Significance
The development and function of the nervous system is profoundly influenced by the interactions of neurons with glial cells and the extracellular matrix (ECM). Mechanisms regulating the maturation of oligodendroglial progenitor cells (OPCs) into myelinating cells to facilitate central nervous system myelination play an essential role in circuit plasticity, including motor learning. Our work has identified that THAP1, a transcription factor whose loss of function leads to the neurodevelopmental movement disorder DYT6 dystonia, drives OPC development by regulating the breakdown of glycosaminoglycans (GAGs), essential components of the ECM. GAGs produced and secreted by OPCs act in an autocrine fashion to inhibit their lineage progression, highlighting a unique mechanism regulating the pool of mature oligodendrocytes.
Keywords: CNS myelination, extracellular matrix, neurodevelopmental disease
Abstract
Mechanisms controlling myelination during central nervous system (CNS) maturation play a pivotal role in the development and refinement of CNS circuits. The transcription factor THAP1 is essential for timing the inception of myelination during CNS maturation through a cell-autonomous role in the oligodendrocyte lineage. Here, we demonstrate that THAP1 modulates the extracellular matrix (ECM) composition by regulating glycosaminoglycan (GAG) catabolism within oligodendrocyte progenitor cells (OPCs). Thap1−/− OPCs accumulate and secrete excess GAGs, inhibiting their maturation through an autoinhibitory mechanism. THAP1 controls GAG metabolism by binding to and regulating the GusB gene encoding β-glucuronidase, a GAG-catabolic lysosomal enzyme. Applying GAG-degrading enzymes or overexpressing β-glucuronidase rescues Thap1−/− OL maturation deficits in vitro and in vivo. Our studies establish lysosomal GAG catabolism within OPCs as a critical mechanism regulating oligodendrocyte development.
The oligodendrocyte (OL) lineage plays a critical role in central nervous system (CNS) development and function (1, 2). The OL lineage in its mature cell state is responsible for wrapping axons with an insulating myelin sheath that supports rapid neurotransmission (3). Developmental myelination during a critical period of postnatal maturation plays an important role in the establishment of neural circuits (4–6), which are disrupted in neurodevelopment disease (7). Regulated myelination in the adult CNS is also increasingly recognized to play an essential role in circuit plasticity, including motor learning (1, 8–10). Mechanisms regulating OL maturation are therefore central to the development of neuronal circuits, as well as their plasticity and maintenance.
OL differentiation, leading to myelination, is regulated by cell-intrinsic pathways (11) and extrinsic signals that emanate from axons and the surrounding extracellular matrix (ECM) (12, 13). The ECM is a complex three-dimensional structure surrounding cells, providing both a physical scaffold and a growth factor organizing center, orchestrating signaling cascades that regulate mobility, development, and function (12, 14, 15). The ECM is composed of glycosaminoglycans (GAGs), a class of long unbranched mucopolysaccharides, GAG-modified proteoglycans, and fibrous proteins (e.g., collagen and elastin) (16). Considerable literature demonstrates that GAGs in their free form and as proteoglycans (e.g., chondroitin sulfate proteoglycan; CSPG) inhibit OL maturation both in vitro and in vivo, demonstrating that GAGs powerfully regulate OL development (17–23).
GAG and proteoglycan content is determined through a balance of synthesis, secretion, and catabolism (16, 24, 25). In addition to the extracellular degradation of ECM components by secreted proteases (26), receptor and cell surface proteoglycans and GAGs are endocytosed and enzymatically catabolized in lysosomes (27, 28). The failure of intracellular GAG catabolism results in mucopolysaccharidoses, a class of lysosomal storage disorders (27, 29). Despite their importance, the sources and cellular mechanisms regulating GAG content and composition in the CNS are ill defined. Most studies on the cellular origins of ECM have focused on CSPG synthesis from astrocytes (30, 31). The study of ECM metabolism within the OL lineage itself has been limited (24, 32–34), despite the likely importance of these processes for OL biology in normal development and disease states.
We demonstrated a cell-autonomous role for the transcription factor THAP1 in regulating the maturation of OL progenitor cell (OPC) to mature myelinating OLs (35). Loss-of-function mutations in the THAP1 gene cause the neurodevelopmental disorder DYT6 dystonia, directly implicating these events in dystonia pathogenesis (36). Conditional deletion of Thap1 from the OL lineage significantly impairs CNS myelination by retarding the transition of OLs to mature myelin-producing cells, without overtly altering the number of OPCs (35). The marked delay in myelination corresponds temporally to the development of motor dysfunction in the Thap1 mutants. The mechanism whereby THAP1 regulates OL lineage maturation was not defined in this study.
Here, we identify a direct role for THAP1 in regulating GAG catabolism and demonstrate that dysregulation of GAG catabolism within OPCs impairs their maturation. Thap1−/− OPCs autoinhibit their own maturation by accumulating and secreting excess GAGs. Liquid chromatography tandem mass spectrometry (LC/MS) analyses of OPC-conditioned media demonstrate that they predominantly secrete chondroitin 4-sulfate, which is significantly increased in Thap1−/− cells. THAP1 regulates GAG catabolism by binding to and regulating the GusB (MPS7) gene encoding the lysosomal, GAG-catabolizing enzyme β-glucuronidase. Treatment with GAG-degrading enzymes or overexpressing β-glucuronidase rescues the Thap1−/− OPC deficits in OL maturation and CNS myelination, establishing that excess GAGs mediate maturation delay in the Thap1−/− oligodendroglial lineage. These findings establish GAG catabolism as a major cell-intrinsic regulatory pathway driving OL maturation.
Results
THAP1 Regulates ECM Pathway Composition during OL Development.
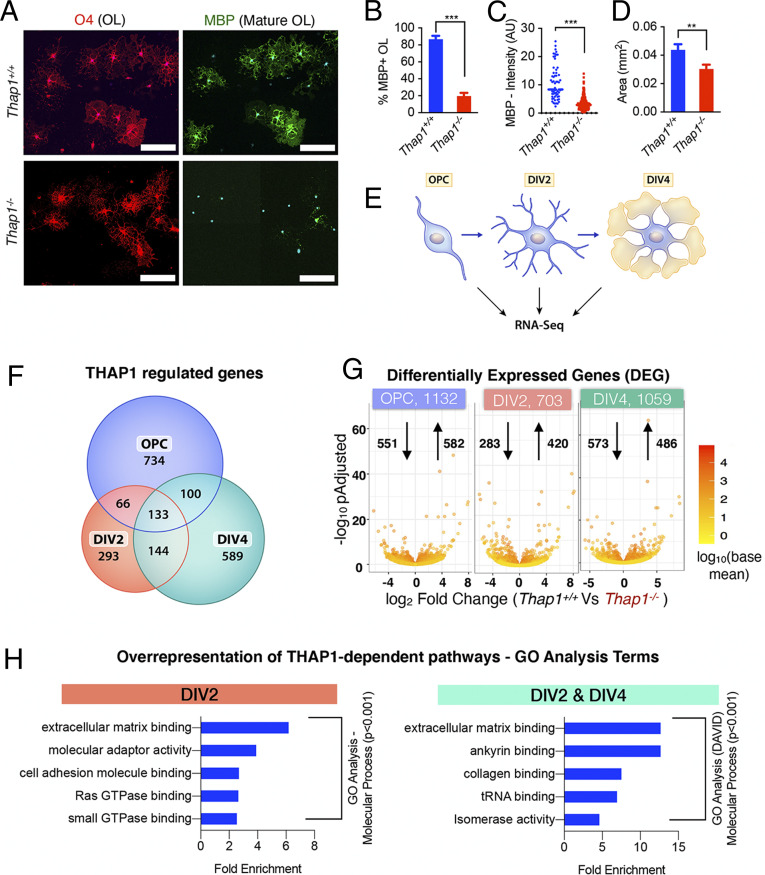
We demonstrated previously that THAP1 plays a cell-autonomous role in regulating myelination during early postnatal CNS development (35). To begin to examine the mechanisms responsible for this role, we performed RNA sequencing (RNAseq) at multiple time points on cultured control (Thap1+/+) and Thap1−/− (derived from Thap1flx/−; Olig2-Cre+) OPC under differentiating conditions. Significantly fewer Thap1−/− OPC matured to express myelin basic protein (MBP+) following 4 days in vitro (DIV4) of treatment with 40 μg/mL T3 thyroid hormone (Fig. 1 A and B). MBP+ Thap1−/− OLs are also significantly smaller (Fig. 1 A and D) and exhibit less intense MBP staining than controls (Fig. 1 A and C). We isolated total messenger RNA (mRNA) from progenitor (OPC) and differentiating OLs (DIV2 and DIV4) from three clonal lines each for Thap1+/+ and Thap1−/− genotypes (schematic, Fig. 1E). We used DESeq2 for the RNAseq data derived from 18 samples to identify THAP1-dependent, differentially expressed genes (DEGs), requiring a greater than log2 1.5-fold change and P < 0.05 significance by the Benjamini–Hochberg false discovery procedure. These analyses identified 1,132 (OPC), 703 (DIV2), and 1,059 (DIV4) DEGs (Fig. 1G and Dataset S1).
Fig. 1.
THAP1 loss disrupts the ECM transcriptome. (A–D) Loss of THAP1 causes maturation deficits in differentiating OPC. (A) Representative images of control (Thap1+/+) and THAP1 null (derived from Thap1flx/−;Olig2-Cre+) OL differentiated (+T3) for 4 d (scale bar, 100 μm.) Cultures are stained for O4, MBP, and DAPI. (B) Quantification of the percentage of O4+ cells expressing MBP. The bar graph shows mean values ± SEM Thap1+/+ = 86.78% ± 3.98; Thap1−/− = 19.58% ± 3.88; t test: t(4) = 12.07; P = 0.0003; and n = 3 clonal lines (100 cells per clone) per genotype. (C) Mean intensity of MBP staining in O4+ cells, represented as arbitrary units (AUs), normalized to background. Each data point in the scatter plot represents MBP+ staining intensity for individual cells. Thap1+/+ = 10.76 AU ± 0.76; Thap1−/− = 3.32 AU ± 0.108; t test: t(427) = 18.21; P < 0.0001; and n = 3 clonal lines (>100 cells per clone) per genotype. (D) Average area of O4+ cells. The bar graph shows mean values ± SEM Thap1+/+ = 0.043 mm2 ± 0.00402; Thap1−/− = 0.030 mm2 ± 0.00309; t test: t(633) = 2.639; P = 0.008; and n = 3 clonal lines (>100 cells per clone) per genotype. (E) Schematic illustrating RNAseq analyses. Total RNA was extracted from three independent clonal lines, each of Thap1+/+ and Thap1−/− cells, at three different time points (OPC, DIV2, and DIV4) and analyzed using Illumina Hiseq (Materials and Methods). (F) Venn diagram depicting the overlap of Thap1+/+ versus Thap1−/− DEGs from OPC, DIV2, and DIV4. (G) Number of DEGs at each time point analyzed. The x-axis = log2 fold change (magnitude of change for the DEG) in Thap1−/−. The y-axis = −log10 adjusted P value depicts the statistical significance of the change. The number of up- and down-regulated genes are indicated by arrows. (H) The DEG at either DIV2 (703) or both DIV2 and DIV4 (277) were used for enrichment analysis of GO terms to identify overrepresented biological pathways using both GENEONTOLOGY and DAVID. Graphs show the most significantly overrepresented GO terms (P < 0.001, sorted as fold overrepresented) in OL from DIV2 (GENEONTOLOGY, GO–Molecular Function) and from the DIV2 and DIV4 overlapping group (DAVID, GO–Molecular Function). tRNA, transfer RNA.
