Significance
PIP4Ks are druggable lipid kinases critical in cancer biology whose function in human immunity remains unknown. Here, we show that PIP4Ks specifically control the growth and activity of a subset of human immune cells called Tregs, isolated from the blood of healthy donors. Tregs function to exquisitely control the strength of the immune response. If the immune response is too strong, this can trigger autoimmune disease insurgence, while weak responses can lead to increased infections or enable tumor cell growth. Being able to selectively control Treg activity would impact the strength of immune responses and ultimately how we treat human diseases. Accordingly, we show that a drug-like inhibitor of PIP4K can be used to control Treg cell activity.
Keywords: phosphoinositide kinases, T-regulatory cells, immunosuppression, UHRF1, Phosphatidylinositol 5-phosphate 4-kinase
Abstract
Regulatory T cells (Tregs) play fundamental roles in maintaining peripheral tolerance to prevent autoimmunity and limit legitimate immune responses, a feature hijacked in tumor microenvironments in which the recruitment of Tregs often extinguishes immune surveillance through suppression of T-effector cell signaling and tumor cell killing. The pharmacological tuning of Treg activity without impacting on T conventional (Tconv) cell activity would likely be beneficial in the treatment of various human pathologies. PIP4K2A, 2B, and 2C constitute a family of lipid kinases that phosphorylate PtdIns5P to PtdIns(4,5)P2. They are involved in stress signaling, act as synthetic lethal targets in p53-null tumors, and in mice, the loss of PIP4K2C leads to late onset hyperinflammation. Accordingly, a human single nucleotide polymorphism (SNP) near the PIP4K2C gene is linked with susceptibility to autoimmune diseases. How PIP4Ks impact on human T cell signaling is not known. Using ex vivo human primary T cells, we found that PIP4K activity is required for Treg cell signaling and immunosuppressive activity. Genetic and pharmacological inhibition of PIP4K in Tregs reduces signaling through the PI3K, mTORC1/S6, and MAPK pathways, impairs cell proliferation, and increases activation-induced cell death while sparing Tconv. PIP4K and PI3K signaling regulate the expression of the Treg master transcriptional activator FOXP3 and the epigenetic signaling protein Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1). Our studies suggest that the pharmacological inhibition of PIP4K can reprogram human Treg identity while leaving Tconv cell signaling and T-helper differentiation to largely intact potentially enhancing overall immunological activity.
CD4+ T cells orchestrate the activation of the immune system as well as suppressing uncontrolled immune reactions (1). CD4+ T cells with distinct effector or regulatory functions are characterized by well-defined cytokine profiles and transcriptional programs giving rise to T-helper (Th) cells, follicular T cells (Tfh), and regulatory T cells (Treg) (2, 3). Tregs are critical for the maintenance of immunological self-tolerance and negative control of immune responses to nonself-antigens (4), and their loss leads to severe autoimmune diseases including type I diabetes, gastritis, and thyroiditis (4, 5). Circulating Tregs are generated in the thymus through positive selection of cells presenting high-affinity MHC class II restricted self-peptides (2, 3). Additionally, peripheral Tregs can be generated through differentiation of CD4+ conventional T cells (Tconv) preferentially in peripheral lymphoid tissues (6, 7). Tregs express high levels of the Treg lineage specification transcription factor FOXP3 (Forkhead box P3) (5, 8–10), which controls both their function and identity. Deletion of FOXP3 in Tregs leads to loss in suppressive activity (11), while its heterologous overexpression confers suppressive activity to Tconvs (8–10). FOXP3 controls the expression of human Treg signature genes including IL2RA (which encodes CD25, the IL-2 receptor alpha chain that enhances survival and IL-2 uptake), CTLA4, and TNFRSF18 (GITR) (12). In peripheral blood, Tregs represent ∼1 to 5% of CD4+ T cells, while in tumors they may represent up to 20 to 30% of the CD4+ T cell repertoire (13). Tumor infiltration by Tregs is often associated with poor prognosis, and Treg depletion can boost antitumor immune responses (13, 14). Thus, suppressing Treg function while maintaining Tconv activity could boost immune cancer cell killing and increase the capacity of immunotherapy in cancer treatment.
Polyphosphoinositide lipid messengers impact a wide array of cell processes including receptor-mediated control of cell proliferation and survival, migration, and transcription. While studies have shown that genetic and pharmacological inhibition of the PI3K pathway in mice and in human patients reduces Treg suppressive activity (15–17), much less is known about the role of other PPIns pathways. The PIP4K family of lipid kinases phosphorylate PtdIns5P to generate PtdIns(4,5)P2 (18). The three isoforms of PIP4Ks, 2A, 2B, and 2C, localize predominantly in the cytoplasm (19), the nucleus (19–22), and endomembranes (23), respectively, and have redundant and nonredundant roles in vivo (19, 24, 25). PIP4K2A−/−_PIP4K2B+/− mice suppress tumor development in p53-null mice (24), and PIP4K2C knockout mice develop characteristics of hyperinflammation (25). Moreover, a human SNP near the PIP4K2C gene associates with increased autoimmune diseases (26). These data suggest that PIP4K are intimately linked with immune responses.
Here, we reveal that PIP4K2B and PIP4K2C are required for human Treg cell proliferation, survival, and immunosuppressive functions. Transcriptome and cell biological analyses revealed that PIP4K depletion reduces PI3K/AKT signaling and proliferation and increases activation-induced cell death in Tregs while leaving Tconv cell activation intact. Reduced Treg function was associated with reduced expression of the essential Treg epigenetic regulator Ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) and decreased expression levels of FOXP3. Moreover, pharmacological inhibition of PIP4K2C phenocopies its knockdown. These data identify the importance of PIP4K signaling in human Treg immunosuppressive function and illustrate that their inhibition can reprogram Treg cells while sparing Tconv cell signaling and Th cell differentiation.
Materials and Methods
Materials.
Antibodies and short hairpin RNA (shRNA) sequences are listed in SI Appendix, Tables S1, S2, and S4 and qRT-PCR probes in SI Appendix, Table S3.
Isolation of T Cells.
Naïve, Treg, and Tconv cells were isolated from buffy coats of healthy donors obtained from Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Ca’Granda Ospedale Maggiore Policlinico in Milan and de-identified before collection and manipulation. Peripheral blood mononuclear cells (PBMCs) were purified on a Ficoll-Paque PLUS (GE Healthcare), and CD4+ T cells were purified (CD4+ T cells isolation kit, Miltenyi). Subsequently, Naïve (CD4+, CD62l+, CD45RO−), naïve Treg (CD4+, CD25+, CD127−, CD45RO−, CD62l+), or Tconv cells (CD4+, CD25−, CD127+, CD45RO−, CD62l+) were isolated by fluorescence-activated cell sorting (FACS) (BD FACSAria).
Primary T Cell Stimulation, Expansion, and Differentiation.
Naïve T cells were expanded using anti-CD3/CD28–coated magnetic Dynabeads (Life Technologies) for 5 d. Freshly isolated naïve Treg cells were expanded using rapid expansion protocol (REP) (27). For Treg differentiation, naïve T cells were T cell receptor (TCR) stimulated for 5 d together with IL-2 and TGFβ1 (5 ng/mL, Sigma Aldrich). For Th1/Th2 polarization, naïve T cells were TCR stimulated and cultured with IL-2, IL-12 (10 ng/mL, Miltenyi), and anti–IL-4 (2 μg/mL, Miltenyi) or with IL-2, IL-4 (10 ng/mL, Miltenyi), anti–IFN-γ (2 μg/mL, Miltenyi), and anti-IL12 (2 μg/mL, Miltenyi), respectively, for 8 d.
Immunostaining.
