Significance
Polyhydroxyalkanoates (PHAs) are bacterial polyesters considered as sustainable and environmentally friendly substitutes for petroleum-based plastics. Despite great advances in PHA research, spatiotemporal characteristics of PHA granule formation and distribution in a live bacterial cell are not well understood. Here, we report the results of a three-dimensional (3D) analysis of live bacterial cells accumulating poly(3-hydroxybutyrate) (PHB), the best-studied member of PHAs. Optical diffraction tomography was applied for visualizing individual cells and in vivo PHB granules and quantifying their physical properties such as weight, volume, density, and localization by measuring 3D refractive index distributions. Comparative analyses of native and recombinant PHB-producing strains revealed distinctive characteristics of PHB accumulation, providing insights into spatiotemporal localization of PHA granules in vivo.
Keywords: polyhydroxyalkanoate, biopolymer, optical diffraction tomography, microorganism, three-dimensional imaging
Abstract
Polyhydroxyalkanoates (PHAs) are biodegradable polyesters that are intracellularly accumulated as distinct insoluble granules by various microorganisms. PHAs have attracted much attention as sustainable substitutes for petroleum-based plastics. However, the formation of PHA granules and their characteristics, such as localization, volume, weight, and density of granules, in an individual live bacterial cell are not well understood. Here, we report the results of three-dimensional (3D) quantitative label-free analysis of PHA granules in individual live bacterial cells through measuring the refractive index distributions by optical diffraction tomography (ODT). The formation and growth of PHA granules in the cells of Cupriavidus necator, the best-studied native PHA producer, and recombinant Escherichia coli harboring C. necator poly(3-hydroxybutyrate) (PHB) biosynthesis pathway are comparatively examined. Through the statistical ODT analyses of the bacterial cells, the distinctive characteristics for density and localization of PHB granules in vivo could be observed. The PHB granules in recombinant E. coli show higher density and localization polarity compared with those of C. necator, indicating that polymer chains are more densely packed and granules tend to be located at the cell poles, respectively. The cells were investigated in more detail through real-time 3D analyses, showing how differently PHA granules are processed in relation to the cell division process in native and nonnative PHA-producing strains. We also show that PHA granule–associated protein PhaM of C. necator plays a key role in making these differences between C. necator and recombinant E. coli strains. This study provides spatiotemporal insights into PHA accumulation inside the native and recombinant bacterial cells.
Due to our increasing concerns on the accumulation of nondegradable plastic wastes, polyhydroxyalkanoates (PHAs), a family of bacterial polyesters, have emerged as a promising alternative to synthetic plastics. PHAs have been attracting much attention because they are biodegradable and possess material properties that are comparable to those of popular synthetic plastics, such as polypropylene and polyethylene (1–4).
PHAs are synthesized by numerous bacteria as an energy, carbon, and redox storage material under unbalanced growth conditions in the presence of an excess carbon source. PHAs exist in the form of insoluble granules in the cytoplasm. PHA granules in vivo are amorphous polymers bound by several different enzymes and proteins, such as PHA synthase, PHA depolymerase, regulatory proteins, and other granule-associated proteins, known as phasin and PhaM (5, 6). Through the actions of these enzymes and proteins, microorganisms synthesize and degrade PHAs in response to the environmental condition. For example, Cupriavidus necator (also known as Ralstonia eutropha), which is the model organism for studying poly(3-hydroxybutyrate) (PHB) biosynthesis, accumulates PHB under an unfavorable growth condition, such as a nitrogen- or phosphorus-deficient condition, to store the excess carbon sources inside cells (1, 2). Cells degrade PHB and utilize it for growth and maintenance when needed under balanced growth conditions (1, 2).
It has always been amazing to observe cells accumulating such large amounts of polymers inside the cells. Several techniques have been used to observe the dynamic behavior and characteristics of PHA granules in vivo. Fluorescence microscopy (7–9), transmission electron microscopy (TEM) (10, 11), and electron cryotomography (12) have provided information on the shape and localization of PHA granules in bacteria, such as C. necator, Rhodospirillum rubrum, and recombinant Escherichia coli expressing the heterologous PHA biosynthesis genes, under various conditions. However, these studies have provided only two-dimensional image information and/or showed a nonnative state of the cells and PHA granules due to the pretreatment process used (more explanations are provided in SI Appendix, Text S1). Although several models for granule formation have been proposed based on these observations (6, 12–14), many of the details for the formation and characteristics of in vivo PHA granules are still unknown.
Here, we present the results of three-dimensional (3D) quantitative analyses for PHA granules in individual live bacterial cells by measuring the refractive index (RI) distribution using optical diffraction tomography (ODT). Formation of PHB granules in the cells of C. necator and recombinant E. coli harboring C. necator PHB biosynthesis genes was monitored with culture time to understand the spatiotemporal formation, growth, and distribution of PHB granules during cell growth. Based on the reconstructed 3D RI, the volumes and weights of cells and PHB granules, PHB content, and PHB granule density could be quantified at a single-cell level. Also, the variations of PHB formation in the individual cells could be obtained through statistical analyses. In addition, we present real-time movies of PHB accumulation in living C. necator and recombinant E. coli cells. These results reveal the distinctive characteristics of in vivo PHA granule formation and distribution in native and nonnative PHA producers.
Results and Discussion
Identification of PHB Granules via Optical Diffraction Tomography.