Analyses of the RNAseq data using estimated sample distance (Heat map; SI Appendix, Fig. S1A) and principal component analysis (SI Appendix, Fig. S1B) demonstrated that, within the same genotype, differentiating OLs (DIV2 and DIV4) were distinctly different from OPC. Thus, we focused on DIV2 and DIV4 cells, identifying two groups of THAP1-dependent DEGs: those in immature (DIV2) cells and those persisting through maturation (i.e., differentially expressed at both DIV2 and DIV4) (Fig. 1F). There were 703 DEG at DIV2 and 277 DEG present at both DIV2 and DIV4 (Fig. 1F). These two groups of DEG were used for enrichment analysis of Gene Ontology (GO) terms to identify overrepresented, biological pathways using both GENEONTOLOGY (http://geneontology.org) and DAVID (https://david.ncifcrf.gov/home.jsp) (Dataset S2). GO enrichment analyses (see Materials and Methods) demonstrated significant overrepresentation of ECM-related pathways in both groups of genes (Fig. 1H, SI Appendix, Fig. S1C and Dataset S2). Specific pathways identified were 1) GAG metabolism, 2) ECM binding (including members of integrin and collagen families), 3) signaling components (Rho GTPases), and 4) cytoskeletal polymerization and filopodium assembly (Fig. 1H and SI Appendix, Fig. S1C and Dataset S2). These analyses suggested strongly that ECM pathways are dysregulated in the Thap1−/− oligodendroglial lineage.
Accumulation of GAG in Thap1−/− OLs.
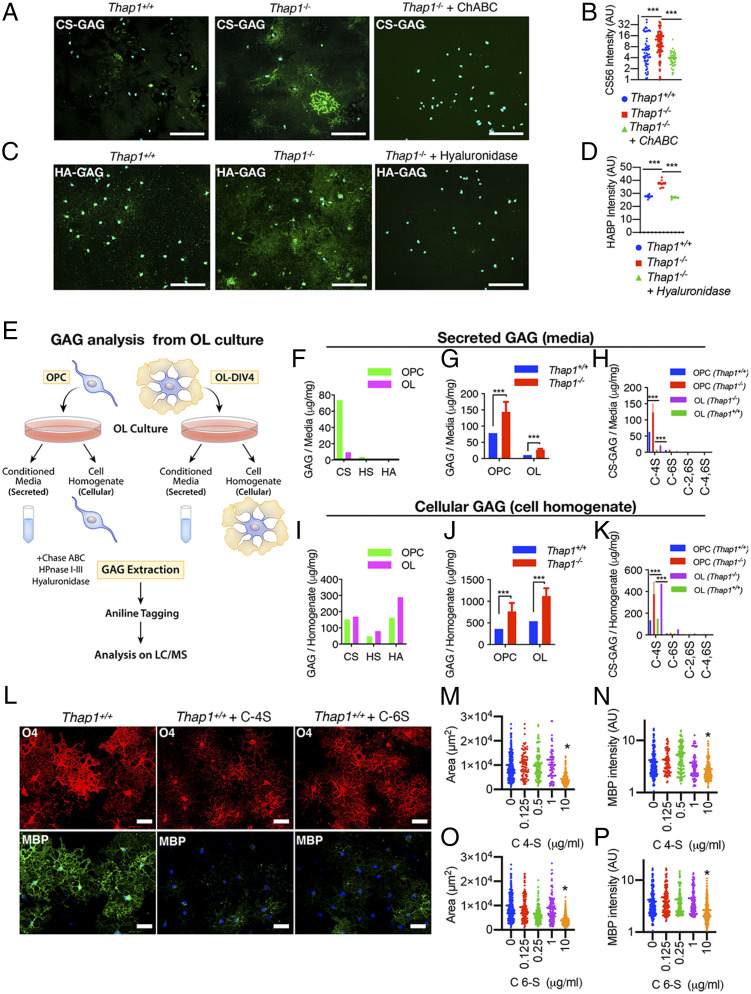
Guided by the strong representation of ECM and particularly GAG metabolism pathways, we explored whether Thap1−/− cultures exhibited abnormalities in the levels of GAGs. We focused on two species most closely linked to OL biology: chondroitin sulfate (CS) and hyaluronate (HA) (17, 18, 20). Immunostaining of nonpermeabilized mature (DIV4) cultures demonstrated that Thap1−/− cells exhibited significantly increased CS-GAG (CS56 and 2H6 antibodies; Fig. 2 A and B and SI Appendix, Fig. S2 A and B) and HA-GAG (biotinylated hyaluronic acid binding protein; Fig. 2 C and D), with prominent accumulation around smaller OL processes (SI Appendix, Fig. S2C). Staining specificity was confirmed by treating the cultures with chondroitinase ABC (ChABC) or hyaluronidase, which removed the relevant signals (Fig. 2 A–D). Thap1−/− cultures contained a greater number of positive cells, as well as a significantly greater intensity of staining per cell (Fig. 2 A–D). The measurement of bulk GAG content using the carbazole assay confirmed significantly increased GAGs in Thap1−/− cultures (SI Appendix, Fig. S2D). These findings are consistent with the RNAseq data indicating that THAP1 participates in a GAG metabolism–related pathway.
Fig. 2.
Thap1 null OLs accumulate and secrete excess GAGs. (A–D) Representative images and the corresponding quantification for Thap1+/+ and Thap1−/− OL cells (DIV4) immunostained with CS-56 (CS-GAG) (A and B), and biotinylated hospital-acquired bacterial pneumonia (HA-GAG) (C and D) under nonpermeabilizing conditions. Thap1−/− OL exhibit a significant accumulation of CS-GAG (Thap1+/+ = 6.68 ± 1.13; Thap1−/− = 12.60 ± 1.29; Thap1−/− + ChABC = 3.93 ± 0.286; and one-way ANOVA F(2,205) = 14.59, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001); n = 2 clonal lines (50 cells per clone) per genotype and HA-GAG (Thap1+/+ = 27.64 arbitrary units [AU] ± 0.38; Thap1−/− = 37.45 AU ± 2.68; Thap1−/− + hyaluronidase = 26.77 AU ± 1.16; and one-way ANOVA F(2,21) = 75.59, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value = 0.0004 and < 0.0001); and n = 2 clonal lines (10 images per clone). (E) Schematic illustrating the study design for mass spectrometry analyses of CS, HS, and HA GAGs using GRIL LC/MS analysis. (F and I) Total CS, HS, and HA GAGs (microgram/milligram/mg) secreted in the media (F) or from cell homogenate Thap1+/+ OPC and OL (I) (see SI Appendix, Fig. S2 C and D for GAG content). (G, H, J, and K) Abnormal accumulation of GAGs in Thap1−/− cultures (n = 2 independent clonal lines simultaneously isolated and processed). (G and J) Total GAGs (microgram/milligram) (CS, HA, and HA-GAGs combined) secreted in the media (G) or from cell homogenate from Thap1+/+ and Thap1−/− OPC and OL (J) (see SI Appendix, Fig. S2 C and D for GAG content). (H and K) CS-GAGs (microgram/milligram) and the composition of mono (C-4S and C-6S) and bisulfated (C-2,6S and C-4,6S) CS-GAGs secreted in the media (H) or from cell homogenate from Thap1+/+ and Thap1−/− OPC and OL (K) (see SI Appendix, Fig. S2 E and F for GAG content). (L–P) C-4S and C-6S inhibit OL differentiation. (L–P) Representative images (L) and corresponding quantification (M–P) of differentiated wild-type OPC without (L, Left column) or with C-4S (10 μg/mL; Middle column) or C-6S (10 μg/mL; Right column) GAG applied to the culture media from DIV2 to DIV5 (scale bar, 50 μm.) (M and O) Area (square micrometer; y-axis) of O4-stained OL at DIV5 for differentiated wild-type OPC treated with C-4S or C-6S DIV2 to DIV5 (0 to 10 μg/mL; x-axis); >250 cells from five different images were quantified for each condition. One-way ANOVA C-4S = F(4,545) = 24.554, P < 0.0001; C-6S = F(4,776) = 43.31, P < 0.0001; and Dunnett’s multiple comparisons test: adjusted P value = < 0.0001). Each data point in the scatter plot represents the area measured for individual cells. (N and P) Mean intensity of MBP staining (AU; y-axis) at DIV5 normalized to background for wild-type OPC treated with C-4S or C-6S (0 to 10 μg/mL; x-axis). One-way ANOVA C-4S = F(2,687) = 13.61, P < 0.0001; and C-6S = F(4,919) = 12.41, P < 0.0001 Dunnett’s multiple comparisons test: adjusted P value = 0.0004 and < 0.0001. Each data point in the scatter plot represents MBP+ staining intensity for individual cells.
Chondroitin-4-Sulfate Is the Major GAG Elevated in Thap1−/− OL.