PBMC were washed with phosphate-buffered saline (PBS), and cell surface antigens were stained at 37 °C using specific conjugated antibodies. For intracellular staining, cells were treated with Fixation/Permeabilization Buffer (eBioscience) and then incubated with specific conjugated antibodies diluted in Permeabilization Buffer (Life Technologies) and analyzed by FACS (Canto II, BD Biosciences).
Molecular Cloning.
pLKO_1 vectors targeting specific genes were purchased from Sigma Aldrich (sh_Mission). Puromycin resistance sequence was substituted with sequences encoding either GFP or mCherry. pCDH vectors were purchased from System Bioscience, and PIP4K2B or PIP4K2C coding sequences were inserted between XbaI/BamHI restriction sites. Plasmids are available upon request.
Lentiviral Production and Primary T Cell Transduction.
Lentiviral particles were produced in Hek293T cells, mixed with Polybrene (8 μg/mL), and used to transduce 5 × 105 TCR-stimulated naïve T, Tconv, or Treg cells by spinoculation for 20 min at 2,000 rpm. Cells were seeded in Roswell Park Memorial Institute 1640 10% fetal bovine serum supplemented with IL-2 or treated to induce differentiation into Treg or Th1/Th2 cells. GFP/mCherry-positive cells were FACS sorted.
PIP4K, PI3K, and mTORC1 Inhibition.
Naïve T, Treg, and Tconv cells were treated with NIH-12848 (10:30 μm), CAL-101 (10 μm), and rapamycin (100 nm) to inhibit PIP4K2C, PI3Kδ, and mTORC1 signaling, respectively. Treg differentiation studies used naïve T cells freshly isolated from peripheral blood, directly treated with inhibitors as above, and differentiated into Treg cells. For analysis of UHRF1 or FOXP3 levels, Tregs expanded by REP were treated with NIH-12848 or CAL-101 for 24 or 48 h and analyzed by RT-qPCR or FACS. shRNA silenced cells were treated with inhibitors and TCR restimulated for 48 h. Finally, for analysis of cell viability, cell proliferation, and suppressive capacity of Tregs, cells were treated with the indicated doses of NIH-12848 for 48 h. Dimethyl sulfoxide (DMSO) was used as vehicle control.
Tconv Cell Suppression Assay.
Naïve T cells were labeled with Cell Trace (Violet/CFSE, Thermo Fisher) and cocultured with or without Treg cells at specified target-to-effector ratios. Cells were stimulated with CD3/CD28 (1 cell: 0. 1 bead) without IL-2 for 4 d (28). Proliferation was quantitated by the dilution of Cell Trace/CFSE using FACS and analyzed using FlowJo 8/10.
Interleukin Production Assay.
Interleukin production was assessed in the presence of brefeldin-A after stimulation with Phorbol 12-myristate 13-acetate (50 ng/mL) and ionomycin (1 μM) (29) using FACS.
Proliferation Assay.
Cell Trace–labeled (Violet/CSFE, Thermo Fisher) primary Treg or Tconv cells were stimulated with CD3/CD28 (1 cell: 0. 1 bead) for 4 d. Proliferation was monitored by the dilution of Cell Trace/CFSE using FACS.
Cell Cycle Analysis.
Cells were stimulated for 3 d, washed in PBS, and fixed in cold EtOH 70% overnight. Cells were washed with PBS and stained with propidium iodide (Abcam) and analyzed by FACS.
Cell Viability Analyses.
Cells were resuspended in Annexin V resuspension buffer and stained with Annexin V-APC (BioLegend) for 20 min at room temperature and then directly analyzed by FACS. Live/dead cell staining (Thermo Fisher) was used following manufacturer’s instructions.
qRT-PCR.
Cells were lysed, and RNA was extracted using a commercial kit (Invitrogen). Complementary DNA was synthesized using a Reverse-Transcription Kit (Life Technologies). qRT-PCR was performed using SYBR Green (Life Technologies) and the GAPDH gene used as housekeeping control.
Western Blotting.
Cell lysates were separated on bis-Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, electroblotted to nitrocellulose, blocked in PBS–Tween-20 (0.1%) dried milk (5%), and then incubated with the primary antibodies. After washing, blots were incubated with appropriate secondary antibodies conjugated to horseradish peroxidase or fluorescent dyes and visualized by chemiluminescence or Licor imaging, respectively. For analysis of signaling pathways, Treg and Tconv cells expanded through REP were transduced to silence PIP4K or treated with inhibitors (48 h) and restimulated with anti-CD3/CD28 Dynabeads for 1 h.
RNA Sequencing.
RNA was extracted and sequenced by Novogene (paired-end 150 base pairs [bp]). The reads were clipped to a max score and to a length of 120 bp using Trimmomatic and aligned to the human reference genome (GRCh38) using HISAT2. Gene-level quantitation and differential expression was determined using FeatureCounts and DESeq2. An Excel file is provided which contains a summary of the donors and shRNA (Dataset S1A), expression data, statistics for gene set enrichment analysis, comparison tables for, and the normalized count data (Dataset S1). Principal component analysis (PCA) and gene ontology analysis was carried out using prcomp and ClusterProfiler, respectively, and heat maps were generated in Pheatmap. Volcano plots were generated using the R package DeBrowser.
Statistical Analysis.
In all figures, n refers to cells isolated from different donors. Statistical analysis was performed employing the two-way unpaired Student’s t test or paired Student’s t test using Prism 9. *P < 0.05, **P < 0.01, ***P < 0.001 unless otherwise stated.
Results
PIP4K2B or 2C depletion impairs FOXP3 up-regulation and Treg differentiation of stimulated CD4+ T cells while sparing Th-specific transcription factors.
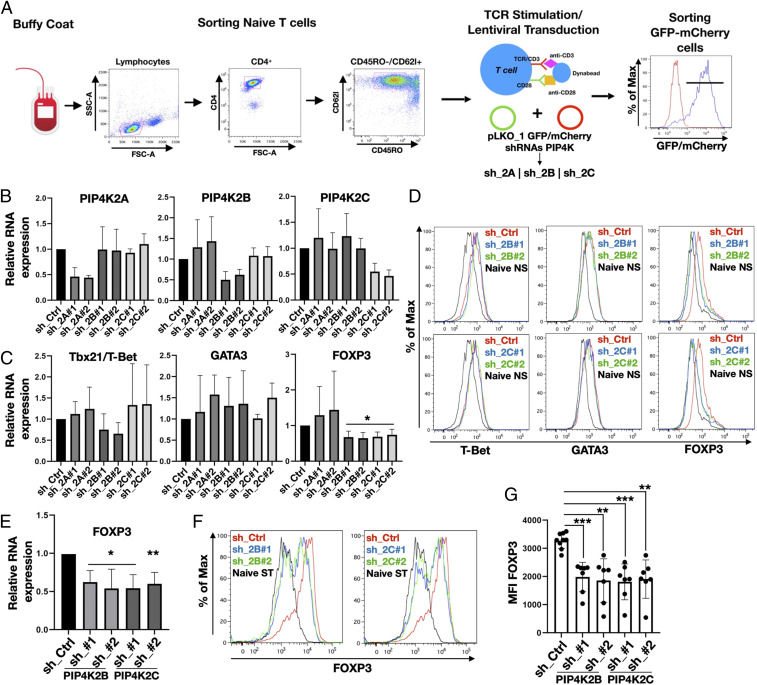
Antigen-MHC activation of naïve CD4+ T cells in the presence of specific cytokines induces their differentiation into distinct populations of T cells controlling immune responses to diverse organisms and foreign bodies (1). IFN-γ and IL-12 drive Th1 differentiation, which express the lineage specification factor T-bet (T-box transcription factor TBX21) (30). Th2 differentiation is mainly driven by IL-4, which up-regulates the transcription factor GATA3 (GATA binding protein), while IL6, IL21, IL23, and TGF-β drive Th17 differentiation and the expression of ROR-γτ (retinoic acid receptor-related orphan receptor gamma-T) transcription factor (31, 32). Although Tregs mainly differentiate in the thymus after antigen priming, TGF-β can induce peripheral Treg cell commitment by triggering FOXP3 expression in naïve T cells (33). Since in vitro TCR-mediated stimulation of naïve T cells induces transient expression of these master transcription factors (34, 35), we analyzed if PIP4K depletion (sh_2B/sh_2C) impacted on their expression. Naïve CD4+ T cells were sorted by FACS, stimulated with anti-CD3/CD28 coated beads, and transduced with lentiviruses to silence different PIP4K isoforms, and GFP/mCherry-positive cells were selected by FACS. Up to 90% of Tconv cells were positively transduced (Fig. 1A), and qRT-PCR (Fig. 1B) revealed sh-mediated isoform selective knockdown of PIP4K gene expression by 60 to 85% compared to the control samples (sh_Ctrl).
