Significance
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer and is one of the most deadly cancer types, with a 5-y survival rate below 10%. One reason for this high mortality rate is that PDAC cells have an enhanced ability to survive and proliferate despite existing in nutrient-deprived environments. Understanding the metabolic rewiring that enables nutrient scavenging and rapid metabolic processing of PDAC cells can provide new strategies to develop effective treatment options for this intractable disease. In this study, convergent findings reveal that UGP2 has a central role in the growth and metabolism of PDAC cells through the regulation of glycogen synthesis and protein N-glycosylation, highlighting therapeutic possibilities for this deadly cancer.
Keywords: PDAC, UGP2, UDP-glucose, glycogen, N-glycosylation
Abstract
UDP-glucose pyrophosphorylase 2 (UGP2), the enzyme that synthesizes uridine diphosphate (UDP)-glucose, rests at the convergence of multiple metabolic pathways, however, the role of UGP2 in tumor maintenance and cancer metabolism remains unclear. Here, we identify an important role for UGP2 in the maintenance of pancreatic ductal adenocarcinoma (PDAC) growth in both in vitro and in vivo tumor models. We found that transcription of UGP2 is directly regulated by the Yes-associated protein 1 (YAP)–TEA domain transcription factor (TEAD) complex, identifying UGP2 as a bona fide YAP target gene. Loss of UGP2 leads to decreased intracellular glycogen levels and defects in N-glycosylation targets that are important for the survival of PDACs, including the epidermal growth factor receptor (EGFR). These critical roles of UGP2 in cancer maintenance, metabolism, and protein glycosylation may offer insights into therapeutic options for otherwise intractable PDACs.
Pancreatic ductal adenocarcinomas (PDACs) are mostly refractory to existing therapies and show a high mortality rate (1). As PDAC cells grow and proliferate to expand their biomass, they must adapt to thrive in an increasingly dense, nutrient-limited, and hypoxic tumor microenvironment. Cancer cells can adapt by rewiring their metabolism via stimulating nutrient uptake pathways and upregulating rate-limiting enzymes involved in anabolic pathways (2, 3). For example, the increase in glycolytic flux driven by metabolic reprogramming has been shown to enhance cancer cell survival (4, 5). Understanding the strategies used by cancer cells to obtain a survival advantage can be used against these cells to target metabolic pathways that can selectively inhibit the survival or proliferation of cancer cells.
Glucose is well-known to be an essential nutrient in most types of cells but, beyond its function as a nutrient, glucose is also a key substrate in many anabolic pathways. The use of glucose in these pathways depends on its activation to uridine diphosphate (UDP)-glucose catalyzed by UDP-glucose pyrophosphorylase 2 (UGP2), the only enzyme capable of converting glucose 1-phosphate to UDP-glucose in mammalian cells (6). UGP2 is upregulated in some cancers, including PDACs, and its expression is correlated with an increased rate of progression and poor prognosis (7–11). However, the direct role of UGP2 in growth and maintenance of PDAC has not been explored and the mechanisms of regulation and action of UGP2 remain unknown.
Here, we demonstrate that UGP2 is a key mediator required for the survival and proliferation of PDAC cells in vitro and in vivo. We identify the transcriptional regulator Yes-associated protein 1 (YAP) as an important upstream activator of UGP2 expression. We further characterize two regulatory functions of UGP2 in PDAC cells: first, that UGP2 regulates glycogen synthesis which impacts survival in nutrient-starved conditions and, second, that UGP2 is critical for an array of protein N-glycosylation modifications, including regulation of key sites on the epidermal growth factor receptor (EGFR) that lead to impaired downstream signaling. These findings reveal metabolic pathways that are critical for PDAC cells and provide potential clinical targets for further investigation.
Results
UGP2 Is Critical for PDAC Cell Survival and Proliferation In Vitro and In Vivo.
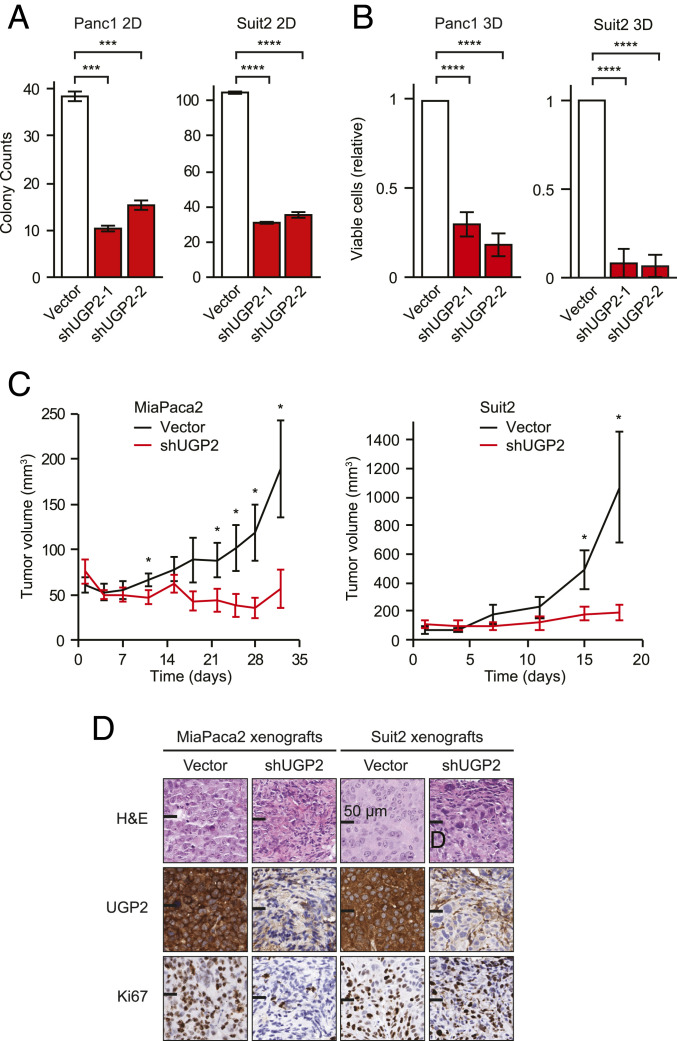
UGP2 is an important enzyme for multiple metabolic pathways in cells; however, it has not been well-studied for its role in the survival or proliferation of cancer cells. We first sought to examine whether UGP2 expression correlated with prognosis in PDAC patients. We found that high expression of UGP2 correlated with worse clinical prognosis in a panel of 177 PDAC patient samples (12) (SI Appendix, Fig. S1A). Multivariate analyses indicated that UGP2 expression correlates strongly with poor prognosis in PDAC tumors, particularly in early-phase and low-grade tumors (SI Appendix, Fig. S1 B and C). To ascertain whether UGP2 is functionally required for the survival and proliferation of PDAC cells, UGP2 expression was depleted in multiple PDAC cell lines (SI Appendix, Fig. S1D). Knockdown of UGP2 led to significant growth inhibition in both two-dimensional (2D) and three-dimensional (3D) culture conditions in Panc1 and Suit2 (Fig. 1 A and B and SI Appendix, Fig. S1 E and F). To further test the extent to which UGP2 was required for tumor growth in vivo, MiaPaca2 and Suit2 cell lines with stable knockdown of UGP2 or empty vector controls were xenografted onto the opposite flanks of nude mice. Knockdown of UGP2 halted tumor growth in both MiaPaca2 and Suit2 xenografts over the course of several weeks (Fig. 1C and SI Appendix, Fig. S1G) and displayed a reduced end-point tumor size (SI Appendix, Fig. S1H). We further confirmed that tumors with knockdown of UGP2 expressed lower UGP2 protein levels and displayed a reduced proliferative index shown by Ki67 staining (Fig. 1D).
