Significance
The ability to detect precancerous clones and reconstruct cancer evolution is important for early cancer detection and improving prevention and treatment strategies. We present PolyG-DS, a sequencing method that combines the unique properties of polyguanine tracts (PolyGs) for cell lineage tracing with ultrahighaccuracy duplex sequencing (DS). PolyG-DS enables accurate and reproducible PolyG genotyping, providing high sensitivity for the detection of low-frequency alleles in mixed populations. This translates into an improved ability to identify clonal expansions within normal tissue, with potential application to detect cancer progression in preneoplastic diseases such as ulcerative colitis. Because PolyG-DS is driver mutation agnostic, it provides a universal, cost-effective approach for assessing tumor evolution across cancer types.
Keywords: cancer evolution, preneoplastic, phylogenetic reconstruction, carcinogenic fields, somatic evolution
Abstract
Polyguanine tracts (PolyGs) are short guanine homopolymer repeats that are prone to accumulating mutations when cells divide. This feature makes them especially suitable for cell lineage tracing, which has been exploited to detect and characterize precancerous and cancerous somatic evolution. PolyG genotyping, however, is challenging because of the inherent biochemical difficulties in amplifying and sequencing repetitive regions. To overcome this limitation, we developed PolyG-DS, a next-generation sequencing (NGS) method that combines the error-correction capabilities of duplex sequencing (DS) with enrichment of PolyG loci using CRISPR-Cas9–targeted genomic fragmentation. PolyG-DS markedly reduces technical artifacts by comparing the sequences derived from the complementary strands of each original DNA molecule. We demonstrate that PolyG-DS genotyping is accurate, reproducible, and highly sensitive, enabling the detection of low-frequency alleles (<0.01) in spike-in samples using a panel of only 19 PolyG markers. PolyG-DS replicated prior results based on PolyG fragment length analysis by capillary electrophoresis, and exhibited higher sensitivity for identifying clonal expansions in the nondysplastic colon of patients with ulcerative colitis. We illustrate the utility of this method for resolving the phylogenetic relationship among precancerous lesions in ulcerative colitis and for tracing the metastatic dissemination of ovarian cancer. PolyG-DS enables the study of tumor evolution without prior knowledge of tumor driver mutations and provides a tool to perform cost-effective and easily scalable ultra-accurate NGS-based PolyG genotyping for multiple applications in biology, genetics, and cancer research.
Cancers evolve through mutation, selection, and clonal expansion (1). While the mutational landscape of cancers has been extensively characterized (2), the evolutionary process that leads to malignancy remains poorly understood (3). Significant evidence indicates that cancer evolution starts many years prior to diagnosis (4), with clonal expansions preceding morphological changes (5). While these findings open a promising window for early cancer detection and prevention (6), the identification of early clonal expansions is challenging, even with modern next-generation sequencing (NGS) technologies, because mutant clones are often small and the mutations driving expansions are not always known. In addition, clonal expansions in healthy tissues are being increasingly recognized as a common feature of normal aging (7, 8), challenging our understanding of the boundaries between age-related and cancer-related, somatic evolution. Given the critical importance of somatic evolution in cancer and aging (6, 9), there is an urgent need for improved methods to sensitively detect and quantify clonal expansions and to determine their malignant potential by elucidating their phylogenetic relationship to tumors.
Polyguanine tracts (PolyGs) are homopolymeric repeats of guanine nucleotides that are highly prone to mutation (as high as 10−4 mutations per base per cell division in humans) due to slippage errors of polymerases during replication (10–13). As cells divide, they accumulate random insertions and deletions (indels) in PolyGs throughout the genome, which creates a fingerprint of their evolutionary history, encoded in the unique mutational profile of each cell (14). These unique fingerprints make PolyG genotyping an ideal system for detecting early clonal expansions and elucidating cancer evolutionary trajectories. Specifically, PolyG profiling offers four main advantages over gene sequencing to trace the evolution of cancer cells: 1) the rate of PolyG mutations is several orders of magnitude higher than that of nonrepetitive regions (10, 13), which yields more data per locus, leading to higher sensitivity to identify genetic relationships; 2) the inference of phylogeny is more accurate because PolyG mutations are thought to evolve neutrally, whereas driver mutation-based lineage mapping may be biased by positive selection (or negative selection during treatment) of markers being tracked; 3) PolyG mutations arise ubiquitously throughout the body, thus serving as universal lineage markers of cell replication that are agnostic of tissue type or prior knowledge of driver mutations; and 4) cell phylogenies can be resolved by analyzing only a small subset of PolyG loci (15, 16). Together, these features make PolyG genotyping a practical and cost-effective alternative to whole exome or whole genome sequencing for the study of tumor evolution. PolyGs have been used to trace the origin of colorectal metastases (15, 16) and to quantify their genetic diversity (17); to detect preneoplastic clonal expansions in ulcerative colitis (18, 19); and to build cell fate maps of mouse development (14, 20, 21).
In prior studies, PolyG genotyping was performed by fragment length analysis (PCR followed by capillary electrophoresis) (15–19). While this approach is quite effective, its scalability is limited. In general, fragment analysis is low throughput, labor intensive, and requires more input DNA for every marker added. Most importantly, it has limited resolution for the detection of subclonal variants because of artificial alleles introduced during PCR (11). These artificial alleles, often referred to as “stutter,” obscure true genotypes and limit the ability of the method to detect low-frequency alleles in DNA mixtures. Several technical and analytical approaches have been developed to leverage modern NGS tools for high-throughput and error-corrected genotyping of other forms of short tandem repeats (STRs) (22–25). While NGS overcomes some of the disadvantages of fragment analysis, genotyping of the most highly mutable, monomeric repeats (i.e., PolyG tracts) remains challenging because of the compounding errors introduced by PCR, cluster generation, sequencing, and alignment. These errors compromise genotyping accuracy and limit the detection of low-frequency mutations.
To address these limitations, we developed PolyG duplex sequencing (PolyG-DS), a method for accurate PolyG genotyping based on CRISPR-DS (26). CRISPR-DS combines the error-correction capabilities of duplex sequencing (DS) (27, 28) with target enrichment by CRISPR-Cas9 digestion to increase efficiency. DS employs double-strand molecular barcodes, which facilitates error correction by allowing the comparison of the independent sequences obtained from the complementary strands of DNA of each original molecule. This approach removes artificial mutations introduced during PCR and sequencing, thus producing highly accurate genotypes that can be used to trace cell lineage and to detect low-frequency alleles in genetic mixtures with high sensitivity. We illustrate the broad applicability of the technology for the detection of clonal expansions and reconstruction of tumor evolution.