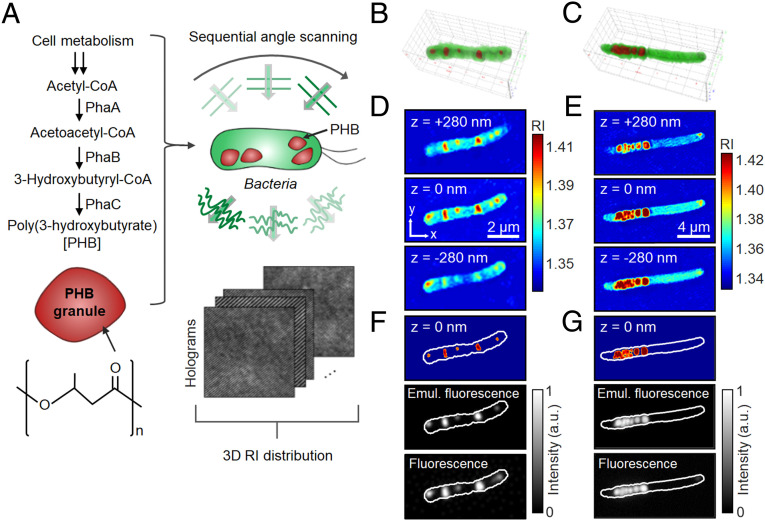
Cells of C. necator and recombinant E. coli accumulating PHB were subjected to ODT analysis (Fig. 1A and SI Appendix, Materials and Methods and Figs. S1 and S2 and Table S1). For the accumulation of PHB, C. necator was first cultured in a nutrient-rich medium, washed with distilled water, and transferred to a nitrogen-limiting medium containing 20 g/L of fructose. On the other hand, recombinant E. coli was cultured under a nutrient-rich condition in the presence of 20 g/L of glucose (SI Appendix, Materials and Methods). In the reconstructed 3D RI distributions of C. necator and recombinant E. coli cells, distinctively high RI regions inside each cell were observed as PHB granules (Fig. 1 B–E). To verify that the high RI regions correspond to the accumulated PHB, the RI tomogram was compared with the fluorescence image of the same cell labeled with Nile Red, which has routinely been used for PHA staining (7). For better comparison, fluorescence-like images were computationally generated from 3D RI distribution by assuming that the granules stained homogeneously (Fig. 1 F and G). When the threshold RI for PHB was set above 1.39, the emulated fluorescence images from 3D RI distribution were consistent with the granule location, morphology, and the signal intensity observed with the actual fluorescence images obtained using Nile Red. Based on the comparison of 3D RI tomograms and conventional fluorescence images, it could be concluded that the regions of RI > 1.39 in the 3D RI distribution correspond to PHB granules. In this study, all quantifications for PHA (PHA granule volume, weight, density, and polymer content) were performed on the region of RI > 1.39.
Fig. 1.
ODT analysis for bacterial cells accumulating PHB. (A) Schematic illustration of the ODT analysis. PHB biosynthesis pathway and the chemical structure (Left). Two-dimensional holograms of a bacterial cell are recorded at various illumination angles to reconstruct the 3D RI distribution of the cell (Right). (B and C) 3D-rendered images of the reconstructed RI distribution of C. necator and recombinant E. coli cells, respectively. (D and E) Cross-sectional images of the corresponding 3D RI distributions. (F and G) PHB identification based on the RI threshold (RI > 1.39) (Top), emulated fluorescence-like images obtained by 3D convolution-based imaging processing (Middle), and wide-field fluorescence images of Nile Red–stained cells (Bottom).
In addition to PHB, bacterial cells accumulating different types of PHAs, poly(3-hydroxybutyrate-co-3-hydroxyvalerate), poly(3-hydroxybutyrate-co-lactate), and medium-chain-length PHA, were subjected to ODT analysis. For all the three types of PHAs, PHA granules were identified as distinctively high RI regions as well as the PHB granules (SI Appendix, Fig. S3). Although we focus on the PHB-producing bacterial cells here, the ODT analyses can be widely used for the cells accumulating various types of PHAs.
Quantitative Characterization of Bacterial Cells and In Vivo PHB Granules.
To understand the formation, growth, and distribution of PHB granules during cell growth, the characteristics of individual cells and cytoplasmic PHB were quantitatively analyzed according to culture time. C. necator and recombinant E. coli cells were cultured for 24 h and 48 h, respectively, to reach the maximum PHB accumulation and subjected to ODT at 8 h intervals (Figs. 2 and 3). Representative 3D RI distributions of C. necator and recombinant E. coli cells during culture time are presented in Figs. 2 A and B and 3 A and B. For both C. necator and recombinant E. coli, the changes of cell morphologies and accumulation of PHB granules according to the culture time were clearly observed.
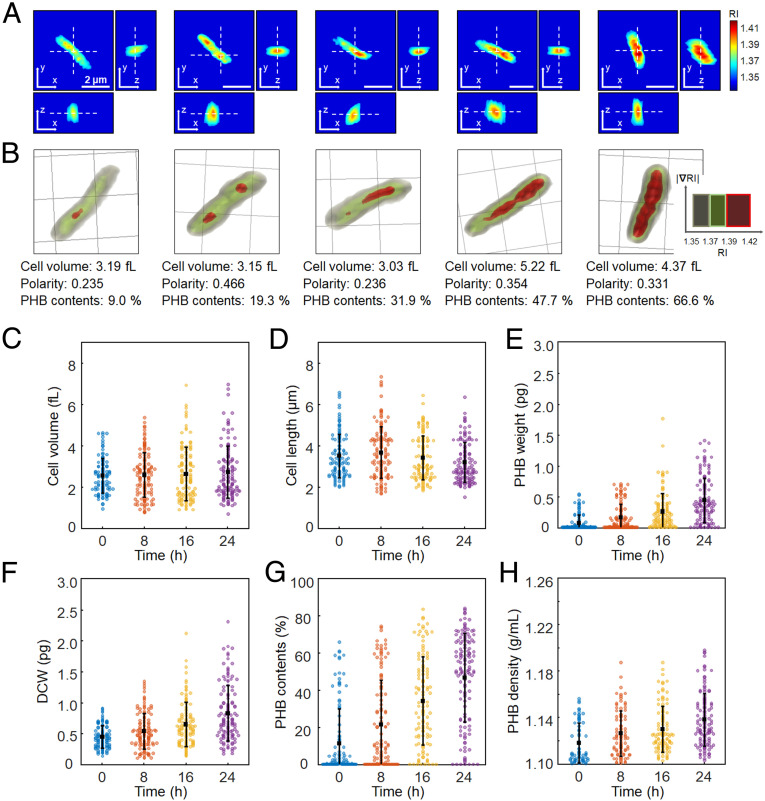
Fig. 2.
