Significance
Streptomyces genomes harbor a trove of biosynthetic gene clusters (BGCs) that encode for drug-like molecules. However, only a fraction of these readily yield expected products. To investigate why this is, we used polycyclic tetramate macrolactam (PTM) antibiotic production as a model system. By comparing the genomes and PTM production profiles of several closely related Streptomyces griseus clade members, we uncovered two mechanisms that differentiate more robust producers from weaker ones. The first involves small insertion–deletion lesions in PTM BGC promoters that significantly modulate production. The second mechanism involves biosynthetic pathway interactions, in which robust PTM producers unexpectedly benefit from griseorhodin coproduction, and weaker producers lack the pathway. We highlight comparative metabologenomics as a powerful approach to understand antibiotic crypticity.
Keywords: Streptomyces griseus, cryptic metabolism, regulation, metabologenomics, tetramic acid
Abstract
Streptomyces genomes harbor numerous, biosynthetic gene clusters (BGCs) encoding for drug-like compounds. While some of these BGCs readily yield expected products, many do not. Biosynthetic crypticity represents a significant hurdle to drug discovery, and the biological mechanisms that underpin it remain poorly understood. Polycyclic tetramate macrolactam (PTM) antibiotic production is widespread within the Streptomyces genus, and examples of active and cryptic PTM BGCs are known. To reveal further insights into the causes of biosynthetic crypticity, we employed a PTM-targeted comparative metabologenomics approach to analyze a panel of S. griseus clade strains that included both poor and robust PTM producers. By comparing the genomes and PTM production profiles of these strains, we systematically mapped the PTM promoter architecture within the group, revealed that these promoters are directly activated via the global regulator AdpA, and discovered that small promoter insertion–deletion lesions (indels) differentiate weaker PTM producers from stronger ones. We also revealed an unexpected link between robust PTM expression and griseorhodin pigment coproduction, with weaker S. griseus–clade PTM producers being unable to produce the latter compound. This study highlights promoter indels and biosynthetic interactions as important, genetically encoded factors that impact BGC outputs, providing mechanistic insights that will undoubtedly extend to other Streptomyces BGCs. We highlight comparative metabologenomics as a powerful approach to expose genomic features that differentiate strong, antibiotic producers from weaker ones. This should prove useful for rational discovery efforts and is orthogonal to current engineering and molecular signaling approaches now standard in the field.
Many therapeutics derive from natural products and their synthetic analogs (1). Historically, Streptomyces and related actinobacteria were heavily screened for these molecules, which resulted in numerous essential medicines. These include many clinical antibiotics (2), and there is an urgent need for new anti-infectives to counter increasing drug resistance (3). A massive reservoir of uncharacterized, biosynthetic gene clusters (BGCs) encoding drug-like molecules resides within Streptomyces genomes, which has triggered resurgent interest in these organisms (4). However, a large proportion of these BGCs fail to produce detectable levels of the expected compounds under laboratory conditions. This phenomenon of cryptic or silent metabolism thus poses a significant hurdle to genomics-driven drug discovery (5, 6). Silent BGCs are often thought to be transcriptionally deficient, and synthetic biology, cell signaling, and stress mechanisms are commonly used to activate silent BGCs for molecule discovery (7). Despite decades of research on the regulation of antibiotic production in Streptomyces (8, 9), a deeper mechanistic understanding of Streptomyces silent metabolism is still needed to access the full biosynthetic potential of these organisms to overcome the drug discovery gap (10).
Here, we employed a comparative metabologenomic approach to dissect why certain Streptomyces strains are ready antibiotic producers while others have apparently silent BGCs. Polycyclic tetramate macrolactam (PTM) antibiotics were specifically targeted in this study because they provide an opportune model system for multistrain comparative analyses. This is because PTM BGCs can contain as few as three genes (11), greatly simplifying regulatory studies. Another advantage of PTM BGCs is their relative commonality (12). A survey of bacterial genomes in GenBank, published in June 2016, reported that over 80 PTM BGCs were detected within 669 Streptomyces genomes available on GenBank at the time (13), and this commonality is leverageable for in-depth comparisons between PTM producers. The environmentally and biotechnologically important Streptomyces griseus clade (14) was identified as a particularly advantageous cohort for these comparisons because multiple strains within it had established active or cryptic PTM BGCs (12, 15), plus several additional family members with sequenced genomes and yet unstudied PTM clusters were available from public collections. Finally, PTM biosynthesis is increasingly understood, and this foundational knowledge was necessary to enact a targeted metabolomics approach to document sensitive production differences. PTMs have experienced intense study toward understanding their unusual hybrid nonribosomal peptide/polyketide origins, investigating PTM therapeutic potential, and leveraging their ease of manipulation via synthetic biology (13).
This study was initiated by comparing the genomes and PTM production profiles among a cohort of PTM locus-bearing S. griseus clade strains, which included known examples of both strong and poor PTM producers. This led to the discovery of a subclade that has consistently higher PTM production and PTM BGC promoter strengths compared to the rest of the test strains. Despite the exceptional commonality of PTM biosynthetic loci in Streptomyces bacteria, PTM regulation remains poorly understood. To reveal how promoter sequence variations might underpin the observed PTM phenotype differences, the promoter driving production in the robust producer Streptomyces species (sp.) strain JV180 was thus mapped and compared against the rest of the clade. Overall, many promoter features appeared to be largely conserved within the group, regardless of host–strain PTM capability. Additionally, the well-characterized global regulator AdpA (16) was confirmed to play a direct positive role on PTM locus control through gene deletion, binding site mutation, and in vitro binding experiments. AdpA binding sites were detected in the PTM promoters of all tested clade members, in which they display a contextually unusual arrangement downstream of promoter −10 boxes. Critically, comparisons of strong and weak PTM promoters from several S. griseus clade members identified a 2 to 3 bp indel, located between the −10 box and AdpA operator site, that strongly influences PTM production strength. Yet another PTM control mechanism that differentiates weak from stronger PTM producers was discovered following the mutagenesis of strain JV180’s griseorhodin BGC. Weak S. griseus clade PTM producers natively lack griseorhodin BGCs, and loss of the JV180 cluster severely curtailed PTM production. Further dissection revealed that strain JV180’s PTM production likely benefits from PTM–griseorhodin coexpression via a yet uncharacterized transcriptional mechanism. In sum, this work revealed two atypical mechanisms by which stronger and weaker PTM producers are differentiated, and it highlights the application of targeted comparative metabologenomics to cohorts of related strains to successfully reveal otherwise difficult-to-detect genomic features that tune antibiotic production.
