melibiose, lactose and raffinose; N sources: nitrate, nitrite, l-lysine and cadaverine. Results were read after 1, 2 and 3 weeks of incubation at 15 °C. In addition, YT Biolog plates were used according to the manufacturer’s instructions to test the assimilation capabilities of a larger number of C sources for the above-mentioned strains of C. gelus, isolates of group 2 and for EXF-13111 of group 3. In short, the fungi were grown on PDA for 14 days at 15 °C. Cell aliquots were resuspended in YT fluid, adjusted to reach a turbidity transmittance of about 47 % in case of yeast strains, and unadjusted for filamentous strains. Biolog YT plates were incubated at 15 °C for up to 14 days, and the assimilation was followed by measuring the absorbance at 590 nm (A590) on a CytationI3 Imaging reader, supported by the Gen5 Microplate Reader and Imager Software (BioTek Instruments). Absorbance readings were taken daily. Measurements were compared to the negative control (water) provided on the same plate, as well as to the absorbance in each individual well immediately after inoculation. Values of absorbance on day 0 in the inoculated plates were subtracted from absorbance values on days 7 and 14, and % value towards assimilation of glucose (100 %) was calculated for all the wells with assimilation tests. Calculated values of 50–70 % according to absorbance of glucose on day 7 were interpreted as weak reactions (w), values of 70–100 % (or higher) as positive reactions. After 14 days, values between 50–70 % were interpreted as weak and delayed (wd), and values between 70–100 % (or higher) as delayed reactions. With filamentous representatives of group 3, absorbance values obtained on Tween 80 were taken as 100 % reaction, interpretation of assimilation results was performed as described above. Oxidation reactions on YT plates did not give any positive reaction, and were therefore not considered.
Phylogeny
For phylogenetic analyses, genomic DNA of the isolates was extracted using the PrepMan Ultra reagent (Applied Biosystems) according to the manufacturer instructions. DNA of filamentous cultures was extracted after mechanical lysis in CTAB buffer according to the protocol described by Gerrits van den Ende and de Hoog [40]. DNA regions/genes used in the phylogenetic analyses were the small subunit (SSU) rDNA, partial large subunit rDNA including its D1/D2 domains (LSU), the internal transcribed spacers 1 and 2 including the 5.8S rDNA (ITS), and partial sequences of genes encoding for translation elongation factor one alpha (TEF), cytochrome b (CYTB), RNA polymerase II largest subunit (RPB1) and RNA polymerase II second largest subunit (RPB2). These were amplified and sequenced with the following primer sets: NS1/NS24, NL1/NL4, ITS1/ITS4, EF1-983F/EF1-2218R, E1M4/E2mr3, RPB1-Af/RPB1-Cr, fRPB2-5F/fRPB2-7cR, respectively [27, 41–44]. Alignments of ITS and LSU sequences of strains described in this study and of the most closely related sequences of type strains and other reference strains, found with the blast algorithm [45] in the non-redundant GenBank nucleotide database, were used for phylogenetic analyses of a concatenated dataset. Partial sequences of the four protein-coding genes, TEF, CYTB, RPB1 and RPB2, were selected from available sequences of the Microbotryomycetes published by Wang et al. [27] and Li et al. [19]. Accordingly, taxa such as Cryolevonia and Meredithblackwellia were excluded from analyses. Ribosomal genes were aligned in mega7 using ClustalX [46], corrections were made by hand. Protein-coding genes were initially aligned in mega7 using ClustalX, and then manually corrected according to the amino acid translations. Introns were excluded from phylogenetic analyses. The best model of nucleotide substitution was estimated with jModelTest 2.1.10 [47] and was used as the custom model (model ‘010230’) input to PhyML 3.1 to estimate the phylogenetic trees [48]. aLRT as Chi2-based support was used for calculation of branch supports. The alpha parameter of the gamma distribution of substitution rate categories and the proportion of invariable sites were estimated by PhyML. Additionally, phylogenies were reconstructed from the same alignments using MrBayes 3.2.7 [49]. Two substitution types of the 4by4 model and gamma distributed rates with a proportion of invariable sites (approximated with four categories of gamma distribution) were used for the estimation through 10 million generations (sampling every 100th generation), two runs of 15 chains each, heated at temperature 0.2 (LSU) or 0.1 (ITS, concatenated dataset) and discarding the first 25 % trees from the final consensus tree. The minimum spanning networks were constructed without ties based on a pairwise matrix of bitwise distances derived from the above described alignment of ITS using packages 'ape' and 'poppr' in R [50–52]. For the analysis of the fungal community by amplicon sequencing, the total genomic DNA was extracted in triplicate from filtered biomass and 1 g cryoconite sediment as described by Perini et al. [15, 16]. The PCR amplification of the ITS2 region was performed using the primers ITS4-Fun and 5.8S-Fun [53] in three reactions per sample. The sequencing of the fungal amplicons was performed by Illumina MiSeq version 3. The sequencing data were processed in QIIME2 [54] as described by Perini et al. [15, 16]. Among the set of representative sequences of individual amplicon sequence variants (ASVs) found in the samples, the ones similar to Camptobasidium arcticum, C. gelus and Psychromyces glacialis were identified with stand-alone blast [45]. These sequences were added to the set of sequences recovered from pure cultures and from the databases and aligned in mega7 using ClustalX [46]. The maximum-likelihood tree was estimated using this alignment with PhyML as described above in order to confirm the taxonomic placement of the Camptobasidium and Psychromyces ASVs.
blast searches using ITS and LSU as queries suggested that 32 isolates from our study are identical to Camptobasidium gelus (Pucciniomycotina, Microbotryomycetes, Kriegeriales, Camptobasidiaceae). Twenty-two isolates (group 2) could not be identified to the species level: 21 isolates differed from C. gelus in five positions in LSU and 17 positions in ITS, while EXF-12685, differed from C. gelus in 15 LSU and 45 ITS positions. Sequences of group 3 were almost identical (two and five positions difference in ITS and LSU) to ‘Rhodotorula svalbardensis’, described here as Psychromyces glacialis (Table 2).
Table 2.
Nucleotide substitutions in ITS and LSU sequences in type strains of Camptobasidium species, Psychromyces glacialis and selected closely related species of genera Cryolevonia, Glaciozyma, Phenoliferia, Rhodotorula and Oberwinklerozyma
Strains: 1, Cr. schafbergensis PYCC 8347T; 2, Cr. giraudoae CRUB 2086T;3, C. hydrophilum CCM 8060T; 4, C. gelus CBS 8941T; 5, C. gelus EXF-12745; 6, C. arcticum EXF-12713T; 7, Camptobasidium sp. EXF-12685; 8, Ps. glacialis EXF-13111T; 9, G. antarctica CBS5942T; 10, G. watsonii CBS10986T; 11, G. martinii CBS 10620T; 12, Ph. psychrophenolica. CBS 10438T;13, ‘R. svalbardensis’ MBL-I; 14, O. yarrowii CBS 7417T.Values above the diagonal are numbers of nucleotide substitutions and sequence similarity (%, in parentheses) in the LSU. Values below the diagonal are numbers of nucleotide substitutions and sequence similarity (%, in parentheses) in the sequences of the ITS.