Quantification of individual C. necator cells accumulating PHB granules. (A) Representative 3D RI tomograms at selected x-y, y-z, and z-x planes and (B) 3D rendering images of C. necator cells with different levels of PHB accumulation. One grid equals 2 μm. The quantified cell volume, polarity, and PHB content (%, wt/wt) are noted below the rendering images. (C) Cell volume (fL). (D) Cell length (μm). (E) PHB granule weight (pg). (F) DCW (pg). (G) PHB content (%, wt/wt). (H) PHB granule density (g/mL). The bars indicate averages and SDs. The numbers of analyzed cells are 121, 126, 120, and 136 for 0, 8, 16, and 24 h, respectively.
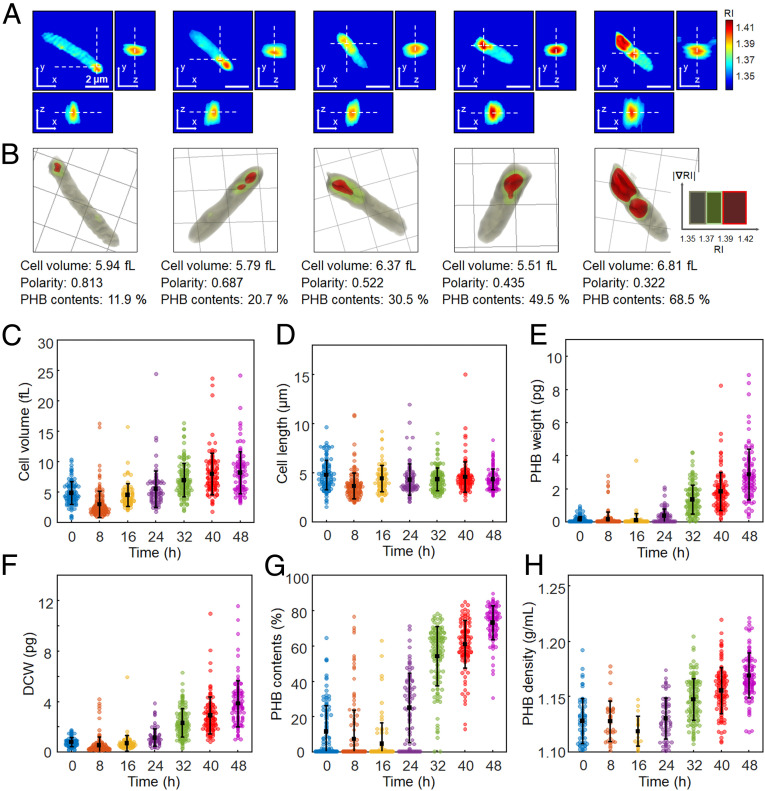
Fig. 3.
Quantification of individual recombinant E. coli cells accumulating PHB granules. (A) Representative 3D RI tomograms at selected x-y, y-z, and z-x planes and (B) 3D rendering images of E. coli cells with different levels of PHB accumulation. One grid equals 2 μm. The quantified cell volume, polarity, and PHB content (%, wt/wt) are noted below the rendering images. (C) Cell volume (fL). (D) Cell length (μm). (E) PHB granule weight (pg). (F) DCW (pg). (G) PHB content (%, wt/wt). (H) PHB granule density (g/mL). The bars indicate averages and SDs. The numbers of analyzed cells are 112, 151, 87, 85, 135, 108, and 103 for 0, 8, 16, 24, 32, 40, and 48 h, respectively.
For statistical analysis, 121, 126, 120, and 136 cells of C. necator were randomly selected at 0, 8, 16, and 24 h of cultivation, respectively. Cell volume and length, dry cell weight (DCW), PHB granule weight, PHB content (the accumulated PHB granule weight to DCW, wt/wt%), and PHB granule density were able to be quantified for the individual cells (Fig. 2). The detailed description on quantification is provided in SI Appendix, Materials and Methods. The average volume of C. necator cells showed an increasing trend with culture time, which were 2.52 ± 0.84, 2.56 ± 1.07, 2.61 ± 1.30, and 2.70 ± 1.27 fL (mean ± SD) at 0, 8, 16, and 24 h, respectively (Fig. 2C). The average values of cell length were 3.47 ± 1.05, 3.63 ± 1.25, 3.38 ± 1.06, 3.15 ± 0.96 µm at the same time points, respectively (Fig. 2D). As C. necator cells actively accumulated PHB, the average values of PHB granule weight increased with culture time, which were 0.070 ± 0.130, 0.157 ± 0.211, 0.255 ± 0.289, and 0.438 ± 0.359 pg at 0, 8, 16, and 24 h, respectively (Fig. 2E). According to the changes of accumulated PHB granule weight and cell volume, the average DCW continued to increase; the average values of DCW were 0.44 ± 0.18, 0.53 ± 0.29, 0.64 ± 0.36, and 0.82 ± 0.45 pg at the same time points, respectively (Fig. 2F). The average PHB contents also increased, which were 11.05 ± 18.59 (0 h), 21.07 ± 23.80 (8 h), 33.77 ± 23.72 (16 h), and 46.30 ± 23.81% (24 h) (Fig. 2G). For comparison, PHB contents were also calculated by gas chromatography analysis, which is a prevalent method for determining PHB concentration and content (SI Appendix, Text S2 and Table S2). The average PHB granule densities were 1.118 ± 0.017, 1.126 ± 0.019, 1.129 ± 0.020, and 1.138 ± 0.023 g/mL at 0, 8, 16, and 24 h, respectively (Fig. 2H).