Little is known regarding the contribution of the OL lineage to the ECM, such as its ability to produce and secrete GAGs or the identity of those produced. Several species of GAGs (CS, HS, and HA) regulate CNS development, including OL lineage maturation (13, 17, 18, 20, 37). To determine if GAG species accumulate within THAP1 mutant OL and to characterize their identity, we employed glycan reductive isotope labeling (GRIL), combined with LC/MS, to quantify CS, HS, and HA GAGs (38) in cell homogenates (intracellular) and supernatant (secreted) from DIV4 Thap1+/+ and Thap1−/− cultures (Fig. 2E, schematic). These analyses demonstrated that there are striking changes in the levels of secreted GAGs during OL maturation (Fig. 2F and SI Appendix, Fig. S2E). Media from Thap1+/+ OPC cultures exhibited approximately eightfold higher GAG levels than in mature OLs, over 95% of which is CS-GAG (Fig. 2F and SI Appendix, Fig. S2E). In contrast, while significant differences in the composition of GAG were observed in cell homogenates (Fig. 2I and SI Appendix, Fig. S2F), the relative differences were far less than those observed in the media. THAP1 depletion significantly increased both secreted and cellular GAG (CS, HS, and HA combined) levels (Fig. 2 G and J and SI Appendix, Fig. S2 E and F). These differences were primarily driven by increases in CS-GAG, the dominant GAG species observed (Fig. 2F and SI Appendix, Fig. S2E). Levels of HA- and HS-GAGs were also significantly increased in Thap1−/− cultures (SI Appendix, Fig. S2 E and F).
The CS-GAG family includes many species with differing amounts and patterns of sulfation that can be detected and resolved by LC/MS. In Thap1+/+ cultures, we found that the dominant form of CS-GAG secreted by OL cultures is chondroitin-4-sulfate (“C-4S”), constituting 85 to 90% of CS-GAG secreted by OL cultures (Fig. 2 H and K and SI Appendix, Fig. S2 G and H). Chondroitin-6-sulfate (“C-6S”) was the only other abundant species (∼10% of CS-GAGs) with negligible amounts of non- and bisulfated species (“C-2,6S” and “C-4,6S”) (Fig. 2 H and K and SI Appendix, Fig. S2 G and H). Consistent with the immunostaining data (Fig. 2 A and B), C-4S and C-6S were significantly increased in Thap1−/− cultures (Fig. 2 H and K and SI Appendix, Fig. S2 G and H). Application of these CS-GAG species (0.125 to 10 μg/mL of C-4S or C-6S from DIV2 to DIV5) to differentiating Thap1+/+ OPC cultures significantly altered OL lineage progression (Fig. 2 L–P). Both C-4S and C-6S significantly impaired the progression to mature MBP+ OLs at 10 μg/mL (∼50% decrease in MBP intensity; ANOVA; P < 0.0001; Fig. 2 L, N, and P) and caused mature cells to occupy a smaller area (∼60% decrease in area; ANOVA; P < 0.0001; Fig. 2 L, M, and O). These data demonstrate that the specific species of CS-GAG elevated in Thap1−/− cultures impair oligodendroglial maturation.
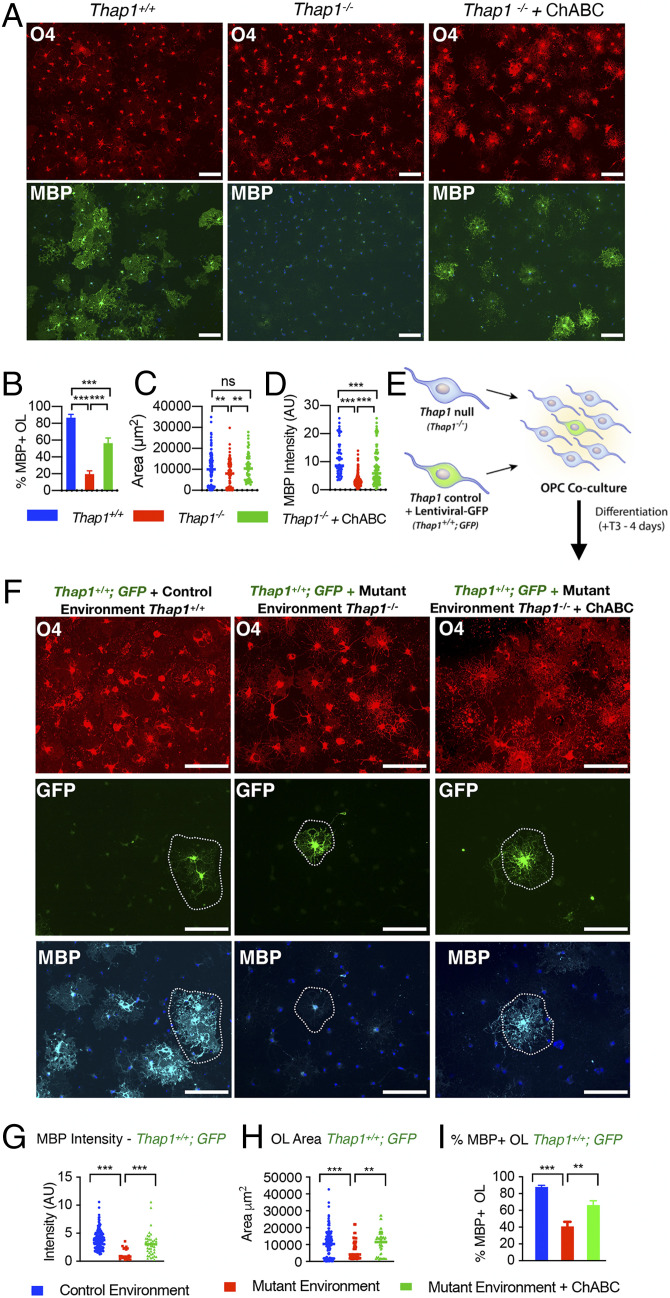
Accumulation of Secreted CS-GAG Contributes to Thap1−/− Maturation Deficits and Exerts an Inhibitory Paracrine Effect.
To further explore the hypothesis that increased CS-GAG secretion is responsible for the maturation impairments of Thap1−/− cultures, we tested the ability of the CS-GAG–degrading enzyme ChABC to rescue this phenotype. Consistent with our hypothesis, treatment of Thap1−/− cultures with 0.1 U/mL ChABC significantly increased the proportion of myelinating OL (approximately threefold increase of MBP+ cells), the intensity of MBP staining (approximately threefold increase), and OL size (approximately twofold increase in cell surface area) (Fig. 3 A–D). Since Thap1−/− OLs secrete excess CS-GAGs (Fig. 2), we hypothesized that they may exert noncell autonomous paracrine effects on neighboring cells. We tested this idea by exploring the ability of Thap1+/+ OPC (GFP labeled with lentivirus; “LV-GFP”) to mature when cocultured with unlabeled Thap1−/− or Thap1+/+ OL (Fig. 3E and SI Appendix, Fig. S3A; cultures contain one Thap1+/+;LV-GFP cell: ∼10 Thap1−/− or Thap1+/+ OL). Thap1−/− OLs significantly impaired the maturation of Thap1+/+;LV-GFP cells (Fig. 3 F–I). Compared to OPCs cultured in the presence of Thap1+/+ OLs, significantly fewer Thap1+/+;LV-GFP OPCs cultured in the presence of Thap1−/− OLs matured into MBP+-expressing OLs (Fig. 3 F and I; ∼50% lower than percentage of MBP+ OLs). LV-GFP+ cells cocultured with Thap1−/− OLs also expressed significantly less MBP (Fig. 3 F and G; approximately fourfold lower that MBP intensity) and covered a significantly smaller area (Fig. 3 F and H; ∼30% smaller area). This inhibitory paracrine effect was rescued by the addition of ChABC to the media (Fig. 3 F–I and SI Appendix, Fig. S3B). Considered together, these data are consistent with a model whereby Thap1−/− OLs inhibit the maturation of surrounding cells through a CS-GAG inhibitory paracrine signal.
Fig. 3.
Excess glycosaminoglycan secretion by Thap1 null cells impairs oligodendroglial maturation via a noncell autonomous mechanism. (A–D) Treatment with ChABC rescues maturation defects in Thap1−/− OL. (A) Representative images (scale bar, 100 μm) of Thap1+/+ and Thap1−/− OL differentiated for 4 d (+T3) either untreated or with ChABC from DIV2 to DIV4 (0.1 U/mL). Cultures are stained for O4, MBP, and DAPI. (B) Quantification of the percentage of O4+ cells expressing MBP. Bar graph shows mean values ± SEM Thap1+/+ = 85.31% ± 4.14; Thap1−/− = 20.81% ± 4.76; and Thap1−/− + ChABC = 56.29% ± 6.31. One-way ANOVA F(2,27) = 47.94, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001; n = 2 clonal lines (>100 cells per clone). (C) Average area of O4+ cells. Each data point in the scatter plot represents the area measured for individual cells. Thap1+/+ = 10,163 μm2 ± 675.9; Thap1−/− = 7,667 μm2 ± 549.5; and Thap1−/− + ChABC = 11,401 μm2 ± 968.3. One-way ANOVA F(2,309) = 6.35, P = 0.002, Dunnett's multiple comparisons test: adjusted P value = 0.009; n = 2 clonal lines (>100 cells per clone). (D) Mean intensity of MBP staining in O4+ cells, represented as arbitrary units (AUs), normalized to background. Each data point in the scatter plot represents MBP+ staining intensity for individual cells. Thap1+/+ = 11.07 AU ± 0.772; Thap1−/− = 3.22 AU ± 0.124; and Thap1−/− + ChABC = 7.86 AU ± 0.63. One-way ANOVA F(2,524) = 134.7, P < 0.0001, Dunnett's multiple comparisons test: adjusted P value < 0.0001; n = 2 clonal lines (>100 cells per clone). (E) Schematic illustrating coculture experimental paradigm to test for paracrine effects of Thap1 null (Thap1−/−) OL cocultured with LV-GFP–labeled control (Thap1+/+;GFP) OPC at 10:1 ratio in differentiation media for 4 d (+T3). (F) Representative images (scale bar, 100 μm) of Thap1+/+;GFP OL cultured in control or mutant environments for 4 d (+T3). Cultures are stained for O4, MBP, and DAPI. (G) MBP+ staining intensity of Thap1+/+;GFP OL. Each data point in the scatter plot represents MBP+ staining intensity for individual cells. Thap1+/+;GFP + control environment = 3.90 AU ± 0.146; Thap1+/+;GFP + mutant environment = 1.286 AU ± 0.227; Thap1+/+;GFP + mutant environment + ChABC = 3.05 AU ± 0.348; one-way ANOVA F(2,197) = 20.26, P < 0.0001, Dunnett's multiple comparisons test: adjusted P value < 0.0001. (H) Area of Thap1+/+;GFP OL. Each data point in the scatter plot represents the area measured for individual cells. Thap1+/+;GFP + control environment = 10,163 μm2 ± 675.9; Thap1+/+;GFP + mutant environment = 7,146 μm2 ± 1,220; Thap1+/+;GFP + mutant environment + ChABC = 9,151 μm2 ± 1,160; one-way ANOVA F(2,266) = 6.84, P = 0.0013, Dunnetts’s multiple comparisons test: adjusted P value < 0.0049. (I) Bar graph shows mean values ± SEM Thap1+/+;GFP + control environment = 87.91% ± 2.09; Thap1+/+;GFP + mutant environment = 40.99% ± 5.48; and Thap1+/+;GFP + mutant environment + ChABC = 66.2% ± 5.22. One-way ANOVA F(2,3.24) = 27.91, P = 0.009, Dunnett’s multiple comparisons test: adjusted P value < 0.001. All quantifications are derived from single clone of Thap1+/+;GFP cocultured in n = 2 mutant clonal lines (>100 cells per clone).