Fig. 1.
Silencing of PIP4K2B or PIP4K2C impairs FOXP3 expression in TCR-stimulated naïve T cells and Treg differentiation. (A) Graphical representation of isolation/stimulation and transduction of CD4+ naïve T cells. (B) PIP4Ks were silenced using different shRNAs targeting PIP4Ks isoforms (sh_2A, sh_2B, sh_2C). mRNA expression was analyzed using qRT-PCR and compared to control (sh_Ctrl). (C) qRT-PCR analysis of expression of transcription factors for Th1 (Tbx21/T-bet), Th2 (GATA3), and Treg (FOXP3) in cells depleted of PIP4Ks (n ≥ 3). (D) FACS analyses of T-bet, GATA3, and FOXP3 protein levels after PIP4K2B and PIP4K2C silencing (representative of n = 5). (E) PIP4Ks were silenced in naïve T cells differentiated into Tregs, and FOXP3 gene expression was assessed by qRT-PCR (n = 4). (F) Protein levels of FOXP3 in differentiated Tregs were analyzed by FACS. (G) Mean fluorescence intensity (MFI) quantitation of FOXP3 staining from independent experiments shown in F) (n ≥ 7). The charts represent means with SD. Statistical analyses were performed using unpaired Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. The number of replicates (n) refers to individual healthy donors.
Surprisingly, knockdown of PIP4K2B or PIP4K2C impaired FOXP3 messenger RNA (mRNA) and protein accumulation (Fig. 1 C and D) but not T-bet or GATA3 (Fig. 1 C and D) and slightly increased ROR-γτ expression (SI Appendix, Fig. S1A). Depletion of PIP4K2B or 2C in Tconv cells did not strongly affect lineage-specific cytokine synthesis such as IL-4 (Th2) or IFN-γ (Th1) (SI Appendix, Fig. S1 B and C), confirming a lack of impact on Th polarization.
As PIP4K signaling impacts on FOXP3 expression in naïve T cells, we tested how their depletion might regulate in vitro induction of Treg cells. Differentiation of naïve T cells toward Tregs can be induced in vitro through TCR stimulation in the presence of TGFβ1 and IL-2 (33), leading to strong up-regulation of FOXP3 expression (Fig. 1F). Strikingly, accumulation of both FOXP3 mRNA (Fig. 1E) and protein (Fig. 1 F and G) were impaired by depletion of PIP4K2B or 2C. A reduction in the accumulation of FOXP3 mRNA and protein was observed using four different shRNA-targeting constructs per isoform (SI Appendix, Fig. S1 D–F). The depletion of PIP4K2A had no effect on FOXP3 accumulation (SI Appendix, Fig. S1G). On the contrary, Th1 (driven by T-bet) and Th2 (driven by GATA3) differentiation was not changed by PIP4K2B or 2C depletion (SI Appendix, Fig. S2 A–D) and did not alter IFN-γ synthesis in Th1 cells (SI Appendix, Fig. S2E) but did slightly reduce IL-4 synthesis in Th2 cells (SI Appendix, Fig. S2F). These data suggest that depletion of PIP4Ks leave Th1 or Th2 differentiation pathways intact but impair in vitro Treg induction. To better define how PIP4K2B and 2C impact on FOXP3 expression and Treg function, we investigated their impact on CD4+FOXP3+ Tregs isolated from the peripheral blood of healthy donors.
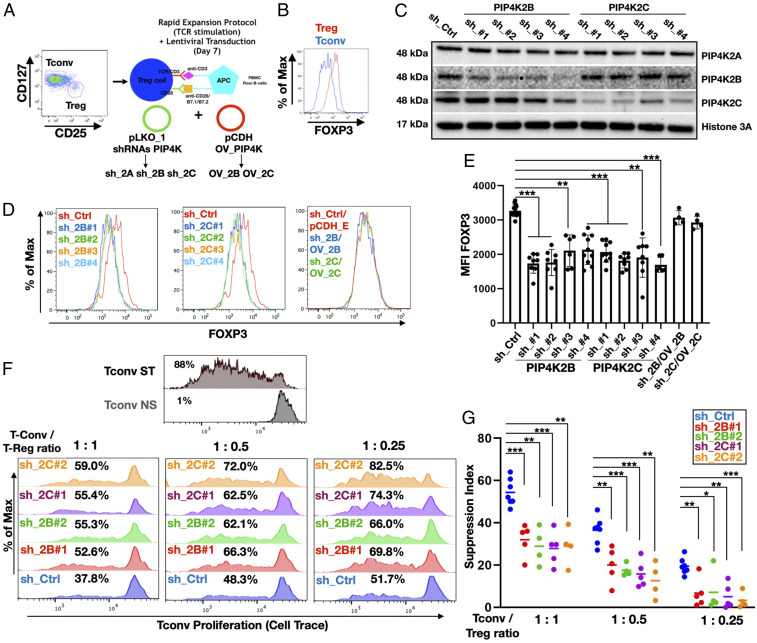
PIP4K2B and PIP4K2C Are Required for Treg Cell–Mediated Immunosuppression.
Circulatory Tregs can be subdivided in different subsets (3) depending on the expression of FOXP3 and CD45RA markers (7). Naïve Tregs (herein referred to as Tregs) isolated by FACS (CD4+/CD25high/CD127−/CD45RO−) showed high FOXP3 expression compared to conventional T cells (CD4+/CD25low/CD127+/CD45RO−) (Fig. 2 A and B). The depletion of PIP4K2B or 2C in expanded Tregs using four different shRNAs per isoform (Fig. 2C) impaired FOXP3 protein expression (Fig. 2 D and E) and reduced TCR-activated FOXP3 induction (SI Appendix, Fig. S3 A and B). PIP4K2A depletion did not change FOXP3 protein levels (SI Appendix, Figs. S2B and S3C). Conversely, PIP4K2B or 2C overexpression slightly increased FOXP3 expression in stimulated naïve T cells and in vitro–differentiated Treg cells (SI Appendix, Fig. S3 D and E) and in Treg cells (SI Appendix, Fig. S3 F–H). Moreover, exogenous overexpression of PIP4K2B or 2C rescued decreased FOXP3 levels in Treg cells after depletion of endogenous PIP4Ks (Fig. 2 D and E and SI Appendix, Fig. S3 G and H). Interestingly, combined depletion of PIP4K2B and 2C (SI Appendix, Fig. S3I) more strongly decreased FOXP3 expression compared to depletion of either PIP4K alone (SI Appendix, Fig. S3J). Continuous FOXP3 expression maintains Treg immunosuppressive activity (36), and therefore, we assessed if PIP4K depletion in Tregs impacts on their immunosuppressive function. CD3/CD28 stimulated strong proliferation in Tconv cells, which was dose dependently inhibited by control Tregs. However, knockdown of either PIP4K2B or 2C in Tregs reduced their capacity to suppress Tconv cell proliferation. Strikingly, combined knockdown of PIP4K2B and 2C more strongly attenuated Treg suppressive activity (Fig. 2 F and G and SI Appendix, Fig. S4 A and B). In contrast, PIP4K2B or 2C overexpression in Tregs increased Treg immunosuppressive activity (SI Appendix, Fig. S4C). Tregs express many proteins involved in suppressive activity and also up-regulate the expression of particular proteins within specific physiological and tumor-induced microenvironments (37, 38). We assessed the expression of some of these in response to PIP4K depletion but found only the expression of CTLA4, a FOXP3 target, was decreased by PIP4K2B/2C depletion (SI Appendix, Fig. S5A). CTLA4 is critical for Treg suppressive function (39), and FOXP3 knockdown reduced CTLA4 expression (SI Appendix, Fig. S5B). These data show that PIP4K2B and 2C impact on FOXP3 and CTLA4 expression to control Treg immunosuppressive activity.