For recombinant E. coli, 112, 151, 87, 85, 135, 108, and 103 cells were randomly selected for ODT analysis at 0, 8, 16, 24, 32, 40, and 48 h of cultivation, respectively (Fig. 3). After the initial 8 h of culture, the average volumes of the recombinant E. coli cells continued to increase, which were 4.71 ± 1.89, 2.85 ± 2.21, 4.38 ± 1.90, 5.39 ± 3.00, 6.85 ± 2.75, 7.88 ± 3.42, and 8.07 ± 3.49 fL at 0, 8, 16, 24, 32, 40, and 48 h, respectively (Fig. 3C). The average cell volume at 48 h was increased by 183% of the average value at 8 h. On the other hand, the cell lengths showed almost constant average values with no significant change with culture time. The average values of cell length were 4.71 ± 1.51, 3.57 ± 1.31, 4.33 ± 1.35, 4.22 ± 1.61, 4.25 ± 1.17, 4.49 ± 1.57, and 4.26 ± 1.07 µm at the same time points, respectively (Fig. 3D). The recombinant E. coli cells became thicker with culture time. The cross-sectional area of the cells was significantly increased; the average values of the cross-sectional area at 8 and 48 h were 1.16 and 2.41 µm2, respectively. As expected from the increase of cell volume, the average values of DCW also increased with culture time after 8 h, which were 0.73 ± 0.36, 0.46 ± 0.68, 0.65 ± 0.62, 1.10 ± 0.68, 2.25 ± 1.13, 2.86 ± 1.46, and 3.78 ± 1.83 pg at 0, 8, 16, 24, 32, 40, and 48 h, respectively (Fig. 3F). The change of the DCW is more remarkable compared to the change of cell volume because the PHB granule weights were rapidly increased after 16 h; the average weights of PHB granules were 0.117 ± 0.187, 0.124 ± 0.443, 0.079 ± 0.409, 0.336 ± 0.411, 1.330 ± 0.869, 1.811 ± 1.151, and 2.830 ± 1.519 pg at the same time points, respectively (Fig. 3E). The average PHB contents also gradually increased with time after 16 h: 11.23 ± 14.85, 7.10 ± 16.37, 4.39 ± 11.72, 24.63 ± 19.59, 53.80 ± 16.75, 60.65 ± 13.64, and 72.83 ± 9.68% at 0, 8, 16, 24, 32, 40, and 48 h, respectively (Fig. 3G). The average values of PHB granule density were also increased after 16 h, which were 1.127 ± 0.020, 1.127 ± 0.018, 1.118 ± 0.013, 1.130 ± 0.019, 1.147 ± 0.019, 1.155 ± 0.021, and 1.168 ± 0.020 g/mL at the same time points, respectively (Fig. 3H).
One unanticipated finding was the decreased average values of PHB granule weight and content at the early stage of culture (0 to 16 h). Through the real-time analysis of the E. coli cells (Movie S1 and further discussed in Real-Time Analysis), we observed that the E. coli cells did not actively accumulate PHB granules at that time point while cells grew in number by cell division, resulting in the decrease of the average values.
Morphological Characterization of In Vivo PHB Granules.
In addition to the PHB granule corresponding to the region of RI above 1.39, we focused on the RI region below 1.39 to observe the granule formation in more detail. It seems that the RI region of 1.37 to 1.39 also represents PHB, distinguishable from the cytoplasmic space (Figs. 2B and 3B and SI Appendix, Fig. S4), as an RI region of above 1.37 gradually began to appear in cells and became granules as time goes by (SI Appendix, Fig. S4). Thus, we assume that the RI region of 1.37 to 1.39 represents the insoluble polymeric chains that did not yet form into the granule and also the relatively mobile surface region of the PHB granule in which several enzymes and proteins are associated. It was further verified by comparing the ODT results with the fluorescence microscopy analysis of the in vivo PHB granules labeled with superfolder green fluorescent protein (sfGFP) (SI Appendix, Text S3 and Fig. S5).
It was found that the distributions of RI below 1.39 were significantly different between the two strains. For C. necator cells, the RI region of 1.37 to 1.39 (the green-colored region in Fig. 2B) was distributed widely around the PHB granules that covered most of the cytoplasm, while the RI region of 1.37 to 1.39 for recombinant E. coli cells (the green-colored region in Fig. 3B) was shown in a relatively small region near the PHB granules (SI Appendix, Fig. S6). Such RI distributions reveal that the in vivo PHB morphology and distribution are different between C. necator and recombinant E. coli cells. In C. necator cells, PHB exists in the form of granules and the mobile polymeric chains which are relatively loosely distributed (Fig. 2B). In contrast, PHB in E. coli cells tends to be concentrated, which mostly leads to the formation of granules (Fig. 3B). This difference is reflected as an observation that the E. coli cells contained more PHB granules of higher density compared with C. necator cells as discussed above.
Correlative Analysis.