Increased ECM in the CNS from OL-Specific Thap1 Deletion.
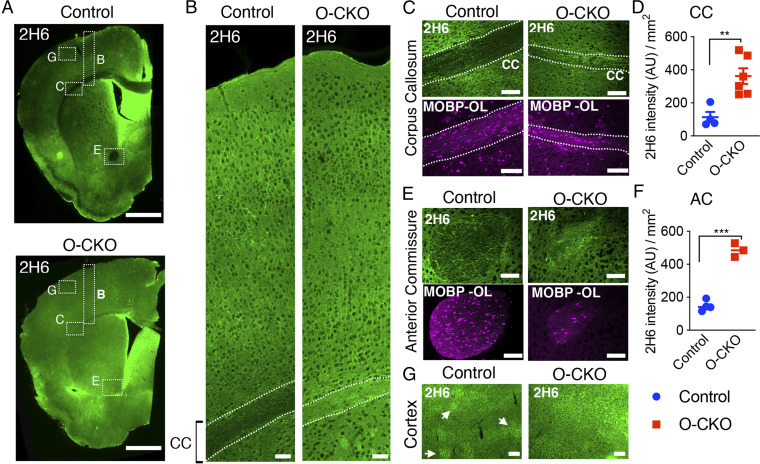
Based on these in vitro data, we assessed CS-GAGs in the brains of THAP1 mutant mice. We characterized the normal trajectory of CS-GAGs in control (Thap1+/flx), juvenile (P14), and adult (P80) mouse CNS by immunostaining with the 2H6 antibody. There was robust CS-GAG immunoreactivity in the juvenile (P14) brains, whereas staining was markedly and subsequently reduced in the adult (P80) CNS (SI Appendix, Fig. S4). In contrast to the homogenous expression in juveniles, the adult CNS showed a distinct pattern of CS-GAG staining, including minimal expression in the corpus callosum (CC) and anterior commissure (AC) white matter tracts (Fig. 4A and SI Appendix, Fig. S4), and CS-GAG structures previously identified as DACS (dandelion clock-like structure) in gray matter regions, including the cortex (Fig. 4G) (39).
Fig. 4.
Increased ECM in the CNS from OL-specific Thap1 deletion. (A) Representative image (scale bar, 1 mm) demonstrating C-4S GAG (2H6) staining for control (Thap1flx/+) (Top) and O-CKO (Thap1flx/−;Olig2-Cre+) (Bottom) mouse P80 tissue. Dashed boxes highlight the CC, AC, and cortex, which are analyzed in control and O-CKO insets. (B) Representative image (scale bar, 100 μm) of the cortical thickness spanning all layers, from CC to pial surface, stained for 2H6. (C–F) Representative images (scale bar, 100 μm) and corresponding quantification of 2H6 staining in CC (C and D) (control = 114.0 arbitrary units (AU) /mm2 ± 31.26; O-CKO = 362.4 AU /mm2 ± 47.48; t test; t(8) = 3.86, P = 0.0048; n = 3 for control and 4 for O-CKO) and AC (E and F) (control = 147.7 AU /mm2 ± 16.12; O-CKO = 485.5 AU /mm2 ± 23.84; t test; t(5) = 12.24, P < 0.0001; (n = 3 for control and 4 for O-CKO). (G) Representative images (scale bar, 100 μm) of 2H6-stained cortex. Arrow heads indicate DACS structures formed in control P80 tissue, which are not observed in O-CKO tissue because of the increased overall expression.
We next assessed CS-GAGs in adult (P80) CNS from OL-conditional Thap1 knockout mice (“O-CKO”; Thap1flx/−;Olig2-Cre). Strikingly, O-CKO CNS shows increased staining for CS-GAG in the white matter tracts analyzed: CC (approximately fourfold increase in CS-GAG intensity compared to littermate control Thap1+/flx mice; t test; P < 0.001 Fig. 4 B–D) and AC (approximately fivefold increase in CS-GAG intensity; t test; P < 0.001; Fig. 4 E and F). Cortical CS-GAGs were also significantly increased and DACS appeared to be disrupted (Fig. 4G). These data indicate that OL-specific Thap1 deletion disrupts levels of CS-GAG in vivo in the CNS.
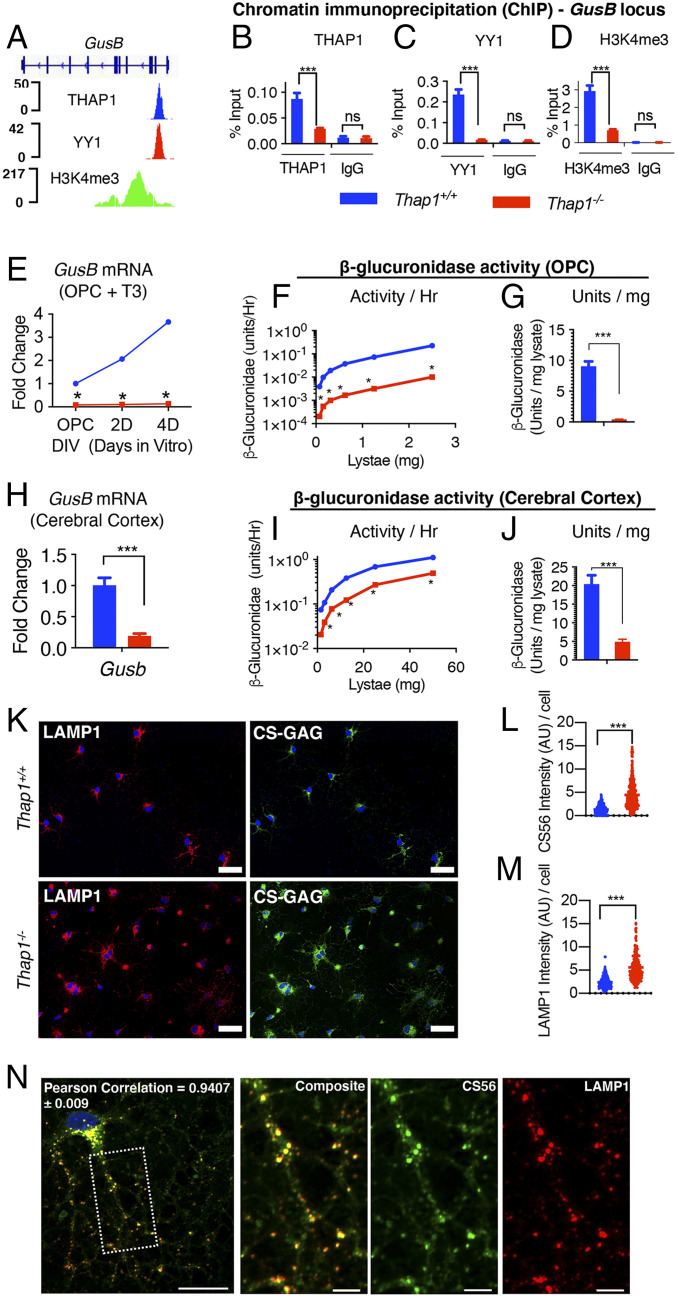
THAP1 Binds to and Regulates the GusB (MPS VII) Gene Encoding the GAG-Degrading Enzyme β-Glucuronidase.
To mechanistically dissect the connection between THAP1 and GAG metabolism, we examined our RNAseq analyses (DIV2 and DIV2 and DIV4 common; Fig. 1F and Dataset S1). We identified a set of 42 genes (Dataset S3) relevant to GAG and proteoglycan metabolism that were differentially regulated in Thap1−/− cultures (Dataset S2). We focused our attention on genes that are 1) directly involved in GAG metabolism and 2) direct targets of THAP1. These criteria identified the Gusb gene encoding β-glucuronidase as a strong THAP1 candidate target. β-glucuronidase is an essential lysosomal enzyme that participates in the catabolism of multiple species of GAG polysaccharides and proteoglycans. Recessively inherited loss-of-function mutations in GUSB causes the mucopolysaccharidosis Sly’s syndrome (MPS VII) (40). Prior chromatin immunoprecipitation (ChIP) sequencing data from human K562 cells demonstrates that THAP1 is bound to the GUSB promoter (Fig. 5A; Encyclopedia of DNA Elements dataset). We confirmed that THAP1 plays a critical role in the transcription of the GusB locus in the OL lineage (Fig. 5 B–E). Quantitative ChIP analyses in DIV4 OL demonstrated that THAP1 is bound to the GusB promoter in the OL lineage (Fig. 5B). Consistent with that finding, GusB expression is significantly and profoundly reduced in Thap1−/− cultures (OPC through DIV4; Fig. 5E). We previously demonstrated that most THAP1 targets are also bound by YY1, a transcription factor known to regulate OL maturation, and that YY1 DNA binding is significantly reduced in Thap1−/− OLs (35). Analyses of the GusB locus demonstrate that it is bound by YY1 in a THAP1-dependent manner (Fig. 5C). H3K4me3, an epigenetic modification associated with active transcription, is significantly reduced at the GusB locus in Thap1−/− OL, further connecting THAP1 to GusB function (Fig. 5D).
Fig. 5.