Fig. 1.
UGP2 is critical for PDAC cell survival and proliferation in vitro and in vivo. (A) Colony counts of Panc1 and Suit2 cells grown in two-dimensional culture conditions for 10 d; n = 3; ***P < 0.001, ****P < 0.0001. (B) Relative viability of Panc1 and Suit2 cells grown in three-dimensional culture conditions for 14 d, measured by CellTiter-Glo; n = 4; ****P < 0.0001. (C) Tumor volumes of MiaPaca2 and Suit2 cells with shUGP2 or empty vector control xenografted on opposite flanks of nude mice; n = 5; *P < 0.05. Error bars represent SEM. (D) Representative immunohistochemical staining of paired opposite-flank tumors with shUGP2 or vector control at end point, stained as indicated. H&E, hematoxylin and eosin. (Scale bars, 50 μm.)
UGP2 Is a Target Gene of the YAP–TEAD Complex.
Further pursuing these indications that UGP2 is required for PDAC growth and maintenance, we sought to understand how UGP2 expression is regulated in these contexts. As the vast majority of PDACs exhibit KRAS mutations and an elevated RAF–MEK–ERK pathway, we first tested the association between mutant KRAS and UGP2 expression. We found that enforced overexpression of the oncogenic KRAS mutants G12D or G12V drives an increase in messenger RNA (mRNA) and protein levels of UGP2 in the nontumorigenic cell line MCF10A (SI Appendix, Fig. S2 A and B). Furthermore, these mutant KRAS-overexpressing cells were more sensitive to UGP2 knockdown in both 2D and 3D culture conditions (SI Appendix, Fig. S2 C–E). However, inhibition of the RAF–MEK–ERK pathway by treatment with trametinib, a MEK inhibitor, did not affect UGP2 expression (SI Appendix, Fig. S2F), suggesting that UGP2 is regulated by a pathway independent of RAF–MEK–ERK signaling downstream of KRAS.
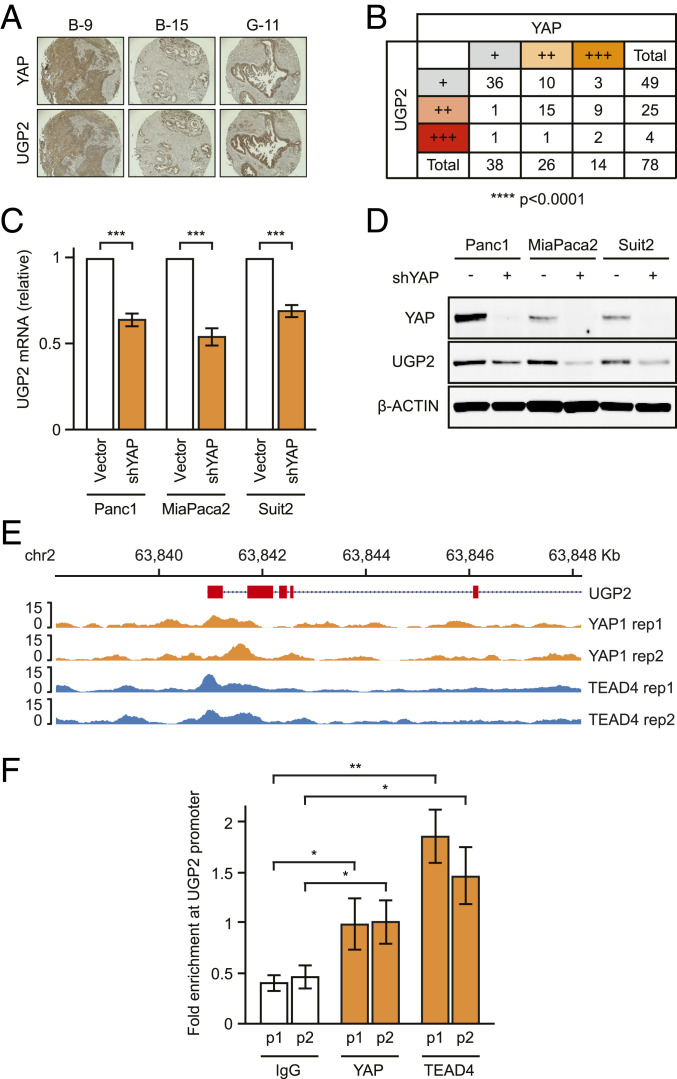
In particular, YAP, a transcriptional coactivator, has been shown to be required for PDAC progression in murine models (13). Similar to UGP2, YAP expression is a negative prognostic marker in PDAC, especially in early-phase tumors (SI Appendix, Fig. S3 A–C). The mRNA expression levels of YAP and UGP2 were also positively correlated in a panel of 178 PDAC patient samples from The Cancer Genome Atlas (SI Appendix, Fig. S3D). To further examine the relationship between YAP and UGP2 in PDACs, immunohistochemistry was employed to probe a tissue microarray panel of 78 PDAC patient samples. Protein levels of YAP and UGP2 showed similar expression patterns and positively correlated with each other through blinded scoring analysis (Fig. 2 A and B). We then tested whether YAP can directly regulate UGP2 expression in PDAC cells. Knockdown of YAP in PDAC cells resulted in decreased UGP2 mRNA and protein levels (Fig. 2 C and D and SI Appendix, Fig. S3E), which was recapitulated by using multiple short hairpins with different targeting sequences (SI Appendix, Fig. S3F). Depletion of YAP was also able to partially reverse UGP2 expression induced by mutant KRAS in MCF10A cells (SI Appendix, Fig. S3G). Conversely, overexpression of YAP increased UGP2 expression in the nontumorigenic cell lines MCF10A and 293T (SI Appendix, Fig. S3 H and I). Introduction of the constitutively active YAP 5SA mutant, in which five inhibitory phosphorylation sites are removed (14), further increased UGP2 expression more so than wild-type YAP, whereas the inactive dominant-negative S94A mutant (15) suppressed UGP2 expression (SI Appendix, Fig. S3I).