|
Species |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
1 |
4 (99.2) |
10 (97.9) |
4 (99.2) |
4 (99.2) |
7 (98.5) |
12 (97.5) |
22 (95.4) |
19 (96.0) |
20 (95.8) |
25 (94.8) |
13 (97.3) |
22 (95.4) |
22 (95.2) |
|
|
2 |
14 (97.3) |
17 (96.9) |
12 (97.9) |
12 (97.9) |
15 (97.4) |
19 (96.7) |
30 (94.4) |
26 (95.5) |
28 (95.1) |
35 (93.7) |
15 (97.4) |
29 (94.9) |
26 (95.3) |
|
|
3 |
53 (88.8) |
65 (87.0) |
8 (98.5) |
8 (98.5) |
12 (97.8) |
11 (98.0) |
26 (95.1) |
26 (95.2) |
28 (94.9) |
30 (94.3) |
18 (96.7) |
25 (95.4) |
27 (94.9) |
|
|
4 |
53 (88.9) |
71 (86.1) |
26 (94.9) |
0 (100) |
5 (99.1) |
15 (97.4) |
30 (94.4) |
27 (95.2) |
29 (94.9) |
33 (94.1) |
18 (96.9) |
28 (95.1) |
24 (95.7) |
|
|
5 |
53 (88.9) |
71 (86.1) |
26 (94.9) |
0 (100) |
5 (99.1) |
15 (97.4) |
30 (94.4) |
27 (95.3) |
29 (94.9) |
33 (94.1) |
18 (96.9) |
28 (95.1) |
24 (95.7) |
|
|
6 |
49 (89.7) |
67 (86.8) |
16 (96.8) |
17 (96.8) |
17 (96.8) |
14 (97.6) |
35 (93.5) |
26 (95.5) |
28 (95.1) |
32 (94.2) |
19 (96.7) |
33 (94.3) |
29 (94.8) |
|
|
7 |
55 (88.7) |
68 (86.7) |
40 (91.9) |
45 (91.3) |
45 (91.3) |
40 (92.2) |
31 (94.2) |
28 (95.1) |
26 (95.5) |
36 (93.5) |
21 (96.3) |
29 (94.9) |
35 (93.7) |
|
|
8 |
54 (88.5) |
71 (85.6) |
56 (88.6) |
57 (89.0) |
57 (89.0) |
54 (89.5) |
50 (90.2) |
33 (93.1) |
33 (93.9) |
40 (92.3) |
22 (95.9) |
2 (99.6) |
32 (93.9) |
|
|
9 |
67 (86.1) |
81 (84.0) |
69 (85.9) |
81 (84.3) |
81 (84.3) |
77 (85.09 |
81 (84.3) |
83 (84.1) |
7 (98.8) |
10 (98.2) |
24 (95.8) |
32 (94.4) |
31 (94.4) |
|
|
10 |
61 (87.2) |
77 (84.7) |
75 (84.8) |
88 (83.1) |
88 (83.1) |
81 (84.4) |
81 (84.3) |
79 (84.9) |
40 (92.6) |
14 (97.5) |
26 (95.5) |
32 (94.4) |
33 (94.1) |
|
|
11 |
61 (87.0) |
77 (84.4) |
77 (84.2) |
82 (84.0) |
82 (84.0) |
81 (84.1) |
67 (86.9) |
78 (84.9) |
47 (91.1) |
52 (90.2) |
33 (94.1) |
40 (92.8) |
36 (93.3) |
|
|
12 |
65 (86.2) |
81 (83.6) |
70 (85.6) |
71 (86.1) |
71 (86.1) |
67 (86.8) |
70 (86.4) |
49 (90.4) |
82 (84.0) |
94 (81.8) |
89 (82.5) |
20 (96.5) |
20 (96.4) |
|
|
13 |
52 (88.9) |
69 (86.0) |
53 (89.2) |
56 (89.2) |
56 (89.2) |
50 (90.3) |
49 (90.4) |
5 (99.1) |
79 (84.9) |
76 (85.5) |
75 (85.4) |
46 (91.0) |
30 (94.6) |
|
|
14 |
93 (81.0) |
107 (79.2) |
69 (86.1) |
73 (86.0) |
73 (86.0) |
69 (86.7) |
73 (85.9) |
64 (87.5) |
82 (84.0) |
90 (82.6) |
83 (83.7) |
74 (85.6) |
61 (88.1) |
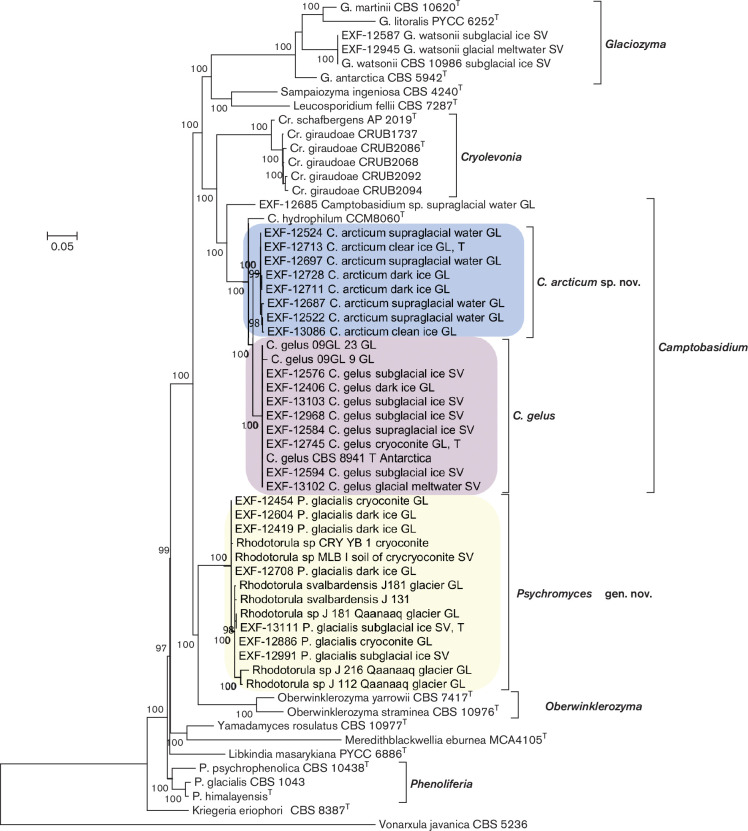
Phylogenetic analyses of concatenated sequences of the ITS and the LSU placed representatives of the newly sampled strains into well-separated lineages of the Microbotryomycetes. Group 1 isolates were identified as Camptobasidum gelus, recently described based on one strain isolated from Antarctica and two from Greenland [18]. Phylogenetic sister group relatedness of group 2 isolates and C. gelus was highly supported (branch support BS=100 %). The phylogenetic analysis confirmed that group 3 isolates belong to ‘Rhodotorula svalbardensis’/ Psychromyces glacialis. They clustered together with other sequences identified as Rhodotorula species. As indicated in Fig. 1, genera Glaciozyma (grouping together with Sampaiozyma and Leucosporidium), Camptobasidium and Cryolevonia form a highly supported monophyletic group (BS=100 %) that is a sister of a joint clade accommodating Psychromyces and Oberwinklerozyma. Genera such as Phenoliferia, Yamadamyces and Meredithblackwellia formed an unresolved, paraphyletic assemblage near the base of the tree, but their relatedness with Oberwinklerozyma, Psychromyces, Camptobasidum, Cryolevonia and Glaciozyma was highly supported (BS=100 %).
Fig. 1.
Phylogenetic tree based on alignment of the LSU rDNA and complete ITS including 5.8S rDNA, estimated by maximum likelihood. The best model of nucleotide substitution was estimated with jModelTest, other parameters (alpha parameter of the gamma distribution of substitution rate categories, proportion of invariable sites) were estimated within PhyML. aLRT as Chi2 based support was used for calculation of branch supports.
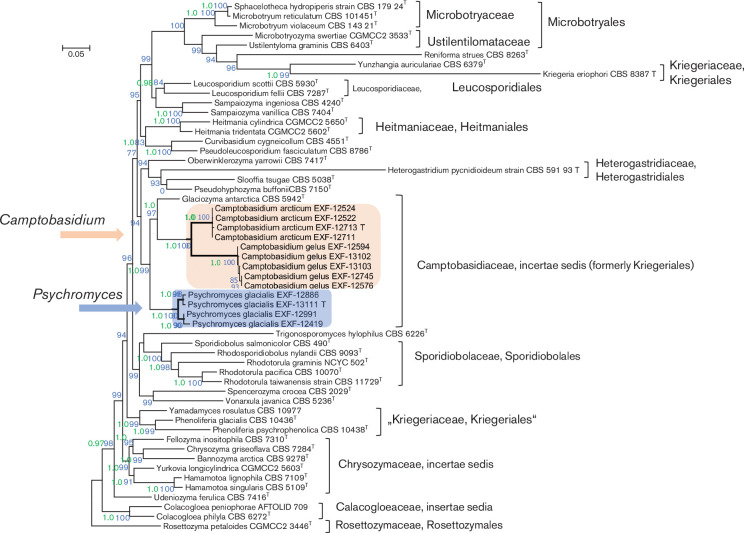
Further phylogenetic analyses were based on concatenated sequences of rDNA loci (SSU, ITS, LSU) and partial sequences of the four protein-encoding genes (TEF, CYTB, RPB1 and RPB2). The complete alignment consisted of 9938 nt. Group 1 introns encountered in SSU sequences and introns in protein encoding genes were excluded from further analyses. The 5297 nucleotides long alignment used for analyses consisted of 1696 nt from SSU, 590 from LSU, 161 from 5.8S rDNA, 372 from CYTB, 746 from TEF, 722 from RPB1 and 1010 from RPB2. Two group 1 introns (covering positions 1514–1992 in EXF-12711, 12522 and positions 2256–2690 in strains EXF-12524, 12713, 12711 of the compiled alignment) were encountered in SSU sequences of C. arcticum. Camptobasidium gelus and C. arcticum had three introns in TEF sequences, while there were only two in Psychromyces glacialis. No introns were encountered in RPB2 and CYTB sequences. CYTB sequences could not be amplified for C. gelus, and RPB1 not for P. glacialis. The obtained phylogenetic tree (Fig. 2) confirmed close relatedness of Camptobasidium and Psychromyces that clustered together with Glaciozyma (BS=99 %; BI-PP, 1.00). We therefore propose inclusion of Psychromyces into the Camptobasidiaceae. The analyses suggested that Kriegeriales sensu stricto, represented by Kriegeria eriophori, is relatively closely related to Psychromyces, Camptobasidum, Glaciozyma and others, while taxa formerly included in the Kriegeriales, such as Phenoliferia and Yamadamyces, appeared distantly related (Fig. 2). Monophyly of Psychromyces, Camptobasidum, Glaciozyma and K. eriophori is, however, not supported due to the phylogenetic interference of orders, such as Heitmaniales, Leucosporidiales and Heterogastridiales. Accordingly, classification of Psychromyces, Camptobasidum, Glaciozyma in the Kriegeriales is not supported in the present study, which is why the Camptobasidiaceae is for the time being considered as an incertae sedis within the Mycobotryomycetes (Fig. 2). It is to be emphasized, however, that the downloaded amino acid sequences of K. eriophori (CBS 8387) were partly difficult to align with numerous other included taxa (RPB1, RPB2) and that several codons could not be translated into amino acids at all (CYTB). It is clear that these numerous apomorphies are responsible for the long terminal branch that K. eriophori obtained in analyses of the non-translated DNA sequences (Fig. 2).