Results
Development of PolyG-DS for Ultra-Accurate Sequencing of PolyGs.
Homopolymers have historically been challenging to accurately genotype with NGS, both because of polymerase slippage errors during amplification and sequencing and because of difficulties with sequence alignment when working with randomly fragmented, DNA-derived reads. To overcome these challenges, we used CRISPR-Cas9–targeted fragmentation to isolate PolyG sequences prior to DS library construction, as previously described in CRISPR-DS (26) (Fig. 1A). The targeted digestion creates fragments with invariant starting points, which facilitates sequence alignment by anchoring the read outside the repetitive region and enables the ligation of adapters with double-stranded molecular barcodes for DS. While PCR-based amplicon targeting also generates invariant read starting points, it does not allow for double-strand barcoding. Thus, we chose CRISPR-Cas9 for target enrichment and developed a purpose-built analytical pipeline for PolyG genotyping based on DS error correction (Fig. 1B) (SI Appendix, Methods).
Fig. 1.
Overview of PolyG-DS. (A) CRISPR-Cas9 target enrichment. gRNAs are designed to excise PolyG sites into fragments of ∼260 bp. Size selection using 0.5× solid-phase, reversible immobilization (SPRI) beads enables the recovery of the homogenously sized, excised fragments in the solution. Fragments are then ligated with DS adapters, which contain a double-stranded molecular barcode comprised of 10 bp random nucleotides and a 3′-dT overhang. (B) Error correction by DS. Molecules with same barcode are grouped and the most common genotype is selected as the single-strand consensus sequence (SSCS) call for each strand of DNA. SSCS calls are then compared, and a duplex consensus sequence (DCS) call is generated only if the two calls agree. (C) PolyG profiling enables the detection of mutant alleles and phylogenetic reconstruction. Precancerous and cancerous cells accumulate passenger PolyG mutations as they clonally expand. For each sample, the fraction of mutant PolyG markers (markers with a genotype significantly different between a given sample and a control sample by Fisher exact test) informs of the level of clonal expansion. In addition, genotype changes can be used to estimate genetic distance (Jensen-Shannon Distance, JSD) and build phylogenetic trees.
The excision of PolyG sequences with CRISPR-Cas9 in short fragments of homogeneous length enables a high degree of enrichment via size selection with solid-phase, reversible immobilization (SPRI) beads prior to adapter ligation and PCR. This approach improves the recovery rate of DS by 10-fold relative to ultrasonic fragmentation, reduces the required DNA input to as little as 10 ng, and decreases the time and cost of library preparation (26). While PolyG tracts are typically short sequences (<30 bp), we designed guide RNAs (gRNAs) (SI Appendix, Table S1) to excise PolyGs in fragments of ∼260 bp to enable sufficient sequencing for the alignment of unique flanking regions, while fitting into a 300-cycle Illumina run with full traversal of the PolyG sequence. For hybrid capture, we designed two biotinylated probes per fragment, maximizing specificity by basic local alignment search tool (BLAST) (29) and avoiding secondary structures (SI Appendix, Table S2 and Methods). We focused our PolyG locus selection on sets of PolyGs previously tested for fragment analysis (16, 19). The final PolyG target panel included 19 PolyGs, comprising 4,988 bp in total, which represents 0.0002% of the human genome. CRISPR-based size selection alone, in the absence of hybridization capture, yielded an ∼0.4% on-target rate, corresponding to an enrichment of ∼2,400×-fold, which is consistent with prior data (26). After hybridization capture, the percentage of on-target reads increased to an average of 91.6%, demonstrating the efficiency of the dual-enrichment approach.
DS of PolyG tracts required the development of a specialized analytical pipeline (Fig. 1B and SI Appendix, Fig. S1). Most algorithms developed for STR genotyping for forensics or microsatellite instability analysis consider the number of repeats but not the actual sequence variation (30). PolyGs are a special class of STRs because they are monomeric, highly mutable, and highly polymorphic, not just in number of repeats but also in sequence (e.g., monomers are sometimes interrupted by a different nucleotide), which provides an extra layer of data but also complexity. To leverage the informativity of all PolyG alleles, we utilized the lobSTR algorithm, specifically designed to genotype STRs (24), to obtain the sequence and length of the PolyG tract in each read. We then adapted the DS pipeline (28) to obtain a consensus genotype based on comparison of the tract sequence among reads sharing the same molecular barcode (Fig. 1B). The consensus algorithm first compares the genotypes of all the sequencing reads corresponding to the same strand of a DNA molecule and selects the most common genotype as the single-strand consensus sequence (SSCS) call. Then the two SSCS calls that correspond to the complementary DNA strands of original duplex DNA molecules are compared and, if their sequence agrees, a duplex consensus sequence (DCS) call is produced (Fig. 1B and SI Appendix, Fig. S2).
While ultrasensitive PolyG genotyping is useful for multiple applications, we focus here on the detection and tracing of precancerous and cancerous clones (Fig. 1C), because this is an area of high clinical interest and one for which we previously demonstrated the value of PolyG profiling using capillary electrophoresis (15–19). The samples used in the study, and their sequencing metrics, are listed in SI Appendix, Table S3.
PolyG-DS Yields Highly Accurate and Reproducible PolyG Genotypes.
PolyG tracts are prone to accumulating errors during PCR amplification and sequencing, which hampers the detection of subclonal mutations. We first tested whether DS was proficient at eliminating these errors by comparing the genotypes obtained with raw, SSCS, and DCS calls in the same sample. We observed that low-frequency, artifactual alleles were common in raw and SSCS calls but were efficiently eliminated at the DCS level (Fig. 2A). Raw genotypes for all the PolyGs tested typically contained hundreds of low-frequency allele sequences, likely derived from PCR, cluster generation, sequencing by synthesis, and/or alignment artifacts. The number of alleles was reduced by an order of magnitude in SSCS calls and to single digits in DCS calls (SI Appendix, Fig. S3A). It should be noted that while a single human diploid cell has only one or two different alleles, human biopsies containing thousands of cells might have additional low-frequency alleles that reflect the heterogeneous evolutionary history of different cell populations. While all PolyGs experienced a reduction in the number of alleles from SSCS to DCS calls, this reduction was significantly larger for longer PolyGs (P = 0.009 Spearman correlation, SI Appendix, Fig. S3B). This result is consistent with higher chances of polymerase slippage in the first PCR cycle in longer than in shorter PolyGs. First cycle errors cannot be corrected with SSCS, but they are corrected with DCS, which makes PolyG-DS especially suitable for accurate PolyG genotyping, particularly for longer PolyGs.