Results
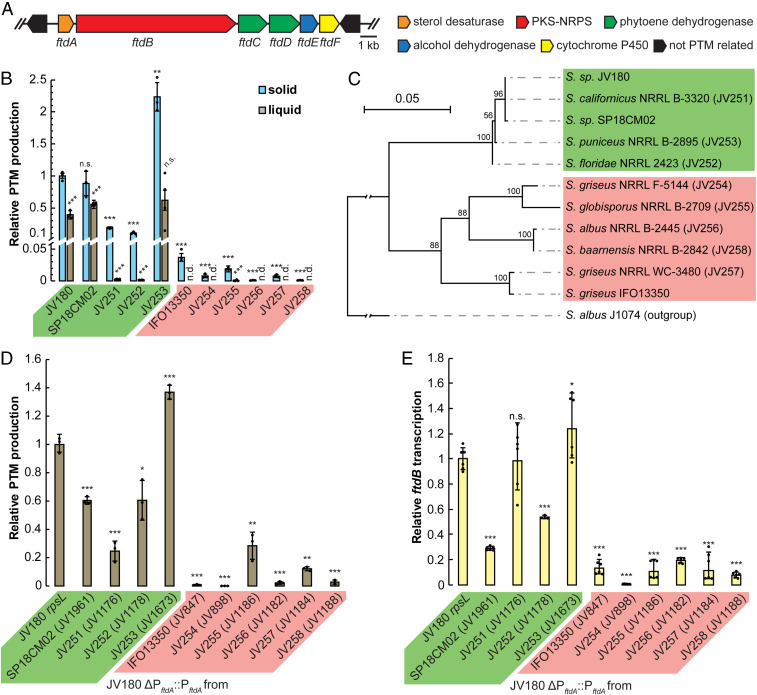
The S. griseus Clade Is an Ideal Model Group to Compare and Reveal Genetic Underpinnings of PTM Crypticity.
Several features of the S. griseus clade make the group attractive for comparison-based approaches to reveal BGC-silencing mechanisms. Clade member Streptomyces sp. strain JV180 readily produces PTM compounds (12, 17), while S. griseus subspecies griseus strain IFO13350 harbors a silent PTM BGC (15, 18). Nevertheless, promoter refactoring of the IFO13350 PTM BGC and its expression in a heterologous host successfully yielded PTMs with 5/5-carbocyclic ring systems, proving the functionality of its encoded enzymes. The PTM production status of most other clade members remained unknown and, to benefit from the increased analytical depth afforded by expanded cohort sizes, several additional clade members having PTM loci in their sequenced genomes (19) were obtained from public strain repositories (JV251 to JV258). To complete the cohort, the environmental clade member Streptomyces sp. strain SP18CM02, whose genome was recently reported by our group (17), (SI Appendix, Table S1) was also included. The phylogenetic relationships of these strains were inferred through multilocus phylogeny (14) (SI Appendix, Fig. S1). Importantly, the PTM BGCs of these strains appeared to be orthologous. This was based on several observations, including that their PTM BGCs share identically ordered biosynthetic genes (ftdA, ftdB, ftdC, ftdD, ftdE, and ftdF), and the chromosomal regions that immediately flank their respective PTM BGCs also have identical gene content (see Fig. 1A for a PTM BGC diagram representative of all strains studied here and the figure legend for additional detail). Finally, the PTM enzymes encoded within each studied BGC also shared high pairwise identities to those of strain JV180 (SI Appendix, Table S4).
Fig. 1.
Differences in S. griseus clade PTM production correlate with putative, PTM-promoter region sequence variation. (A) In all tested S. griseus clade strains, the PTM locus is comprised of a conserved set of genes, ftdA to ftdF. The non-PTM upstream and downstream genes in all tested strains, respectively, encode a cysteine desulfurase (open reading frame [ORF] −1) and mandelate racemase (ORF +1). (B) PTM production is most robust on solid media (blue bars, n = 3) versus when strains are grown in liquid (brown bars, n = 4) media. n.d. = not detected. (C) Neighbor-joining tree of the 500-bp upstream of each strain’s ftdA gene. This tree indicates that PftdA sequences of JV180-like strains form a separate group from the other (IFO13350 like) PftdAs. The analogous PftdA region of the Streptomyces albus strain J1074 was used as an outgroup. Sequences were aligned with Multiple Sequence Comparison by Log Expectation, and the tree was built using the Kimura substitution model in CLC Main Workbench. Branches supported by less than 50% of 500 bootstrap simulations were collapsed. (D) PTM production resulting from non-JV180 PftdA regions after introduction into JV180. The x-axis labels indicate which strains were used to source each heterologously tested promoter. The strain numbers in parentheses specify which strains in SI Appendix, Table S1 were used in each experiment (n = 3). (E) Relative ftdB transcript abundance from JV180 promoter–swapped strains in D (n = 6, except for JV1178 where n = 3). From these data, a clear trend emerges, in which strains and their PftdA region promoters sourced from the subclade that includes JV180 (A–D, green highlights) yield higher PTM production and transcription, compared to the rest of the tested S. griseus clade strains (A–D, red highlights). The statistical significance in the differences in PTM production or transcription were calculated by two-tailed Student’s t test, relative to JV180 or its rpsL mutant, JV307. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. = not significant. Error bars represent SD.
To assess and compare PTM production in these strains, the robust PTM-producing strain JV180 served as an archetype. Strain JV180’s PTM production was extensively analyzed using approaches similar to those reported for clifednamide-type PTM analyses in other Streptomyces by our group (20). Putative PTM peaks were identified using 13C5-labeled ornithine precursor incorporation and diagnostic daughter ion production following collision-induced dissociation in liquid chromatography–coupled tandem mass spectrometry (LC-MS/MS). These target the conserved ornithine-derived tetramate region, which has two possible hydroxylation states (SI Appendix, Figs. S2 B–E and S3). As expected, all PTM peaks identified through these methods were absent when the JV180 PTM BGC was deleted (ΔftdABCDEF; Fig. 1A and SI Appendix, S2A), yielding a high-confidence set of S. griseus clade PTM congeners for quantitative production comparisons.
The remaining S. griseus clade strains were then grown under several conditions, and their extracts were analyzed via LC-MS/MS and ultraviolet (UV) spectrometry for PTM production (see Materials and Methods). As expected from orthologous PTM BGCs, all producing strains gave PTM signals that largely overlapped with those established in JV180, but strain-to-strain differences in quantity and relative PTM congener ratios were observed. From these comparative PTM production data, an interesting trend emerged. Strains JV180, SP18CM02, and JV251 to JV253 displayed robust PTM production, while the remaining strains generally showed little production (Fig. 1B and SI Appendix, Figs. S4–S6). We noted that these strains belong to a distinct subclade on the S. griseus clade phylogenetic tree (SI Appendix, Fig. S1, group VI in green), suggesting they might share a conserved genetic basis for increased PTM production versus the other studied clade members. Interestingly, these comparative production analyses revealed a clear bias for higher PTM production on solid media over shake flask cultures for all tested strains (Fig. 1B). The strongest PTM producers continued to have the highest production in liquid media as well. Throughout, to obtain consistent transcriptional information, liquid media was used. Furthermore, in agreement with prior publications, PTM production could be detected by UV absorbance in JV180 (12), while strain IFO13350 produced insufficient PTMs for detection using this method (SI Appendix, Fig. S7A). In contrast, our PTM-targeted LC-MS/MS analyses revealed detectable production in all tested clade members, but the amounts varied widely by strain. This suggests that prior efforts to characterize weak PTM producers may have overlooked scant actual production due to inherent UV limitations, overcome here by MS/MS methodology (SI Appendix, Fig. S7 B and C).