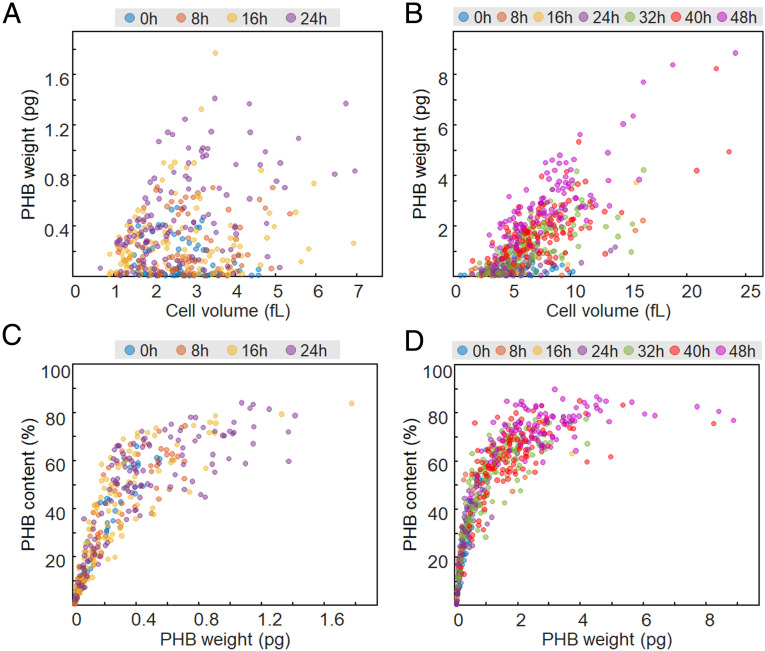
To examine and capture the distinctive features of individual cells and accumulated PHB granules, correlative analyses were performed using the individual cell data obtained. Given that C. necator and recombinant E. coli cells showed the increase of average cell volume with culture time (Figs. 2 and 3), the dependence of cell size on PHB accumulation was evaluated by making the scatter plots of PHB weight (or PHB content) versus cell volume (Fig. 4 A and B).
Fig. 4.
Scatter plots of (A and B) PHB weight versus cell volume and (C and D) PHB content versus PHB granule weight for C. necator and recombinant E. coli cells, respectively.
Differently from C. necator (Fig. 4A), the scatter plot of PHB weight versus cell volume for recombinant E. coli cells (Fig. 4B) revealed a partially linear relationship with the correlation coefficients of 0.72, 0.86, 0.52, 0.67, 0.70, and 0.83 for 8, 16, 24, 32, 40, and 48 h, respectively. These results imply that in vivo PHB accumulation affects the volume of the E. coli cell. The cross-sectional area of the E. coli cells showed a tendency to increase as PHB granules were accumulated that was prominent for the cells with more than 50% of PHB content (SI Appendix, Fig. S7).
Interestingly, the scatter plots for PHB content versus PHB weight exhibited saturation-like curves for both C. necator and recombinant E. coli cells (Fig. 4 C and D). At low PHB weights, the PHB contents increased with PHB weight, as expected. When the PHB content reached a certain value, however, it no longer increased and plateaued with a further increase of PHB weight. Under the experimental condition we used, PHB contents of 83% (C. necator) and 89% (E. coli) were observed as plateau values. These results may be because bacteria accumulate PHA by maintaining their viability; cells need to retain a certain intracellular space for metabolic and physiological activities other than PHB accumulation.
PHB Granule Localization Analysis.
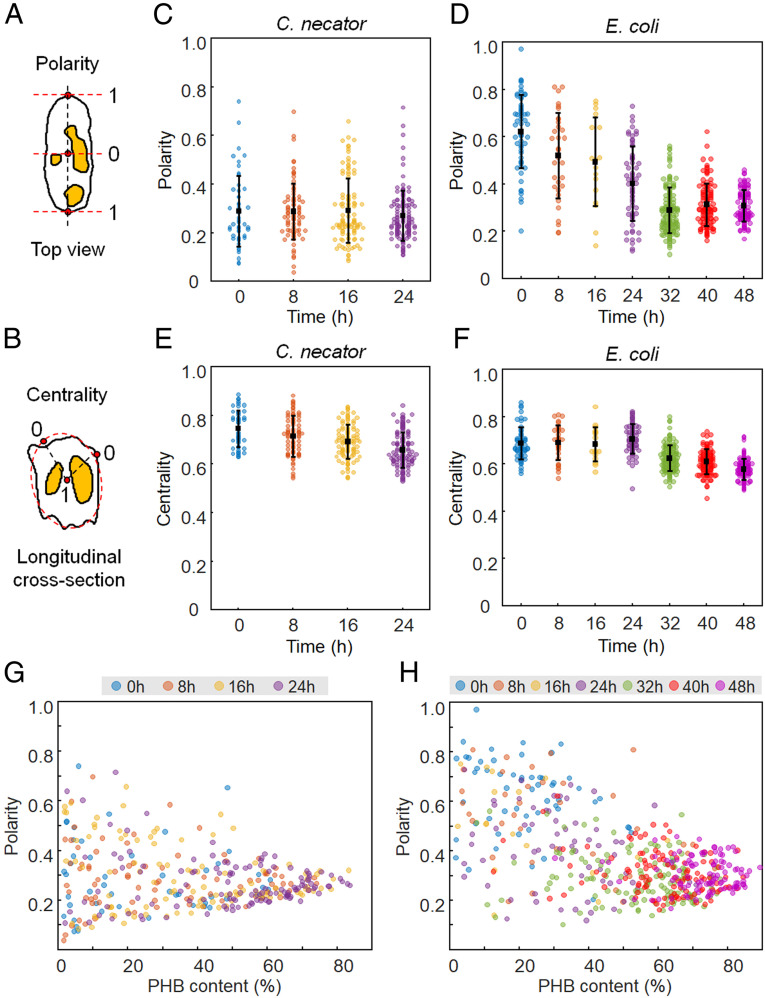
In addition to quantitative analysis, the spatial distribution of PHB granules in individual cells was investigated based on the 3D RI distribution. The relative positions of the in vivo PHB granules were presented with two parameters, polarity and centrality (Fig. 5), as previously described (12). Polarity is defined as the ratio of the axial distance between the cell center and the PHB granule to the distance between the cell center and cell pole, corresponding to a half of the cell length (Fig. 5A). Centrality is defined as the ratio of the distance of the PHB granule from the cell membrane to the distance between the long center axis and the cell membrane at each cross-section (Fig. 5B). Polarity and centrality have a range of 0 to 1 and higher polarity and centrality indicate that the PHB granules are located near the pole and the long center axis of the cell, respectively. Details of localization analysis are described in SI Appendix, Materials and Methods.