Fig. 2.
Phylogenetic tree based on alignment of the SSU, LSU, 5.8S rDNA, TEF, CYTB, RPB1 and RPB2 estimated by maximum-likelihood and Bayesian analyses. The tree includes representatives of Psychromyces glacialis gen. nov., sp. nov., Camptobasidium arcticum sp. nov., C. gelus and representative species of genera of Microbotryomycetes. In the maximum-likelihood estimation, the best model of nucleotide substitution was estimated with jModelTest, other parameters (alpha parameter of the gamma distribution of substitution rate categories, proportion of invariable sites) were estimated within PhyML. aLRT as Chi2-based support was used for calculation of branch supports. In estimation by MrBayes, a total 10 million generations were calculated and the first 25 % were discarded before the shown consensus tree was calculated from trees sampled every 100 generations. Posterior probabilities higher than 0.9 (in green) and maximum-likelihood bootstrap value from 1000 bootstrap replicates larger than 70 % (in blue) are shown near the nodes.
To analyse the effect of protein-encoding gene sequences on the dataset analysed in Fig. 2 and to allow comparison with already published trees that are based only on nc rDNA data, protein-encoding genes were excluded from the dataset and the remaining alignment was analysed separately (Fig. S1). This analysis also addressed more Mycobotryomycetes as included in Fig. 1 and supported some of the conclusions made on the basis of concatenated dataset presented in Fig. 2. Relatedness of Camptobasidium, Psychromyces and Glaciozyma is almost equally highly supported (BS=92 %), while the monophyly of the so defined Camptobasidiaceae with Kriegeria sensu stricto is not supported. Monophyly of Kriegeria eriophori with Phenoliferia and Yamadamyces (Fig. S1, BS=89 %) is, however, not in congruence with inferences made on the basis of the dataset including protein-encoding genes (Fig. 2). The inconsistent clustering of K. eriophori is most likely caused by numerous apomorphies that resulted in the above-discussed alignment problems and a long terminal branch for K. eriophori in Fig. 2.
Camptobasidium
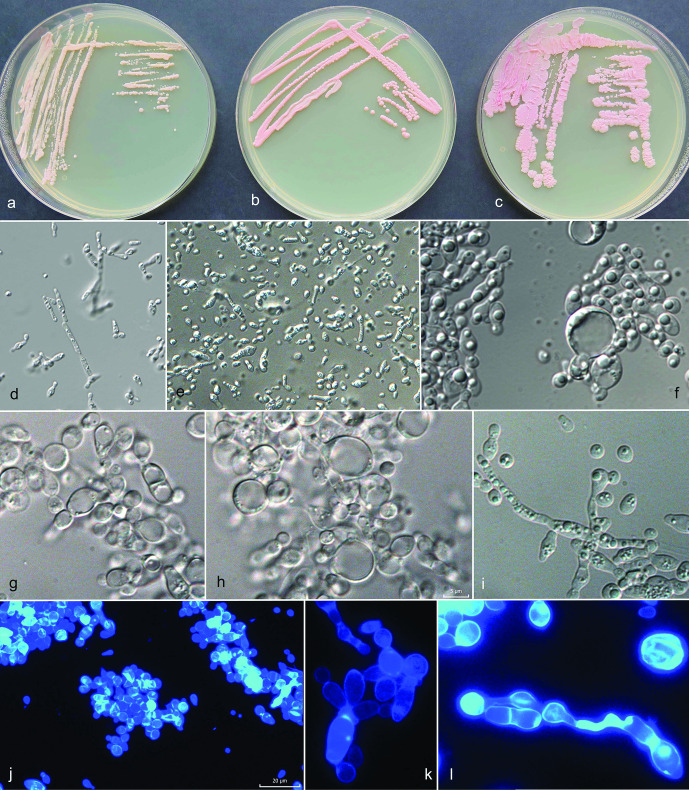
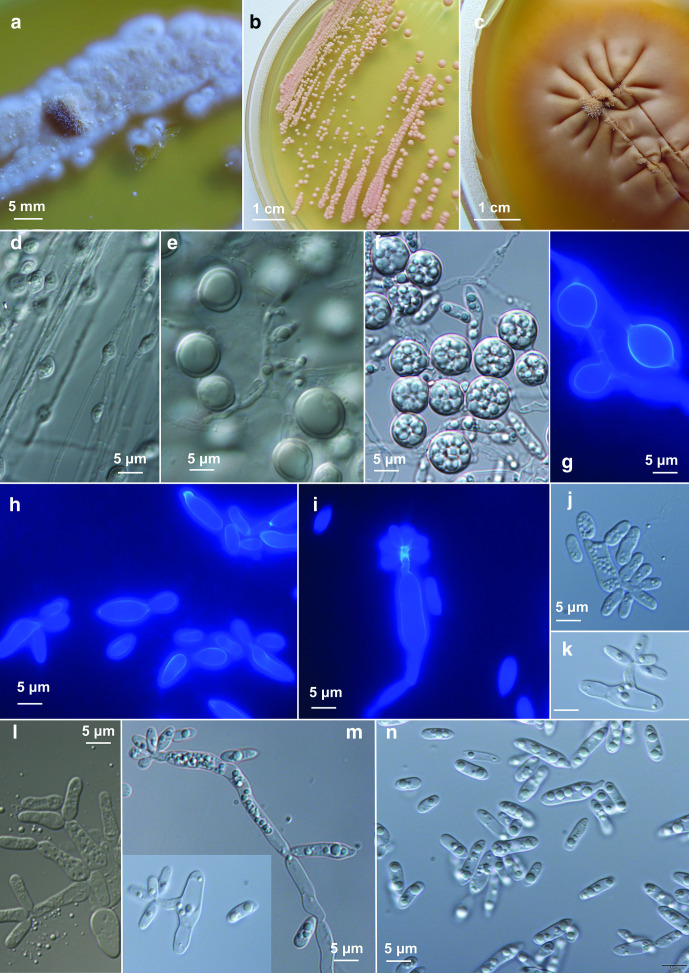
The maximum-likelihood analysis of selected ITS and LSU sequences of Camptobasidium strains supported the recognition of three species: 31 strains were identified as C. gelus and 22 strains as two yet undescribed species, of which one (EXF-12685) is no longer alive. The other species is here newly described as C. arcticum. The ITS/LSU sequences of C. arcticum strain EXF-12713 differed from the those of C. gelus CBS 8941 by 17/5 nt (99/96,9 % similarity). Strain EXF-12685 was most closely related to C. hydrophilum CCM8060 (11 nt difference in LSU, 98 % similarity; 40 nt difference in ITS, 91.9 % similarity), while it differed from C. arcticum strain EXF-12713 by 14 nt in the LSU and 42 nt in the ITS sequences. A similar comparison with C. gelus strain EXF-12745 showed a difference of 2.6 % (15 nt) in LSU, and 8.6 % (45 nt) in ITS (Table 2). Since EXF-12685 did not survive deep-freezing preservation, it could not be taxonomically considered. The representatives of other closely related species listed in Table 2 had lower similarities and the number of differing nucleotides in ITS was for all drastically larger as in LSU, as already described in other studies [27]. When comparing ITS sequences almost no variability was noted in C. gelus, while some were noted within C. arcticum (Fig. S2). The type strain of C. gelus, CBS 8941 (AY040665) from Antarctica, strain BL58-2 (AB474396) isolated from Russian glacier ice core in Siberia [55] and EXF-12745 from cryoconite in Greenland, had identical ITS sequences. The Antarctic yeast strain CBS 8941, now recognized as C. gelus, was noticed as a potentially new phylogenetic lineage already by Wang et al. [23] on the level of LSU sequences. It was then selected as the type of C. gelus by de Garcia et al. [11], who described this species based on two additional strains from Greenland. The same authors [23] addressed the separate position of CRUB 1733 (GenBank FJ841888), which was recently described as the new species Cryolevonia giraudoae by de Garcia et al. [11]. The genus Cryolevonia, closely related to Camptobasidium (Fig. 2), was described and typified with Cr. schafbergensis by Pontes et al. [17]. Classification of these psychrophilic yeast species was based only on ITS /LSU data [11, 17]. The type species of the genus Camptobasidium, C. hydrophilum, is strictly filamentous and forms peculiar anamorphic Ingoldian spores [56], and no yeast phase, which is the only morphology observed in C. gelus. Analyses of ribosomal DNA sequences placed two of our three newly discovered phylogenetic groups in Camptobasidium; however, ribosomal DNA sequences are only restrictively used for phylogenetic analyses of the Microbotryomycetes [23]. No household gene sequences could be generated or are accessible for the type species, C. hydrophilum, which is why monophyly of all Camptobasidium species cannot be confirmed yet on the basis of all loci selected here. Camptobasidium cultures are characterized by pink to reddish-grey colonies [11, 56], in our case studied on PDA (Figs 3 and 4). According to our observation, strains identified as C. gelus were typically purplish white (14A2), pastel pink (11A3), greyish rose (11B3) or reddish grey (11B2), with either glistening or ridged, membranous surface appearance, raised with entire or undulating margin (Fig. 4a–d). De Garcia et al. [11] described Camptobasidium gelus as ovoidal to ellipsoidal yeasts multiplying by multilateral budding. According to the provided micrograph [11], they also observed larger cells containing large vacuoles, as well as smaller storage granules, which were observed also in our isolates (Fig. 4f, j and m). The original description of C. gelus states that hyphae or pseudohyphae are not formed, however we observed the presence of weak pseudomycelium on PDA and in Dalmau plate culture on CMA in our isolates (Fig. 4e, l). Individual yeast cells of C. gelus isolates from this study were oblong, 5.5±2 (mean±SD; min-max: 3–9)×3.5±1 (mean±SD; min-max: 2–5) µm when one-celled, up to 13 µm long when two-celled, while C. arcticum forms slightly smaller cells, measuring 5±2 (mean±SD; min-max: 3–9)×3±0.6 (mean±SD; min-max: 2–4) µm due to developed pseudomycelium less glistening colonies.