PTM Promoter Sequence Heterogeneity Contributes to Differences in Antibiotic Production.
Because the most robust PTM production was found in strains of the JV180 subclade, we hypothesized that comparing these genomes against other S. griseus clade strains could reveal specific differences that underpin the observed production disparities. PTM BGC transcriptional differences were immediately targeted as a potential mechanism. While weak transcription is often implicated in the cryptic biosynthesis literature, this assertion is often speculative or left without mechanistic investigation (21). The possibility of PTM biosynthetic enzyme defects in some strains was ruled out as a potential cause. This is because all tested strains have high–PTM BGC protein sequence identity (SI Appendix, Table S4), and previous transcriptional refactoring experiments were able to successfully activate the cryptic PTM BGC of strain IFO13350 in heterologous hosts, as a demonstration of BGC functionality (15, 18).
Possible strain-linked differences in PTM BGC transcription were initially explored by comparing the presumed PTM promoter regions of each strain tested here. S. griseus clade PTM BGCs have a simple and conserved gene arrangement with minimal intergenic gaps, suggesting a single operon (SI Appendix, Fig. S8A). In JV180, PCR amplification of complementary DNA (cDNA) intergenic junctions confirmed this supposition (SI Appendix, Fig. S8B).
Phylogenetic analysis of the ∼500 nt located upstream of the first gene in each studied PTM BGC, ftdA, formed two groups: one by JV180-like strains and one by the remaining strains (these sequences are hereafter referred to as JV180- or IFO13350-like PftdAs, respectively; Fig. 1C and SI Appendix, Fig. S9). To test if the sequence differences in PftdA regions are transcriptionally relevant, we leveraged the strong PTM producer JV180. JV180 represents a uniform background to eliminate complicating, metabolic, and genetic variables inherent to strain-to-strain comparisons. After replacing the native promoter region of JV180 with the corresponding region of the other 10 S. griseus clade strains in this study, we generally observed higher PTM production and ftdB transcription from JV180-like PftdAs than IFO13350-like PftdAs (Fig. 1 D and E). This supports the idea that stronger PTM producers might result from inherently stronger promoters.
Surmising that the basic transcriptional machinery of all S. griseus clade members should be highly similar, we also hypothesized that if JV180-like PftdAs are stronger than IFO13350-like PftdAs, then introducing the former promoter type into strain IFO13350’s PTM BGC should increase PTM production and transcription. However, exhaustive attempts to introduce the necessary plasmids to engineer increased production in IFO13350 (and other group member JV257; SI Appendix, Fig. S1), failed by both conjugation and protoplast transformation. Instead, the effects of PftdA sequence variation were assayed in strain IFO13350 using an xylE (22) reporter gene chromosomally integrated into the ΦC31 site. Controls testing the efficacy of colorimetric XylE assays in both strains JV180 and IFO13350 were successful when using the strong, constitutive promoter PermE*, but both PftdA_JV180 and PftdA_IFO13350 failed to drive observable XylE in both strains. However, qRT-PCR assays to detect xylE transcripts were successful in making the desired comparison, confirming PftdA_JV180 to be stronger than PftdA_IFO13350 in both JV180 and IFO13350 hosts (SI Appendix, Fig. S10). Together, the above data supported a model in which PftdA sequence variations might significantly influence PTM production or silence. While BGC promoter heterogeneity is a concept that remains underexplored in the biosynthetic literature, the idea of tuning BGC outputs through promoter strength is grounded in numerous studies in which silent BGCs can be activated by replacing their native promoters with stronger ones (see recent review in ref. 23).
Mapping Promoter Regions of the JV180 PTM BGC Enables Structure–Function Comparisons.
Existing PTM regulatory knowledge is sparse (24, 25), including within the S. griseus clade. Our above results suggested that S. griseus clade PTM regulation involves cis-regulatory elements located directly upstream of ftdA (∼500 nt), although we could not rule out additional influences from trans-regulatory elements. To define the PTM promoter architecture of these strains, the transcriptional start site (TSS) of strain JV180 was mapped via circular rapid amplification of cDNA ends. The putative TSS was a cytosine residue 195-nt upstream of the predicted start codon of ftdA (SI Appendix, Fig. S11). Likely −10 and −35 boxes were assigned based upon established spacing (26, 27) (Fig. 2A). The high-sequence conservation between the −35 box and the TSS (82.4 to 100% pairwise sequence identity) suggested that all examined S. griseus clade strains share the same core promoter (−35, −10, and TSS).
Fig. 2.
Mapping of PftdA and identification of cis-regulatory elements by deletion analysis. (A) Diagram of the JV180 PftdA region, with the zones deleted upstream of PftdA diagrammed below. (Inset) Alignment of the core promoter sequence (note: the sequences are identical in JV180-like PftdAs). (B) Relative PTM production from JV180 mutants, with deletions in the upstream region of PftdA (n = 3). (C) Relative ftdB transcript abundance from JV180 mutants, with deletions in the upstream region of PftdA (n = 6). (D) Diagram of the PftdA UTR downstream of the transcript start site. Regions deleted downstream of PftdA are diagrammed below. (Inset) Cartoonized representation of the JV180 PftdA UTR secondary structure predicted by mFold. (E) Relative PTM production from PftdA UTR truncation mutants (n = 3). (F) Relative ftdB transcript abundance from PftdA UTR truncation mutants (n = 6). The statistical significance in the differences in PTM production or transcription were calculated by two-tailed Student’s t test, relative to the JV180 rpsL mutant, JV307. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. = not significant. Error bars represent SD.
To better resolve PftdA promoter architecture and probe for the presence of cis-regulatory signatures upstream of the predicted −35 boxes, a series of nested deletions in this region were created in JV180 (Fig. 2A). PTM transcription and production in these deletions were largely unaffected, except where the putative −35 box was disrupted (Fig. 2 B and C, Δ−528_−31). This confirmed the position of the JV180 −35 box and suggested this region lacks any critical, regulatory residues. Furthermore, we observed highly variable sequence conservation within this region upstream for all studied S. griseus clade strains (38.2 to 97.8% pairwise sequence identity). This contrasts with the stricter sequence conservation seen in the core promoter region (−35, −10, and TSS).