THAP1 directly binds to and regulates the GusB gene encoding the GAG-catabolizing lysosomal enzyme β-glucuronidase. (A) Genome browser track (using Integrative Genomics Viewer) showing ChIP sequencing signals for THAP1, YY1, and H34me3 at the GusB locus in K652 cells (Encyclopedia of DNA Elements dataset). (B–D) Quantitative ChIP demonstrating THAP1-dependent binding of YY1 and H3K4me3 methylation at the GusB promoter region in OL (+T3 − DIV4). Binding, represented as percentage input (y-axis), is demonstrated for THAP1 (B) (x-axis) (Thap1+/+ = 0.087% ± 0.008; Thap1-/- = 0.02% ± 0.001; t test: t(4) = 6.85; and P = 0.02); YY1 (C) (x-axis) (Thap1+/+ = 0.23% ± 0.017; Thap1-/- = 0.018% ± 0.0009; t test: t(2) = 12.85; and P = 0.006); and H3K4me3 (D) (x-axis) (Thap1+/+ = 2.93% ± 0.219; Thap1-/- = 0.707% ± 0.036; and t test: t(2) = 10.04; P = 0.0098) and their respective isotype control IgG (Goat IgG or Rabbit IgG; x-axis) at the GusB promoter region. The bar graph shows mean values ± SEM. Percentage input represents the final amount of immunoprecipitated chromatin/gene from corresponding Thap1+/+ and Thap1-/- OL cells (Materials and Methods); final values represent the average of three technical repeats from a single clonal population per genotype. (E–J) THAP1 loss significantly reduces GusB mRNA expression and β-glucuronidase enzyme activity in OLs and in CNS tissue. (E) GusB mRNA expression (qRT-PCR) from Thap1+/+- and Thap1-/--differentiating OL (OPC, DIV2, and DIV4). GusB expression was normalized to Rpl19 expression and is represented as fold change (y-axis), with respect to Thap1+/+. Line graphs show mean values ± SEM. Final values represent the three technical repeats from a single clonal population per genotype. (F and G) Deficient β-glucuronidase enzyme activity in Thap1-/- OPC. The line graph shows the mean value of β-glucuronidase activity (y-axis = units/hour) (F) for Thap1+/+ and Thap1-/- OPC lysate (0 to 3 μg; x-axis) derived from fluorometry assay. The bar graph shows mean ± SEM values of normalized β-glucuronidase activity units/lysate (milligram) (G) for OPC (Thap1+/+ = 9.05 U/mg ± 0.803; Thap1-/- = 0.396 U/mg ± 0.038; and t test: t(10) = 10.76; p < 0.0001); final values represent an average of three readings from a two clonal population per genotype. (H) GusB mRNA expression (qRT-PCR) from control (Thap1+/flx) and THAP1 N-CKO (Thap1flx/−;Nestin-Cre+) CNS (cerebral cortex, age P21). GusB expression was normalized to Rpl19 expression and is represented as fold change (y-axis) with respect to control. Line graphs show mean values ± SEM; n = 3. (I and J) Deficient β-glucuronidase enzyme activity in THAP1 N-CKO CNS. The line graph shows the mean value of β-glucuronidase activity (y-axis = units/hour) (I) for control (Thap1+/flx) and THAP1 N-CKO (Thap1flx/−;nestin-Cre+) cerebral cortex lysate (0 to 60 μg; x-axis) derived from fluorometry assay. (J) Bar graph shows mean ± SEM values of normalized β-glucuronidase activity units/lysate (milligram) (J) for CNS (control = 20.43 U/mg ± 2.35; N-CKO = 4.90 U/mg ± 0.68; and t test: t(10) = 6.34; p < 0.0001). Final values represent an average of three readings from a two clonal population per genotype. Two-way ANOVA demonstrates a main effect of genotype (P <0.01) for GusB mRNA (E and H) expression and β-glucuronidase activity (F and J) in OPC (E and F) or cerebral cortex (H and I). * represents time points in which significant differences exist using post hoc Sidak’s multiple comparison test. (K) Representative images (scale bar, 50 μm) derived from images obtained as a tiled scan and corresponding quantification (L and M) of Thap1+/+ and Thap1-/- OL cells (DIV4) immunostained under conditions of membrane permeabilization for CS-56 (CS-GAG; Thap1+/+ = 1.34 ± .072; Thap1-/- = 4.44 ± 0.16; t test: t(530) = 14.51; and P < 0.0001) or LAMP1 (Thap1+/+ = 2.36 arbitrary units [AU] ± 0.082; Thap1-/- = 5.57 AU ± 0.20; t test: t(412) = 15.27; and P < 0.0001). n = 2 clonal lines (100 cells per clone) per genotype. Each data point in the scatter plot represents CS-56 or LAMP1 staining intensity for individual cells. (N) Representative image (scale bar, 20 μm) of CS-GAG (CS-56) colocalization with lysosomes (LAMP1) in OL (DIV4). The region, the dashed box, is represented in a magnified view for individual channels (CS565 and LAMP1) in the inset (scale bar, 5 μm). Pearson’s coefficient value (0.94 ± 0.009) shows significant colocalization (R = 0.94 ± 0.009) of CS565 and LAMP1.
Consistent with significantly reduced GusB expression in Thap1−/− OL (Fig. 5E), we also find reduced β-glucuronidase enzymatic activity in THAP1 mutant cells and CNS tissue, as measured by fluorometry (MUG assay). In Thap1+/+ cultures, β-glucuronidase activity was comparable in OPC and OL (DIV4) (SI Appendix, Fig. S5 A and B). Thap1−/− OLs exhibited a nearly 20-fold decrease of β-glucuronidase activity (Fig. 5 F and G). In Thap1−/− cortical tissue, GusB mRNA was decreased by ∼80% (Fig. 5H), and β-glucuronidase activity was reduced by ∼75% (Fig. 5 I and J). Several other brain regions assessed (cortex, striatum, CC, and cerebellum) showed similar reductions in β-glucuronidase activity (SI Appendix, Fig. S5C). β-Glucuronidase is a lysosomal enzyme, suggesting that loss of its activity should cause lysosomal accumulation of CS-GAGs. Indeed, assessing intracellular CS-GAG by staining permeabilized cells demonstrated significantly higher levels in Thap1−/− OL cultures (Fig. 5 K–M). Colabeling cells for CS-GAG and the pan-lysosomal marker LAMP1 confirmed that the intracellular CS-GAG accumulation is almost exclusively localized to lysosomes (Fig. 5N and SI Appendix, Fig. S5D). Considered together, with the effects of THAP1 at the GusB locus, these data indicate a direct and essential role for THAP1 in regulating levels of CNS β-glucuronidase.
GUSB Overexpression Rescues THAP1 Mutant GAG-Homeostatic and Oligodendroglial Maturation Defect In Vitro and In Vivo.
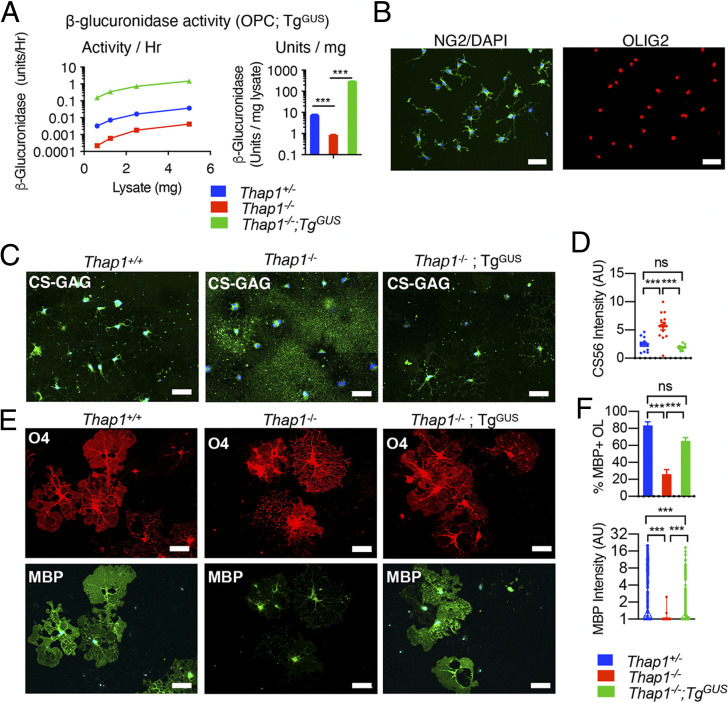
To formally test the necessity of reduced β-glucuronidase for THAP1-related GAG homeostatic and oligodendroglial lineage defects, we pursued rescue studies with a GUSB transgene [TgGUS; (41)]. Prior work demonstrated that the TgGUS does not produce a phenotype when in the hemizygous state on a wild-type background (41). We ingressed TgGUS onto the CNS conditional Thap1 null background (Thap1flx/−;Nestin-Cre). We used these mice to generate OPCs from subventricular zone–derived neural stem cells from Thap1+/+ and Thap1−/− CNS [as previously described (35)], with or without the TgGUS transgene. We confirmed that the TgGUS transgene significantly increased β-glucuronidase activity in Thap1−/− OPCs (Fig. 6A) and that Thap1−/−;TgGUS OPCs normally express the OPC-specific markers OLIG2 and CSPG4 (NG2) (Fig. 6B).
Fig. 6.
β-glucuronidase overexpression rescues the Thap1 null GAG homeostatic and OL maturation defects. (A) β-glucuronidase activity in OPC lysate from Thap1+/+, Thap1−/−, and Thap1−/−;TgGUS genotypes (mean ± SEM of normalized β-glucuronidase activity; Thap1+/+ = 7.48 U/mg ± 0.193; Thap1−/− = 0.87 U/mg ± 1.29; Thap1−/−; TgGUS = 304.1 U/mg ± 1.6; and one-way ANOVA F(2,149972) = 34628.96, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001). (B) Representative images (scale bar, 50 μm) of Thap1−/−;TgGUS OPC-expressing NG2 (CSPG4) and pan-OL lineage marker OLIG2. (C and D) β-glucuronidase overexpression prevents CS-GAG accumulation in Thap1−/− OL. Representative images (C) (scale bar, 50 μm) derived from images obtained as a tiled scan and corresponding quantification (D) for Thap1+/+, Thap1−/−, and Thap1−/−;TgGUS OL (+T3 − DIV5) immunostained with CS-56 (CS-GAG) under nonpermeabilized conditions. Each data point in the scatter plot represents CS-56 staining intensity for multiple cells normalized to the area in the field of view. Thap1+/+ = 2.46 arbitrary units [AU] ± 0.35; Thap1−/− = 5.7 AU ± 0.67; Thap1−/−;TgGUS = 1.909 AU ± 0.13; and one-way ANOVA F(2,34) = 19.95, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001. (E and F) β-glucuronidase overexpression rescues maturation defects in Thap1−/− OL. (E) Representative images (scale bar, 50 μm) for Thap1+/+, Thap1−/−, and Thap1−/−;TgGUS OL differentiated for 5 d (+T3). Cultures were stained with O4, MBP, and DAPI. (F) Quantification of the percentage of O4+ cells expressing MBP and intensity of MBP staining. (F, Upper graph) Percentage of O4+ cells expressing MBP. The bar graph shows mean ± SEM values. The y-axis; Thap1+/+ = 83.48% ± 5.08; Thap1−/− = 26.17% ± 5.33; Thap1−/−;TgGUS = 65.18 ± 4.09; and one-way ANOVA F(2,49) = 23.27, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001. (F, Lower graph) Mean intensity of MBP staining normalized to background. Each data point in the scatter plot represents MBP+ staining intensity for individual cells. Thap1+/+ = 4.81 AU ± 0.24; Thap1−/− = 1.005 ± 0.004; Thap1−/−;TgGUS = 1.933 ± 0.128; and one-way ANOVA F(2,1195) = 127.8, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001. ns, not significant.