Fig. 5.
In vivo localization of PHB granules. Diagrams for calculation of (A) polarity and (B) centrality. PHB granules are indicated in yellow. (C and D) Polarity distributions of C. necator and recombinant E. coli cells, respectively. (E and F) Centrality distributions of C. necator and recombinant E. coli cells, respectively. (G and H) Scatter plots of polarity versus PHB content (%, wt/wt) for C. necator and recombinant E. coli cells, respectively.
The average polarity values for C. necator cells were 0.29 ± 0.15, 0.29 ± 0.12, 0.29 ± 0.13, and 0.27 ± 0.10 at 0, 8, 16, and 24 h, respectively. On the other hand, recombinant E. coli cells exhibited higher average polarity values of 0.62 ± 0.16, 0.52 ± 0.18, 0.49 ± 0.19, 0.40 ± 0.16, 0.29 ± 0.10, 0.31 ± 0.09, and 0.31 ± 0.07 at 0, 8, 16, 24, 32, 40, and 48 h, respectively (Fig. 5 C and D). Compared with C. necator, recombinant E. coli cells showed a tendency to localize PHB granules near the cell poles, especially at the early period of cultivation.
To further examine the localization preference of PHB granules, correlative analyses between PHB content and polarity were performed (Fig. 5 G and H). Both scatter plots for C. necator and recombinant E. coli cells show that the polarity values have a wide range of 0 to 0.8 at low PHB contents and converge into a relatively narrow range of 0.3 to 0.4 as PHB contents increase. Considering the cells with low PHB contents (e.g., below 5%) were in the early phase of PHB granule formation, the wide range of polarity implies that the formation of PHB granule can occur anywhere in the cytoplasm. In addition, it is obvious that the polarity of PHB granules decreased as PHB granules occupied more volume within the cell. Interestingly, the polarity for recombinant E. coli cells seems to be negatively correlated with PHB content while the scatter plot of C. necator does not exhibit any noticeable correlation between polarity and PHB content. To examine whether the correlation is indeed significant, new scatter plots were made to contain only the cells, which are longer than the average length of the analyzed cell (SI Appendix, Fig. S8 A and B). In the case of recombinant E. coli, the negative correlation between PHB content and polarity is more clearly observed while the plot for C. necator still does not show any clear correlation. The decreasing polarity with increasing PHB content indicates that the PHB granules tend to be localized to the poles even though PHB granules can be formed anywhere in cell cytoplasm. This was further investigated through real-time monitoring, which is discussed in Real-Time Analysis.
The average centrality values for C. necator were 0.75 ± 0.07, 0.71 ± 0.08, 0.69 ± 0.07, and 0.66 ± 0.07 at 0, 8, 16, and 24 h of cultivation, respectively (Fig. 5E). Recombinant E. coli showed the average centrality values of 0.69 ± 0.07, 0.69 ± 0.07, 0.68 ± 0.07, 0.71 ± 0.06, 0.62 ± 0.05, 0.61 ± 0.05, and 0.58 ± 0.04, respectively (Fig. 5F). The two bacterial species showed similar results that PHB granules localization was biased toward the cell center rather than the cell membrane (SI Appendix, Fig. S8 C and D). Some studies have reported the temporal image of PHA granules near the membrane, leading to the membrane budding model for PHA formation (15–17). However, except for the PHB granules at the poles in recombinant E. coli cells, most PHB granules in C. necator and recombinant E. coli were localized in the middle between the cell center and membrane.
Real-Time Analysis.
In addition to the statistical analyses of data obtained at each sampling time, real-time analyses tracking live individual cells were performed for several hours to understand more detailed spatiotemporal changes of PHB granules in vivo. Time-lapse movies showing the growth of cells and PHB granules could be obtained for C. necator and recombinant E. coli cells (Movies S1–S4). The dynamic changes of cells and in vivo PHB granules shown in the time-lapse movie were also quantitatively analyzed at the individual cell level (SI Appendix, Text S4 and Fig. S9).
The movies show the actual in vivo dynamics of PHB granule synthesis and degradation in living cells, even considering the possible existence of small optical perturbations caused by the movement of cells. As shown in Movies S2–S4 and SI Appendix, Fig. S10, the PHB granule localization during the cell division process is distinctively different between the two species. The PHB granules in C. necator cells were generally localized throughout the cytoplasm (Movie S2 and SI Appendix, Fig. S10). The daughter cells generated by cell division could divide up the PHB granules.
In addition to the Movie S2 monitoring the C. necator cells cultured in the nutrient-limited medium for boosting PHB accumulation, we also obtained a time-lapse movie (Movie S3) for C. necator cells cultured in a nutrient-rich medium to investigate the different characteristics caused by the medium condition. Given that this condition is more suitable for cell growth, the C. necator cells in Movie S3 showed more active cell division with larger cells than the cells shown in Movie S2. During cell growth, PHB granules in C. necator cells seem to be constantly appearing and disappearing. The PHB granules were localized throughout the cytoplasm and the daughter cells generated by cell division contained a similar amount of PHB granules.
In contrast, most recombinant E. coli cells in Movie S4 showed locally asymmetric accumulation of PHB granules that corresponds to the high-polarity values. Interestingly, most PHB granules were localized near only one pole of the cells rather than both poles. The uneven distribution of PHB granules is closely related to the process of inheritance of PHB granules during cell division of E. coli. Movie S4 shows that recombinant E. coli cells elongated toward the opposite side of the accumulated PHB granules and then were divided into two cells. Thus, one daughter cell contained the majority of PHB granules while the other cell had almost no PHB granules. It should be noted that not all PHB granules are formed at the polar region in E. coli; some PHB granules are formed in the middle of the cells. However, after cell division of the E. coli cells containing PHB granules in the middle, the PHB granules tend to be localized near a new pole of the daughter cell; this results in eventual localization of PHB granules near the polar region in E. coli (SI Appendix, Fig. S11). It corresponds well to the statistical analyses that recombinant E. coli cells contain more PHB granules of higher polarity and show the negative correlation between polarity and PHB content.