Fig. 2.
PolyG-DS accuracy and reproducibility. (A) Allele distribution for a representative PolyG analyzed without error correction (raw), with SSCS correction, and with DCS correction. The true allele (black bar) is obscured by artificial alleles (gray bars) when analyzing without error correction or with SSCS correction. DCS correction removes artificial alleles and reveals the true genotype of the sample. (B) Technical reproducibility of a sample analyzed in two independent experiments. In addition to homozygous and heterozygous alleles, PolyG-DS identifies a subset of low-frequency alleles (<0.1) likely to correspond to subclonal populations. The allele frequencies of the two replicates are highly correlated (Pearson test R2 = 0.995, P < 0.001).
We next tested the reproducibility of DCS PolyG profiling by comparing the allele frequencies obtained from two technical replicates using 100 ng of the same DNA extracted from colon stroma (Fig. 2B). A subset of PolyGs produced few DCS calls, which was mostly due to difficulties in lobSTR genotyping. To ensure high-quality genotype comparisons in this and subsequent experiments, we implemented a minimum of 40% genotyped raw reads and 25 DCS calls for each PolyG and sample to be analyzed (SI Appendix, Methods and Figs. S4 and S5). In this reproducibility experiment, 12 PolyGs met these criteria in both replicates and their allele frequencies were calculated and compared between the two samples. We observed excellent allele frequency concordance for near-homozygous and heterozygous PolyG alleles, as expected, but also for low-frequency alleles (<0.1), which are likely to represent subclonal populations present in this sample. We also demonstrated that the allele frequency was reproducible when decreasing the input DNA to 25 ng (R2 = 0.987) and 10 ng (R2 = 0.961) (SI Appendix, Fig. S6). Overall, these studies demonstrate the power of DS to eliminate artifactual alleles in PolyG NGS genotyping and to achieve high reproducibility in allele frequency quantification for DNA input as low as 10 ng.
PolyG-DS Provides High Sensitivity for Detecting Low-Frequency Alleles in Population Mixtures.
High accuracy in PolyG genotyping is expected to result in high sensitivity for the detection of mutations at low-variant allele frequencies. To estimate the sensitivity of our method, we mixed two DNA samples from different individuals (Samples A and B, colon stroma) at decreasing proportions of Sample A into B, ranging from 0.5 to 0.001 (Fig. 3A). This spike-in experiment simulates the challenge of detecting small subclones within samples and enables a rigorous assessment of sensitivity by comparing observed and expected allele frequencies. A total of 12 PolyGs produced successful genotypes across all samples (Fig. 3B and SI Appendix, Fig. S7A). While some PolyGs had overlapping alleles between the two samples, the presence of multiple alleles within each sample, the variability in allele frequency, and the four different mixing conditions in the experimental design provided a large number of data points (184) to compare observed and expected frequency. Both values were highly correlated, even when including low-frequency (<0.1) alleles (Pearson correlation, R2 = 0.99, P < 0.0001, Fig. 3C). We observed 50 out of 54 alleles (93% sensitivity) expected to be present at a frequency <0.1 and 22 out of 24 alleles (92% sensitivity) expected to be present at a frequency <0.01. To gain further insight into the question of whether low-frequency alleles corresponded to true alleles or artifactual alleles, we tracked one variant that was present in Sample A at a very low frequency (0.006; SI Appendix, Fig. S7B). We observed that this variant appeared in the 0.5 and 0.1 mixtures close to the expected frequency, supporting the true nature of this allele. Collectively, these results confirm the genotyping accuracy of our method and demonstrate its high sensitivity for low-frequency allele detection.
Fig. 3.
PolyG-DS sensitivity. (A) Spike-in experimental design. Sample A was spiked in Sample B at different proportions from 0.5 to 0.001. (B) Example of consistent allele frequency between observation (O) and expectation (E) for a representative PolyG. Insert shows the magnification of low-frequency alleles. (C) For all PolyGs and four sample mixtures, observed allele frequencies are plotted against expected allele frequencies, showing high concordance. Expected allele frequency levels are color coded. P value corresponds to Pearson correlation. (D) Heatmap of the genetic distance to Sample B, quantified using JSD based on DCS calls. Average JSD for each sample is indicated at the bottom of the panel. (E) PolyG-DS resolution for subclonal detection at raw, SSCS, and DCS level. As the proportion of Sample A decreased in mixture samples, the average JSD to Sample B also decreased (AVE: average). The dashed rectangle compares the JSD of the two last dilutions with the background level from the replication of Sample B. (F) Background noise measured by the JSD of raw, SSCS, and DCS genotypes of two technical replicates of Sample B.
We next calculated the Jensen–Shannon distance (JSD; SI Appendix, Methods), which has previously been used to quantify genetic divergence across a panel of PolyG tracts (16, 19), between each sample and Sample B (Fig. 3D). The average JSD recapitulated the genetic composition of each sample: the higher the proportion of Sample A in the mixture, the higher the genetic distance to Sample B (Fig. 3E). The genetic distance of the 0.5 and 0.1 mixtures was similar between raw, SSCS, and DCS calls, indicating that artifactual alleles in raw and SSCS calls (Fig. 2A) did not interfere with the detection of variants at larger allele frequencies (Fig. 3E). However, for the 0.01 and 0.001 mixtures, the JSD calculated with raw and SSCS calls was at the same level as the background JSD, indicating no ability to detect genetic differences in those mixtures when using raw and SSCS calls. The technical background was estimated by measuring the JSD between replicate samples (B to B) (Fig. 3F). For DCS calls, the background was ∼50% lower than for raw and SSCS calls, which enabled the resolution to distinguish the JSD of the 0.01 and 0.001 mixtures above the background (Fig. 3E). Consistently, phylogenetic trees built with this data demonstrated that DCS calls, but not raw or SSCS calls, resolved the low-frequency mixtures (0.01 and 0.001) with high-bootstrap confidence (SI Appendix, Fig. S8). These results demonstrate the benefit of using duplex error correction over single-stranded or no correction to detect variants at low-allele frequencies with maximal resolution.