We began assessing the effects of increasing β-glucuronidase by measuring CS-GAG levels in Thap1−/− OPCs. Immunostaining Thap1−/−;TgGUS OL (+T3-DIV4) for CS-GAG under nonpermeabilizing conditions demonstrated that the TgGUS transgene completely reversed the CS-GAG accumulation phenotype (Fig. 6 C and D). At DIV4, Thap1−/−;TgGUS cultures also exhibited a significantly greater number of myelinating OL (percentage of MBP+ OL; Fig. 6 E and F) and increased MBP staining (MBP+ intensity; Fig. 6 E and F) compared to Thap1−/− OL.
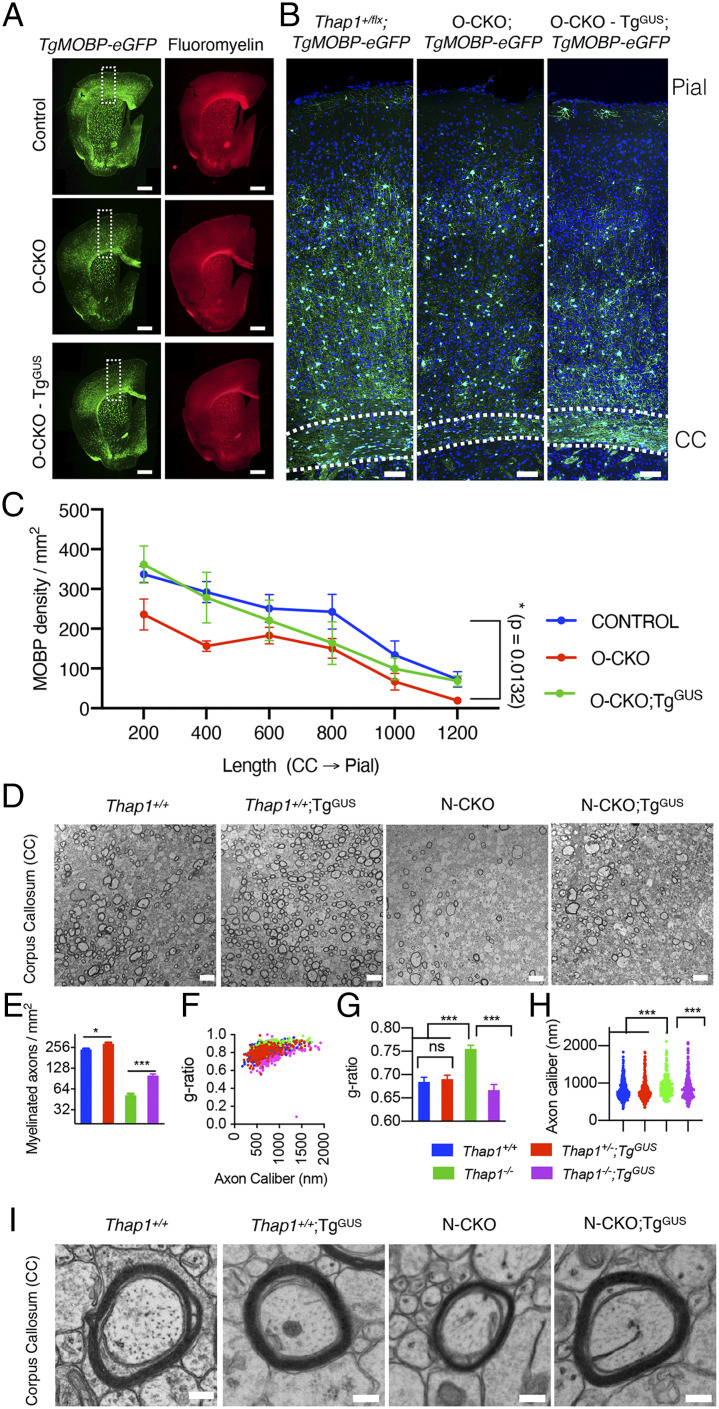
Having confirmed in vitro rescue we next tested the ability of the TgGUS transgene to rescue THAP1-related myelination defects in vivo. We confirmed that the TgGUS allele increases β-glucuronidase activity in all the CNS regions tested (SI Appendix, Fig. S6). To directly quantify mature myelinating OL (MOBP-OL), we ingressed the BAC TgMOBP-eGFP transgene into our Thap1 conditional lines (42). This transgene expresses eGFP under the control of the myelin-associated OL basic protein regulatory elements, thereby selectively labeling mature OLs (43). We assessed THAP1-dependent changes in MOBP-OL density by comparing the number of eGFP+ cells in the cerebral cortex costained with fluoromyelin in O-CKO (O-CKO;TgMOBP-eGFP) and control (Thap1+/flx;TgMOBP-eGFP) P80 mice (Fig. 7A). O-CKO mice had significantly fewer MOBP-OL across the entire cortical thickness from the CC to the pial surface, with the most prominent reduction in the OL dense layer proximal to CC (layer VI) (Fig. 7 B and C). GUSB overexpression (TgGUS) rescued these OL defects, with a comparable number of mature OL observed in the cortex of O-CKO + GusB mice (O-CKO;TgGUS;TgMOBP-eGFP), relative to similar regions of control CNS (Fig. 7 B and C). These findings demonstrate in vivo that the deficit in mature OLs in O-CKO mice is rescued by β-glucuronidase overexpression.
Fig. 7.
β-glucuronidase overexpression rescues the myelination defect in the THAP1 null CNS. (A) Representative images (scale bar, 500 μm) of a coronal hemisection derived from stitching multiple images (Image J) demonstrating MOBP-eGFP and fluoromyelin staining for P80 tissue derived from control (Thap1flx/+;TgMOBP-eGFP), O-CKO (Thap1flx/−;Olig2-Cre+;TgMOBP-eGFP), and O-CKO;TgGUS (Thap1flx/−;Olig2-Cre+;TgGUS;TgMOBP-eGFP). Dashed boxes indicate the corresponding cortical regions analyzed for MOBP-OL number. Representative images (B) (scale bar, 100 μm) and corresponding quantification (C) of MOBP-OL counts (MOBP-OL/square millimeters; measured through the thickness of the cortex from CC through the pial surface) from P80 control, O-CKO, and O-CKO-TgGUS. Two-way ANOVA post hoc analysis shows the main effect of genotype, control versus O-CKO P = 0.0132; control versus O-CKO;TgGUS, and P = 0.156; n = 3 for control, 4 for O-CKO, and 3 for O-CKO-TgGUS. (D) Representative transmission electron microscopy (EM) images (scale bar, 4 μm) of the genu of CC at P21 from control (Thap1+/+;Cre negative) or N-CKO (Thap1flx/−;Nestin-Cre+) with or without TgGUS. (E–H) Quantification of EM images (n = 3 per genotype) demonstrates that β-glucuronidase overexpression (N-CKO; TgGUS) significantly increases myelination in the THAP1 null CNS. (E) Density of myelinated axons (number of axons/square millimeters). Mean ± SEM; control = 238.09 ± 12.58, control;TgGUS = 289.85 ± 10.36; N-CKO = 51.74 ± 2.70; N-CKO;TgGUS 100.71 ± 4.790; and one-way ANOVA F(3,6) = 182.7, P < 0.0001, Dunnett’s multiple comparisons test: adjusted P value < 0.0001. (F and G) The g-ratio represented in relation to axon caliber (F) and by genotype (G, mean ± SEM; control = 0.684 ± 0.009, control;TgGUS = 0.690 ± 0.0087; N-CKO = 0.755 ± 0.0082; N-CKO;TgGUS = 0.666 ± 0.0119; and one-way ANOVA F(3,8) = 15.68, P = 0.001, Dunnett’s multiple comparisons test: adjusted P value < 0.004). (H) Axon caliber (each data point represents axon caliber [nanometer] of individual axons; control = 813.2 nm ± 14.76, control;TgGUS = 825.9 nm ± 19.54; N-CKO = 935.0 ± 21.42; N-CKO;TgGUS = 897.4 ± 21.08; and one-way ANOVA F(3,1047) = 8.613, P = 0.001, Dunnett’s multiple comparisons test: adjusted P value < 0.004). n = 3 per genotype. (I) Representative EM images (scale bar, 200 nm) demonstrating the rescue of myelin thickness (g-ratio) observed in N-CKO with β-glucuronidase overexpression (N-CKO; TgGUS).
We hypothesized that the β-glucuronidase rescue of mature OL number would also enable the rescue of compact myelin ultrastructure deficits in Thap1 null mice (35). Our prior work demonstrated that conditional Thap1 deletion from either the entire CNS (Thap1flx/−;Nestin-Cre; “N-CKO”) or OL lineage (O-CKO) cause hypomyelination and myelin ultrastructural defects in the genu of CC (35). Consistent with the cell-autonomous role of THAP1 in the OL lineage, N-CKO CNS showed no changes in the density of other glial cell types—astrocytes and microglia in CC (SI Appendix, Fig. S7 A and B). N-CKO;TgGUS mice exhibited a twofold increase in the density of myelinated axons, compared to N-CKO mice without the transgene (Fig. 7 D and E). TgGUS did not fully restore myelinated axon density to control levels, however. The abnormally high g-ratio characteristic of N-CKO mice, in contrast, was rescued completely by the restoration of β-glucuronidase (Fig. 7 D, F, G, and I). N-CKO mice exhibit a modest but significant increase in the size of the myelinated axons, which was also rescued by the TgGUS transgene (Fig. 7 D and H). These findings establish β-glucuronidase activity as a key mediator of THAP1-related CNS pathology.