Examination of Other Bacterial Strains.
To investigate whether the differences in PHA granule localization of C. necator and recombinant E. coli are observed in other species, we additionally analyzed the cells of Pseudomonas putida, which is a native MCL-PHA producer, and also recombinant Klebsiella pneumoniae expressing C. necator phaCAB genes. Interestingly, we observed that PHA granule localization was similar between two native PHA-producing strains (C. necator and P. putida) and also between two nonnative PHA-producing strains (recombinant E. coli and recombinant K. pneumoniae). Similar to C. necator cells, P. putida cells exhibited no correlation between the polarity and PHA content (SI Appendix, Fig. S12), and PHA granules were distributed throughout the cell cytoplasm (Movie S5 and SI Appendix, Fig. S13). On the other hand, the recombinant K. pneumoniae cells showed a negative correlation between the polarity and PHB content (SI Appendix, Fig. S14) and pole-biased localization of PHB granules with asymmetric inheritance as observed in the recombinant E. coli cells (Movie S6 and SI Appendix, Fig. S15). It is especially notable that the distributions of RI region of 1.37 to 1.39 in P. putida (SI Appendix, Fig. S13) and recombinant K. pneumoniae (SI Appendix, Fig. S15) cells were similar to those for C. necator and recombinant E. coli cells, respectively. The RI regions of 1.37 to 1.39 in C. necator and P. putida cells were distributed widely throughout the cytoplasm compared with the recombinant E. coli and K. pneumoniae cells in which the RI regions were mostly observed near the PHB granules.
We assumed that the differences in localization of PHA granules between native and nonnative PHA-producing strains might be attributed to the existence of the PHA granule-associated proteins and enzymes found in native strains including C. necator and P. putida. To support the hypothesis, ODT analyses were performed for the recombinant E. coli cells additionally expressing the genes of C. necator PhaP1 (phaP1) or PhaM (phaM), which plays a major role in determining granule morphology and distribution (11, 18–20) (SI Appendix, Figs. S16–S19). In the case of expressing phaP1, the size of PHB granules became smaller and the number of granules increased (SI Appendix, Fig. S20). Nevertheless, the localization characteristics were similar to the control strain (E. coli expressing C. necator phaCAB genes only) (SI Appendix, Fig. S16). On the other hand, phaM-expressing E. coli cells behaved similarly to the C. necator cells in that the granule polarity was relatively low and did not exhibit any distinct correlation with PHB content (SI Appendix, Fig. S18). In addition, the distribution of the RI region of 1.37 to 1.39 for phaM-expressing E. coli cells (SI Appendix, Fig. S19) became wider than the distributions for control E. coli and phaP1-expressing E. coli cells, which is similar to the C. necator cells. The broad distribution of the RI region of 1.37 to 1.39 might be due to the PhaM that places PHB polymer chains at the nucleoid periphery as PhaM can bind both PHB and nucleoid (11, 18).
Based on these results, we have concluded that the localization characteristics of PHA are strikingly different between native and nonnative PHA-producing strains. In native PHA producers, the granule-associated protein, in particular, PhaM for the case of C. necator strongly influences in vivo PHA granule formation and localization. Since PhaM controls the formation and localization of PHA granules during cell division by interacting with PHA granules and bacterial nucleoid (11, 18), it contributes to the relatively equal distribution of accumulated PHB to the daughter cells; C. necator might exhibit relatively narrow cell-to-cell variations in cell morphology and PHB accumulation compared with the recombinant E. coli cells. On the other hand, PHA granules synthesized in the nonnative PHA-producing strains have been processed differently. Our observation on PHB granule localization in recombinant E. coli and K. pneumoniae cells is similar to the previous studies which have shown that protein aggregates and inclusion bodies are also localized with polar-bias and show asymmetric inheritance related to nucleoid exclusion (21, 22). In terms of the nucleoid exclusion against the PHB granules in the recombinant bacterial cells, the localization of PHB granules is likely to be governed by the competition with the chromosome for intracellular space to ensure proper cell division.
Conclusion
This study presents 3D imaging and quantitative analysis of PHA granules in living bacteria using ODT analysis. The two representative PHA producers, C. necator and recombinant E. coli harboring C. necator PHB biosynthesis genes were selected for the analysis. Through ODT analysis, in vivo PHB granules could be identified as the distinctively high RI (RI > 1.39) region. Based on the RI distribution, cell volume and length, DCW, PHB granule weight, PHB content, granule density, and localization were determined for a single cell in a spatiotemporal manner. In addition, the growth of PHB granules inside the living cells could be monitored for over 8 h. Through such studies, the distinguishing characteristics of PHB accumulation in the two strains were presented. As a native PHA producer, C. necator has several PHA granule-associated proteins such as PHA synthase, PHA depolymerase, regulatory enzymes, and amphiphilic proteins (5, 6). ODT analysis was able to capture the dynamics of PHB synthesis and degradation in detail in a native PHB producer. Also, unique spatiotemporal characteristics of PHB accumulation in nonnative PHB producer, recombinant E. coli, were observed. These differences were further examined through the ODT analyses for the other native and nonnative bacterial strains, P. putida and recombinant K. pneumoniae. As a result, we have demonstrated the distinctive features for PHA granule formation and distribution in native and nonnative producers. It was found that C. necator PhaM is a key enzyme for determining the morphology and localization of PHA granule in vivo based on the observation of the recombinant E. coli additionally expressing phaM that behaved similarly to C. necator.