Fig. 2.
UGP2 is a target gene of YAP. (A) Representative tissue microarray images stained for YAP or UGP2. B-9, B-15, and G-11 indicate sample codes. (B) Scoring of tissue microarrays containing 78 pancreatic ductal adenocarcinoma samples immunohistochemically stained for YAP or UGP2. +, ++, and +++ represent combined scoring of automated area × intensity measurements. ****P < 0.0001 by χ2 test. (C) qPCR for UGP2 mRNA in Panc1, MiaPaca2, and Suit2 cells with shYAP or empty vector. ***P < 0.001, error bars represent SEM. (D) Immunoblot on lysates from Panc1, MiaPaca2, and Suit2 cells stably expressing shYAP or empty vector, probed as indicated. (E) ChIP-seq for YAP1 and TEAD4 at the UGP2 locus. (F) Targeted PCR using two sets of primers (p1 and p2) in the UGP2 promoter region of Panc1 cells after cross-linking and pull-down assays using antibodies against either YAP, TEAD4, or control immunoglobulin G (IgG). Fold enrichment is percent input target gene/percent input actin. *P < 0.05, **P < 0.01, error bars represent SEM.
YAP is a transcriptional coactivator lacking intrinsic DNA-binding domains and requires DNA-binding partners, such as TEA domain transcription factor (TEAD) proteins, for transcriptional activation (16, 17). To test whether the YAP–TEAD complex regulates UGP2 transcriptional expression by directly binding at the UGP2 locus, chromatin immunoprecipitation sequencing (ChIP-seq) was performed for YAP and TEAD4. As expected, high occupancy of YAP–TEAD4 was observed at the promoters of well-established YAP–TEAD binding sites including cellular communication network factor 2 (CCN2) (15, 16) (SI Appendix, Fig. S3J). Furthermore, we observed binding of both YAP and TEAD4 at the UGP2 promoter region across biological replicates (Fig. 2E) and specific binding of YAP and TEAD4 was confirmed using ChIP-qPCR primer pairs targeted to the UGP2 promoter (Fig. 2F). These results support a model wherein transcription of UGP2 is positively regulated by direct binding of the YAP–TEAD complex to the UGP2 locus.
Effects of UGP2 Loss on Glycogen Synthesis.
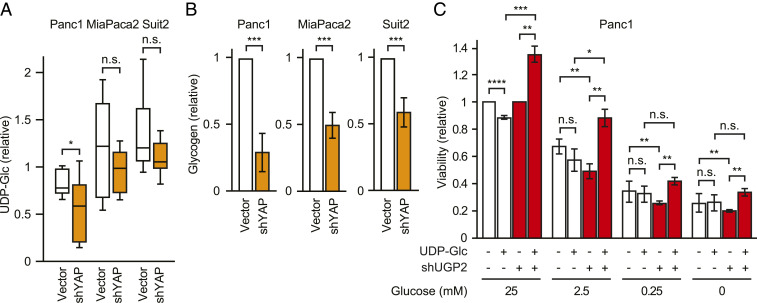
As mentioned above, UGP2 is an enzyme that generates UDP-glucose from glucose 1-phosphate. An important cellular process that utilizes UDP-glucose as a direct precursor is glycogen synthesis (18). Glycogen is a multibranched polysaccharide that serves as an important form of energy for proliferating cancer cells, especially in low-glucose or hypoxic conditions (19, 20). In fact, hypoxic conditions have been shown to increase glycogen levels (21), UGP2 expression (22), and YAP activity (23). In accordance with these results, we found that knockdown of YAP, which decreases UGP2 expression, led to a decrease in intracellular UDP-glucose and glycogen levels in PDAC cells (Fig. 3 A and B). Conversely, the overexpression of YAP or mutant KRAS G12D and G12V led to increased intracellular glycogen levels in MCF10A cells (SI Appendix, Fig. S4). To investigate the effect of UGP2 on cell survival in response to glucose starvation, we assessed the viability of UGP2-depleted Panc1 cells in standard tissue-culture media with 25 mM glucose or reduced glucose concentrations of 2.5, 0.25, or 0 mM. Knockdown of UGP2 caused a stronger growth inhibitory effect in the low-glucose conditions (Fig. 3C). Furthermore, the supplementation of UDP-glucose rescued proliferation of UGP2-depleted cells but not control cells in different concentrations of glucose, including 25, 2.5, 0.25, and 0 mM (Fig. 3C). These data suggest that loss of UGP2 creates a scarcity of UDP-glucose that inhibits survival in low-glucose contexts.
Fig. 3.