Fig. 2.
PIP4K2B and PIP4K2C regulates FOXP3 expression and suppressive capacity of human Treg cells. (A) Graphical representation of isolation/stimulation and lentiviral transduction of Treg cells. (B) FACS analysis of FOXP3 expression in Tregs compared to Tconv cells. (C) Western blotting analysis of PIP4K2B and PIP4K2C silencing in Treg cells using indicated shRNAs. Empty pLKO_1 was used as control (sh_Ctrl). (D) FACS analyses of FOXP3 levels in Treg cells after silencing of PIP4K2B or PIP4K2C obtained as in C and Treg where PIP4Ks expression was rescued in the cells (PIP4K2B: sh_2B/OV_2B and PIP4K2C: sh_2C/OV_2C). (E) Mean fluorescence intensity (MFI) quantitation of FOXP3 staining of experiments in D (n ≥ 6 for sh_2B and sh_2C, n = 4 for sh_2B/OV_2B and sh_2C/OV_2C). (F) Suppression assay showing Tconv proliferation upon coculture with Treg cells depleted of PIP4K2B or PIP4K2C at different Treg/Tconv ratios. Tconv not stimulated (NS) or TCR stimulated (ST) were employed as negative and positive controls to gate proliferating cells. (G) Suppression index (diminished proliferation of Tconv cells) quantitation of experiments represented in F (n ≥ 4). The charts represent means with SD. Statistical analyses were performed using unpaired Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. The number of replicates (n) refers to individual healthy donors.
Depletion of PIP4Ks Reprogram the Treg Transcriptome.
PIP4K2B or 2C were depleted using two different shRNA in Tregs and Tconvs isolated from two different donors (SI Appendix, Materials and Methods). PIP4Ks were not differentially expressed in Tconv compared to Tregs (SI Appendix, Fig. S6A), and PIP4K depletion reduced FOXP3 expression in Tregs (SI Appendix, Fig. S6B). The transcriptome of these cells was analyzed using RNA sequencing (RNA-seq). After mapping reads to the genome, quantitation at the gene level was assessed and differentially expressed genes extracted using DESeq2. Volcano plots show PIP4K2B depletion changed the expression of 2,569 genes (false discovery rate [FDR] < 0.05 and absolute log2 fold change [LFC] >0.6), while PIP4K2C depletion changed 524 genes (FDR < 0.05 and absolute LFC >0.6) (Fig. 3A). The volcano plots also highlight how more genes were significantly changed in Tregs compared to Tconv cells after depleting PIP4K (Dataset S1 E–H). The differentiation and function of Tconvs was largely unchanged after depleting PIP4K, and accordingly, knockdown of PIP4K2B or 2C in Tconv cells changed only 135 and 66 genes, respectively (FDR = 0.05 and absolute LFC > 0.6, Dataset S1B and Fig. 3B). As depletion of either PIP4K2B or 2C decreased FOXP3 expression and inhibited Treg suppressive activity, we searched for genes that were coregulated in both knockdowns. There was a highly significant degree of overlap of genes regulated in Tregs by both PIP4K2B and PIP4K2C (230 genes: representation factor 3.4 and P < 2.34 × 10−71) (SI Appendix, Fig. S6C). Given this, we reanalyzed the RNA-seq data combining both sh_2B- and sh_2C-depleted Treg samples to generate a PIP4K2B_2C-regulated gene set (Treg_shPIP4K-DEG: 219 genes up and 200 genes down: p-adjusted < 0.05 and absolute LFC > 0.6). The deregulation of Treg_shPIP4K-DEG by depletion of either PIP4K2B (Treg:log(sh_2B/sh_Ctrl)) or 2C (Treg:log(sh_2C/sh_Ctrl)) in Tregs was highly correlated (Fig. 3C, R = 0.93). However, this gene set was only weakly deregulated by PIP4K2B or 2C depletion in Tconv cells (Fig. 3C). Strikingly, genes differentially expressed in Tconv after knockdown of PIP4K were not changed in Tregs (SI Appendix, Fig. S6D). PCA using Treg_shPIP4K-DEG genes show that PC1 and PC2, which account for 90% of the variation, strongly distinguish control, and PIP4K depleted Tregs as expected (Fig. 3D). Surprisingly, Treg_shPIP4K-DEG also strongly distinguishes Tconv cells from Tregs. Moreover, the PCA confirmed the lack of a strong effect of PIP4K depletion in Tconv, as all Tconv samples clustered closely together regardless of PIP4K depletion. To further explain the PCA, we generated heat maps to visualize the expression of Treg_shPIP4K-DEG in different T cell groups. The expression profile of Treg_shPIP4K-DEG was clustered into three groups, cluster 1, cluster 2, and cluster 3 (Fig. 3 E, Left), and the average expression of genes within each cluster was quantitated (Fig. 3E, Right). Cluster 1 genes were significantly up-regulated in Tregs after depletion of either PIP4K2B or 2C and were more highly expressed in Tconv compared to Treg regardless of whether PIP4K were depleted. Cluster 2 genes were significantly down-regulated in Tregs after depletion of PIP4K and had decreased expression in Tconv compared to Treg. In Tconv, cluster 2 gene expression was not further changed by depletion of PIP4K. Cluster 3 genes were decreased in Tregs after knockdown of either PIP4K2B or 2C, but their expression in Tconv was similar to control Tregs and did not show any change upon knockdown of PIP4K. The altered expression of some of the deregulated genes was verified using qRT-PCR of RNA extracted from both Treg and Tconv cells from different patient donors using qRT-PCR (SI Appendix, Fig. S6E). For example, the expression of HMOX1 and BAIAP3 is strongly up-regulated by either PIP4K2B or 2C depletion in Tregs and slightly up-regulated in Tconv cells (cluster 1). PLK1 and FOXP3 are down-regulated mainly in Tregs (cluster 2). PKMYT1, TK1, and IQGAP3 and UHRF1 (see PIP4K, PI3K, and UHRF1 Signaling Controls FOXP3 Expression and Treg Function) are down-regulated in Tregs, while their expression in Tconv mirrors their expression in Treg control cells (cluster 3). Similar down-regulation of Treg_shPIP4K-DEG was observed in an unrelated dataset comprising Tregs and Tconvs isolated from 12 different healthy donors (GSE166866; SI Appendix, Fig. S6F). These data suggest that depletion of PIP4Ks in Tregs reprograms the expression of many genes to mirror their expression in Tconv cells (cluster 1 and 2). The differential effects on gene expression observed on depletion of PIP4K supports our contention of different functional roles for PIP4Ks in Tregs compared to Tconvs. Cluster 3 genes are particularly intriguing, as they are decreased in Tregs upon depletion of PIP4K but are expressed to the same extent as control Tregs. We isolated cluster 3 genes and used them to interrogate an unrelated RNA-seq dataset comprising human Tconv and Tregs before and after T cell stimulation (CD3/CD28) isolated from four patient donors (Gene Expression Omnibus GSE138603). Cluster 3 genes were more highly expressed in Tconv compared to Tregs in the basal state. Surprisingly, cluster 3 genes are strongly up-regulated in response to T cell activation (Fig. 3F) in both Tconv and Tregs. Gene ontology analysis showed that cluster 3 genes were highly enriched for genes involved in the cell cycle, chromosome segregation, and mitosis (Fig. 3G). Cluster 1 and 2 genes were enriched for autophagy and translation terms, respectively (SI Appendix, Fig. S6 G and H). These data suggest that PIP4K depletion specifically decreases the expression of a subset of T cell activation–dependent genes in Tregs without impacting on their expression in Tconvs.