Fig. 3.
Morphology of Camptobasidium arcticum. (a–c) Cultures of Camptobasidium arcticum on PDA in 9 cm Petri dishes after 2 months of incubation at 15 °C: (a) EXF-12713T, (b) EXF-12689, (c) EXF-13086. Micromorphology of cells in water: (d) EXF-12689 on PDA after 14 days, (e) after 2 months, (f) EXF-12713T on PDA after 14 days, (g, h) on OA after 2 months, (i) EXF-12689 on OA forming pseudohyphae, (j–l) EXF-12713T on OA stained with calcofluor white. Scale bar indicated on fig. (j) (20 µm) is valid also for figures (d) and (e) — ×400 magnification; scale bar indicated on fig. (h) (5 µm) is valid also for figures (f), (g), (i), (k), (l) – ×1000 magnification.
Fig. 4.
Morphology of Camptobasidium gelus. (a)–(d) Cultures of C. gelus on PDA in 9 cm Petri dishes after 2 months of incubation at 15 °C. (a) EXF-12597, (b) EXF-13102, (c) EXF-12968, (d) EXF-12745. Micromorphology of cells in water, (e, f) EXF-12745 on PDA, 14 days, (g) EXF-12745 on OA, 28 days, (h) EXF-12968 on PDA and (i) on OA, (j) EXF-12576 on PDA, (k, l) EXF-12745 on OA stained with calcofluor white. (k) EXF-12586 on PDA. Scale bar indicated on fig. (k) (20 µm) is valid also for figures (e) and (h) – ×400 magnification; scale bar indicated on fig. (l) (5 µm) is valid also for figures (f), (g), (i), (j), (l), (m) – ×1000 magnification.
Sexual reproductive structures were neither observed in single colonies or in mixed cell assays of C. gelus, nor in C. arcticum. No true mycelium and no teliospores were observed. The sexual morph is only known for C. hydrophilum from water-submerged cultures and is characterized by recurved, 1–3 septate metabasidia showing sporogenous loci on their convex sides and bearing rarely single basidiospores, mostly in groups of up to 20 that are sessile on inconspicuous denticles [56]. Authors also reported typically broadly fusiform or elliptical chlamydospores.
All Camptobasidium strains in our study showed slow colony development suggesting either oligotrophicity or auxotrophy, thus perhaps the need of a particular nutrient, mediating substance or partner relationship. Also, the closely related and psychrophilic Cryolevonia schafbergensis was described as a species with salient physiological characteristics: not growing at 18 °C, growing on a medium containing 16 % NaCl. These characteristics allowed the distinction between C. giraudoae [11] and Camptobasidium species (Table S2).
Differences in assimilation profile among closely related species are listed in Table 3, and within Camptobasidium and Psychromyces in Table S2. Assimilation of raffinose and sucrose was evident in all Camptobasidium species, but also in Psychromyces glacialis. Assimilation abilities of C. gelus isolates were mostly in accordance with de Garcia et al. [11]. Our isolates variably assimilated l-sorbose, d-xylose and nitrite, but were not able to assimilate melibiose and polyols (i-erythritol, xylitol, d-mannitol, d-sorbitol, adonitol) (Table S2). The newly proposed C. arcticum differs from C. gelus in the ability to assimilate melezitose, cellobiose, methyl β-d-glucoside and in the inability to assimilate nitrite. The additional differences in assimilation of other, not often used C sources were noticed in C. arcticum: the ability to assimilate gentiobiose, maltotriose and dextrin. It variably assimilated nitrate and showed weak or no assimilation of l-lysine. In some cases, differences in assimilation were observed when comparing results of classical tests in liquid media with the results of Biolog YT. As an example, Camptobasidium gelus showed assimilation of maltose and glycerol in classical tests, but no optical density changes were detected on YT plates (Table S2).
Table 3.
Differences and similarities in the assimilation capacity of selected carbon and nitrogen sources among Glaciozyma, Cryolevonia, Camptobasidium and Psychromyces species
Species: 1, Glaciozyma antarctica [62]; 2,Glaciozyma watsonii [24]; 3, Glaciozyma martini [24]; 4, Glaciozyma litorale [25]; 5, Cryolevonia schlafbergensis [17]; 6, Cryolevonia giraudoae [11]; 7, Camptobasidium hydrophilum [11]; 8, Camptobasidium gelus [11]; 9, Camptobasidium gelus (this study); 10, Camptobasidium arcticum (this study); 11, Psychromyces glacialis–yeast phase (this study); 12, Psychromyces glacialis–filamentous phase (this study); 13, ‘Rhodotorula svalbardensis' [57]. +, Positive; –, negative; w, weak positive; d, delayed (slow); wd, weak and delayed; v, variable (–/+/w/d); na, data not available.
|
Source |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
l-Arabinose |
– |
v |
w/– |
– |
– |
– |
w |
– |
– |
– |
– |
– |
– |
|
d-Arabinose |
– |
– |
– |
– |
– |
– |
w |
– |
– |
– |
– |
– |
+ |
|
α-d-Glucose |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
d-Galactose |
v |
+ |
+ |
– |
– |
v |
na |
v |
v (–,w,d) |
– |
d |
– |
– |
|
Sucrose |
v |
– |
– |
– |
+ |
– |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Raffinose |
– |
– |
– |
– |
– |
v |
+ |
+ |
+ |
d |
+ |
+, –*, d |
+ |
|
d-Ribose |
– |
v |
– |
– |
– |
– |
na |
– |
v (d,w,+) |
– |
– |
– |
– |
|
d-Sorbitol |
v |
+ |
w/+ |
na |
na |
w |
na |
+ |
–* |
–* |
wd* |
–* |
na |
|
Lactose |
– |
– |
w/– |
– |
– |
– |
+ |
– |
w/– |
v |
– |
– |
+ |
|
Trehalose |
v |
w/– |
w/– |
v,d |
– |
– |
– |
– |
v |
w/+ |
+ |
d |
na |
|
Maltose |
v |
v |
v |
– |
+ |
v |
w |
w |
w/+, –* |
+ |
d |
+, w*, d |
+ |
|
Melezitose |
– |
– |
– |
– |
+ |
+ |
+ |
– |
– |
+ |
+ |
+ |
+ |
|
Melibiose |
– |
– |
– |
– |
– |
– |
– |
+ |
– |
v, –* |
– |
– |
– |
|
Methyl α-d-glucoside |
– |
– |
– |
– |
– |
– |
w |
– |
–* |
–* |
–* |
–* |
na |
|
d-Xylose |
v |
v |
w/– |
– |
– |
– |
|
– |
w/d |
v (–,w,d) |
wd |
– |
– |
|
d-Glucitol |
v |
na |
w/+ |
– |
v |
na |
na |
+ |
–* |
–* |
wd* |
–* |
na |
|
Cellobiose |
v |
– |
– |
v,d |
– |
w |
na |
w |
– |
+ |
w,–* |
w/+ |
+ |
|
d-Glucosamine |
na |
– |
– |
– |
– |
– |
na |
– |
–* |
–* |
–* |
–* |
na |
|
Glycerol |
v |
– |
v |
d |
d |
+ |
– |
+ |
v, –* |
– |
– |
– |
– |
|
Nitrate |
+ |
w/+ |
+ |
na |
+ |
+ |
– |
+ |
+ |
v |
+ |
+ |
+ |
|
Nitrite |
+ |
– |
w |
na |
+ |
+ |
– |
+ |
w/– |
– |
+ |
w/d |
+ |
|
l-Lysine |
na |
v |
d/– |
na |
– |
+ |
na |
d |
+ |
w/– |
w |
w/– |
na |
|
Tween 80 |
na |
na |
na |
na |
na |
na |
na |
na |
v (–,w,+) |
– |
+ |
+ |
na |
*Results obtained by Biolog YT plates only.