UGP2 and YAP regulate glycogen metabolism in PDAC cells. (A) UDP-glucose levels of Panc1, MiaPaca2, and Suit2 cells with shYAP or empty vector. *P < 0.05 by ANOVA; n.s., not significant. (B) Cellular glycogen levels in Panc1, MiaPaca2, or Suit2 cells with shYAP or empty vector control. **P < 0.01, ***P < 0.001. Error bars represent SEM. (C) Relative viability of Panc1 cells with vector control or shUGP2 grown with or without 100 μM UDP-glucose for 7 d. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
UGP2 Regulates Protein Glycosylation in PDAC Cells.
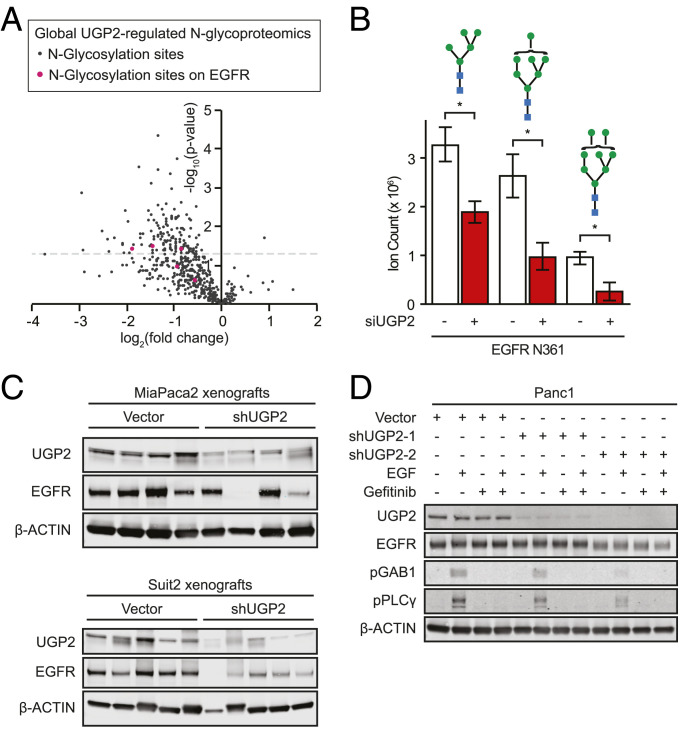
Another prominent pathway that uses UDP-glucose as a substrate is the asparagine (N)-linked glycosylation pathway (24). As the specific modifications that UGP2 regulates in PDAC cells have not been previously elucidated, we performed unbiased global N-glycoproteomic analysis of all asparagine-linked glycan modifications in Suit2 cells following 48 h of UGP2 knockdown using small interfering RNA (siRNA) (SI Appendix, Fig. S5A). A large-scale decrease in glycan modifications was observed relative to cells treated with scrambled siRNA controls (Fig. 4A). Knockdown of UGP2 significantly decreased the incidence of 141 N-glycosylation modifications spread across 89 proteins (SI Appendix, Table S1). Total global proteomics, performed in parallel, revealed that the vast majority of the observed decreases in N-glycosylation were not explained by corresponding changes in total protein levels (SI Appendix, Fig. S5 B and C and Table S1) or global changes in the relative frequency of glycan-chain architectures (SI Appendix, Fig. S5D). Thus, UGP2 knockdown in PDAC cells rapidly reduces N-glycosylation modifications in a distinctive constellation of sites across multiple proteins.
Fig. 4.
UGP2 regulates N-glycan modifications on proteins including EGFR. (A) Unbiased quantitative N-glycoproteomic analysis identified changes in protein modifications in Suit2 cells upon knockdown of UGP2 relative to nontargeting control siRNAs at 48 h; red dots represent modifications on EGFR. (B) Quantification of glycan modifications on EGFR Asn361 in Suit2 cells with siUGP2 or nontargeting control siRNAs at 48 h. *P < 0.05. Blue squares represent N-acetylglucosamines; green circles represent mannoses, error bars represent SEM. (C) Immunoblot of MiaPaca2 and Suit2 xenograft end-point tumor lysates, probed as indicated. (D) Immunoblot of Panc1 cells stably expressing shUGP2-1, shUGP2-2, or empty vector, pretreated with 10 μM gefitinib for 1 h and/or stimulated with 100 ng/mL EGF for 10 min, probed as indicated.
Of the 89 proteins with N-glycan modifications significantly regulated by UGP2, EGFR was among the top proteins whose N-glycan modifications were most frequently and consistently decreased upon knockdown of UGP2. EGFR is a membrane-embedded growth factor receptor whose activation leads to signaling through the RAS–RAF–MEK–ERK growth axis, upon which many PDACs are dependent. N-glycosylation of EGFR is required for its trafficking, efficient ligand binding, protein stability, and receptor activation (25). We identified a UGP2-regulated N-glycosylation site on the extracellular ligand-binding domain of EGFR at Asn361, where three different N-glycan modifications were significantly decreased by knockdown of UGP2 (Fig. 4B). Decrease of an additional N-glycan modification to EGFR was observed at Asn528, although the UGP2-induced change did not reach statistical significance at this alternate site (SI Appendix, Fig. S5E). While the amount of total EGFR protein remained stable upon transient 48-h siRNA-based perturbation of UGP2 (SI Appendix, Fig. S5F), immunoblots for EGFR in cells with long-term stable short hairpin RNA (shRNA) knockdown of UGP2 showed a decrease in total EGFR protein (SI Appendix, Fig. S5G). Similarly, in the majority of xenografted tumors, knockdown of UGP2 resulted in decreased EGFR protein as shown in both immunoblots of tumor lysates and immunohistochemistry on tumor sections (Fig. 4C and SI Appendix, Fig. S5H). These data suggest that UGP2 loss leads to decreases in N-glycosylation of multiple proteins including EGFR. Furthermore, phosphorylation of established EGFR targets GAB1 (26) and PLCγ (27) was decreased in cells with UGP2 knockdown and this effect was ablated by pretreatment with the EGFR inhibitor gefitinib (Fig. 4D and SI Appendix, Fig. S5I). Thus, loss of UGP2 can regulate EGFR expression and downstream biological signaling.