PolyG-DS Detects Preneoplastic Clonal Expansions and Elucidates Their Phylogeny.
To prove the value of PolyG-DS for detection of subclonal mutations in vivo, we analyzed nondysplastic colonic biopsies from patients with ulcerative colitis, an inflammatory bowel disease that predisposes to colorectal cancer. By comparing capillary electrophoresis PolyG profiles in colonic epithelium and stroma, we previously showed that PolyG mutant clones in nondysplastic colonic biopsies distinguish the patients that have progressed to high-grade dysplasia (HGD) or cancer (18, 19). Conceptually, the presence of these clones indicates that the cells have gained the ability to expand abnormally (although not apparent histologically) and represent potentially precancerous populations, which is consistent with the well-stablished role of carcinogenic fields in this disease (31–34). To directly compare our method with the prior method, we analyzed a subset of 13 nondysplastic colonic biopsies previously profiled by capillary electrophoresis and confirmed that biopsies from patients with HGD or cancer elsewhere in their colon harbored a higher burden of PolyG mutant clones than biopsies from patients dysplasia free (Fig. 4A), replicating prior findings (19). As in fragment analysis by capillary electrophoresis, mutations were identified by comparing PolyG genotypes between colonic epithelium and stroma (SI Appendix, Methods). However, the increased sensitivity of DS enabled the identification of a larger fraction of mutations in patients with HGD or cancer, including the detection of mutations in a biopsy previously determined to have no clones by fragment analysis (Fig. 4A). PolyG-DS also revealed some mutations in patients without dysplasia, consistent with recent reports of clonal evolution in ulcerative colitis (34–36). To explore clonal expansion heterogeneity across biopsies, for one patient without dysplasia and two patients with HGD or cancer, we analyzed three independent nondysplastic colonic samples (SI Appendix, Fig. S9). None of the samples from the patient without dysplasia had mutant PolyG genotypes, while two out of the three biopsies from each patient with HGD/cancer had mutant genotypes (SI Appendix, Fig. S9). This rate of mutation in nondysplastic biopsies of patients that had progressed to cancer is consistent with our prior study and with the finding that, in these patients, clonal expansions occur as large patches across the colon (19). From a clinical perspective, we postulate that patients with more extensive clonal expansions might be at a high risk of developing HGD or cancer in the future. PolyG-DS provides a fast, easy, and sensitive approach to analyze clonal expansions in multiple nondysplastic samples, facilitating future studies in larger cohorts of patients with ulcerative colitis.
Fig. 4.
PolyG-DS detects preneoplastic clonal expansions and elucidates their phylogeny. (A) Identical colon biopsies from patients with ulcerative colitis with and without dysplasia and cancer were analyzed by PolyG-DS and capillary electrophoresis (from ref. 19), and the fraction of mutant genotypes was quantified in both cases. PolyG-DS showed higher sensitivity to identify mutant genotypes than capillary electrophoresis. (B) Proximal colon diagram from a patient with ulcerative colitis that underwent colectomy. Colon samples (represented by squares) were collected at evenly spaced intervals within an alphanumeric grid (diagram based on ref. 19). Histological diagnoses are color-coded. The four samples analyzed by PolyG-DS are indicated. (C) Heatmap showing the genetic distance (JSD) from the four colonic epithelial samples to stroma. JSD was calculated by comparing PolyG genotypes obtained with DCS calls. (D) Example of a mutant allele shared between normal tissue (NEG) and dysplastic tissue (LGD and HGD1). The allele frequency increased with the degree of pathological changes. (E) Phylogenetic reconstruction of preneoplastic progression in ulcerative colitis. The phylogenetic tree was built with UPGMA. Bootstrap values (n = 1,000) are shown for each branch. HGD1 and LGD are derived from same normal tissue (NEG), while HGD2 followed a different pathway of progression. NEG: Negative for Dysplasia. LGD: Low Grade Dysplasia. HGD: High Grade Dysplasia.
Next, we aimed to determine whether PolyG-DS could resolve the clonal relationships among dysplastic biopsies. First, we tested the method in a simple scenario including cancer, nondysplastic, and stroma control biopsies and demonstrated that PolyG-DS profiling accurately resolved the cancer lineage using phylogenetic reconstruction based on the JSD and unweighted pair group method with arithmetic mean (UPGMA) (37) (SI Appendix, Fig. S10 and Methods). We then used the same approach to reconstruct the evolutionary history of four adjacent colon biopsies procured from a colectomy specimen from a patient with ulcerative colitis (Fig. 4B). Pathological assessment indicated that one biopsy was negative for dysplasia, one exhibited low-grade dysplasia (LGD), and two contained HGD (HGD1 and HGD2). In our prior study using fragment analysis by capillary electrophoresis, we identified mutations shared by HGD1, LGD, and negative for dysplasia biopsies but not HGD2 (19). Our analysis here identified multiple markers that supported those findings (Fig. 4 C and D) and elucidated the evolutionary path of these biopsies (Fig. 4E). The phylogenetic tree structure indicated that HGD1, LGD, and the negative biopsy evolved as a clonal patch, whereas HGD2 was unrelated. These findings are consistent with the notion of clonal mosaicism and synchronous clonal expansions in ulcerative colitis (33) and illustrate the utility of our method to study the evolutionary trajectories of dysplasia and cancer.
PolyG-DS Detects Age-Related PolyG Mutations Previously Identified by Capillary Electrophoresis.