Discussion
Our studies establish THAP1 as a transcriptional regulator of OL maturation via its control of the GAG metabolism. We demonstrate that THAP1 directly binds to and regulates the GusB gene encoding β-glucuronidase, a lysosomal, GAG-degrading enzyme. THAP1 loss dramatically decreases GusB mRNA and β-glucuronidase enzymatic activity, causing CS-GAG lysosomal accumulation and increasing secretion. Multiple lines of evidence support CS-GAG accumulation as the key mechanism responsible for delaying the maturation of the Thap1−/− OL lineage. Preventing abnormal accumulation of GAGs rescues the Thap1−/− OL maturation defect, whereas administering GAG species that accumulate in Thap1−/− cultures (C-4S and C-6S) impairs wild-type OL myelination. Moreover, overexpressing GUSB rescues GAG accumulation and OL maturation and ameliorates CNS myelination defects in Thap1−/− mice. Considered together, our findings identify a cell-autonomous mechanism of OL lineage regulation and provide insight into the molecular pathophysiology of THAP1 loss of function, the cause of DYT6 dystonia.
The role of CS-GAGs (and CSPGs) in regulating OL differentiation has been well studied (13, 17–23). CSPGs inhibit OL differentiation by stimulating receptor-type protein–tyrosine phosphatase (RPTP) subtypes RPTPζ and RPTPσ, thereby activating Rho-associated kinase, leading to growth cone collapse (44, 45). Studies exploring the cellular source of CSPGs have largely focused on astrocytes (46–48). Here, we demonstrate that the OL lineage itself is a functionally important source of CS-GAGs that are autoinhibitory to OL lineage progression. Several prior studies support this model, including the observation of CSPG production from OL cultures (20, 32–34, 49). Additionally, reducing CSPG signaling via the application of ChABC, inhibition of CSPG production, or inactivation of CSPG receptor PTPRZ promotes OL differentiation in vitro (20, 50). Our mass spectrometry analyses indicate that, within the OL lineage, CS-GAGs are uniquely secreted by OPCs. OL lineage progression is accompanied by a precipitous drop in CS-GAG levels. This drop appears to be an essential step in OL maturation. Our studies suggest that the drop in CS-GAGs is not likely from increased catabolism, as the β-glucuronidase activity remains comparable through maturation (SI Appendix, Fig. S5 A and B). Rather, the expression of genes involved in CS-GAG production (identified from GO terms) decrease through OL differentiation.
We utilized LC/MS studies to analyze three GAGs (CS, HS, and HA) with well-established roles in CNS development. We discovered that C-4S is the major GAG species secreted (∼85% of total CS-GAG) by the OL lineage (Fig. 3). Differentially sulfated CS-GAGs mediate unique biological activities (51). The discovery of C-4S production within OPCs and its autoinhibitory activity on these cells raises important questions about CS-GAG metabolism in normal development and in the context of brain disease and repair. Despite the biological activity conferred by the GAGs in their free form and as proteoglycans, few studies have explored GAG metabolism or secretion in the OL lineage. Rather, secreted proteoglycans (e.g., aggrecan family proteins) have been the major focus of studies exploring GAG/proteoglycan significance in injury and development.
How does THAP1 loss of function cause GAG accumulation? We demonstrate that GusB is the critical downstream effector for this phenotype. GusB encodes β-glucuronidase, a lysosomal enzyme that catalyzes the terminal stages of GAG catabolism by hydrolyzing glucuronide or glucuronic acid from multiple GAG species (CS-, HS-, and HA-GAGs) (52, 53). The complete loss of GUSB causes MPS VII (40), a lysosomal storage disease with marked GAG elevations (53, 54), lysosomal abnormalities (55, 56), and multiorgan defects, including neurodevelopmental, cognitive, and motor dysfunction (57, 58). Importantly, and consistent with our findings, MPS VII patients and mouse models (GusBmps) (59) exhibit reductions in CNS myelination and altered expression of myelin component genes (60). Myelin abnormalities also occur in other MPS syndromes (61, 62). Most studies of MPS VII pathogenesis in the CNS have focused on the GusB function and regulation in neurons (63–66). In contrast, our studies constitute the effort of defining the role of GusB in the glial lineage. Klotho, which codes for a type I membrane protein with glucuronidase activity and is related to β-glucuronidase, has been shown to promote the maturation of OL lineage (67). However, Klotho is not a resident of lysosomes, has not been implicated in any MPS, and it is unknown if it plays any role GAG metabolism. The restoration of β-glucuronidase restores GAG homeostasis in the OL lineage, rescues in vitro maturation of Thap1−/− OLs, and significantly improves Thap1−/− CNS myelination. The lack of a complete rescue of hypomyelination or clasping behavior (preliminary observations) in Thap1 Nestin-CKO from GUSB overexpression indicates the possibility of a critical role of other THAP1 targets or perhaps the inability of GUSB-mediated GAG catabolism to compensate for the loss of other ECM target genes identified in our RNAseq studies. Nevertheless, these observations establish the lysosomal catabolism of GAGs as an essential element of THAP1 CNS function.
Most DYT6 dystonia mutations in THAP1 are predicted to cause LOF, including early stop mutations (68). Our prior studies demonstrate that THAP1 has a prominent role in CNS myelination during juvenile development, the period when dystonia emerges in most DYT6 subjects (35). These observations highlight a critical role for lysosomal GAG catabolism during CNS maturation and implicate this pathway in the neurodevelopmental disease DYT6 dystonia. While the experiments presented here focus exclusively on GAG effects within and on the OL lineage, considerable data suggest that GAG abnormalities impact neural plasticity and function (69). These noncell autonomous effects, the impact of potentially abnormal GAG metabolism within neurons themselves and the impact of delayed OL maturation on neural function, are important future directions in the study of DYT6 pathogenesis, the role of ECM abnormalities in brain injury and disease, and fundamental understanding of neuronal–glial interactions.
Materials and Methods
Generation and Maintenance of Mice.
Animal research was conducted in accord with the NIH laboratory animal care guidelines and with the Institutional Animal Care and Use Committee at the University of Michigan. Thap1 floxed mice generation, characterization, and genotyping have been previously described (35) and detailed in SI Appendix, SI Materials and Methods. Nestin-Cre+ and TgGUS (Stock # 006558) mice were purchased from Jackson Laboratory. The Olig2-Cre mouse line used in this study has been previously described (70). The breeding strategy used to generate and derive all primary OPC cells and conditional null mice is described in SI Appendix, SI Materials and Methods.
GRIL LC/MS Analyses.
The equal density (2 × 104 cells/cm2) of Thap1+/+ and Thap1−/− OPC was plated on a 100-mm culture dish, coated with poly-L-Ornithine (10 μg/mL) and laminin (5 μg/mL), and supplemented with PDGF-α (20 μg/mL) every day for 72 h for OPC cultures. For OL cultures, the similar density of OPC was plated on three 10-cm dishes, coated with poly-L-Ornithine (10 μg/mL) and poly-d-Lysine (10 μg/mL), and differentiated for 96 h with T3 (20 μg/mL). We simultaneously collected 1) media supernatant and concentrated it 10-fold using Amicon Centrifugal Filters (3 kDa molecular weight cutoff) and stored at −80 °C until further use and 2) the cells were scrapped, collected, washed with PBS, concentrated into a cell pellet, and stored at −80 °C until further use. GAG extract; enzymatic depolymerization of CS, HS, and HA chains; and further LC/MS analyses using C18 reversed phase column were conducted, as previously described in ref. 38 and detailed in SI Appendix, SI Materials and Methods.
Statistics.
All data are reported as mean ± SEM. All statistical tests reported (Student’s t tests and one-way or two-way ANOVAs) were performed using Graphpad Prism software.
Supplementary Material
Acknowledgments
We thank Haoran Huang and Stephanie Mrowczynski for technical assistance. We thank Cathy Collins and Jay Li for critical reading of the manuscript. We are grateful to the staff of the University of California San Diego Glycoanalytics Core for GRIL LC/MS analysis. We thank Dr. Herbert Geller for helpful suggestions. We thank the staff of University of Michigan’s Core Facilities (DNA Sequencing Core, Microscopy and Image Analysis Laboratory, and Unit of Laboratory Animal Medicine) and Hong Yi (Electron Microscopy Core, Emory University). This research was supported, in part, by Grants 1R01NS109227 and NINDS to W.T.D.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.R.C. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100862118/-/DCSupplemental.
Data Availability
RNAseq data generated for this study and ChIP sequencing data for THAP1, YY1, and H3K4me3 (generated by the Encyclopedia of DNA Elements Project Consortium) have been deposited in Gene Expression Omnibus (GSE161556, GSM803408, GSM803446, and GSM788087). All other study data are included in the article and/or supporting information.