The –10 boxes and TSS residues of all examined strains are perfectly conserved, but several single-nucleotide polymorphisms (SNPs) differentiate the −35 boxes of the IFO13350-like PftdAs (Fig. 2A, red box) from the strictly conserved −35 boxes of the JV180 group (Fig. 2A, green box). It is known that changes in bacterial −35 boxes can greatly affect promoter strength (28, 29) and could thus affect metabolite production. Therefore, a panel of JV180 mutants carrying each observed −35 box SNP in the IFO13350 group was created and tested for PTM production and transcription (SI Appendix, Fig. S12A). Overall, the IFO13350 group −35 box SNPs failed to significantly change PTM production and only slightly decreased transcription in the chimeric JV180 hosts (SI Appendix, Fig. S12 B and C). These differences were subtle compared to the far more substantial ones seen in our ∼500-nt replacements upstream of the ftdA gene (Fig. 1 D and E), indicating that PftdA strength differences must originate through mechanisms other than −35 box differences.
AdpA Positively Regulates S. griseus Clade PTM BGCs.
Many Streptomyces messenger RNAs (mRNAs) contain long 5′ untranslated regions (5′ UTRs) (26, 27), and these can contribute to regulatory tuning. The DNA regions encoding 5′ UTRs can modulate gene expression through direct regulator binding (30), and their corresponding mRNAs can further modulate expression via riboswitches (31) or other RNA secondary structures (32). A series of ∼20 nt deletions were constructed (Fig. 2D) across the 195-nt 5′ UTR region of JV180 to probe contributions to PTM regulation. This revealed multiple lesions with strongly decreased PTM production and transcript levels, while others had little effect (Fig. 2 E and F). mFold (33) modeling to reveal possible 5′ UTR mRNA secondary structures across S. griseus clade strains yielded several energetically favored outputs, and we surmise some of the deleterious, mutational effects seen in JV180 might stem from the disruption of these types of structures (Fig. 2D, inset and SI Appendix, Fig. S13–17).
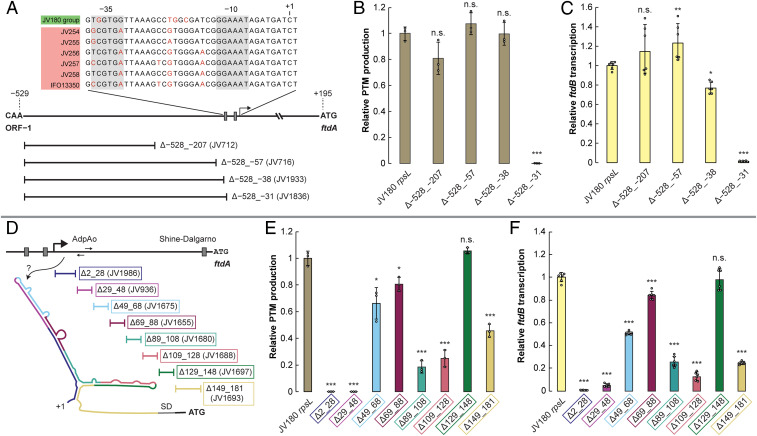
While S. griseus clade PftdA 5′ UTR folding remains to be further explored, the above deletions were particularly useful for identifying a critical region that we subsequently characterized as an AdpA binding site (Fig. 2 D–F, Δ29_48 and SI Appendix, Fig. S9). AdpA is a global regulator that is well studied in strain IFO13350, such that it is known to bind >500 chromosomal sites via its weak consensus sequence TGGCSNGWWY (34). S. griseus AdpA is involved in the hierarchical control of morphological differentiation, the production of streptomycin and other antibiotics, and several other important processes (see review in ref. 16). Prior chromatin immunoprecipitation sequencing and RNA sequencing data indicated that AdpA might bind upstream of IFO13350's PTM BGC (35), but because prior efforts to elicit PTM production from wild-type (WT) IFO13350 were unsuccessful (15, 18), the biological significance of AdpA’s interaction with the gene cluster was tenuous. Our searches for potential AdpA consensus motifs within the PftdA regions of the studied S. griseus clade strains revealed an imperfect inverted repeat 29- to 48-nt downstream of the TSS in the JV180 PftdA (within the 5′ UTR region) that was conserved in all S. griseus clade strains examined (SI Appendix, Fig. S9). Because our nested deletions revealed that this sequence is essential for JV180 PTM production, it renewed the notion that AdpA might positively regulate PTM expression.
Several experiments were carried out to investigate whether AdpA regulates PftdA, including adpA deletion and complementation analysis, mutating the putative AdpA binding site in PftdA, and in vitro binding assays. As expected from prior adpA studies in strain IFO13350 (36), deletion of the JV180 ortholog (ΔadpA) led to the loss of morphological differentiation and pigmentation (SI Appendix, Fig. S18). PTM production and BGC transcription were also abrogated in JV180, and these defects were rescued by ectopically expressing either native adpA, its IFO13350 ortholog (97% amino acid identity versus JV180 AdpA), or by replacing PftdA with the strong, constitutive promoter PermE* to drive PTM BGC expression independent of AdpA (Fig. 3 A and B and SI Appendix, Fig. S19). These results were consistent with AdpA being a transcriptional activator for PftdA. However, in bacterial regulation, regulators binding downstream of the RNA–polymerase complex typically cause transcriptional downshifts (37, 38). Thus, the location of the putative AdpA operator site downstream of the conserved –10 box is unusual for a transcriptional activator. Importantly, another AdpA-activated promoter with a downstream operator site controls S. griseus IFO13350s adsA, encoding an extracytoplasmic function sigma factor (30). This precedent thus supports the idea that AdpA could positively regulate PTM expression, despite its atypical, putative binding arrangement in PftdA.
Fig. 3.
AdpA is required for PTM expression and directly binds PftdA in vivo and in vitro. (A) Relative PTM production from JV180, its ΔadpA mutant, and complementation of PTM production by adpA or replacement of PftdA with the constitutive PermE* (n = 3). (B) Relative ftdB transcript abundance from JV180, its ΔadpA mutant, and complementation of PTM transcription by adpA or replacement of PftdA with the constitutive PermE* (n = 6). n.d. = not detected. (C) Alignment of the PftdA AdpA binding sites, with nonconserved nucleotides highlighted in red (note: the sequences are identical in the JV180-like PftdAs). (D) Relative PTM production from JV180 PftdA AdpA binding site mutants (n = 3). (E) Relative ftdB transcript abundance from JV180 PftdA AdpA binding site mutants (n = 6). Electrophoretic mobility shift assays for His-tagged JV180 AdpA interaction with the (F) JV180 PftdA binding site, (G) IFO13350 PftdA binding site, or (H) a JV180 PftdA binding site containing a transversion mutation in one of the nucleotides (red, underlined) directly interacting with AdpA, which was necessary for protein–DNA interaction. The lane marked + is a positive control using a WT JV180 probe sequence. The statistical significance in the differences in PTM production or transcription were calculated by two-tailed Student’s t test, relative to the JV180 rpsL mutant, JV307. *P < 0.05, **P < 0.01, and ***P < 0.001. Error bars represent SD.