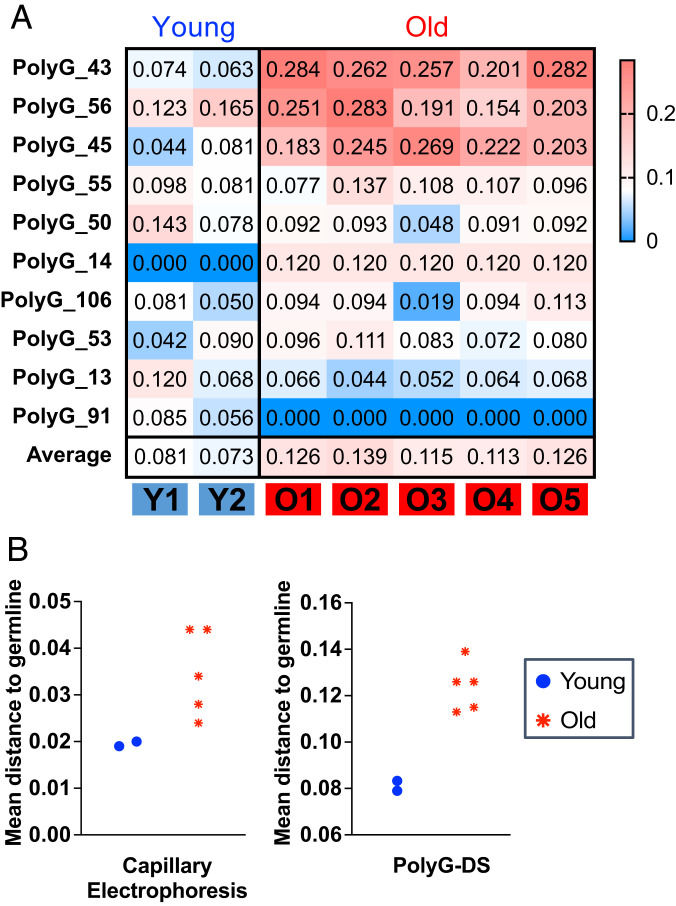
To further compare PolyG-DS to fragment analysis by capillary electrophoresis and to highlight the ability of this method to detect PolyG mutations that accumulate in an age-dependent manner as cells proliferate, we next analyzed DNA from a previously established collection of clonal organoids derived from single normal intestinal epithelial stem cells procured from young (4 y-old) and older (66 y-old) individuals (38). Prior profiling of these samples by capillary electrophoresis demonstrated that organoids derived from older stem cells had more PolyG mutations than organoids derived from younger stem cells, quantified as the genetic distance to a polyclonal control sample representative of the germline genotype (16). Using a subset of the same samples, PolyG-DS revealed multiple markers with higher genetic distances in older than younger organoids (Fig. 5A). When the average genetic distances between young and old were compared, the results closely resembled prior findings by capillary electrophoresis (Fig. 5B), further demonstrating the value of PolyG-DS as an NGS alternative to fragment analysis for PolyG profiling.
Fig. 5.
PolyG profiling of clonal organoids derived from single, normal, and intestinal stem cells from a child (age = 4) and an older individual (age = 66). (A) Heatmaps showing the genetic distance (JSD) between intestinal organoids from each individual and a germline control sample (a polyclonal mixture of intestinal cells from the same individual). JSD was calculated by comparing PolyG genotypes obtained with DCS calls. Only PolyG markers that yielded data for all samples were included. The average JSD for each sample is indicated at the bottom of the panel. (B) Comparison of mean distance to germline calculated from capillary electrophoresis data obtained in prior study (16) and from PolyG-DS.
PolyG-DS Accurately Reconstructs Ovarian Cancer Evolution.
To expand the range of applications of PolyG-DS, next we explored the value of our method to study the evolutionary history of high-grade serous carcinoma (HGSC), a cancer type that originates in the fallopian tube and disseminates locally, following tumor trajectories that are not well understood (39). From a patient who underwent debulking surgery for HGSC (pT3cN1), we obtained two primary tumor biopsies from bilateral fallopian tubes, two primary tumor biopsies from bilateral ovaries, one omental metastatic biopsy, and two normal biopsies from the peritoneum (Fig. 6A and SI Appendix, Methods). We measured the JSD of each sample relative to a normal peritoneal control sample to build a marker-level genetic distance heatmap (Fig. 6B) and used pairwise distances between samples and the UPGMA method to reconstruct their phylogeny (Fig. 6C). The samples clustered in branches that had high-confidence bootstrap but showed no obvious relationship with their anatomical location (e.g., samples from opposite sides clustered together). These results are consistent with prior findings indicating extensive mixing of cells within the peritoneal cavity during metastatic spread (40). Nevertheless, we considered the possibility that the tree structure could be affected by variable tumor purity. Previous studies that leveraged PolyG genotyping for phylogenetic reconstruction (16, 17, 19) had relied on samples with consistently high purity, achieved by microdissection of areas with high–tumor cell content under the microscope. Here, manual dissection of pure tumor was intentionally not performed to represent the usual clinical collection and to challenge our method to detect PolyG mutations in the presence of contaminating normal tissue. This is also the reason that, here, we chose to employ UPGMA instead of the previously used neighbor-joining algorithm for phylogenetic reconstruction, as we found that the ultrametric trees produced by UPGMA were very robust to varying levels of normal admixture. The UPGMA algorithm effectively scales branch lengths, such that all samples have the same distance from the root. This overcomes the problem of artifactually shortened branch lengths due to impurity and results in the correct tree topology.
Fig. 6.
Reconstruction of ovarian cancer evolution using PolyG-DS. (A) Anatomical diagram of tumor location. RFT, primary tumor in the right fallopian tube; ROV, primary tumor in the right ovary; LFT, primary tumor in the left fallopian tube; LOV, primary tumor in the left ovary; MT, metastasis in omentum; and NP, normal peritoneum. Two independent normal peritoneal biopsies were collected (NP1 and NP2). (B) Heatmap of genetic distance to normal tissue (NP1). (C) The phylogenetic tree was built with UPGMA. Confidence values for each interior branch were calculated from 1,000 bootstrap replicates.
To investigate tumor purity, first we performed histological examination of adjacent formalin-fixed, paraffin-embedded (FFPE) tissue, which confirmed variable tumor content (SI Appendix, Fig. S11A). Then, we performed TP53-targeted standard DS and used the mutant allele frequency (MAF) of the TP53 driver mutation to correct the PolyG genotypes of each biopsy by normalizing their known proportion of normal cells (SI Appendix, Methods). TP53 mutations are an early and ubiquitous event in HGSC (41–44) and are typically accompanied by the loss of the second allele (45, 46), which makes them ideal surrogate markers of tumor purity. Deep DS of TP53 (>1,000×; SI Appendix, Methods) revealed the driver tumor mutation (chr17: g.7674241C > T, p.S241F) at variable frequencies across samples, in general agreement with pathological estimates of tumor purity (SI Appendix, Fig. S11A) and in close correlation with TP53 loss of heterozygosity (R2 = 0.999, P < 0.001; SI Appendix, Fig. S11B). These results confirm the loss of the second allele and further validate the use of the driver TP53 mutation MAF to estimate tumor content for PolyG profile correction. Genetic distances calculated on corrected profiles yielded a tree that closely replicated the topology of the tree obtained without correction (SI Appendix, Fig. S11C). This result demonstrates that PolyG-DS coupled with the UPGMA method can resolve phylogenies in samples with variable levels of normal tissue contamination. While this application of PolyG-DS requires more extensive testing with larger numbers of samples and patients, this example demonstrates the versatility of the assay and its potential for high-throughput testing of tumor evolution in research and clinical settings.