Fig. 3.
PIP4K2B and PIP4K2C regulate Treg cells transcriptomic profile. (A) Volcano plots (log fold change against the −log of the adjusted P value) of expressed genes in Tregs after depletion of PIP4K2B (Left) or PIP4K2C (Right). The numbers and dots in blue and red refer to genes that are significantly down-regulated or up-regulated, respectively (P < 0.05 and absolute log2fold change > 0.6). (B) Same as A, except PIP4K depletions were in Tconv cells. In both A and B, the x- and y-axis have the same minimum and maximum to illustrate the difference in effects of PIP4K depletions in Tregs compared to Tconv. (C) Scatter plots to show the changes in the expression of genes from the Treg_shPIP4K-DEG gene set after depletion of PIP4K in either Tregs or Tconvs as indicated. (Left) Depletion of PIP4K2B (log(sh_2B/sh_Ctrl) plotted on the x-axis and PIP4K2C depletion (log(sh_2C/sh_Ctrl) on the y-axis. (Middle) Depletion of PIP4K2B in Tregs against depletion of PIP4K2B in Tconv cells. (Right) Depletion of PIP4K2C in Tregs against depletion of PIP4K2C in Tconv cells. The data depict the average of three RNA-seq data sets for each knockdown in Treg and two in Tconvs. (D) The Treg_shPIP4K-DEG gene set was used for PCA. (E, Left) The Treg_shPIP4K-DEG (P-adjusted < 0.05) was used to generate a heat map with scaled rows and three hierarchical clusters. (Right) Gene sets from each cluster were extracted, and the average expression of genes within each cluster in Tregs and Tconv before and after depletion of PIP4Ks were quantitated. (F) Genes from cluster 3 (E) were used to interrogate their expression in an unrelated dataset comprising Tconv and Tregs before (Tconv_NS, Treg_NS) and after CD3/CD28 activation (Tconv_ST, Treg_ST) isolated from four separate donors. The heat map shows the average expression of each gene within cluster 3, while the graph depicts the average expression of all genes from cluster 3. The data illustrate that cluster 3 genes are up-regulated upon T cell activation. (G) Gene ontology enrichment analysis showing that cluster 3 genes are highly enriched for genes related to cell cycle, mitosis, and chromosome segregation. Box plots follow standard Tukey representation. Statistics are derived using Krushkal–Wallis hypothesis testing with post hoc–paired testing using Benjamini–Hochberg probability adjustments.
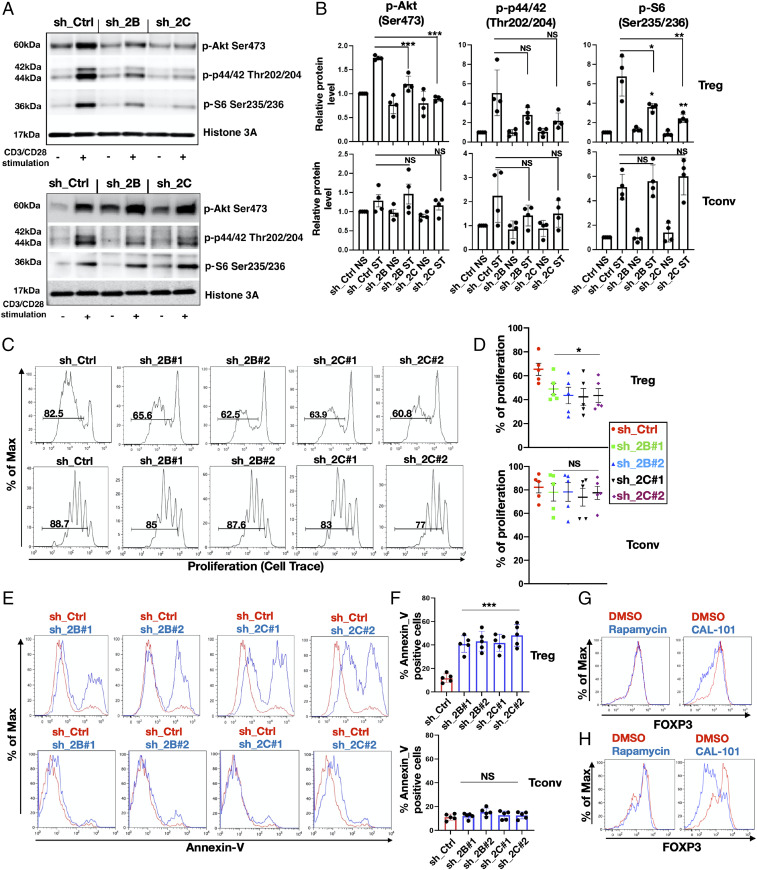
PIP4K Specifically Control PI3K/mTORC1 Pathways in Activated Treg Cells.
Since RNA-seq analysis indicated that the expression of a subset of T cell activation–dependent genes involved in the cell cycle and mitosis were deregulated in Tregs but not in Tconv (cluster 3), we investigated if and how PIP4Ks might differentially control specific signaling pathways stimulated by T cell activation. PIP4K2B and 2C impact on PI3K, mTORC1 signaling (24, 25, 40), and MAPK signaling (21, 24). Stimulation with CD3/CD28 activated the PI3K/mTOR and MAPK pathways in both Tregs and in Tconvs with similar time courses (SI Appendix, Fig. S7 A and B). PIP4K2B or 2C depletion in Tregs strongly attenuated CD3/CD28 activation of Akt, S6, and ERK (Fig. 4A and quantitated in Fig. 4 B, Top), while their depletion in Tconv had little impact (Fig. 4A and quantitated in Fig. 4 B, Lower). PI3K and mTORC1 signaling positively contribute to activated T cell viability and proliferation (41) but have different roles in T cells (17, 42, 43). We next analyzed how PIP4K2B or 2C depletion impacts on proliferative capacity and cell viability of Tregs and Tconvs. Tregs exhibited decreased proliferation when PIP4K2B or 2C was depleted (Fig. 4 C and D, Top), which was not observed in Tconv cells (Fig. 4 C and D, Lower) and was characterized by decreased cell numbers in S and G2/M phases of the cell cycle (SI Appendix, Fig. S7C) and decreased Ki67 staining (SI Appendix, Fig. S7D). Moreover, PIP4K2B or 2C depletion stimulated TCR activation–induced cell death in Tregs (Fig. 4 E and F, Top) but not in Tconvs (Fig. 4 E and F, Lower). The silencing of PIP4K2A had no effects on Treg cell proliferation or viability (SI Appendix, Fig. S7 E–G). These data confirm a stronger role for PIP4K2B and 2C in controlling signaling, proliferation, and viability in Tregs compared to Tconvs.