AdpA–DNA cocrystallization studies indicate that the protein binds target operators as a homoduplex that recognizes a highly variable motif, containing four core invariant guanosine and cytosine nucleotides (Fig. 3C, bold residues in gray box) (34). In all S. griseus clade PftdA promoters studied here, these invariant residues were perfectly conserved, and introducing a transversion mutation at any one of these nucleotides abrogated JV180 PTM production and transcription (Fig. 3 C–E). This additional evidence further suggests that the identified region acts as a functional AdpA operator. Exchanging the JV180 AdpA operator site with that from IFO13350 was PTM proficient, although it showed a slight decrease in PTM titer and ftdB transcription (Fig. 3 C–E). AdpA–PftdA interactions were further examined via electrophoretic mobility shift assays using operator sequences from JV180 and IFO13350, plus the critical point mutants that disrupt essential DNA–AdpA interactions. As expected, recombinant histidine-tagged AdpA bound the JV180 and IFO13350 PftdA AdpA binding sites (Fig. 3 F and G) but failed to shift operators with the in vivo tested point mutations (Fig. 3H and SI Appendix, S20). Together, our in vivo and in vitro data strongly suggest that AdpA directly binds PftdA in both JV180 and IFO13350 but also indicate that the native SNPs (Fig. 3C, red residues) in the nonessential residues of the AdpA operator sites are not the main cause of PTM expression differences seen between PftdA_JV180 and PftdA_IFO13350 (Fig. 3 D and E).
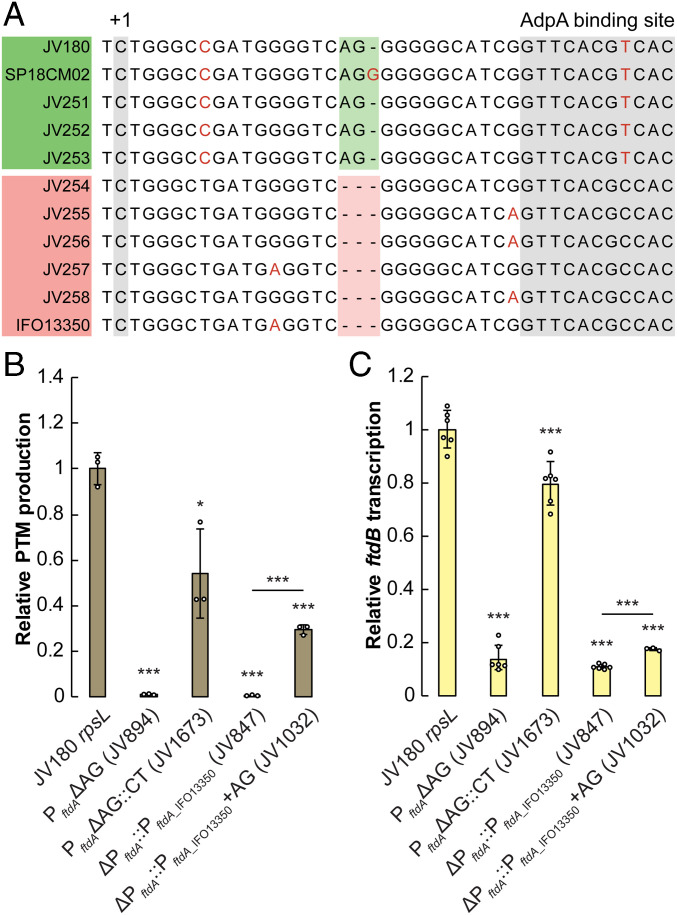
Comparative Promoter Analyses Reveal an Indel “Switch” that Tunes PftdA Strength and PTM Production.
Aside from the AdpA operator, the JV180 PftdA 5′ UTR truncation experiments revealed that the 28 nucleotides between the TSS and the AdpA binding site are also critical for PTM expression (Fig. 2 B–F, Δ2_28). Nucleotide alignments between promoters in this region revealed generally high conservation, except for two nucleotides (AG) that are present in all JV180-like PftdAs but are missing from IFO13350-like PftdAs (Fig. 4A and SI Appendix, Fig. S9). The effects of this indel on PTM regulation were tested by deleting this AG dinucleotide from JV180 and by introducing the dinucleotide at the corresponding position in PftdA_IFO13350 (PftdA_IFO13350+AG). PftdA_IFO13350+AG was tested heterologously in strain JV180. Strikingly, the dinucleotide deletion led to strongly reduced PTM production and transcription, while the amended PftdA_IFO13350+AG insertion variant led to a substantial increase in JV180 PTM production and transcription, compared to the WT PftdA_IFO13350 sequence (Fig. 4 B and C). To test if the indel’s effect was sequence specific, a JV180 transversion (ΔAG::CT) mutant was created, which exhibited reduced PTM production and transcription compared to WT but was much less deleterious than ΔAG (Fig. 4 B and C). This region is seemingly prone to sequence plasticity within the S. griseus clade; the recently isolated strain SP18CM02 contains an additional guanosine in this region compared to other JV180-like PftdAs (Fig. 4A). Despite having otherwise high, overall identity to all other JV180-like PftdAs, PftdA_SP18CM02 drove slightly less transcription and resultant PTM production when heterologously introduced into JV180 (Fig. 1 D and E). Together, these data suggest that, of the natural sequence variants in this region, having the AG dinucleotide is important for PTM production. These data clearly reveal this indel region as a key factor in modulating natural S. griseus clade PftdA strength. Further work is necessary to discern how this indel region modulates promoter strength, possibly via mechanisms such as perturbed AdpA–RNA polymerase interactions or recruitment of another yet unknown, regulatory component.
Fig. 4.
A conserved AG(G) indel tunes strong and weak PftdAs. (A) Alignment of the PftdA UTR section containing the conserved AG(G) indel between the TSS and the AdpA binding site. Nonconserved nucleotides are indicated in red. (B) Relative PTM production from JV180 PftdAΔAG mutants and JV180 ΔPftdA::PftdA_IFO13350 ± AG mutants (n = 3). (C) Relative ftdB transcript abundance from JV180 PftdAΔAG mutants and JV180 ΔPftdA::PftdA_IFO13350 ± AG mutants (n = 6, except for JV1032 where n = 3). The statistical significance in the differences in PTM production or transcription were calculated by two-tailed Student’s t test, relative to the JV180 rpsL mutant, JV307, or otherwise indicated. *P < 0.05, **P < 0.01, and ***P < 0.001. Error bars represent SD.