Discussion
PDACs are extremely intractable solid tumors that experience a hypoxic and nutrient-deprived microenvironment and often undergo a metabolic switch to support increased glycolytic flux for their growth (28). These differential metabolic settings may provide novel opportunities to selectively target cancers via their metabolic pathways. For example, l-asparaginase, an enzyme that induces asparagine deprivation, has been successfully used to treat acute lymphoblastic leukemia and is now undergoing clinical trials for PDAC (28). An emerging paradigm in the field of cancer metabolism is that the network of metabolic reactions, once considered separate from other cellular networks, is in fact closely integrated with key oncogenic signaling pathways. How metabolism is reprogrammed in cancers and how to exploit metabolic changes, such as rate-limiting pathways or metabolites, for therapeutic benefit are leading questions in the field. Additionally, the catalytic nature of enzymes involved in metabolic pathways makes them amenable for small-molecule targeting.
Employing in vitro 2D and 3D models as well as in vivo tumor xenograft models, we identified UGP2 as a regulator of glycogen synthesis and protein N-glycosylation that is critical for PDAC cell growth, exposing a vulnerability that could be exploited for therapeutic interventions. While embryonic deletion of UGP2 is predicted to be lethal in mice (29), the involvement of UGP2 in genetic disease suggests that loss of UGP2 function may be tolerable in specific postnatal tissues (30). Furthermore, we identified the YAP–TEAD complex as an important transcriptional regulator of UGP2 expression in PDAC. YAP, a bona fide oncogene, has recently been identified as an essential mediator of oncogenic KRAS signaling during PDAC progression (13) as well as a marker for poor prognosis in PDAC patients (31). YAP can compensate for the inactivation of oncogenic KRAS in several cancer types (32, 33), suggesting interplay between these major signaling pathways. Downstream target genes of YAP include cyclins and growth factors that are involved in proliferation, but YAP activation also induces stem cell properties, drug resistance, and metastasis, suggesting the presence of additional targets that may affect various cellular processes. Data shown in this study provide potential mechanisms for UGP2 as an effector of YAP activity to regulate growth and metabolism in PDAC cells. However, observations showing that almost complete YAP depletion leads to only a partial down-regulation of UGP2 expression suggest that there may be additional UGP2 regulators in PDAC cells and this will be of interest for future studies.
An intriguing question is how ubiquitous UGP2 dependency is across different cancer types. In the Dependency Map (DepMap) portal (34, 35), cell lines from multiple tissue types display a dependency on UGP2 for their growth, suggesting that the requirement for UGP2 may extend beyond PDACs. In fact, UGP2 was observed to be persistently upregulated both at the mRNA and protein levels with increased metastatic potential and was found to play an essential role in promoting migration and metastasis in hepatocellular carcinoma cells (11). However, in the case of lung cancer, UDP-glucose has been shown to impair metastasis through the regulation of Snail, an epithelial–mesenchymal transition factor (36). Interestingly, UGP2 is codependent with multiple genes which have a role in protein glycosylation, including phosphoglucomutase 3, phosphomannomutase 2, and mannose phosphate isomerase, highlighting the biological complexity of this pathway.
The product of UGP2, UDP-glucose, can act as a substrate for multiple metabolic pathways, such as glycogen synthesis and protein N-glycosylation (37). UGP2 synthesizes UDP-glucose from glucose 1-phosphate, converted from the glycolytic intermediate glucose 6-phosphate, and is the sole enzyme responsible for this reaction in mammalian cells. UDP-glucose can then be incorporated into branch particles to form glycogen and the reverse process can occur rapidly to be utilized as fuel in cells. PDACs are known to exist in severe hypoxic and nutrient-deprived conditions, and restrained glycogen breakdown by glycogen phosphorylase inhibitor treatment was shown to inhibit proliferation and induce apoptosis in PDAC cells (38). Similarly, we observed that cells with UGP2 knockdown showed worse survival in low-glucose conditions compared with control cells and that their proliferation could be rescued by the supplementation of UDP-glucose. These data suggest that the availability of UDP-glucose, through UGP2 activity, is a limiting factor for the survival and proliferation of PDAC cells.
Unbiased global N-glycoproteomics identified 89 proteins with N-glycosylation sites significantly reduced by knockdown of UGP2 in PDAC cells and understanding which of these posttranslational modifications are functionally significant remains an important step for subsequent studies. UGP2 loss led to decreased glycosylation of several proteins with well-established roles in PDAC biology, such as EGFR (SI Appendix, Table S1). Depletion of UGP2 also led to a subsequent decrease in total EGFR protein, which some PDAC cell lines depend upon for growth (34, 35), although the exact mechanism by which this decrease occurred remains unclear. UGP2 depletion also impaired downstream EGFR signaling, indicating that these glycosylation moieties may regulate EGFR function. As differential glycosylation can drive changes in protein behaviors including degradation, localization, and signaling (39), glycan modifications may be important for promoting EGFR stability or inhibiting its degradation. Also, the proximity of Asn361 to the ligand-binding site suggests that the UGP2-regulated EGFR modifications could affect the ability of EGF ligands to bind and initiate downstream signaling cascades. The role of UGP2 in cancer progression in vivo warrants further investigation as an intrinsic mechanism and as a node for discovery of novel pharmacological interventions.
Materials and Methods
Tissue Culture and Xenografts.
All cell lines were confirmed by short tandem repeat analysis and were regularly screened for mycoplasma. Panc1, MiaPaca2, Suit2, and HEK293T were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) with 10% fetal bovine serum (Atlanta Biologicals). MCF10A cells were grown in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/mL EGF (PeproTech), 0.5 mg/mL hydrocortisone (Sigma), 100 ng/mL cholera toxin (Sigma), and 10 μg/mL insulin (Sigma). Prior to experiments, 2 μg/mL puromycin (Invitrogen; A11138-02) was used to select for enhanced green fluorescent protein or KRAS G12D or G12V. Blasticidin (Fisher; AAJ61883WDL) (4 μg/mL) was used to select for the shRNA of interest or the empty vector pLKO. Three-dimensional cultures were seeded using 20% Matrigel (Corning; 354234) on low-attachment 96-well plates (Corning; 3474) for 2 wk. Cell viability was measured using the CellTiter-Glo luminescent cell-viability assay (Promega; G7570). Cellular glycogen was measured using the Glycogen Assay Kit (Abcam; ab65620). Metabolomics including UDP-glucose levels were measured by the Precision Metabolomics liquid chromatography–mass spectrometry (LC-MS) global metabolomics platform (Metabolon).