Taken together, this study presents a real-time quantitative analysis of PHA accumulation in live individual cells to understand PHA granule formation, growth, and distribution together with cell division by tracking the changes of cell volume and length, PHA content, and PHA granule localization for hours. The strategy presented in this study will be useful for understanding the spatiotemporal accumulation of other types of PHAs as well.
Recently, several studies have suggested that the asymmetric segregation of protein aggregates is considered an important basis of bacterial cellular aging and rejuvenation (23, 24). The recombinant E. coli cells accumulating PHB can also serve as a model for such studies, providing new perspectives on bacterial aging.
Last but not least, the ODT analysis reported here can facilitate the development of efficient PHA-producing strains. As several excellent studies have been performed on manipulating cellular morphology and cell division patterns to improve PHA production (25, 26), the ODT technology can assist the strain development by monitoring the detailed behavior of the cells accumulating PHA granules according to different genetic modifications and fermentation conditions.
Materials and Methods
All the materials and methods conducted in this study are detailed in SI Appendix, Materials and Methods: bacterial strain and cultivation, gas chromatography analysis, sample preparation for optical imaging, ODT, wide-field fluorescence analysis, quantification of cell parameters, and polarity and centrality analyses.
Supplementary Material
Acknowledgments
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (Grants NRF2012M1A2A2026556 and NRF-2012M1A2A2026557) and the Bio and Medical Technology Development Program (Grant 2021M3A9I4022740) from the Ministry of Science and ICT (MSIT) through the National Research Foundation of Korea to S.Y.L. This work was also supported by the Korea Advanced Institute of Science and Technology Cross-Generation Collaborative Laboratory project.
Footnotes
Competing interest statement: Y.P. has financial interests in Tomocube Inc., a company that commercializes optical diffraction tomography and quantitative phase imaging instruments.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103956118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Anderson A. J., Dawes E. A., Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54, 450–472 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S. Y., Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49, 1–14 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Chen G. Q., A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 38, 2434–2446 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Choi S. Y., et al., Metabolic engineering for the synthesis of polyesters: A 100-year journey from polyhydroxyalkanoates to non-natural microbial polyesters. Metab. Eng. 58, 47–81 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Jendrossek D., Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J. Bacteriol. 191, 3195–3202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jendrossek D., Pfeiffer D., New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ. Microbiol. 16, 2357–2373 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Peters V., Rehm B. H., In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol. Lett. 248, 93–100 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Mravec F., et al., Accumulation of PHA granules in Cupriavidus necator as seen by confocal fluorescence microscopy. FEMS Microbiol. Lett. 363, fnw094 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Jendrossek D., Fluorescence microscopical investigation of poly(3-hydroxybutyrate) granule formation in bacteria. Biomacromolecules 6, 598–603 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Tian J., Sinskey A. J., Stubbe J., Kinetic studies of polyhydroxybutyrate granule formation in Wautersia eutropha H16 by transmission electron microscopy. J. Bacteriol. 187, 3814–3824 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahl A., Schuth N., Pfeiffer D., Nussberger S., Jendrossek D., PHB granules are attached to the nucleoid via PhaM in Ralstonia eutropha. BMC Microbiol. 12, 262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeby M., Cho M., Stubbe J., Jensen G. J., Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J. Bacteriol. 194, 1092–1099 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerngross T. U., Reilly P., Stubbe J., Sinskey A. J., Peoples O. P., Immunocytochemical analysis of poly-beta-hydroxybutyrate (PHB) synthase in Alcaligenes eutrophus H16: Localization of the synthase enzyme at the surface of PHB granules. J. Bacteriol. 175, 5289–5293 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobes G. A., et al., Growth and kinetics of in vitro poly ([R]‐(–)‐3‐hydroxybutyrate) granules interpreted as particulate polymerization with coalescence. Macromol. Rapid Commun. 21, 77–84 (2000). [Google Scholar]
- 15.Obruca S., et al., Novel unexpected functions of PHA granules. Appl. Microbiol. Biotechnol. 104, 4795–4810 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Stubbe J., Tian J., Polyhydroxyalkanoate (PHA) hemeostasis: The role of PHA synthase. Nat. Prod. Rep. 20, 445–457 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Jendrossek D., Selchow O., Hoppert M., Poly(3-hydroxybutyrate) granules at the early stages of formation are localized close to the cytoplasmic membrane in Caryophanon latum. Appl. Environ. Microbiol. 73, 586–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bresan S., Jendrossek D., New insights into PhaM-PhaC-mediated localization of polyhydroxybutyrate granules in Ralstonia eutropha H16. Appl. Environ. Microbiol. 83, e00505–e00517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pötter M., Müller H., Steinbüchel A., Influence of homologous phasins (PhaP) on PHA accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology (Reading) 151, 825–833 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Neumann L., et al., Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J. Bacteriol. 190, 2911–2919 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coquel A. S., et al., Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput. Biol. 9, e1003038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neeli-Venkata R., et al., Robustness of the process of nucleoid exclusion of protein aggregates in Escherichia coli. J. Bacteriol. 198, 898–906 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindner A. B., Madden R., Demarez A., Stewart E. J., Taddei F., Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 105, 3076–3081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Proenca A. M., Rang C. U., Buetz C., Shi C., Chao L., Age structure landscapes emerge from the equilibrium between aging and rejuvenation in bacterial populations. Nat. Commun. 9, 3722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S. Y., Suppression of filamentation in recombinant Escherichia coli by amplified FtsZ activity. Biotechnol. Lett. 16, 1247–1252 (1994). [Google Scholar]
- 26.Jiang X. R., Chen G. Q., Morphology engineering of bacteria for bio-production. Biotechnol. Adv. 34, 435–440 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.