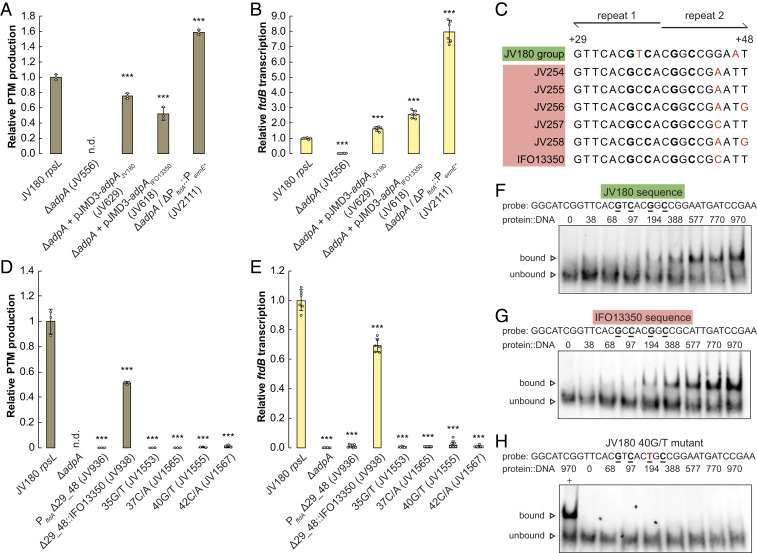
Discovery of an Unexpected Griseorhodin–Biosynthetic Interaction that Strengthens PTM Production in Strain JV180.
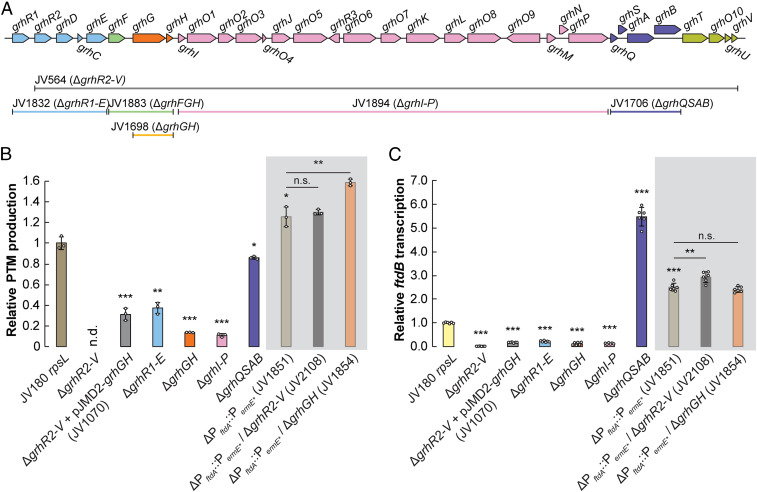
Our data thus far illustrated how leveraging within-clade comparative metabologenomics can assist regulatory region mapping and how small easily overlooked nucleotide changes in these regions can tune antibiotic production. The use of comparative genomics to understand Streptomyces antibiotic production, particularly at the species level, is a relatively recent development in natural products functional genomics. Studies in this area tend to focus on BGC conservation and differentiation (e.g., comparison of the Streptomyces albus clade; see ref. 39). From these comparisons, we anticipated that our S. griseus clade strains would share several BGCs (beyond PTMs) and that some antibiotic BGCs in these strains would not be conserved clade wide. We found that our S. griseus clade strains share a core set of 13 conserved BGCs. Some BGCs were found in only a few strains, and several BGCs were unique by strain (SI Appendix, Fig. S21). Strikingly, strain IFO13350 is one of the oldest known producers of streptomycin (40), but all members of the JV180 group were found to lack this BGC. Likewise, we noted that the JV180 group strains produced red pigments that are absent from the other studied S. griseus clade strains (SI Appendix, Fig. S22). Through comparative BGC analysis and subsequent cluster deletion in strain JV180, we attributed this pigment to the production of griseorhodin polyketide congeners (41) (Fig. 5A, ΔgrhR2-V and SI Appendix, S23 A–C). Unexpectedly, this griseorhodin BGC deletion mutant exhibited several additional phenotypes, including abrogated PTM production, down-regulated PTM BGC transcription, and reduced sporulation (Fig. 5 B and C and SI Appendix, S24).
Fig. 5.
Deletions in the JV180 griseorhodin (grh) BGC often negatively impact PTM production and transcription by PftdA. (A) Diagram of the JV180 grh BGC, with gene ranges deleted in various mutants mapped below as corresponding, colored bars. (B) Relative PTM production from various grh mutants in JV180 rpsL (white background) and JV180 ΔPftdA::PermE* background strains (gray background, n = 3). n.d. = not detected. (C) Relative ftdB transcript abundance from various grh mutants in JV180 rpsL and JV180 ΔPftdA::PermE* background strains, as noted in B (n = 6). The statistical significance in the differences in PTM production or transcription were calculated by two-tailed Student’s t test, relative to the JV180 rpsL mutant, JV307, or otherwise indicated. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. = not significant. Error bars represent SD.
The loss of PTM production in the JV180 ΔgrhR2-V mutant was wholly unexpected, and the mutant’s pleiotropic phenotypes suggested that griseorhodin might act as a signaling molecule. To test this, four adjacent genes encoding griseorhodin polyketide synthase (PKS) assembly-line enzymes were deleted (ΔgrhQSAB) (Fig. 5A). This was done to specifically eliminate griseorhodin production, while leaving all other grh BGC genes intact. Interestingly, this mutant was PTM proficient, arguing against the signaling idea. To further probe how the griseorhodin BGC exerts its influence, we tested a griseorhodin-enriched JV180 growth extract (see Materials and Methods and SI Appendix, Fig. S25A) and authentic γ-rubromycin (42) (a griseorhodin analog) for their ability to chemically complement the ΔgrhR2-V strain. Neither sample could restore PTM production in the ΔgrhR2-V mutant in flask cultures (SI Appendix, Fig. S25B), and disk diffusion tests on agar plates failed to restore sporulation in the diffusion zone (SI Appendix, Fig. 25C). These experiments together ruled out the griseorhodin signaling hypothesis.