Psychromyces
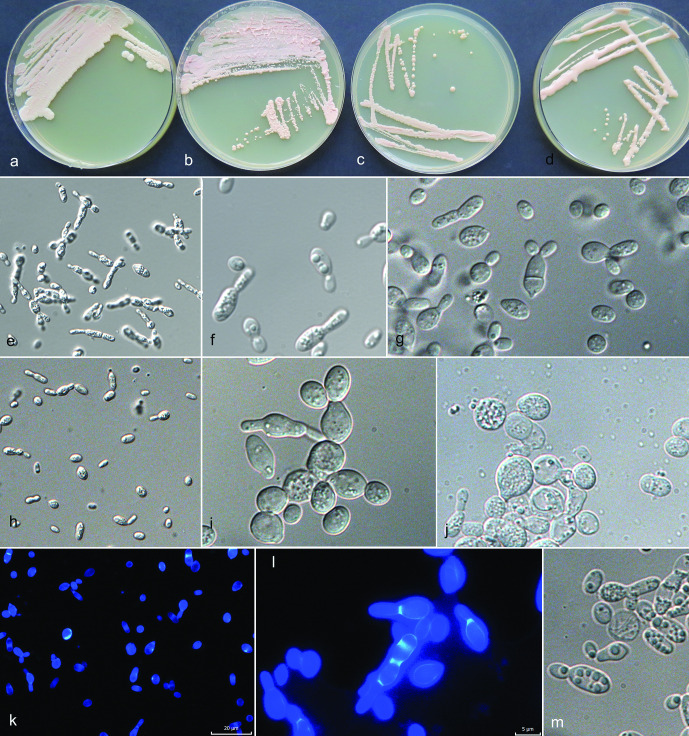
The maximum-likelihood analysis of selected ITS and LSU sequences grouped together dimorphic and filamentous isolates of group 3 with isolates named Rhodotorula sp. and Rhodotorula svalbardensis that were all isolated from cold environments of Svalbard and Greenland [12, 57]. The strongly supported sister group relationship of this group 3 to Oberwinklerozyma was suggested on the basis of analyses of nc rDNA sequences (BS=100 %) (Fig. 1). The concatenated seven-gene analysis justified and confirmed the isolated position of the genus (BS=100 %), and placed it as sister clade to the clade comprising Camptobasidium and Glaciozyma (Fig. 2). These results suggest that the clade should be placed in the family Camptobasidiaceae. We propose here a new genus, Psychromyces, for this clade for reasons described below. Bayesian phylogenetic inference analysis based on ITS sequences revealed this clade as unresolved (Fig. S2). A number of six nucleotide substitutions out of 526 distinguish two groups of strains, represented by EXF-13111 and EXF-12419 (Fig. S5). Exploration of the clade with additional phylogenetic marker genes (CYTB, TEF, RPB2) did not result in any better phylogenetic resolution. Accordingly, only a single species, P. glacialis, could be recognized. Strains with highly similar or identical ITS and LSU sequences were found also in other studies of Arctic glacial environments and supported cladification into two phylogenetically unresolved or paraphyletic subgroups within Psychromyces. Strains EXF-13111, 126476, 12545, 12886, 12991 belong to one, and strains EXF-12419, 12623, 12398, 12718, 13156, 124554, 12890, 12708, 13154, 12420, 12604, 12984, 12626 to the other group (Figs S2b and S5). Strains from Qaanaaq (represented by KY782280, KY782283) and Russell glaciers (KY782281, KY782282), both from Greenland (Singh et al., unpublished), and a strain from cryoconite sediment from Midtre Lovénbreen glacier in Svalbard (KC333170) [12] clustered with EXF-13111 on the basis of ITS sequences. The second group is represented by strain EXF-12419. Its ITS sequence is almost identical to the one of the strains MLB-I (JF805370) and CBS 12863 (=JCM 19699, JCM 19700, MTCC 10952) (AB734690), of which the latter originated from cryoconite soil sampled from the Midtre Lovénbreen glacier, Svalbard [57].
Singh et al. [57] erected the name ‘Rhodotorula svalbardensis’ for strains MLB-I and CBS 12863; however, the name Rhodotorula svalbardensis is invalid because two gatherings, strains MLB-I (CCP-II) and CRY-YB-1, were assigned as type [58], Shenzhen Code, Art. 40.7). Colony characteristics and microscopic characters described by Singh et al. [57] clearly suggested conspecificity of strains MLB-I (CCP-II), CRY-YB-1 and several strains we obtained from Svalbard and Greenland. However, although our strains were isolated as yeast colonies, only a single strain, EXF-13111, retained its ability to grow as a yeast after 2 years of preservation at −80 °C, while the other 16 strains were revived as purely filamentous after deep freezing. Singh et al. [57] described and illustrated ellipsoidal vegetative cells, 5.8–8.3×4.2–7.5 µm (mean 7×5.9 µm), occurring individually or in groups. According to our observation, these thick-walled cells most likely represent teliospores. In our strains these structures were typically globose to subglobose and had a diameter of 3–4 µm in young cultures; however, depending on the culture condition and age of culture, also spores with a diameter of up to 11 µm and of variable shapes were seen. Sometimes they contained oil droplets. In an attempt to induce their germination, the spores were cut out, placed into sterile H2O and kept at 5 °C. After an incubation period of 6 months, they were placed on water agar, however, no germination was observed. Chlamydospores are known in some species of basidiomycetous yeasts, for example, in genera Mrakia, Sporidiobolus, Tilletiaria, Fellomyces and Rhodosporidium, where they can appear in old cultures [59]. They were also observed in Camptobasidium hydrophylum [56]. Singh et al. [57] described, however, not illustrated, unilaterally or occasionally multilaterally budding cells. Yeast colonies were observed only in the strain EXF-13111. The yeast phase was maintained with the subculturing of yeast colonies, however after approx. 1 month of incubation, the colonies started developing hyphae and became strictly filamentous. The observed yeast cells were oblong and measured 7.5±3 (mean±SD; min-max: 5–12) µm×3.5±0.5 (mean±SD; min-max: 3–4) µm. Budding was uni-, bi- or multilateral and occurred solitarily directly on the mother cell or on sympodially proliferating, up to 5 µm long stalks. Daughter yeasts formed on terminal or lateral sympodially proliferating stalks have been so far described for certain Bullera species [59]. Some of the yeast cells in EXF-13111 prolonged to more than 20 µm long stalks, and formed pseudomycelium-like structures. Strains described by Singh et al. [57] formed up to 1.45 µm wide, septate hyphae. The presence of hyphae, with and without clamps, was observed in some strains in our study. Typically, the width of clamped mycelium was 2.5 µm in strain EXF-12419, while hyphae without clamps were narrower (up to 2 µm) and densely septated. In strain EXF-13111, non-clamped mycelium had a width of approx. 1.5 µm. If filamentous colonies were subcultured, they remained filamentous.
The most obvious difference between Psychromyces and other related species was its ability to assimilate Tween 80 (reaction on YT plate), which indicates lipolytic capacity. Camptobasidium gelus isolates varied in assimilation of Tween 80 (negative, positive, week reactions), while C. arcticum was unable to assimilate it. The assimilation ability of Tween 80 is in agreement with Singh’s et al. [57] observation of high lipase activity. In contrast, amylase activity reported by the same authors [57] was not detected by the classical assimilation tests performed in this study. The assimilation profile was similar to the closely related genus Camptobasidium: sucrose and raffinose were assimilated, as for all species of Camptobasidium; however, Psychromyces showed also the ability assimilate trehalose (Tables 3 and S2). It assimilated melesitoze and cellobiose, as C. arcticum. According to Singh et al. [57], Psychromyces glacialis is well-adapted to life in glacial environments, in particular to low temperatures. It can synthesize antifreeze proteins, modulate its membrane lipid composition by increasing the unsaturated fatty acids content and produce extracellular enzymes, particularly amylase, cellulase, protease and catalase at 4 °C [57].
Ecology
Ice samples retrieved from the Greenland Ice Sheet harboured a high abundance of yeasts, e.g. up to 900 c.f.u. ml−1 in dark ice, and contained a large number of yeast taxa. The majority of the taxa belonged to the Microbotryomycetes (Basidiomycota), including Camptobasidium gelus, the here proposed C. arcticum, formerly classified as Glaciozyma antarctica-like, Psychromyces glacialis, formerly classified as ‘Rhodotorula svalbardensis’, Phenoliferia glacialis, Sporobolomyces ruberrimus, and other yet-undescribed basidiomycetous yeasts [15]. A study of glacial environments on Svalbard resulted in the isolation of some of the same taxa, such as C. gelus and Psychromyces glacialis, but also revealed additional species: Glaciozyma watsonii, Leucosporidiella muscorum, Phenoliferia glacialis and P. psychrophenolica, with values up to 5 c.f.u. ml−1 [16]. The phylogenetic and phenotypic analysis of both sets of glacial isolates supported the description of two novel species, herewith proposed as Camptobasidium arcticum and Psychromyces glacialis.
High-throughput amplicon sequencing and analysis of ITS2 sequences from total environmental DNA [15, 16] revealed that species of the Microbotryomycetes commonly and abundantly occur in supra- and subglacial environments. The abundance of this group was up to 96 % in supraglacial water, and clear and dark ice (Greenland Ice Sheet) and up to 75 % in subglacial ice (Svalbard). ITS2 sequences identical to those of Camptobasidium gelus EXF-12745, Camptobasidium arcticum EXF-12713, Camptobasidium sp. EXF-12685 and Psychromyces glacialis EXF-13111 were found in total environmental DNA extracts (Fig. S3). Accordingly, these species appeared to commonly occur in diverse glacial environments and different geographical locations. The genus Camptobasidium has a circumpolar distribution and it also occurs in Antarctica [18]. The genus Psychromyces has so far only been recorded in Arctic glacial environments, both in Greenland and Svalbard. None of the species has been found so far in uppermost layers of the glacier consisting of fresh snow.