For each cell line, five nude mice (NCr-Foxn1nu from Taconic) were injected with 3 million cells expressing either LKO or shUGP2 resuspended in 70% phosphate-buffered saline (PBS), 30% Matrigel on opposite flanks. Subsequent tumors were monitored by caliper two or three times per week beginning 1 d after injection. Sample sizes required for statistically meaningful results were determined based on standards in the field and minimization of animal use. All animal studies and end points were consistent with University of California San Francisco (UCSF) Institutional Animal Care and Use Committee guidelines. At end point, tumors were weighed and cut into portions that were snap-frozen for immunoblots or fixed for histology using 24 h of immersion in 10% neutral buffered formalin. Tumor lysates were prepared using a Fisher PowerGen homogenizer for 30 to 60 s in cell lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 1 mM ethylenediaminetetraacetic acid, 1 mM ethylene glycol tetraacetic acid, 10% glycerol, 50 mM NaF, 5 mM NaPPl, 1 mM phenylmethanesulfonyl fluoride, 0.5 mM dithiothreitol [DTT], PPase inhibitor, protease inhibitor). Lysates were incubated on ice for 30 min and spun at 13,000 rpm at 4 °C; then the supernatants were frozen at −80 °C for immunoblot applications.
Immunoblots and Immunohistochemistry.
Lysates were diluted with 4× NuPage buffer and run on Invitrogen NuPage 4 to 12% Bis-Tris gels. The antibodies employed were KRAS (Sigma; 3B10-2F2), UGP2 (Santa Cruz Biotechnology; sc-377089), YAP (Cell Signaling; 14074), EGFR (Cell Signaling; 4267), p-PLCγ (Cell Signaling; 3233), p-GAB1 (Cell Signaling; 3234), and B-ACTIN (Sigma; A5441). Immunohistochemistry of xenografts was performed by HistoWiz using antibodies against EGFR (EP38Y) and UGP2 (Santa Cruz Biotechnology; sc-377089; 1:200). Immunohistochemistry of historic deidentified PDAC samples used antibodies against YAP1 (Cell Signaling; 14074) and UGP2 (Santa Cruz Biotechnology; sc-377089). To analyze tissue microarray staining, blinded data extraction for mean intensity and percent area was conducted using inForm software from PerkinElmer. Normalization and χ2 statistical analysis were conducted using Matlab.
RNA Interference.
shRNAs targeting YAP and UGP2 were purchased from Sigma. The shRNA constructs were packaged as lentiviruses using third-generation packaging systems with standard protocols. The YAP shRNA sequences were 5′-CCCAGTTAAATGTTCACCAAT-3′ (TRCN0000107265) and 5′-GCCACCAAGCTAGATAAAGAA-3′ (TRCN0000107266). The UGP2 shRNA sequences were 5′-CCACAGCATCATCACATGAAT-3′ (TRCN0000037840) and 5′-GCTAGTTTCTTACAATGAAAT-3′ (TRCN0000435330). For siRNA experiments, Suit2 cells were grown in 2D culture on 15-cm plates (Corning; 430599) and transfected with pools of nontargeting siCtrl or siUGP2. siRNAs in the control nontargeting pool (D-001810-10-05) and UGP2 ON-TARGETplus SMARTpool (L-007739-00-0050) were purchased from Dharmacon. The UGP2 siRNA sequences in the pool were 5′-GAGCUAGAAUUAUCUGUGA-3′, 5′-UAGCAAAGGACGUGUCUUA-3′, 5′-ACAAACAACCUAUGGAUUU-3′, and 5′-UAAUAUAUCUUCCGUGUUG-3′. Transfections were performed using DharmaFECT 1 Transfection Reagents (Dharmacon; T-2001). After 48 h, triplicate cell pellets were washed twice in PBS, harvested, counted, pelleted, and frozen in ∼10 million cell aliquots for downstream immunoblot, N-glycomic, N-glycoproteomic, and total proteomic analyses.
Chromatin Immunoprecipitation.
Five million cells were seeded on 10-cm plates in triplicate. Cross-linking was performed by adding 37% (weight/volume [vol]) formaldehyde to a final concentration of 1.42% dropwise to the plate at room temperature. Cells were incubated with rotation for 15 min at room temperature. Then the formaldehyde was quenched with ice-cold 125 mM glycine for 5 min at room temperature. Cells were scraped, washed twice in ice-cold PBS, and frozen on dry ice. Pellets were resuspended in 1 mL ChIP buffer (330 μL Triton buffer + 660 μL sodium dodecyl sulfate buffer + 10 μL protease inhibitor). Global ChIP-seq and UGP2 promoter ChIP-PCR were performed using antibodies against YAP1 (Novus Biologicals; NB110-58358) and TEAD4 (Abcam; ab58310). ChIP-PCR was performed with two independent sets of PCR primers targeting the UGP2 locus: forward primer 5′-AGAGGTTGGTGGGTGGTTTG-3′ reverse primer 5′-ATACGCGTCTGGAACGTCA-3′; forward primer 5′-TTGTGTGTACGTGGTTTGCG-3′, reverse primer 5′-AATGACCTCCAGCTTCTTCGG-3′.
Glycomics, Glycoproteomics, and Total Proteomics.