To continue probing the grh locus for key PTM-influencing genes outside of grhQSAB, multiple groups of genes were deleted from the BGC (grhR1-E, grhFGH, and grhI-P; Fig. 5A). All three mutations caused a complete loss of red griseorhodin pigmentation and showed reductions in PTM production and transcription (Fig. 5 B and C and SI Appendix, Figs. S23D and S26B). Homology-based annotations of the genes in these regions (41) led us to focus on a subset, which might affect PTM biosynthesis through transcriptional or metabolic mechanisms. Genes grhR2 and grhR3 encode transcriptional regulators, which could influence regulatory cross-talk, grhF encodes a phosphopantetheinyl transferase (essential for posttranslational modification of NRPS and PKS enzymes), and grhGH encode β and ε subunits of acetyl-CoA carboxylase (ACC). ACC enzyme complexes are essential for malonyl-CoA production, a common precursor for fatty acid, griseorhodin, and PTM biosynthesis. PTM production in JV180 ΔgrhR2-V was not complemented by grhR2, grhR3, or grhF, but grhGH was able to restore some PTM production and transcription (Fig. 5 B and C and SI Appendix, Fig. S26A). Interestingly, expressing either grhG or grhH alone was sufficient to restore PTM production, similar to grhGH when expressed together (SI Appendix, Fig. S26A). A ΔgrhGH mutant was thus created, leading to partial griseorhodin pigmentation and reduced PTM production comparable to the initial ΔgrhFGH mutant (SI Appendix, Figs. S23D and S26B). How the ΔgrhR1-E and ΔgrhI-P mutations caused decreased PTM production remains unclear. Because these lesions led to the loss of putative regulatory genes (grhR2 and grhR3; see ref. 41), their phenotypes could be entangled with concomitant grhGH down-regulation.
In addition to the griseorhodin BGC’s grhGH, several other Streptomyces PKS clusters are known to harbor additional, nonhousekeeping copies of ACC genes. These include cpkKL in the coelimycin BGC of Streptomyces coelicolor (43) and jadN in the jadomycin BGC of Streptomyces venezuelae (44), and it can be reasoned that these ACC copies likely assist polyketide biosynthesis through increased malonyl-CoA. Because JV180 PTM production decreased in the ΔgrhR2-V and ΔgrhGH mutants but not the ΔgrhQSAB mutant (Fig. 5B), we likewise hypothesized that PTM downshifts in these mutants might be caused, at least partially, by reduced, intracellular malonyl-CoA concentration. We thus tested several additional S. griseus clade ACC subunit genes for their ability to rescue PTM production in the JV180 ΔgrhR2-V strain to discern if these effects were specific to grhGH or are more broadly attributable across ACC subunit homologs. The tested ACC genes included housekeeping accBE alleles cloned from JV180 and IFO13350 [function assigned by sequence homology and gene neighborhood synteny to the accBE genes in S. coelicolor (45)] plus the previously uncharacterized, PKS-associated ACC genes SGR3280 to SGR3281 of strain IFO13350, all of which encode ACC β and ε subunits like grhGH. All of these ACC homologs partially restored PTM production in the ΔgrhR2-V strain (SI Appendix, Fig. S26A), further supporting a role for malonyl-CoA in the PTM production defects of the ΔgrhR2-V and ΔgrhGH mutants.
To test if JV180 could still produce PTMs independent of the griseorhodin BGC, we constitutively expressed the strain’s PTM BGC by replacing PftdA with PermE*. This ΔPftdA::PermE* promoter replacement in both of the ΔgrhR2-V or ΔgrhGH backgrounds led to increased PTM production and PTM BGC transcription, similar to a JV180 ΔPftdA::PermE* control (Fig. 5 B and C). Our data show that PTM transcription and its production is affected by the lack of grhR2-V and grhGH. It is possible that the griseorhodin BGC may affect the PTM BGC through biosynthetic malonyl-CoA availability. It is also likely that an as of yet undefined transcriptional regulatory interaction also connects the two BGCs. Malonyl-CoA–responsive regulators are well-characterized in other model organisms such as Bacillus subtilis (46), but no such regulators are known in Streptomyces. Further inquiry is underway to characterize this unusual cross-cluster interaction more fully.
Intrigued by the finding that ectopic expression of just grhG or grhH could rescue PTM production in the JV180 ΔgrhR2-V mutant, we tested if the PTM BGCs of weak PTM-producing S. griseus clade strains might be similarly stimulated for production. This was done by heterologously expressing grhG in strains IFO13350, JV254, and JV258 (the grhGH construct had low-conjugation efficiency). This resulted in increased PTM production from these natively low-producing strains on agar media and also increased PTM BGC expression when tested in strain IFO13350, showing that ACC subunit overexpression can stimulate PTM production in natively poor producers (SI Appendix, Fig. S27 A and B). Combined with the above data, these observations support a model in which S. griseus clade PTM production differences have complex origins. In the case of poor PTM producers, promoter indels and insufficient positive BGC interactions likely dampen potential production via low transcription. In contrast, better PTM producers appear to benefit from cross-BGC interactions that increase transcription from a more active PftdA variant (+AG), resulting in PTM production. Strain JV180, and possibly other members of its subclade, do not seem to be bottlenecked for PTM production at the transcriptional level based on several findings throughout this study. This includes the failure of grhG merodiploids to boost WT JV180 PTM production (SI Appendix, Fig. S27A) and evidence from several recombinant strains in which PTM BGC transcription was significantly up-regulated by two- to ninefold, but PTM production increased by only up to 1.4-fold (ΔgrhQSAB, adpA complementation, and ΔPftdA::PermE* strains; Figs. 3 A and B and 5 B and C). The above findings also highlight ACC enzyme overexpression as a way to potentially up-regulate certain silent BGCs, adding to their current use for increasing fatty acid and polyketide titers via malonyl-CoA overproduction (47, 48).
Conclusions
Streptomyces genomes contain many BGCs encoding for drug-like compounds, and to harness their full biosynthetic capacity for discovery, it is crucial to understand what underpins the differences between active and silent BGCs (10). Cryptic metabolism is a well-recognized problem in the field, and poor BGC transcription is often implicated as the predominant mechanism behind biosynthetic silence. However, this supposition is increasingly challenged by a growing body of research that suggests all cryptic clusters aren’t necessarily transcriptionally silent (49). Furthermore, it only addresses how biosynthetic silence could originate in a given organism but does little toward explaining why affected BGCs show little activity to begin with. A common but difficult-to-prove hypothesis is that axenic laboratory growth deprives microorganisms of signals needed to up-regulate quiescent BGCs (21). This idea is based on the complex lifestyles of Streptomyces bacteria, which are known to be heavily influenced by molecular and environmental cues (50). Indeed, chemical elicitor screening, coculturing, and other regulatory manipulation strategies continue to yield new molecules from apparently silent BGCs, bolstering this idea (7, 50, 51). However, the number and types of molecular signals these organisms can sense and respond to must have a finite limit, constrained by the characteristics and capacities of their genetically encoded signal transduction pathways (10). If true, then extracellular signals can only explain one part of antibiotic crypticity, highlighting the need to understand how basic strain-to-strain genetic differences also contribute to the phenomenon.