Discussion
We have shown that by leveraging CRISPR-Cas9–based target enrichment by size selection with DS error correction, we can perform ultra-accurate PolyG genotyping, enabling high resolution for the detection of subclonal variants and cell lineage tracing. By correcting and comparing the genotypes of both strands of DNA independently, PolyG-DS efficiently eliminates artifactual alleles that plague raw sequencing reads. A substantial proportion of these alleles persisted after single-strand error correction, indicating that they probably correspond to stutter in the first PCR cycles and therefore are not removable by single-stranded barcoding methods. Consistently, longer PolyG repeats, which have higher rates of polymerase slippage, exhibited higher reduction in the number of alleles from single-stranded to double-stranded error correction than shorter PolyG repeats. Because of the accuracy of PolyG-DS, genotypes were highly reproducible across replicates, and low-frequency mutations (<0.01) could be detected in spike-in samples according to expectation, demonstrating the high resolution of the assay for PolyG mutation detection.
Our prior studies using PolyG fragment analysis by capillary electrophoresis already demonstrated the value of PolyG genotyping to detect precancerous clonal expansions (18, 19) and trace cancer evolution (15, 16), but this approach is low throughput, time intensive, and cannot detect genetic heterogeneity at low allele frequencies (47). By reanalyzing a subset of samples previously analyzed by capillary electrophoresis and by using a similar number of PolyG markers, we provided a side-by-side demonstration of the increased resolution for subclonal detection enabled by PolyG-DS. An additional advantage of PolyG-DS compared to capillary electrophoresis is that it is highly multiplexable. There are thousands of PolyG tracts in the human genome, and a prior study demonstrated the feasibility of simultaneously interrogating ∼2,500 STR by using CRISPR-Cas9 excision followed by NGS with selective primers (25). With PolyG-DS, such a high number of markers is unlikely to be necessary for most applications, since subclonal resolution can already be achieved with a small and inexpensive subset of markers. We note, however, that our panel included PolyGs that consistently failed in some samples because of sequence complexities that made their genotyping challenging. Thus, it is desirable to test hundreds of markers in a single assay in order to select the best performers and optimize the panel for maximum information with minimal cost. These efforts are currently underway.
While ultra-accurate PolyG genotyping has multiple applications in genetics and biology, here we have focused on the ability of these sequences to inform about preneoplastic and neoplastic progression. PolyG mutations are uniquely suited for this purpose because, unlike cancer driver gene mutations, they are neutral (e.g., not selected) to our knowledge, and the same mutation type (indels) affects all replicating cells. This makes PolyG mutations informative of (pre)neoplastic processes without the limitation of cancer type. In addition, their high mutability and molecular clock nature makes them ideal candidates for phylogenetic reconstruction. We have leveraged these properties to illustrate the value of PolyG-DS in two different settings: the identification of clonal expansions in normal tissue and tracing of cancer evolution.
The study of clonal expansions in morphologically normal tissue has become especially relevant after multiple studies have revealed prevalent small size clonal expansions as an intrinsic component of normal aging across tissues (8, 48). Here, we have replicated prior findings by capillary electrophoresis, demonstrating more PolyG mutations in 66 y-old than in 4 y-old normal intestinal stem cells (16). In addition, we have confirmed the ability of PolyG-DS to detect more frequent clonal expansions in the nondysplastic colonic epithelium of patients with ulcerative colitis that progressed to cancer compared to those without cancer—a finding that we initially reported in two independent studies using capillary electrophoresis (18, 19). While a background level of clonal expansion is expected in noncancerous colonic epithelium of ulcerative colitis patients because of the underlying remodeling of the inflamed mucosa (34–36), different selective pressures enable the later expansion of precancerous clones in the subset of patients who eventually progress to developing colorectal cancer (36). In this context, measuring the “occult” (e.g., not histologically apparent) evolutionary process in nondysplastic tissue might provide venues to predict cancer progression in ulcerative colitis and potentially other preneoplastic diseases (33). PolyG-DS offers a promising tool for this purpose and, more generally, for the quantification of clonal expansions in normal tissue agnostic to specific driver mutations.
Regarding PolyG profiling in tumors, prior studies by fragment analysis already illustrated the value of this approach to trace a cancer’s evolutionary history and to elucidate the pathways of metastatic dissemination, with important implications for the management of advanced disease (15–17). Here, we provide proof of principle of the value of PolyG-DS to reconstruct the evolution of early disease, specifically the dysplastic stages that precede colorectal cancer progression in ulcerative colitis. The analysis of the genetic relationships among histologically normal, dysplastic, and cancerous samples is critical to understanding the evolution of cancer in this disease and to identifying the specific dysplastic lesions that are cancer precursors, which is an unmet clinical need (49). Similar approaches could be employed to study the evolution of precancerous lesions in other cancer types in order to elucidate what lesions are genetically linked to cancer (aggressive) versus those that are unrelated to cancer (indolent) (50, 51). In addition, we have demonstrated that PolyG-DS could be useful to characterize complex cancer evolutionary trajectories, such as the one identified in this study and in prior studies of HGSC evolution (40, 45). Here, we analyzed typical clinical samples without time-intensive tissue processing to determine whether ultra-accurate PolyG genotyping coupled with UPGMA phylogenetic reconstruction was tolerant to normal tissue contamination. UPGMA produces ultrametric trees, which is helpful in avoiding branch-length artifacts caused by severely variable purity. By comparing phylogenetic trees before and after purity correction, we demonstrated that the topology of the trees was highly consistent. However, we have not formally tested how much contamination is tolerable, which would likely depend on the cancer type and the selected panel of PolyG markers. Thus, future studies should approach this issue carefully. Furthermore, we anticipate that as more NGS-based PolyG data become available, we will be able to develop rigorous methods for subclonal reconstruction from these data. Currently, the PolyG allele mixture present in each sample is fed “as is” into distance-based, phylogenetic reconstruction algorithms. While this method produces highly accurate sample trees, it cannot resolve subclones, which is a significant limitation. The digital data generated by PolyG-DS has the potential to illuminate subclonal structure, if used in conjunction with an appropriate evolutionary model (52).