Cell-membrane extraction methods were applied as previously described with slight modifications (40–42). Cell pellets were resuspended in a homogenization buffer containing 20 mM Hepes (pH 7.4), 0.25 M sucrose, and 1:100 protease inhibitors (Calbiochem/EMD Chemicals). Cells were lysed with five alternating on and off pulses in 5- and 10-s intervals using a probe sonicator (Qsonica) followed by centrifugation at 2,000 × g for 10 min and ultracentrifugation at 200,000 × g for 45 min at 4 °C. Pellets were resuspended in 500 μL of 0.2 M Na2CO3 and 500 μL of water followed by two additional ultracentrifugations at 200,000 × g for 45 min at 4 °C. For protein digestion, the cell-membrane fractions were reconstituted in 60 μL of 8 M urea and sonicated for 10 min to homogenize the pellet (40–42). To further the denaturing process, 2 μL DTT was added to each sample and incubated for 50 min at 55 °C, followed by the addition of 4 μL iodoacetamide for 20 min at room temperature in the dark. Ammonium bicarbonate (NH4HCO3) (420 μL) solution was added to adjust the pH and 2 μg of trypsin for protein digestion was added for 18 h at 37 °C. Peptides were desalted by solid-phase extraction (SPE) with C18 cartridges (Sigma) containing 500 mg of materials for proteomic analysis or enriched by SPE using iSPE-HILIC cartridges (The Nest Group) for glycopeptide analysis. Samples were dried in vacuo using a miVac (SP Scientific).
Glycopeptide samples were reconstituted in 10 μL of Nanopure water prior to injection onto a Nanospray Flex ion source Orbitrap Fusion Lumos Tribrid mass spectrometer coupled with an UltiMate WPS-3000RS nanoLC 980 System (Thermo Fisher Scientific) conducted with a binary gradient system where solvent A was composed of 0.1% (vol/vol) formic acid and solvent B was composed of 80% (vol/vol) acetonitrile (43). Samples were separated on an Acclaim PepMap 100 C18 LC column (3 µm, 0.075 × 150 mm; Thermo Fisher Scientific) with the following gradient sequence for separation: 0 to 5 min, 4% B; 5 to 133 min, 4 to 32% B; 133 to 152 min, 32 to 48% B; 152 to 155 min, 48 to 100% B; 155 to 170 min, 100% B; 170 to 171 min, 100 to 4% B; 171 to 180 min, 4% B. Data acquisition was performed with a mass range of m/z 700 to 2,000 in positive-ionization mode. Ionization spray voltage was set to 1.8 kV with a 275 °C ion transfer capillary temperature. Precursor ions were subjected to stepped higher-energy C trap dissociation (30 ± 10%) applied to obtain tandem MS/MS spectra with a minimum mass range set at 120 m/z. Proteomic samples were reconstituted to 60 μL and analysis was performed on the same instrument with the following gradient sequence for separation: 0 to 5 min, 4% B; 5 to 90 min, 4 to 47% B; 90 to 100 min, 47 to 70% B; 100 to 100.5 min, 70 to 100% B; 100.5 to 115.5 min, 100% B; 115.5 to 116 min, 100 to 4% B; 116 to 130 min, 4% B.
Raw files from the glycoproteomic and proteomic analysis were identified from a UniProt FASTA database using Byonic and Byologic software (Protein Metrics) (43). Search parameters were set with a precursor mass tolerance of 10 parts per million (ppm) and quadrupole time-of-flight/higher-energy collisional dissociation (Q-TOF/HCD) fragmentation with mass tolerance of 20 ppm. Fixed modification was assigned to carbamidomethylation at cysteine. Variable modification of deamidated amino acids were assigned to asparagine and glutamine, methylation of lysine and arginine.
Cell-membrane fractions were reconstituted in 100 μL of 100 mM NH4HCO3 with 5 mM DTT (44). Samples were heated for 1 min at 100 °C followed by the addition of 2 μL peptide N-glycosidase F (New England Biolabs) to release the N-glycans. Samples were then incubated in a microwave reactor (CEM) at 20 W, 37 °C for 10 min. The resulting solutions were incubated in a 37 °C water bath for 18 h. Ultracentrifugation at 200,000 × g for 45 min was performed to isolate the N-glycan fractions from the protein precipitates. N-glycans were purified using a porous graphitic carbon (PGC) 96-well SPE plate. Samples were dried in vacuo using a miVac (SP Scientific).
N-glycan samples were reconstituted in 60 μL of Nanopure water prior to injection onto an Agilent 6520 Accurate Mass Q-TOF LC-MS System coupled to a PGC nanochip. N-glycan separation was conducted with a binary solvent system where solvent A was composed of 0.1% (vol/vol) formic acid and 3% (vol/vol) acetonitrile and solvent B was composed of 1% (vol/vol) formic acid and 90% (vol/vol) acetonitrile. The gradient sequence for separation was 0 to 2 min, 3% B; 2 to 20 min, 3 to 16% B; 20 to 40 min, 16 to 72% B; 40 to 42 min, 72 to 100% B; 42 to 52 min, 100% B; 52 to 54 min, 100 to 0% B; 54 to 65 min, 3% B with a constant flow rate of 300 nL⋅min−1. Spectra were collected every 1.5 s in positive-mode ionization with a mass range of m/z 600 to 2,000. The top five most abundant precursor ions in each MS1 spectrum were subjected to collision-induced dissociation fragmentation. The collision equation is Vcollision = 1.8 × (m/z)/100 V − 2.4 V. Data were analyzed using the MassHunter Qualitative Analysis B08 software (Agilent) Find by Molecular Feature function with a mass tolerance of 20 ppm. N-glycans were identified based on an in-house library with accurate mass. Relative abundances were determined based on integrated peak areas for each glycan composition and normalized to the summed peak areas of all detected glycans.
Supplementary Material
Acknowledgments
We thank the UCSF Biorepository & Tissue Biomarker Technology Core for tissue microarray image analysis. HistoWiz, Inc. performed immunohistochemical staining of murine tumors, and UDP-glucose levels were measured by Metabolon, Inc. Patient PDAC mRNA expression data were generated by The Cancer Genome Atlas Research Network. We thank members of the F.M. laboratory for discussions and critical reading of the manuscript. A.L.W. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation through postdoctoral fellowship number DRG-2214-15. Research reported in this publication was supported by the National Cancer Institute of the NIH under Award K99CA226363 (to A.L.W.). Funding for this work was provided by NIH Grant R01GM049077 (to Q.Z. and C.B.L.), Memorial Sloan Kettering Cancer Center Support Grant/Core Grant P30 CA008748 (to M.S.), and UCSF Research Allocation Program and National Research Foundation of Korea Grant NRF-2020R1C1C1013220 (to S.E.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing interest statement: A.L.W. has received research funding from Oncogenuity. M.S. has received research funds from Puma Biotechnology, AstraZeneca, Daiichi-Sankyo, Immunomedics, Targimmune, and Menarini Ricerche. He is on the scientific advisory board of Menarini Ricerche and the Bioscience Institute, a cofounder of medendi.org, and an employee and stockholder of AstraZeneca. F.M. is a consultant for Daiichi-Sankyo, Pfizer, Amgen, BridgeBio, Olema, OPNA-IO, PMV, Quanta, and Remedy Plan and has received research funding from Daiichi-Sankyo.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103592118/-/DCSupplemental.