References
- 1.Bergles D. E., Richardson W. D., Oligodendrocyte development and plasticity. Cold Spring Harb. Perspect. Biol. 8, a020453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida R. G., Lyons D. A., On myelinated axon plasticity and neuronal circuit formation and function. J. Neurosci. 37, 10023–10034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waxman S. G., Bennett M. V., Relative conduction velocities of small myelinated and non-myelinated fibres in the central nervous system. Nat. New Biol. 238, 217–219 (1972). [DOI] [PubMed] [Google Scholar]
- 4.Rice D., Barone S. Jr, Critical periods of vulnerability for the developing nervous system: Evidence from humans and animal models. Environ. Health Perspect. 108 (suppl. 3), 511–533 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields R. D., White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 31, 361–370 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang F., et al., Enhancing oligodendrocyte myelination rescues synaptic loss and improves functional recovery after chronic hypoxia. Neuron 99, 689–701.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Kim S., Pappas S. S., Dauer W. T., CNS critical periods: Implications for dystonia and other neurodevelopmental disorders. JCI Insight 6, e142483 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenzie I. A., et al., Motor skill learning requires active central myelination. Science 346, 318–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao L., et al., Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat. Neurosci. 19, 1210–1217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bacmeister C. M., et al., Motor learning promotes remyelination via new and surviving oligodendrocytes. Nat. Neurosci. 23, 819–831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbaz B., Popko B., Molecular control of oligodendrocyte development. Trends Neurosci. 42, 263–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colognato H., Tzvetanova I. D., Glia unglued: How signals from the extracellular matrix regulate the development of myelinating glia. Dev. Neurobiol. 71, 924–955 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow D. E., Macklin W. B., Inhibitors of myelination: ECM changes, CSPGs and PTPs. Exp. Neurol. 251, 39–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theocharidis U., Long K., Ffrench-Constant C., Faissner A., Regulation of the neural stem cell compartment by extracellular matrix constituents. Prog. Brain Res. 214, 3–28 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Fawcett J. W., Oohashi T., Pizzorusso T., The roles of perineuronal nets and the perinodal extracellular matrix in neuronal function. Nat. Rev. Neurosci. 20, 451–465 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Esko J. D., Kimata K., Lindahl U., “Proteoglycans and sulfated glycosaminoglycans” in Essentials of Glycobiology, Varki A., Ed. et al. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2nd Ed., 2009), pp. 229–248. [PubMed] [Google Scholar]
- 17.Sloane J. A., et al., Hyaluronan blocks oligodendrocyte progenitor maturation and remyelination through TLR2. Proc. Natl. Acad. Sci. U.S.A. 107, 11555–11560 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siebert J. R., Osterhout D. J., The inhibitory effects of chondroitin sulfate proteoglycans on oligodendrocytes. J. Neurochem. 119, 176–188 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Lau L. W., et al., Chondroitin sulfate proteoglycans in demyelinated lesions impair remyelination. Ann. Neurol. 72, 419–432 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Karus M., et al., Regulation of oligodendrocyte precursor maintenance by chondroitin sulphate glycosaminoglycans. Glia 64, 270–286 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Keough M. B., et al., An inhibitor of chondroitin sulfate proteoglycan synthesis promotes central nervous system remyelination. Nat. Commun. 7, 11312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuboyama K., Tanga N., Suzuki R., Fujikawa A., Noda M., Protamine neutralizes chondroitin sulfate proteoglycan-mediated inhibition of oligodendrocyte differentiation. PLoS One 12, e0189164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pu A., Stephenson E. L., Yong V. W., The extracellular matrix: Focus on oligodendrocyte biology and targeting CSPGs for remyelination therapies. Glia 66, 1809–1825 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Song I., Dityatev A., Crosstalk between glia, extracellular matrix and neurons. Brain Res. Bull. 136, 101–108 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Nowicka D., Gręda A., Chondroitin sulfate metabolism in the brain. Acta Neurobiol. Exp. (Wars) 79, 338–351 (2019). [PubMed] [Google Scholar]
- 26.Berezin V., Walmod P. S., Filippov M., Dityatev A., Targeting of ECM molecules and their metabolizing enzymes and receptors for the treatment of CNS diseases. Prog. Brain Res. 214, 353–388 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Winchester B., Lysosomal metabolism of glycoproteins. Glycobiology 15, 1R–15R (2005). [DOI] [PubMed] [Google Scholar]
- 28.Rainero E., Extracellular matrix endocytosis in controlling matrix turnover and beyond: Emerging roles in cancer. Biochem. Soc. Trans. 44, 1347–1354 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi H., Recent trends in mucopolysaccharidosis research. J. Hum. Genet. 64, 127–137 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Jones L. L., Margolis R. U., Tuszynski M. H., The chondroitin sulfate proteoglycans neurocan, brevican, phosphacan, and versican are differentially regulated following spinal cord injury. Exp. Neurol. 182, 399–411 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sandvig A., Berry M., Barrett L. B., Butt A., Logan A., Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: Expression, receptor signaling, and correlation with axon regeneration. Glia 46, 225–251 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Yim S. H., Sherin J. E., Szuchet S., Oligodendrocyte proteoglycans: Modulation by cell-substratum adhesion. J. Neurosci. Res. 34, 401–413 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Courel M. N., et al., Hyaluronectin is produced by oligodendrocytes and Schwann cells in vitro. J. Neurocytol. 27, 27–32 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Garwood J., et al., The extracellular matrix glycoprotein Tenascin-C is expressed by oligodendrocyte precursor cells and required for the regulation of maturation rate, survival and responsiveness to platelet-derived growth factor. Eur. J. Neurosci. 20, 2524–2540 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Yellajoshyula D., et al., The DYT6 dystonia protein THAP1 regulates myelination within the oligodendrocyte lineage. Dev. Cell 42, 52–67.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs T., et al., Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat. Genet. 41, 286–288 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Properzi F., et al., Heparan sulphate proteoglycans in glia and in the normal and injured CNS: Expression of sulphotransferases and changes in sulphation. Eur. J. Neurosci. 27, 593–604 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Lawrence R., et al., Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 283, 33674–33684 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi N., et al., DACS, novel matrix structure composed of chondroitin sulfate proteoglycan in the brain. Biochem. Biophys. Res. Commun. 364, 410–415 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Sly W. S., et al., Beta-glucuronidase deficiency mucopolysaccharidosis. Birth Defects Orig. Artic. Ser. 10, 239–245 (1974). [PubMed] [Google Scholar]
- 41.Kyle J. W., et al., Correction of murine mucopolysaccharidosis VII by a human beta-glucuronidase transgene. Proc. Natl. Acad. Sci. U.S.A. 87, 3914–3918 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong S., et al., A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Lee Y., et al., Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pendleton J. C., et al., Chondroitin sulfate proteoglycans inhibit oligodendrocyte myelination through PTPσ. Exp. Neurol. 247, 113–121 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Kuboyama K., Fujikawa A., Suzuki R., Tanga N., Noda M., Role of chondroitin sulfate (CS) modification in the regulation of protein-tyrosine phosphatase receptor type Z (PTPRZ) activity: Pleiotrophin-PTPRZ-A signaling is involved in oligodendrocyte differentiation. J. Biol. Chem. 291, 18117–18128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKeon R. J., Jurynec M. J., Buck C. R., The chondroitin sulfate proteoglycans neurocan and phosphacan are expressed by reactive astrocytes in the chronic CNS glial scar. J. Neurosci. 19, 10778–10788 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asher R. A., et al., Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J. Neurosci. 20, 2427–2438 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laabs T. L., et al., Inhibiting glycosaminoglycan chain polymerization decreases the inhibitory activity of astrocyte-derived chondroitin sulfate proteoglycans. J. Neurosci. 27, 14494–14501 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Probstmeier R., Stichel C. C., Müller H. W., Asou H., Pesheva P., Chondroitin sulfates expressed on oligodendrocyte-derived tenascin-R are involved in neural cell recognition. Functional implications during CNS development and regeneration. J. Neurosci. Res. 60, 21–36 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Kuboyama K., Fujikawa A., Suzuki R., Noda M., Inactivation of protein tyrosine phosphatase receptor type Z by pleiotrophin promotes remyelination through activation of differentiation of oligodendrocyte precursor cells. J. Neurosci. 35, 12162–12171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Properzi F., et al., Chondroitin 6-sulphate synthesis is up-regulated in injured CNS, induced by injury-related cytokines and enhanced in axon-growth inhibitory glia. Eur. J. Neurosci. 21, 378–390 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Ray J., Scarpino V., Laing C., Haskins M. E., Biochemical basis of the beta-glucuronidase gene defect causing canine mucopolysaccharidosis VII. J. Hered. 90, 119–123 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Heuer G. G., Skorupa A. F., Prasad Alur R. K., Jiang K., Wolfe J. H., Accumulation of abnormal amounts of glycosaminoglycans in murine mucopolysaccharidosis type VII neural progenitor cells does not alter the growth rate or efficiency of differentiation into neurons. Mol. Cell. Neurosci. 17, 167–178 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Kubaski F., et al., Elevation of glycosaminoglycans in the amniotic fluid of a fetus with mucopolysaccharidosis VII. Prenat. Diagn. 37, 435–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogler C., et al., A murine model of mucopolysaccharidosis VII. Gross and microscopic findings in beta-glucuronidase-deficient mice. Am. J. Pathol. 136, 207–217 (1990). [PMC free article] [PubMed] [Google Scholar]
- 56.Bayó-Puxan N., et al., Lysosomal and network alterations in human mucopolysaccharidosis type VII iPSC-derived neurons. Sci. Rep. 8, 16644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomatsu S., Montaño A. M., Dung V. C., Grubb J. H., Sly W. S., Mutations and polymorphisms in GUSB gene in mucopolysaccharidosis VII (Sly Syndrome). Hum. Mutat. 30, 511–519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montaño A. M., et al., Clinical course of sly syndrome (mucopolysaccharidosis type VII). J. Med. Genet. 53, 403–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar M., et al., High-resolution magnetic resonance microscopy and diffusion tensor imaging to assess brain structural abnormalities in the murine mucopolysaccharidosis VII model. J. Neuropathol. Exp. Neurol. 73, 39–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parente M. K., Rozen R., Cearley C. N., Wolfe J. H., Dysregulation of gene expression in a lysosomal storage disease varies between brain regions implicating unexpected mechanisms of neuropathology. PLoS One 7, e32419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seto T., et al., Brain magnetic resonance imaging in 23 patients with mucopolysaccharidoses and the effect of bone marrow transplantation. Ann. Neurol. 50, 79–92 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Reichert R., et al., Neuroimaging findings in patients with mucopolysaccharidosis: What you really need to know. Radiographics 36, 1448–1462 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Levy B., Galvin N., Vogler C., Birkenmeier E. H., Sly W. S., Neuropathology of murine mucopolysaccharidosis type VII. Acta Neuropathol. 92, 562–568 (1996). [DOI] [PubMed] [Google Scholar]
- 64.Sly W. S., Vogler C., Gene therapy for lysosomal storage disease: A no-brainer? Transplants of fibroblasts secreting high levels of beta-glucuronidase decrease lesions in the brains of mice with Sly syndrome, a lysosomal storage disease. Nat. Med. 3, 719–720 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Vogler C., et al., Murine mucopolysaccharidosis type VII: The impact of therapies on the clinical course and pathology in a murine model of lysosomal storage disease. J. Inherit. Metab. Dis. 21, 575–586 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Parente M. K., Rozen R., Seeholzer S. H., Wolfe J. H., Integrated analysis of proteome and transcriptome changes in the mucopolysaccharidosis type VII mouse hippocampus. Mol. Genet. Metab. 118, 41–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen C.-D., et al., The antiaging protein Klotho enhances oligodendrocyte maturation and myelination of the CNS. J. Neurosci. 33, 1927–1939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanchard A., et al., DYT6 dystonia: Review of the literature and creation of the UMD Locus-Specific Database (LSDB) for mutations in the THAP1 gene. Hum. Mutat. 32, 1213–1224 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Lin R., Kwok J. C. F., Crespo D., Fawcett J. W., Chondroitinase ABC has a long-lasting effect on chondroitin sulphate glycosaminoglycan content in the injured rat brain. J. Neurochem. 104, 400–408 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Lin J.-P., Mironova Y. A., Shrager P., Giger R. J., LRP1 regulates peroxisome biogenesis and cholesterol homeostasis in oligodendrocytes and is required for proper CNS myelin development and repair. eLife 6, e30498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data generated for this study and ChIP sequencing data for THAP1, YY1, and H3K4me3 (generated by the Encyclopedia of DNA Elements Project Consortium) have been deposited in Gene Expression Omnibus (GSE161556, GSM803408, GSM803446, and GSM788087). All other study data are included in the article and/or supporting information.