Efficient target enrichment by CRISPR-Cas9 fragmentation depends on DNA quality and therefore the method, as currently developed, is not immediately applicable to FFPE samples. An alternative includes the elimination of CRISPR-Cas9 enrichment in favor of two rounds of hybridization capture (53). This approach, however, requires more DNA, and it is more expensive long term because of the additional cost of two rounds of capture versus the upfront cost of gRNAs. Other approaches to improve the assay for future applications include the implementation of newer algorithms for PolyG genotyping (30), the inclusion of a larger number of markers and selection of the best performers, and the addition of CRISPR-Cas9 guides and hybridization probes for selected target genes to simultaneously screen for mutations in PolyG tracts and candidate cancer drivers.
In summary, we have developed a method to transition PolyG profiling from capillary electrophoresis to an NGS sequencing platform that leverages the advantages of CRISPR-Cas9 target enrichment and error correction by DS. We have demonstrated that the profiling of less than 20 PolyG provides sufficient information to detect subclonal alleles and resolve cancer phylogenies. In addition, the high accuracy and sensitivity of PolyG-DS allows the identification of clonal expansions in histologically normal tissue without prior knowledge of driver mutations. PolyG-DS is high throughput and readily scalable to fit different study needs, providing a comprehensive solution to PolyG profiling for multiple applications in biology and medicine.
Materials and Methods
Samples.
Study samples are listed in SI Appendix, Table S3 and information about collection and processing is provided in SI Appendix, Methods.
PolyG-DS Library Preparation.
CRISPR-Cas9 digestion was performed using gRNAs for 19 PolyGs (Fig. 1A and SI Appendix, Table S1), following published protocols (26) (SI Appendix, Methods). Digested fragments were size selected using 0.5× AMPure XP Beads (Beckman Coulter) and then A-tailed and ligated to DS adapters using the NEBNext Ultra II DNA Library Prep Kit (NEB). We used DS adapters (TwinStrand Biosciences) that contain 10 bp random double-stranded molecular barcodes (also known as tags) and a 3′-dT overhang (Fig. 1A). Ligated fragments were PCR amplified, captured with 120-mer biotinylated oligos (IDT), and indexed by PCR, as previously described (26) (SI Appendix, Methods).
Sequencing.
Samples were visualized on the Agilent 4200 TapeStation to confirm the presence of distinct peaks corresponding to the fragment length of the designed CRISPR-Cas9 cut fragments. Then, samples were quantified using the Qubit dsDNA HS Assay Kit and were sequenced in an Illumina MiSeq using a version 2 300-cycle kit or in an Illumina HiSeq 3000 (Genewiz).
PolyG Genotyping.
Sequencing reads were processed with an in-house pipeline that integrates lobSTR, which is an STR profiler for NGS data (24) (http://lobstr.teamerlich.org), and DS for error correction (28) (SI Appendix, Fig. S1 and https://github.com/risqueslab/PolyG-DS). First, duplex barcodes are extracted from the reads. Second, the lobSTR algorithm aligns PolyG-containing reads to the human reference genome and uses the flanking sequence to identify and extract the PolyG sequence. Third, duplex barcodes are used to identify all the raw reads derived from the same original DNA molecule to determine the consensus PolyG genotype (Fig. 1B and SI Appendix, Fig. S2). As in standard DS (28), consensus making occurs at two levels: SSCS and DCS. However, the consensus is not produced at the nucleotide level but at the PolyG genotype level, which we refer to as “call.” SSCS calls are produced by comparing the PolyG genotypes of all the raw reads sharing the same barcode and selecting the most common one. DCS calls are produced by comparing the PolyG genotypes of the two SSCS with complementary barcodes (which correspond to the two strands of the same original DNA molecule). Only when the two SSCS genotypes agree, a DCS call is made (Fig. 1B and SI Appendix, Fig. S2). For final PolyG profiling, we filtered out DCS calls that contained more than two non-G/C nucleotides or more than two nucleotides that were the reverse compliment of the motif. Details on the output files generated in the analysis are provided in SI Appendix, Methods and Fig. S1. A postprocessing script was used to calculate, for each sample, the percentage of raw reads aligned to each PolyG region using Burrows–Wheeler Aligner mem (54) and the percentage of reads successfully aligned by lobSTR for each PolyG. Several PolyGs failed genotyping in some samples (SI Appendix, Fig. S4), resulting in a low-DCS generation (SI Appendix, Fig. S5). For each sample, we only considered PolyG markers that had at least 40% genotyped reads and that produced a minimum of 25 DCS passing filter (SI Appendix, Methods and Fig. S5). In addition, we required that all the samples compared against each other shared the same informative PolyGs.
Data Analysis.
Statistical and data analyses were performed in Prism (Graphpad) and R (https://cran.r-project.org) using the software packages ape and phangorn (55, 56). We used the JSD to quantify genetic divergence to a reference sample, to build heatmaps, and to calculate pairwise JSD among samples to build phylogenetic trees (16). Because PolyG mutations behave as molecular clocks, we built phylogenetic trees using the UPGMA (37). For a two-sample comparison of the fraction of mutant genotypes, we used Fisher exact test with P < 0.0005 as criteria for mutation. Additional methods and statistical analyses are explained in SI Appendix, Methods.
Software Availability.
Software for PolyG-DS data analysis is available at https://github.com/risqueslab/PolyG-DS.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grant CA160674 to L.A.L. and T.A.B. and NIH Grants CA181308 and CA240885 to R.A.R. We thank Marshall Horwitz, Evan Boyle, Alan Rubin, and Jay Shendure for contributing initial ideas, support, and materials; Enna Hun for procuring ovarian cancer samples; and Jeanne Fredrickson and Ngoc-Han Nguyen for providing experimental assistance.
Footnotes
Competing interest statement: S.R.K., L.A.L., and R.A.R. are consultants and equity holders at TwinStrand Biosciences Inc. J.J.S. is an employee and equity holder at TwinStrand Biosciences Inc. S.R.K., L.A.L., R.A.R., D.N., and J.J.S. are named inventors on patents owned by the University of Washington and licensed to TwinStrand Biosciences Inc. R.A.R. is an equity holder at NanoString Technologies Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023373118/-/DCSupplemental.
Data Availability
Sequencing reads data have been deposited in the Sequence Read Archive (PRJNA674403). All other study data are included in the article and/or supporting information.