Data Availability
All study data reported in this article are included in the main text and/or SI Appendix.
Change History
May 24, 2022: The affiliation of author Sung Eun Kim has been updated.
References
- 1.Ying H., et al., Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 30, 355–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Commisso C., et al., Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamphorst J. J., et al., Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasso C., Jansen G., Giovannetti E., Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit. Rev. Oncol. Hematol. 114, 139–152 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Ying H., et al., Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duggleby R. G., Chao Y. C., Huang J. G., Peng H.-L., Chang H.-Y., Sequence differences between human muscle and liver cDNAs for UDPglucose pyrophosphorylase and kinetic properties of the recombinant enzymes expressed in Escherichia coli. Eur. J. Biochem. 235, 173–179 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Wang L., et al., Expression of UGP2 and CFL1 expression levels in benign and malignant pancreatic lesions and their clinicopathological significance. World J. Surg. Oncol. 16, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jonge H. J. M., et al., Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 25, 1825–1833 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Wang Q., et al., SHP2 and UGP2 are biomarkers for progression and poor prognosis of gallbladder cancer. Cancer Invest. 34, 255–264 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Zeng C., Xing W., Liu Y., Identification of UGP2 as a progression marker that promotes cell growth and motility in human glioma. J. Cell. Biochem. 120, 12489–12499 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Li Y., et al., Multiomics integration reveals the landscape of prometastasis metabolism in hepatocellular carcinoma. Mol. Cell. Proteomics 17, 607–618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy Á., Munkácsy G., Győrffy B., Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11, 6047 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W., et al., Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci. Signal. 7, ra42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B., et al., Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B., et al., TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein C., et al., YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 11, e1005465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L., TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuita J.-C., Alape-Girón A., Thelestam M., Katz A., A point mutation in the UDP-glucose pyrophosphorylase gene results in decreases of UDP-glucose and inactivation of glycogen synthase. Biochem. J. 370, 995–1001 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Favaro E., et al., Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 16, 751–764 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Zois C. E., Favaro E., Harris A. L., Glycogen metabolism in cancer. Biochem. Pharmacol. 92, 3–11 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Pelletier J., et al., Glycogen synthesis is induced in hypoxia by the hypoxia-inducible factor and promotes cancer cell survival. Front. Oncol. 2, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pescador N., et al., Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS One 5, e9644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou T. Y., et al., Inactivation of hypoxia-induced YAP by statins overcomes hypoxic resistance tosorafenib in hepatocellular carcinoma cells. Sci. Rep. 6, 30483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bieberich E., “Synthesis, processing, and function of N-glycans in N-glycoproteins” in Glycobiology of the Nervous System, Advances in Neurobiology, Yu R. K., Schengrund C.-L., Eds. (Springer, New York, 2014), pp. 47–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaszuba K., et al., N-glycosylation as determinant of epidermal growth factor receptor conformation in membranes. Proc. Natl. Acad. Sci. U.S.A. 112, 4334–4339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holgado-Madruga M., Emlet D. R., Moscatello D. K., Godwin A. K., Wong A. J., A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature 379, 560–564 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Meisenhelder J., Suh P. G., Rhee S. G., Hunter T., Phospholipase C-γ is a substrate for the PDGF and EGF receptor protein-tyrosine kinases in vivo and in vitro. Cell 57, 1109–1122 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Cohen R., et al., Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget 6, 16832–16847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian D., et al., Identifying mouse developmental essential genes using machine learning. Dis. Model. Mech. 11, dmm034546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perenthaler E., et al., Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases. Acta Neuropathol. 139, 415–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salcedo Allende M. T., et al., Overexpression of Yes associated protein 1, an independent prognostic marker in patients with pancreatic ductal adenocarcinoma, correlated with liver metastasis and poor prognosis. Pancreas 46, 913–920 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Shao D. D., et al., KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158, 171–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapoor A., et al., Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158, 185–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broad Institute , DepMap Achilles 18Q3 public (2018). https://depmap.org/portal/. Accessed 15 January 2019.
- 35.Meyers R. M., et al., Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., et al., UDP-glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature 571, 127–131 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Führing J. I., et al., A quaternary mechanism enables the complex biological functions of octameric human UDP-glucose pyrophosphorylase, a key enzyme in cell metabolism. Sci. Rep. 5, 9618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee W.-N. P., et al., Metabolic sensitivity of pancreatic tumour cell apoptosis to glycogen phosphorylase inhibitor treatment. Br. J. Cancer 91, 2094–2100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q., et al., Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal. Chem. 91, 5433–5445 (2019). [DOI] [PubMed] [Google Scholar]
- 40.An H. J., et al., Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteomics 11, M111.010660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park D., et al., Characteristic changes in cell surface glycosylation accompany intestinal epithelial cell (IEC) differentiation: High mannose structures dominate the cell surface glycome of undifferentiated enterocytes. Mol. Cell. Proteomics 14, 2910–2921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park D. D., et al., Membrane glycomics reveal heterogeneity and quantitative distribution of cell surface sialylation. Chem. Sci. (Camb.) 9, 6271–6285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q., Xie Y., Xu G., Lebrilla C. B., Identification of potential sialic acid binding proteins on cell membranes by proximity chemical labeling. Chem. Sci. (Camb.) 10, 6199–6209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu G., et al., Unveiling the metabolic fate of monosaccharides in cell membranes with glycomic and glycoproteomic analyses. Chem. Sci. (Camb.) 10, 6992–7002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data reported in this article are included in the main text and/or SI Appendix.