Slow growth of axenic cultures and unsuccessful preservation of circa 15 % of the isolates might indicate that Camptobasidium species, especially C. arcticum, accommodates a complex ecological niche or that it has complex nutritional requirements. It is possible that they grow optimally only in associations with other glacial organisms. Myco- and plant-parasitism is a frequent trait of the Pucciniomycotina, including Microbotryomycetes [60]. Camptobasidium hydrophilum, a close relative of C. arcticum, was identified as a mycoparasite, since it can form coiling hyphae around hyphae of aquatic hyphomycetes in dual cultures [56]. Parasitic life mode at the host–parasite interface is otherwise known to be supported via special interactive organelles, called colacosomes. These structures were so far detected in mycoparasitic Heterogastridiales, but also, occasionally, in non-parasitic Sporidiobolales and Leucosporidiales within Microbotryomycetes [60], implying their loss in saprobic species. The presence of these structures, indicative of mycoparasitism, have not been studied in Camptobasidium and Psychromyces so far, and should be the focus of future studies.
Statistical analysis of ITS2 NGS sequencing data from Greenland glacial environments revealed high co-occurrence of non-identifiable Microbotryomycetes with Phialophora (Chaetothyriales, Ascomycota) and with unidentified Leucosporidiales (Mycrobotryomycetes, Basidiomycota). While in Svalbard glacial samples high co-occurrence was calculated for non-identifiable Microbotryomycetes with an unidentified species of Leucosporidiales, with Didymellaceae (Pleosporales, Ascomycota), with Libkindia masarykiana (incertae sedis, Microbotryomycetes), and with Kriegeriaceae (Kriegeriales, Microbotryomycetes; data not shown). These co-occurrences might indicate similar ecological preferences of these taxa (e.g. for cold aquatic environments) or interactions between the taxa.
The transition from species only showing a yeast phase, a presumable ancestral morphology to species also having a filamentous phase, co-occurs with ecological niche diversification and adaptations to various lifestyles and environments [60]. Exclusively filamentous species in vitro are found only in Heterogastridium, Pycnopulvinus (both Heterogastridiales), some genera of the Microbotryales, Camptobasidium (type species) and Psychromyces. It is unclear whether C. gelus or C. arcticum, showing only yeast growth in vitro, produce hyphae in their natural environment. However, it is rather likely that their life styles in nature are far more complex than can be predicted by in vitro studies. Occurrence of teliospores and clamped mycelium in P. glacialis, for example, implies that this species might produce basidia and basidiospores in nature, although these structures have not yet been observed on agar plates. Similarly, only a single P. glacialis strain displayed dimorphic characters while all other strains were strictly filamentous. Although the ecological role of this species is yet unresolved, numerous adaptations to cold environments have been already recognized [53].
This study shows that the glacial environments of Greenland and Svalbard harbour a high abundance of ecologically highly adapted species of the Microbotryomycetes. Additionally, a large yeast diversity remains uncharacterized, which should be investigated in future studies with novel isolation, cultivation and preservation approaches. These studies should also investigate species interactions, which might be particularly important in environments characterized by extremely low temperatures, lack of water and oligotrophic conditions. The presented results also underline the power of combining culture-dependent and independent strategies to assess yeast diversity and to progress towards unravelling the function of these yeasts in glacial ecosystems.
Description of Camptobasidium arcticum sp. nov. (Perini & Zalar)
Camptobasidium arcticum (arc'ti.cum. L. neut. adj. arcticum, northern, Arctic, referring to the geographical area the species was discovered).
MycoBank number: MB 834188
Streak cultures at 15 °C after 14 days on PDA mucoid, reddish white (8A2, 11A2), made of minute colonies. Colonies at 15 °C after 8 weeks on PDA 2–4 mm, either pastel pink (8A2) to pink (11A5) with glistening or rough surface with a wrinkled structure, almost entire margin in the glistening colony type, margin undulate in rough colony type (Fig. 3a–c). Individual yeast cells on PDA one- or two- celled, oblong, 5±2 (mean±SD; min-max: 3–9)×3±0,6 (mean±SD; min-max: 2–4) µm, often filled with inclusions (storage granules), some cells swell to round, up to 10 µm large structures, containing a single large vacuole (Fig. 3f–h). Yeasts are occurring singly or in clusters. Budding is predominantly uni- and bi-polar (Fig. 3g), but also with several loci at one pole (Fig. 3k). Pseudohyphae observed in some strains on OA (Fig. 3l). Sexual reproductive structures not observed in single or mixed cell assays.
Assimilated compounds: α-d-glucose, sucrose, trehalose, raffinose (delayed), maltose, melezitose, cellobiose, gentiobiose, maltotriose, stachyose (delayed), methyl β-d-glucoside, dextrin (sometimes delayed) and cadaverine (sometimes weak and delayed). Variable: melibiose, lactose, salicin, palatinose, d-xylose, maltitol, nitrate and l-lysine. The following compounds are not assimilated: d-galactose, starch, l-sorbose, l-arabinose, d-arabinose, d-ribose, d-glucosamine and nitrite. Additional data on C source assimilation obtained by Biology YT plates only are given in Table S2.
Does not grow in the presence of 10 %, 15 % NaCl or 50 % glucose.
Growth at 5, 10, 15 °C is positive; no growth is evident at 20 °C. Fermentation abilities absent.
The holotype, EXF-12713H, originated from clear ice in Greenland Ice Sheet, 60 km east of Kangerlussuaq, 67° 04′ 43″ N 49° 20′ 29″ W, in July 2017 by Laura Perini. It is permanently preserved in a metabolically inactive state at the Ex Culture Collection of the Infrastructural Centre Mycosmo (MRIC UL), Slovenia (www.ex-genebank.com) at the Department of Biology, Biotechnical Faculty, University of Ljubljana, Slovenia. Accession numbers of DNA sequences derived from ex-type strain EXF-12713, deposited into the above mentioned Ex Culture Collection: MK454798 (LSU), MN983248 (ITS), MT304813 (SSU), MT260394 (CYTB), MT260390 (TEF), MT260386 (RPB2).
Other examined strains: Greenland, isolated from supraglacial water (EXF-12522, EXF-12524, EXF-12689), from dark ice (EXF-12711), from clear ice (EXF-13086), August 2017, L. Perini.
Camptobasidium arcticum (Fig. 3) differed from C. gelus (Fig. 4) in several phenotypic characteristics, such as colony colour and morphology, colony and cell size. Moreover, the newly proposed C. arcticum differs from C. gelus in the ability of assimilation of melezitose, cellobiose, gentiobiose, maltotriose, methyl β-d-glucoside and dextrin, in the inability of assimilation of nitrite, in variable assimilation of nitrate, and in weak or no assimilation of l-lysine. Camptobasidium arcticum was isolated from all sampled environments in Greenland, with no evidence of a clear preference for a specific habitat. Camptobasidium gelus occurred in cryoconite, dark ice, clear ice, supraglacial water in Greenland and in subglacial ice and glacial meltwater in Svalbard. None of the species were recovered from snow. Camptobasidium arcticum was until now found only in Greenland samples of this study. Three identical LSU rDNA sequences (JQ768846, AB558448, AY040647) were deposited at NCBI. The first sequence (JQ768846) was named Basidiomycota sp. TP-Snow-Y73 by Shao and Ma and was isolated from glacier surface snow of the Tibetan plateau (PR China), but no publication has been linked to the strain. The second sequence (AB558448), named Basidiomycota sp. GU54, was produced by Uetake et al. [61], and was isolated from glacial surface ice and snow of the Gulkana glacier (Alaska).
Description of Psychromyces gen. nov. (Perini & Zalar)
Psychromyces (Psy.chro.my’ces. Gr. masc. ad. psychros cold; Gr. masc. n. mykes a fungus; N.L. masc. n. Psychromyces a fungus from the cold).
Belongs to phylum Basidiomycota, subphylum Pucciniomycotina, class Microbotryomycetes, family Camptobasidiaceae. The genus is circumscribed based on phylogenetic analyses shown in Figs 1 and 2, with close relationships to the genera Camptobasidium, Glaciozyma and Cryolevonia. Cultures pink, dimorphic or filamentous, initially yeast-like, after incubation filamentous with some or no remaining yeast colonies. Hyphae septate, clamp connections absent or present. Oblong intercalary teliospores produced after 1 month of incubation. Sexual reproduction in the form of germinating basidia from teliospores and basidiospores is not known, but clamped hyphae indicate its sexual reproduction. Assimilated C sources: α-d-glucose, sucrose, raffinose and melezitose. Assimilated N sources: nitrate, nitrite and cadaverine. Glucose is not fermented. Urease reaction is positive. Type species is Psychromyces glacialis (Perini & Zalar).
MycoBank accession number: MB 834189.
Type species: Psychromyces glacialis (Perini & Zalar).
Species accepted: Psychromyces glacialis (Perini & Zalar) MB 834190.
Description of Psychromyces glacialis sp. nov. (Perini & Zalar)
Psychromyces glacialis (gla.ci.a’lis. L. masc. adj. glacialis icy, frozen).
MycoBank accession number: MB 834190.