References
- 1.Nowell P. C., The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 2.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium , Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGranahan N., Swanton C., Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 168, 613–628 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Gerstung M.et al., PCAWG Evolution & Heterogeneity Working Group, and PCAWG Consortium , The evolutionary history of 2,658 cancers. Nature 578, 122–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtius K., Wright N. A., Graham T. A., An evolutionary perspective on field cancerization. Nat. Rev. Cancer 18, 19–32 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Fittall M. W., Van Loo P., Translating insights into tumor evolution to clinical practice: Promises and challenges. Genome Med. 11, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risques R. A., Kennedy S. R., Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 14, e1007108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy S. R., Zhang Y., Risques R. A., Cancer-associated mutations but no cancer: Insights into the early steps of carcinogenesis and implications for early cancer detection. Trends Cancer 5, 531–540 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijg J., Dong X., Pathogenic mechanisms of somatic mutation and genome mosaicism in aging. Cell 182, 12–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer J. C., et al., Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum. Mol. Genet. 11, 707–713 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Ellegren H., Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Weber J. L., Wong C., Mutation of human short tandem repeats. Hum. Mol. Genet. 2, 1123–1128 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Whittaker J. C., et al., Likelihood-based estimation of microsatellite mutation rates. Genetics 164, 781–787 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salipante S. J., Horwitz M. S., Phylogenetic fate mapping. Proc. Natl. Acad. Sci. U.S.A. 103, 5448–5453 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naxerova K., et al., Hypermutable DNA chronicles the evolution of human colon cancer. Proc. Natl. Acad. Sci. U.S.A. 111, E1889–E1898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naxerova K., et al., Origins of lymphatic and distant metastases in human colorectal cancer. Science 357, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiter J. G., et al., Lymph node metastases develop through a wider evolutionary bottleneck than distant metastases. Nat. Genet. 52, 692–700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salk J. J., et al., Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm. Bowel Dis. 19, 2593–2602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salk J. J., et al., Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc. Natl. Acad. Sci. U.S.A. 106, 20871–20876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salipante S. J., Kas A., McMonagle E., Horwitz M. S., Phylogenetic analysis of developmental and postnatal mouse cell lineages. Evol. Dev. 12, 84–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salipante S. J., Thompson J. M., Horwitz M. S., Phylogenetic fate mapping: Theoretical and experimental studies applied to the development of mouse fibroblasts. Genetics 178, 967–977 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson K. D., et al., MIPSTR: A method for multiplex genotyping of germline and somatic STR variation across many individuals. Genome Res. 25, 750–761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guilmatre A., Highnam G., Borel C., Mittelman D., Sharp A. J., Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum. Mutat. 34, 1304–1311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gymrek M., Golan D., Rosset S., Erlich Y., lobSTR: A short tandem repeat profiler for personal genomes. Genome Res. 22, 1154–1162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin G., et al., CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat. Commun. 8, 14291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nachmanson D., et al., Targeted genome fragmentation with CRISPR/Cas9 enables fast and efficient enrichment of small genomic regions and ultra-accurate sequencing with low DNA input (CRISPR-DS). Genome Res. 28, 1589–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salk J. J., Schmitt M. W., Loeb L. A., Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 19, 269–285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy S. R., et al., Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc. 9, 2586–2606 (2014). Correction in: Nat. Protoc. 9, 2903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson M., et al., NCBI BLAST: A better web interface. Nucleic Acids Res. 36, W5–W9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gymrek M., A genomic view of short tandem repeats. Curr. Opin. Genet. Dev. 44, 9–16 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Baker A.-M., et al., Evolutionary history of human colitis-associated colorectal cancer. Gut 68, 985–995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker K. T., Salk J. J., Brentnall T. A., Risques R. A., Precancer in ulcerative colitis: The role of the field effect and its clinical implications. Carcinogenesis 39, 11–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi C.-H. R., Bakir I. A., Hart A. L., Graham T. A., Clonal evolution of colorectal cancer in IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 218–229 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Olafsson S., et al., Somatic evolution in non-neoplastic IBD-affected colon. Cell 182, 672–684.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nanki K., et al., Somatic inflammatory gene mutations in human ulcerative colitis epithelium. Nature 577, 254–259 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Kakiuchi N., et al., Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 577, 260–265 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Sokal R. R., Michener C. D., A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38, 1409–1438 (1958). [Google Scholar]
- 38.Blokzijl F., et al., Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soong T. R., Howitt B. E., Horowitz N., Nucci M. R., Crum C. P., The fallopian tube, “precursor escape” and narrowing the knowledge gap to the origins of high-grade serous carcinoma. Gynecol. Oncol. 152, 426–433 (2019). [DOI] [PubMed] [Google Scholar]
- 40.McPherson A., et al., Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat. Genet. 48, 758–767 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Ahmed A. A., et al., Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 221, 49–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network , Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vang R., et al., Molecular alterations of TP53 are a defining feature of ovarian high-grade serous carcinoma: A rereview of cases lacking TP53 mutations in the cancer genome atlas ovarian study. Int. J. Gynecol. Pathol. 35, 48–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn E., et al., TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma—Evidence supporting the clonal relationship of the two lesions. J. Pathol. 226, 421–426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckert M. A., et al., Genomics of ovarian cancer progression reveals diverse metastatic trajectories including intraepithelial metastasis to the fallopian tube. Cancer Discov. 6, 1342–1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labidi-Galy S. I., et al., High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 8, 1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou W., et al., Use of somatic mutations to quantify random contributions to mouse development. BMC Genomics 14, 39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martincorena I., Somatic mutation and clonal expansions in human tissues. Genome Med. 11, 35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gui X., et al., Histological and molecular diversity and heterogeneity of precancerous lesions associated with inflammatory bowel diseases. J. Clin. Pathol. 73, 391–402 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Hewitt K., et al., The evolution of our understanding of the biology of cancer is the key to avoiding overdiagnosis and overtreatment. Cancer Epidemiol. Biomarkers Prev. 29, 2463–2474 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Srivastava S., et al., Cancer overdiagnosis: A biological challenge and clinical dilemma. Nat. Rev. Cancer 19, 349–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caravagna G., et al., Subclonal reconstruction of tumors by using machine learning and population genetics. Nat. Genet. 52, 898–907 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt M. W., et al., Sequencing small genomic targets with high efficiency and extreme accuracy. Nat. Methods 12, 423–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https://arxiv.org/abs/1303.3997. Accessed 25 July 2016.
- 55.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Schliep K. P., phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads data have been deposited in the Sequence Read Archive (PRJNA674403). All other study data are included in the article and/or supporting information.