Fig. 4.
PIP4K2B and PIP4K2C silencing specifically inhibits TCR-dependent signaling and activation of Tregs. (A) PIP4K2B and PIP4K2C were silenced in Tregs (Top) and in Tconv (Bottom) and stimulated or not (CD3/CD28 stimulation) for 1 h. The activation of PI3K, MAPK, and mTORC1 signaling was assessed by Western blotting using the indicated antibodies. (B) Quantitation of experiments shown in A in Tregs (Top) and in Tconv (Bottom) upon PIP4Ks depletion and presented after normalization to sh_Ctrl NS and Histone 3A (n = 4). (C) Proliferation analysis (Cell Trace dye dilution) of Tregs (Top) and Tconv (Bottom) upon PIP4K silencing as indicated. (D) Quantitative data for experiments in G are presented (n = 5). (E) Cell apoptosis (Annexin V staining) in Tregs (Top) and Tconv (Bottom) upon PIP4Ks silencing. (F) Quantitative data are presented for the experiments in E (n = 5). (G and H) PI3K inhibition impacts on FOXP3 levels. Expanded Treg cells (Top) and in vitro–differentiated Treg cells (Bottom) were treated with rapamycin (100 nm) or CAL-101 (10 μM), and the levels of FOXP3 were analyzed by FACS. DMSO was used as vehicle control (n = 3). The charts represent means with SD. Statistical analyses were performed using unpaired Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. The number of replicates (n) refers to individual healthy donors.
PI3K Inhibition Decreases FOXP3 Levels in Tregs.
Since PI3K and mTORC1 signaling were altered by PIP4K, we investigated the relationship between the activation of these pathways and FOXP3 expression by their direct inhibition. Rapamycin treatment attenuated phosphorylation of S6, a downstream target of mTORC1, but not phosphorylation of AKT. Treatment with CAL-101 (PI3Kδ inhibitor) attenuated phosphorylation of both AKT and S6 (SI Appendix, Fig. S7H). The treatment of Tregs with CAL-101 but not rapamycin decreased expression of FOXP3 in Treg cells (Fig. 4G) and decreased FOXP3 accumulation in in vitro–differentiated Tregs (Fig. 4H). CAL-101 also decreased FOXP3 expression after CD3/CD28 stimulation (SI Appendix, Fig. S7I). Interestingly, combined PI3Kδ inhibition with depletion of either PIP4K2B or 2C almost completely attenuated the CD3/CD28-mediated increase in FOXP3 levels (SI Appendix, Fig. S7I). These data suggest that PIP4K utilizes PI3K-dependent and -independent pathways to impact on FOXP3 expression.
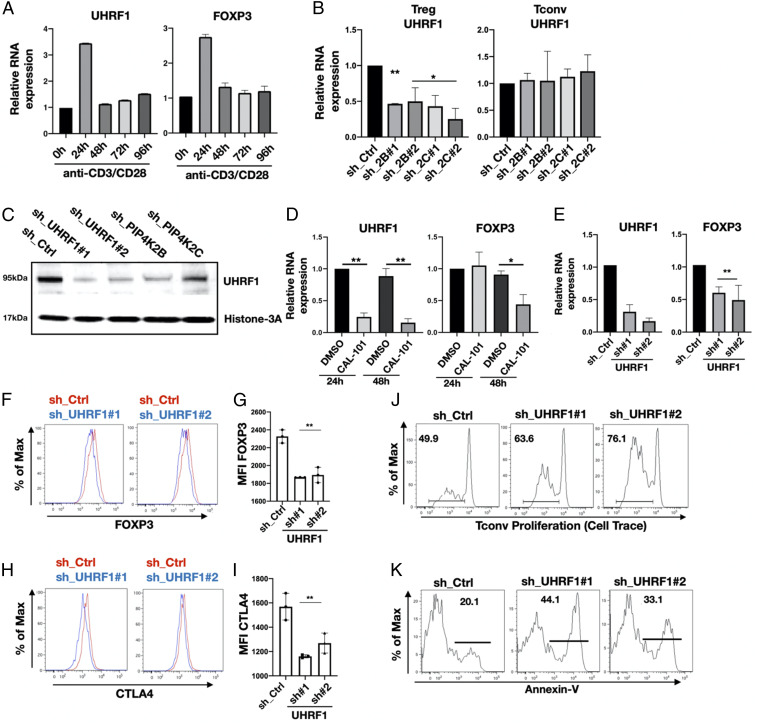
PIP4K, PI3K, and UHRF1 Signaling Controls FOXP3 Expression and Treg Function.
To further delineate how PIP4K signaling impacts on FOXP3 expression and Treg proliferation while sparing Tconv cells, we investigated genes from cluster 3. UHRF1 encodes a nuclear E3-ubiquitin ligase that interacts with and recruits DNA methyltransferases to control DNA methylation (44). UHRF1 is a key regulator of Treg differentiation, maintenance, and activity (45, 46) and is allosterically regulated by PtdIns5P, the substrate for PIP4K (47). Furthermore, the levels of UHRF1 and FOXP3 mRNA changed coordinately after CD3/CD28 stimulation (Fig. 5A), demonstrating that their expressions are both T cell activation dependent. We confirmed that depletion of PIP4K2B or 2C decreased UHRF1 mRNA levels in human Treg cells but not in Tconv cells (Fig. 5B) and decreased UHRF1 protein expression (Fig. 5C). The treatment of Tregs with the PI3Kδ inhibitor CAL-101 strongly inhibited UHRF1 expression and subsequently impaired FOXP3 expression (Fig. 5D). These data suggest that UHRF1 couples PIP4K/PI3K signaling to FOXP3 gene regulation. To directly implicate UHRF1 in the control of FOXP3 expression, we showed that depletion of UHRF1 using two different shRNAs decreased FOXP3 mRNA (Fig. 5E) and protein levels (Fig. 5 F and G) and reduced CTLA4 protein expression (Fig. 5 H and I). Importantly, UHRF1 depletion reduced Treg suppression of Tconv cell proliferation (Fig. 5J) and increased apoptosis in Tregs (Fig. 5K). These data indicate that UHRF1 silencing phenocopies PIP4K depletion in Tregs and that depletion of UHRF1 or PIP4K leads to two differentially regulated phenotypes. The first is decreased expression of FOXP3 and immunosuppression. The second is an increase in cell death and a decrease in cell proliferation. That these phenotypes are not related is illustrated by a lack of cell cycle blockade or increased apoptosis in Tregs after the knockdown of FOXP3, although there is a decrease in CTLA4 levels (SI Appendix, Fig. S8 A and B). Analysis of FOXP3 expression in live or dead cells isolated by FACS clearly demonstrated that reduced FOXP3 expression is not a consequence of PIP4K depletion–induced cell death (SI Appendix, Fig. S8C). These data are consistent with a pathway in which PIP4K2B and 2C impact on PI3K and UHRF1 signaling to control FOXP3 expression and Treg immunosuppressive activity. Additionally, PIP4K and UHRF1 signaling control Treg cell proliferation and apoptosis independently of FOXP3.
Fig. 5.