Streak cultures at 15 °C after 14 days on PDA agar mucoid, orange white (5A2), filamentous, 2–6 mm diameter, margin composed of fine filaments immersed into the medium (Fig. 5a). Synnemata-like dense bundles of hyphae form on initial inoculation points (Fig. 5c). Yeast colonies (Fig. 5b) visible after 3 weeks, attain 2 mm diameter after 7 weeks on PDA and MEA, 0.5 mm on SNA, pastel red (7A4), convex and mat. After 3 months on PDA at 15 °C, the filamentous colonies reach up to 40 mm diameter, become pale red to pastel red (7A3, 8B4), mat. Mycelium in some strains without clamps (EXF-13111), 1.5–2.0 µm wide (Fig. 5d), or with clamps (Fig. S6), 2–4 µm wide, branched (Fig. S6 d–j). After 14 days of incubation, numerous teliospores are produced intercalary (Figs 5d–g and S6 h, j–l). Teliospores oblong, (5.5–) 8×10 (–13) (mean; min-max: 5.5–13) µm. Budding occurring in yeast colonies polar or occasionally multilateral, sessile or on short or long denticles, and with sympodial proliferation. Individual yeast cells on MEA: 8.5±2 (mean±SD; min-max: 5.5–11)×(2.5) 3.5±0.5 (4.5) µm, on PDA: 7.5±3 (mean±SD; min-max: 5–12)×3.5±0.5 (mean±SD; min-max: 3–4) µm. Dalmau plate culture on CMA agar, 7 weeks of incubation: numerous teliospores on densely septated mycelium.
Fig. 5.
Morphology of Psychromyces glacialis. (a, b) cultures of Psychromyces glacialis EXF-13111 at 15 °C: (a) PDA, dimorphic culture after 3 weeks of incubation, (b) PDA, yeast culture grown from single yeast colony after 1 month of incubation, (c) PDA, filamentous culture grown from the margin of filamentous colony after 1 month of incubation; (d–g) teliospores grown on (d) CMA, Dalmau plate after 1 month of incubation in 60 % lactic acid, (e) PDA, margin of filamentous colony after 10 weeks of incubation in 60 % lactic acid, (f) PDA, yeast part of colony after 3 weeks of incubation, mounted in water, (g) PDA, filamentous colony after 3 weeks, coloured with calcofluor white; (h–n) yeast cells, (h, i, m, n) PDA, from dimorphic culture after 3 weeks of incubation, (h–i) mounted in water, (m–n) stained with calcofluor white (l) PDA, dimorphic culture in water after 1 month of growth on PDA, rudimentary pseudohyphae; (i–m) PDA, budding cells on short or long denticles, (n) PDA, yeast cells with inclusions.
Assimilated compounds: α-d-glucose, sucrose, raffinose, trehalose, maltose, melezitose, cellobiose (sometimes weak), nitrate, nitrite and cadaverine. Variable: salicin, l-sorbose, d-xylose, l-arabinose, d-glucosamine and l-lysine. Not assimilated compounds: melibiose, d-galactose, starch, d-arabinose, d-ribose and glycerol. Additional data on C source assimilation obtained by Biology YT plates only are given in Table S2. Does not grow in the presence of 10%, 15 % NaCl, 50 % glucose.
Growth at 5, 10, 15 °C is positive; no growth evident at 20 °C. Fermentation abilities absent.
The holotype, CBS 16467, originated from subglacial ice of Vestre Brøggerbreen glacier (78° 54′ 55″ N 11° 45′ 48″ E) in Norway, Svalbard, in July 2017 by Laura Perini. It is permanently preserved in a metabolically inactive state at the CBS Yeast Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands. The ex-type strain has been deposited in the Ex Culture Collection of the Infrastructural Centre Mycosmo (MRIC UL), Slovenia (www.ex-genebank.com) at the Department of Biology, Biotechnical Faculty, University of Ljubljana, Slovenia, as strain EXF-13111. Accession numbers of DNA sequences derived from type: MT301949 (LSU rDNA), MK671633 (ITS), MT248408 (SSU), MT260392 (CYTB), MT260389 (TEF), MW036268 (RPB2).
Other examined strains: Greenland, Greenland Ice Sheet, 60 km east of Kangerlussuaq, 67° 04′ 43″ N 49° 20′ 29″ W, isolated from dark ice (EXF-12419), isolated from cryoconite (EXF-12886, EXF-12454), July 2017, L. Perini; Norway, Svalbard, Midtre Lovénbreen glacier, isolated from subglacial ice (EXF-12984), SV, Vestre Brøggerbreen glacier, isolated from subglacial ice (EXF-12991), August 2017, L. Perini.
Supplementary Data
Funding information
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 675546. We further acknowledge support from the United Kingdom Natural Environment Research Council Consortium Grant ‘Black and Bloom’ (NE/M021025/1). We acknowledge the financial support from the Slovenian Research Agency to the Infrastructural Centre Mycosmo (MRIC UL), to the research programmes ARRS P1-0170 and ARRS P1-0198, and projects ARRS J4-2549 and J7-1815.
Acknowledgements
The authors are grateful to Barbara Kastelic-Bokal and Mojca Matul for their technical assistance. We thank the Ny-Ålesund UK-NERC station for providing accommodation and laboratory facilities. We sincerely thank Malcom Airey for his logistic support during the Svalbard sampling campaign.
Author contributions
Conceptualization, L.P., C.G., N.G.-C., P.Z.; investigation, L.P., K.A., C.G., P.Z.; supervision, C.G., N.G.-C., P.Z.; validation, C.G., P.Z.; writing – original draft, L.P., P.Z.; writing – review and editing, L.P., C.G., N.G.-C., P. Z.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CBS, Culture Collection of the Westerdijk Fungal Biodiversity Institute; CYTB, cytochrome b; EXF, abbreviation for fungal strains preserved in Ex Culture Collection of the Infrastructural Centre Mycosmo (MRIC UL), Slovenia (www.ex-genebank.com); ITS, internal transcribed spacer; LSU, D1/D2 domain of the large subunit (28S) rRNA gene; RPB1, the largest subunit of DNA polymerase II; RPB2, the second largest subunit of DNA polymerase II; SSU, small subunit rRNA gene; TEF, translation elongation factor 1-α.
Two supplementary tables and six supplementary figures are available with the online version of this article.
References
- 1.Buzzini P, Turk M, Perini L, Turchetti B, Gunde-Cimerman N. Yeasts in polar and subpolar habitats. In: Buzzini P, Lachance MA, Yurkov A, editors. Yeasts in Natural Ecosystems: Diversity. Cham: Springer; 2017. pp. 331–365. [Google Scholar]
- 2.Yurkov A. Temporal and geographic patterns in yeast distribution. In: Buzzini P, Lachance MA, Yurkov A, editors. Yeasts in Natural Ecosystems: Ecology. Cham: Springer; 2017. pp. 101–130. [Google Scholar]
- 3.Gostinčar C, Turk M, Trbuha T, Vaupotič T, Plemenitaš A, et al. Expression of fatty-acid-modifying enzymes in the halotolerant black yeast Aureobasidium pullulans (de Bary) G. Arnaud under salt stress. Stud Mycol. 2008;61:51–59. doi: 10.3114/sim.2008.61.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon C, Wiezer A, Strittmatter AW, Daniel R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl Environ Microbiol. 2009;75:7519–7526. doi: 10.1128/AEM.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pathan AAK, Bhadra B, Begum Z, Shivaji S. Diversity of yeasts from puddles in the vicinity of midre lovénbreen glacier, Arctic and bioprospecting for enzymes and fatty acids. Curr Microbiol. 2010;60:307–314. doi: 10.1007/s00284-009-9543-3. [DOI] [PubMed] [Google Scholar]
- 6.Buzzini P, Margesin R. Cold-adapted yeasts: a lesson from the cold and a challenge for the XXI century. In: Buzzini P, Margesin R, editors. Cold-adapted yeasts: Biodiversity, Adaptation Strategies and Biotechnological Significance. Berlin, Heidelberg: Springer; 2014. pp. 3–22. [Google Scholar]
- 7.Kim HJ, Shim HE, Lee JH, Kang Y-C, Hur YB. Ice-binding protein derived from Glaciozyma can improve the viability of cryopreserved mammalian cells. J Microbiol Biotechnol. 2015;25:1989–1996. doi: 10.4014/jmb.1507.07041. [DOI] [PubMed] [Google Scholar]
- 8.Alcaíno J, Cifuentes V, Baeza M. Physiological adaptations of yeasts living in cold environments and their potential applications. World J Microbiol Biotechnol. 2015;31:1467–1473. doi: 10.1007/s11274-015-1900-8. [DOI] [PubMed] [Google Scholar]
- 9.Butinar L, Spencer-Martins I, Gunde-Cimerman N. Yeasts in high Arctic glaciers: the discovery of a new habitat for eukaryotic microorganisms. Antonie van Leeuwenhoek. 2007;91:277–289. doi: 10.1007/s10482-006-9117-3. [DOI] [PubMed] [Google Scholar]
- 10.Arenz BE, Blanchette RA. Distribution and abundance of soil fungi in Antarctica at sites on the Peninsula, Ross sea region and McMurdo dry Valleys. Soil Biology and Biochemistry. 2011;43:308–315. doi: 10.1016/j.soilbio.2010.10.016. [DOI] [Google Scholar]
- 11.de Garcia V, Zalar P, Brizzio S, Gunde-Cimerman N, van Broock M. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiol Ecol. 2012;82:523–539. doi: 10.1111/j.1574-6941.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- 12.Edwards A, Douglas B, Anesio AM, Rassner SM, Irvine-Fynn TDL, et al. A distinctive fungal community inhabiting cryoconite holes on glaciers in Svalbard. Fungal Ecol. 2013;6:168–176. doi: 10.1016/j.funeco.2012.11.001. [DOI] [Google Scholar]
- 13.Luo B, Sun H, Zhang Y, Gu Y, Yan W, et al. Habitat-specificity and diversity of culturable cold-adapted yeasts of a cold-based glacier in the Tianshan Mountains, northwestern China. Appl Microbiol Biotechnol. 2019;103:2311–2327. doi: 10.1007/s00253-018-9512-5. [DOI] [PubMed] [Google Scholar]