PIP4K depletion decreases UHRF1 levels in Tregs altering immunosuppressive capacity and cell viability. (A) Treg cells were TCR stimulated for the times indicated, and the expression of UHRF1 and FOXP3 was measured by qRT-PCR. (B) Expression levels of UHRF1 in Tregs (Left) and Tconv (Right) cells upon PIP4K silencing as indicated was analyzed using qRT-PCR. The empty pLKO_1 was used as control (sh_Ctrl) (n = 3). (C) Western blotting analysis of the expression levels of UHRF1 in Treg cells depleted of PIP4K or UHRF1 as indicated compared to control cells (sh_Ctrl). (D) RT-qPCR analysis of mRNA levels of UHRF1 (Left) or FOXP3 (Right) in Tregs treated with p110δ inhibitor (CAL-101, 10 μM) for 24 or 48 h (n = 3). DMSO was used as vehicle control. (E) UHRF1 was silenced in Tregs (sh_UHRF1), and the expression of UHRF1 (Left) and of FOXP3 (Right) were measured using qRT-PCR (n = 3). (F–I) UHRF1 was silenced, and protein levels of FOXP3 (F) or CTLA4 (H) were measured by FACS. (G and I) Quantitation of mean fluorescence intensity (MFI) values of FOXP3 and CTLA4 (n = 3). (J) UHRF1 was silenced in Tregs, and their immunosuppressive activity toward T conv cells was assessed (representative of n = 3). (K) UHRF1 was silenced in Tregs, and apoptosis was assessed by Annexin V staining (representative of n = 3). The charts represent means with SD. Statistical analyses were performed using unpaired Student’s t test, *P < 0.05, **P < 0.01. The number of replicates (n) refers to individual healthy donors.
Pharmacological Inhibition of PIP4K2C Phenocopies Gene Knockdown.
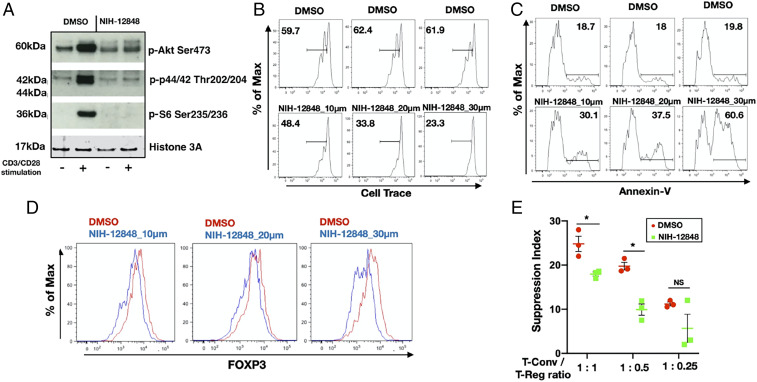
Given the recent interest in PIP4K inhibitors and to test if PIP4K activity underlies suppression of Treg function, we next tested if an irreversible inhibitor targeting PIP4K2C, NIH-12848 (48) might phenocopy PIP4K2C depletion in human Treg cells. NIH-12848 attenuated CD3/CD28-mediated stimulation of phosphorylation of AKT, S6, and MAP kinase in Tregs (Fig. 6A) but had little effect on the same pathways in Tconv (SI Appendix, Fig. S9A). NIH-12848 dose dependently inhibited CD3/CD28-stimulated proliferation (Fig. 6B), increased apoptosis (Fig. 6C), impaired FOXP3 expression in Tregs (Fig. 6D), and suppressed induction of in vitro Treg differentiation from naïve T cells (SI Appendix, Fig. S9B). Importantly, NIH-12848 impaired Treg suppressive activity on Tconv proliferation (Fig. 6E and SI Appendix, Fig. S9C).
Fig. 6.
Pharmacological inhibition of PIP4K2C with NIH-12848 phenocopies PIP4K2C silencing in Treg cells. (A) Treg cells were treated with DMSO or NIH-12848 (20 μM) for 48 h and then TCR stimulated (+) or maintained as controls (−). Lysates were probed with the indicated antibodies by Western blotting (representative of n = 3). Tregs were treated with DMSO or NIH-12848 for 48 h at the concentrations indicated, and cell proliferation (B), apoptosis (C), or FOXP3 levels (D) were measured by FACS (representative of n = 3). (E) Inhibition of PIP4K2C affects Treg cells capacity to suppress Tconv proliferation. Quantitative data for suppressive experiments are presented (n = 3). The charts represent means with SD. Statistical analyses were performed using unpaired Student’s t test, *P < 0.05. The number of replicates (n) refers to individual healthy donors.
Our studies illustrate that pharmacological inhibition or depletion of PIP4K can be used to suppress Treg cell proliferation, survival, and immunosuppressive activity whilst largely sparing Tconv cell signaling and Th differentiation (SI Appendix, Fig. S10).
Discussion
We show that genetic and pharmacological inhibition of two isoforms of PIP4K lipid kinases regulate FOXP3-dependent and -independent pathways to suppress human Treg function in three ways. Firstly, they control signaling through the PI3K, mTORC1/S6, and MAPK pathways. Secondly, they impact on Treg proliferation and apoptosis. Thirdly, they control the expression of the master transcriptional regulator FOXP3 and, more broadly, reprogram Treg cell transcription profiles. Importantly, PIP4K2B or 2C depletion in Tconvs does not attenuate PI3K and mTORC1 signaling or Th differentiation and has little impact on transcriptional output. Moreover, PIP4Ks affect the expression of a subset of T cell–dependent activation genes involved in cell cycle control only in Tregs. PIP4K silencing does reduce FOXP3 expression in activated Tconvs and reduces their differentiation into Treg in vitro. Broadly, PIP4K depletion would be expected to engender a hyperinflammatory environment in vivo, which is observed in PIP4K2C knockout mice (25), and implicate deregulation of Treg cells in the enhanced tumor suppression observed in PIP4K knockout mice (24). Interestingly, the nonredundancy of PIPK2B and 2C in human Treg immunosuppression suggests that Treg activity could be rheostatically controlled by using inhibitors that target one or both of the PIP4K isoforms in combination with clinically relevant inhibitors of PI3Kδ.
PIP4Ks control PtdIns5P and PtdIns(4,5)P2 levels in a kinase-dependent (21, 40) and -independent manner (49). PIP4Ks impact positively and negatively on PI3K/Akt signaling (40, 49–51), but in Tregs, we show that PIP4K2B and 2C depletion inhibits PI3K/mTOR. The decrease in T cell activation signaling imparted by PIP4K silencing in Tregs is reflected by a decreased expression of a subset of activation-dependent genes involved in cell cycle, mitosis, and chromosome segregation. These changes might explain the specific effect of PIP4K depletion on Treg proliferation and apoptosis compared to Tconv. Treg-specific inhibition has also been observed with PI3Kδ inhibitors (16, 17)
PIP4K2B is localized to the cytosol and nucleus (22), suggesting it may directly regulate gene transcriptional output. PIP4K2B controls nuclear PtdIns5P, which binds to nuclear proteins to modulate epigenetic signaling and transcriptional output (20, 52). Moreover, nuclear PtdIns5P allosterically regulates UHRF1 (47), directly linking PIP4K to the regulation of DNA methylation. UHRF1 appears to be a common mediator of PIP4K/PI3K-dependent changes in immunosuppression and proliferation (SI Appendix, Fig. S10). In mice, UHRF1 limits Treg and Tconv to Treg differentiation (53, 54), while UHRF1 (45, 55) and its partner, DNA methylase DNMT1 (56), are essential in Tregs for cell proliferation, FOXP3 and CTLA4 expression, and immunosuppressive activity. In mice, UHRF1 depletion in Tregs is associated with increased incidences of inflammatory diseases and enhanced tumor immunity (45, 55). Interestingly, reducing UHRF1 levels in tumor cells leads to the re-expression of tumor suppressor genes and a reduction of tumor cell growth (57). These data suggest that the antitumor effects of PI3K and PIP4K inhibition observed in vivo might in part be executed through the deregulation of UHRF1 expression in Tregs and perhaps more broadly in tumor cells.