- 14.Duo Saito RA, Connell L, Rodriguez R, Redman R, Libkind D, et al. Metabarcoding analysis of the fungal biodiversity associated with Castaño Overa glacier – Mount Tronador, Patagonia, Argentina. Fungal Ecol. 2018;36:8–16. doi: 10.1016/j.funeco.2018.07.006. [DOI] [Google Scholar]
- 15.Perini L, Gostinčar C, Anesio AM, Williamson C, Tranter M, et al. Darkening of the Greenland ice sheet: fungal abundance and diversity are associated with algal Bloom. Front Microbiol. 2019;10:557. doi: 10.3389/fmicb.2019.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perini L, Gostinčar C, Gunde-Cimerman N. Fungal and bacterial diversity of Svalbard subglacial ice. Sci Rep. 2019;9:20230. doi: 10.1038/s41598-019-56290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pontes A, Ruethi J, Frey B, Aires A, Thomas A, et al. Cryolevonia gen. nov. and Cryolevonia schafbergensis sp. nov., a cryophilic yeast from ancient permafrost and melted sea ice. Int J Syst Evol Microbiol. 2020;70:2334–2338. doi: 10.1099/ijsem.0.004040. [DOI] [PubMed] [Google Scholar]
- 18.de Garcia V, Trochine A, Uetake J, Bellora N, Libkind D. Novel yeast taxa from the cold: description of Cryolevonia giraudoae sp. nov. and Camptobasidium gelus sp. nov. Int J Syst Evol Microbiol. 2020;70:3711–3717. doi: 10.1099/ijsem.0.004223. [DOI] [PubMed] [Google Scholar]
- 19.Li A-H, Yuan F-X, Groenewald M, Bensch K, Yurkov AM, et al. Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud Mycol. 2020;96:17–140. doi: 10.1016/j.simyco.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin M-xiu, Zhou P-jin. Mrakia psychrophila sp. nov., a new species isolated from Antarctic soil. J Zhejiang Univ Sci B. 2007;8:260–265. doi: 10.1631/jzus.2007.B0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas-Hall SR, Turchetti B, Buzzini P, Branda E, Boekhout T, et al. Cold-adapted yeasts from Antarctica and the Italian Alps-description of three novel species: Mrakia robertii sp. nov., Mrakia blollopis sp. nov. and Mrakiella niccombsii sp. nov. Extremophiles. 2010;14:47–59. doi: 10.1007/s00792-009-0286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vishniac HS, Takashima M. Rhodotorula arctica sp. nov., a basidiomycetous yeast from Arctic soil. Int J Syst Evol Microbiol. 2010;60:1215–1218. doi: 10.1099/ijs.0.014910-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q-M, Yurkov AM, Göker M, Lumbsch HT, Leavitt SD, et al. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina . Stud Mycol. 2015;81:149–189. doi: 10.1016/j.simyco.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turchetti B, Thomas Hall SR, Connell LB, Branda E, Buzzini P, et al. Psychrophilic yeasts from Antarctica and European glaciers: description of Glaciozyma gen. nov., Glaciozyma martinii sp. nov. and Glaciozyma watsonii sp. nov. Extremophiles. 2011;15:573–586. doi: 10.1007/s00792-011-0388-x. [DOI] [PubMed] [Google Scholar]
- 25.Kachalkin AV. Yeasts of the White Sea intertidal zone and description of Glaciozyma litorale sp. nov. Antonie van Leeuwenhoek. 2014;105:1073–1083. doi: 10.1007/s10482-014-0165-9. [DOI] [PubMed] [Google Scholar]
- 26.Toome M, Roberson RW, Aime MC. Meredithblackwellia eburnea gen. et sp. nov., Kriegeriaceae fam. nov. and Kriegeriales ord. nov.--toward resolving higher-level classification in Microbotryomycetes . Mycologia. 2013;105:486–495. doi: 10.3852/12-251. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q-M, Groenewald M, Takashima M, Theelen B, Han P-J, et al. Phylogeny of yeasts and related filamentous fungi within Pucciniomycotina determined from multigene sequence analyses. Stud Mycol. 2015;81:27–53. doi: 10.1016/j.simyco.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachalkin AV, Turchetti B, Inácio J, Carvalho C, Mašínová T, et al. Rare and undersampled dimorphic basidiomycetes. Mycol Prog. 2019;18:945–971. doi: 10.1007/s11557-019-01491-5. [DOI] [Google Scholar]
- 29.Box JE, Fettweis X, Stroeve JC, Tedesco M, Hall DK, et al. Greenland ice sheet albedo feedback: thermodynamics and atmospheric drivers. The Cryosphere. 2012;6:821–839. doi: 10.5194/tc-6-821-2012. [DOI] [Google Scholar]
- 30.de Vries RP, Burgers K, van de Vondervoort PJI, Frisvad JC, Samson RA, et al. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl Environ Microbiol. 2004;70:3954–3959. doi: 10.1128/AEM.70.7.3954-3959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nirenberg HI. A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany. 1981;59:1599–1609. doi: 10.1139/b81-217. [DOI] [Google Scholar]
- 32.Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/AEM.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King AD, Hocking AD, Pitt JI. Dichloran-rose Bengal medium for enumeration and isolation of molds from foods. Appl Environ Microbiol. 1979;37:959–964. doi: 10.1128/AEM.37.5.959-964.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hocking AD, Pitt JI. Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Appl Environ Microbiol. 1980;39:488–492. doi: 10.1128/AEM.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beever RE, Bollard EG. The nature of the stimulation of fungal growth by potato extract. Microbiology. 1970;60:273–279. [Google Scholar]
- 36.Küster E. Outline of a comparative study of criteria used in characterization of the actinomycetes. International Bulletin of Bacteriological Nomenclature and Taxonomy. 1959;9:97–104. doi: 10.1099/0096266X-9-2-97. [DOI] [Google Scholar]
- 37.Pollack JD, Benham RW. The chlamydospores of Candida albicans: comparison of three media for their induction. J Lab Clin Med. 1957;50:313–317. [PubMed] [Google Scholar]
- 38.Kornerup A, Wanscher JH. Methuen Handbook of Colour. 2nd ed. London, UK: Methuen; 1967. [Google Scholar]
- 39.Kurtzman CP, Fell JW, Boekhout T, Robert V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a taxonomic study. 5th ed. Amsterdam: Elsevier; 2011. pp. 87–110. [Google Scholar]
- 40.Gerrits van den Ende AHG, de Hoog GS. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana . Stud Mycol. 1999;43:151–162. [Google Scholar]
- 41.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 42.Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear its and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 43.Wang Q-M, Bai F-Y. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rRNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Res. 2008;8:799–814. doi: 10.1111/j.1567-1364.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 49.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamvar ZN, Brooks JC, Grünwald NJ. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet. 2015;6:208. doi: 10.3389/fgene.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 52.R Development Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2020. https://www.r-project.org
- 53.Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, et al. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for illumina amplicon sequencing. Appl Environ Microbiol. 2016;82:7217–7226. doi: 10.1128/AEM.02576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, et al. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints. 2018;Dec 3 doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uetake J, Kohshima S, Nakazawa F, Takeuchi N, Fujita K, et al. Evidence for propagation of cold-adapted yeast in an ice core from a Siberian Altai glacier. J Geophys Res. 2011;116 doi: 10.1029/2010JG001337. [DOI] [Google Scholar]
- 56.Marvanová L, Suberkropp K. Camptobasidium hydrophilum and its anamorph, Crucella subtilis: a new heterobasidiomycete from streams. Mycologia. 1990;82:208–217. doi: 10.1080/00275514.1990.12025866. [DOI] [Google Scholar]
- 57.Singh P, Singh SM, Tsuji M, Prasad GS, Hoshino T. Rhodotorula svalbardensis sp. nov., a novel yeast species isolated from cryoconite holes of Ny-Ålesund, Arctic. Cryobiology. 2014;68:122–128. doi: 10.1016/j.cryobiol.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Turland NJ, Wiersema JH, Barrie FR, Greuter W, et al. International Code of Nomenclature for algae, fungi, and plants (Shenzhen code): adopted by the nineteenth international botanical Congress Shenzhen, China, July 2017. Glashütten: Koeltz botanical books. 2018.
- 59.Kurtzman CP, Fell JW, Boekhout T, Robert V. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam: Elsevier; 2011. [Google Scholar]
- 60.Oberwinkler F. Yeasts in Pucciniomycotina . Mycol Progress. 2017;16:831–856. doi: 10.1007/s11557-017-1327-8. [DOI] [Google Scholar]
- 61.Uetake J, Yoshimura Y, Nagatsuka N, Kanda H. Isolation of oligotrophic yeasts from supraglacial environments of different altitude on the Gulkana glacier (Alaska) FEMS Microbiol Ecol. 2012;82:279–286. doi: 10.1111/j.1574-6941.2012.01323.x. [DOI] [PubMed] [Google Scholar]
- 62.Kurtzman CP, Fell JW, Boekhout T, Robert V. Leucosporidium antarcticum Fell, Statzell, I.L. Hunter & Phaff. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam: Elsevier; 2011. pp. 1487–1488. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.