The discovery that cell autonomous inhibition of PIP4Ks can inhibit tumor cell growth (24, 58, 59) has stimulated the development of PIP4K inhibitors (48, 60). The roles for PIP4K in reprogramming human Treg cells to attenuate their immunosuppressive activity, while sparing Tconv cell signaling and Th differentiation and thereby potentially enhancing tumor immunosurveillance, increases the relevance of these PIP4K inhibitors as anticancer therapeutics.
Supplementary Material
Acknowledgments
We acknowledge the Marie Curie fellowship (nuc-inositide: ID 625639 2015 to 2017), the National Institute of Molecular Genetics, the Malaysian government, and the Biotechnology and Biological Sciences Research Council for funding (BB/P003508/1). We thank Dr. Mariacristina Crosti, Dr. Tiziano Donnarumma, Prof. Klaus Okkenhaug, and Dr. Jonathan H. Clarke for help and reagents and Dr. Paolo Maiuri for scientific support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010053118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Luckheeram R. V., Zhou R., Verma A. D., Xia B., CD4+T cells: Differentiation and functions. Clin. Dev. Immunol. 2012, 925135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Josefowicz S. Z., Lu L. F., Rudensky A. Y., Regulatory T cells: Mechanisms of differentiation and function. Annu. Rev. Immunol. 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M. O., Rudensky A. Y., T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat. Rev. Immunol. 16, 220–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakaguchi S., Yamaguchi T., Nomura T., Ono M., Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Vignali D. A., Collison L. W., Workman C. J., How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottschalk R. A., Corse E., Allison J. P., TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J. Exp. Med. 207, 1701–1711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyara M., et al., Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Walker M. R., et al., Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25− T cells. J. Clin. Invest. 112, 1437–1443 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hori S., Nomura T., Sakaguchi S., Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Fontenot J. D., Gavin M. A., Rudensky A. Y., Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Wildin R. S., et al., X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 27, 18–20 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Chen C., Rowell E. A., Thomas R. M., Hancock W. W., Wells A. D., Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J. Biol. Chem. 281, 36828–36834 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Tanaka A., Sakaguchi S., Regulatory T cells in cancer immunotherapy. Cell Res. 27, 109–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curiel T. J., et al., Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Patton D. T., et al., Cutting edge: The phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J. Immunol. 177, 6598–6602 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Ali K., et al., Inactivation of PI(3)K p110δ breaks regulatory T-cell-mediated immune tolerance to cancer. Nature 510, 407–411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chellappa S., et al., The PI3K p110δ isoform inhibitor idelalisib preferentially inhibits human regulatory T cell function. J. Immunol. 202, 1397–1405 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Rameh L. E., Tolias K. F., Duckworth B. C., Cantley L. C., A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 390, 192–196 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Bultsma Y., Keune W. J., Divecha N., PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem. J. 430, 223–235 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Gozani O., et al., The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114, 99–111 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Jones D. R., et al., Nuclear PtdIns5P as a transducer of stress signaling: An in vivo role for PIP4Kbeta. Mol. Cell 23, 685–695 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Ciruela A., Hinchliffe K. A., Divecha N., Irvine R. F., Nuclear targeting of the beta isoform of type II phosphatidylinositol phosphate kinase (phosphatidylinositol 5-phosphate 4-kinase) by its alpha-helix 7. Biochem. J. 346, 587–591 (2000). [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke J. H., Emson P. C., Irvine R. F., Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am. J. Physiol. Renal Physiol. 295, F1422–F1430 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emerling B. M., et al., Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155, 844–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shim H., et al., Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl. Acad. Sci. U.S.A. 113, 7596–7601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raychaudhuri S., et al., Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 40, 1216–1223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudley M. E., Wunderlich J. R., Shelton T. E., Even J., Rosenberg S. A., Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J. Immunother. 26, 332–342 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collison L. W., Vignali D. A., In vitro Treg suppression assays. Methods Mol. Biol. 707, 21–37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai W., Li H., Song N., Li L., Chen H., Optimal method to stimulate cytokine production and its use in immunotoxicity assessment. Int. J. Environ. Res. Public Health 10, 3834–3842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szabo S. J., et al., A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Yang X. O., et al., T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J., Yamane H., Cote-Sierra J., Guo L., Paul W. E., GATA-3 promotes Th2 responses through three different mechanisms: Induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 16, 3–10 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Chen W., et al., Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuoka K., et al., T-bet upregulation and subsequent interleukin 12 stimulation are essential for induction of Th1 mediated immunopathology in Crohn’s disease. Gut 53, 1303–1308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernández-Hoyos G., Anderson M. K., Wang C., Rothenberg E. V., Alberola-Ila J., GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Williams L. M., Rudensky A. Y., Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 (2007). [DOI] [PubMed] [Google Scholar]
- 37.De Simone M., et al., Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity 45, 1135–1147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miragaia R. J., et al., Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chambers C. A., Kuhns M. S., Egen J. G., Allison J. P., CTLA-4-mediated inhibition in regulation of T cell responses: Mechanisms and manipulation in tumor immunotherapy. Annu. Rev. Immunol. 19, 565–594 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Jones D. R., Foulger R., Keune W. J., Bultsma Y., Divecha N., PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 27, 1644–1656 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Chi H., Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12, 325–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C., Chapman N. M., Karmaus P. W., Zeng H., Chi H., mTOR and metabolic regulation of conventional and regulatory T cells. J. Leukoc. Biol. 97, 837–847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abu-Eid R., et al., Selective inhibition of regulatory T cells by targeting the PI3K-Akt pathway. Cancer Immunol. Res. 2, 1080–1089 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bostick M., et al., UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Obata Y., et al., The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat. Immunol. 15, 571–579 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Helmin K. A., et al., Maintenance DNA methylation is essential for regulatory T cell development and stability of suppressive function. J. Clin. Invest. 130, 6571–6587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gelato K. A., et al., Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 54, 905–919 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Clarke J. H., et al., The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem. J. 466, 359–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D. G., et al., PIP4Ks suppress insulin signaling through a catalytic-independent mechanism. Cell Rep. 27, 1991–2001.e1995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carricaburu V., et al., The phosphatidylinositol (PI)-5-phosphate 4-kinase type II enzyme controls insulin signaling by regulating PI-3,4,5-trisphosphate degradation. Proc. Natl. Acad. Sci. U.S.A. 100, 9867–9872 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramel D., et al., PtdIns5P protects Akt from dephosphorylation through PP2A inhibition. Biochem. Biophys. Res. Commun. 387, 127–131 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Stijf-Bultsma Y., et al., The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell 58, 453–467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohkura N., Kitagawa Y., Sakaguchi S., Development and maintenance of regulatory T cells. Immunity 38, 414–423 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Sun X., Cui Y., Feng H., Liu H., Liu X., TGF-β signaling controls Foxp3 methylation and T reg cell differentiation by modulating Uhrf1 activity. J. Exp. Med. 216, 2819–2837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helmin K. A., et al., Maintenance DNA methylation is essential for regulatory T cell development and stability of suppressive function. J Clin. Invest. 130, 6571–6587, 10.1172/JCI137712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L., et al., Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood 121, 3631–3639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alhosin M., et al., Signalling pathways in UHRF1-dependent regulation of tumor suppressor genes in cancer. J. Exp. Clin. Cancer Res. 35, 174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jude J. G., et al., A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene 34, 1253–1262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitagawa M., et al., Dual blockade of the lipid kinase PIP4Ks and mitotic pathways leads to cancer-selective lethality. Nat. Commun. 8, 2200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manz T. D., et al., Discovery and structure-activity relationship study of (Z)-5-methylenethiazolidin-4-one derivatives as potent and selective pan-phosphatidylinositol 5-phosphate 4-kinase inhibitors. J. Med. Chem. 63, 4880–4895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.