Toward this, we leveraged the unusual commonality of PTM BGCs in Streptomyces to compare several highly related S. griseus clade strains to discover genomic features that differentiate strong and weak producers. These efforts led to a cohort of sequence-defined PTM promoters, confirmation that the global regulator AdpA acts directly on S. griseus clade PTM production by binding PftdA promoter regions in an atypical way, and that 2- to 3-bp lesions between −10 promoter regions and AdpA operator sites can cause substantial differences in transcription strength and biosynthetic output. To our knowledge, this type of inquiry is largely absent from the biosynthetic–regulatory literature, but similar promoter heterogeneity, with resultant-tuned transcription, has been documented to control phase variation in bacterial pathogenicity (52). Furthermore, because the AG indel discovered here resides in a polyguanosine-rich region, poly-N strand slippage (52) or a similar mechanism might plausibly explain how these promoter variants arise in S. griseus clade member populations. As more Streptomyces are sequenced and characterized, it is likely that additional promoter region sequence variations will emerge as drivers of silent metabolism in other biosynthetic pathways. In proof, scientists working at Warp Drive Bio, Inc. patented a method for activating silent BGCs encoding rapamycin-like antibiotics by “reversing” naturally occurring short indels within LAL family regulator binding sites (53, 54). Those regulatory indels were discovered through large-scale actinomycete genome sequence and molecule production comparisons at that company. Such knowledge is thus highly desirable to streamline targeted discovery efforts and direct new rational engineering approaches to activate select silent BGCs.
Finally, we also found that, in strain JV180, a BGC encoding griseorhodin production was required for both PTM production and transcription. Part of this relationship seems to implicate malonyl-CoA, whose biosynthesis is encoded for by grhGH, but further research is clearly needed to reveal a definitive mechanism. The deletion of unwanted BGCs is commonly used to engineer “clean” chassis strains (55) or to uncover the metabolites of BGCs (43, 56). In light of this, our discovery of PTM dependence on griseorhodin was surprising and provides a rare, cautionary example of how genome minimization can have unintended consequences on broader strain-wide BGC function. Together, our results show that the seemingly simple, monocistronic PTM BGC of strain JV180 appears to be under highly complex control, adding to a growing body of similarly complex BGC regulatory circuits and mechanisms in other Streptomyces (57, 58). Much remains to be learned about these types of regulatory networks in Streptomyces, and continued inquiry is required for a more comprehensive understanding to support rational drug discovery and production.
Materials and Methods
Strains, Plasmids, Primers, Enzymes, Chemicals, and General Methods.
Strains, plasmids, and primers are described in SI Appendix, Tables S1–S3. Strains IFO13350 and JV251 to JV258 were obtained from the Agricultural Research Service Culture Collection (NRRL). All primers were purchased from Integrated DNA Technologies. All restriction enzymes, Taq polymerase, and T4 ligase were purchased from New England BioLabs. PCR was generally carried out using KOD Hot Start DNA Polymerase (EMD Millipore) in FailSafe PCR 2× PreMix G (Epicentre). Taq polymerase was used for colony PCR to verify cloning and genome editing. Streptomyces genomic DNA was prepared for PCR by grinding a colony in DMSO, as described by Van Dessel et al. (59). Most media components and chemicals were purchased from Sigma-Aldrich or Thermo Fisher Scientific unless specified. γ-rubromycin was purchased from Abcam. 13C5-L-ornithine was purchased from Cambridge Isotope Laboratories. Standard protocols for manipulating Escherichia coli were based on those of Sambrook et al. (60). Streptomyces cultures were routinely propagated on International Streptomyces Project-2 medium (ISP2) agar and Trypticase Soy Broth (Difco) at 28 °C. Glass beads were added to liquid cultures to disrupt mycelial clumps.
Streptomyces Conjugations.
Conjugations were performed using JV36 as the general E. coli donor, as previously described (61). Streptomyces sp. strain JV180 spores were collected from lawns plated on 8,340 agar [1% Proflo cottonseed meal (ADM); 2% d-mannitol; 0.1% yeast extract; 0.01% KH2PO4; 0.01% MgSO4-7H2O; 0.002% CaCl2-2H2O; 0.2% (volume/volume) R2 trace elements solution (62); 2.067% MES hemisodium salt; 2% agar; and pH 6.5] using sterile cotton swabs into TX Buffer (63). Strain JV254 and JV258 spores were collected from lawns plated on Soya Flour Mannitol (62) and ISP-S (61), respectively. Exconjugants were selected with 50 µg/mL colistin and 50 µg/mL apramycin. Successful integrations by ΦC31 integrase–based vectors were verified by colony PCR, as previously described (20).
Marker-Less Gene Deletion/Promoter Replacement.
All gene deletions and PftdA mutants were constructed using double homologous recombination, as previously described (20). To avoid undesired recombination between the WT and the mutant PftdA sequences, an intermediate ΔPftdA::tsr mutant was constructed, with specifics provided the SI Appendix, Supplementary Methods. This ΔPftdA::tsr mutant was used as the parent strain for the construction of most PftdA mutants. Some plasmids for genome editing were cloned using overlap extension PCR instead of Gibson assembly, as described previously (64) (SI Appendix, Table S2).
PTM Production and Analysis.
Strains were streaked from −80 °C glycerol stocks onto ISP2 plates and incubated at 28 °C for 2 to 3 d. A plug was cut from the plate and used to inoculate 3 mL Trypticase Soy Broth in 24-well deep well plates, which were shaken at 300 rpm at 28 °C. One 4-mm glass bead was added per well to disrupt mycelial clumps. After 2 d of growth, 200 µL cultures were either inoculated into 20 mL ATCC-MOPS (adapted from ATCC172: 1% dextrose; 2% soluble starch, 0.5% yeast extract; 0.5% N-Z amine; 0.63% MOPS; and pH 7.2) in a 125-mL flask with 6-mm glass beads for disrupting clumps or plated on 8,340 agar overlaid with cellophane and incubated at 28 °C. Flask cultures were shaken at 250 rpm. After 4 d of growth, 1 mL was collected in RNAlater for protein and RNA analyses (see qRT-PCR), and the rest of the cultures were extracted twice with equal volumes of ethyl acetate. Solid media (plate) cultures were incubated for 6 d, and the mycelia and spores were collected from the cellophane for protein extraction by trichloroacetic acid, as described by Bose and Newman (65) and quantification by Bradford assay. The remaining agar was diced and extracted with ethyl acetate by soaking overnight.
The ethyl acetate extracts were dried at low pressure and resuspended in 500 µL LC-MS grade methanol and syringe filtered before LC-MS analysis. PTM analysis was performed using a Phenomenex Luna C18 column (75 × 3 mm, 3 µm pore size) installed on an Agilent 1260 Infinity HPLC connected to an Agilent 6420 Triple-Quad mass spectrometer. For each run, 10 µL sample was injected and the chromatography conditions were as follows: T = 0, 5% B; T = 4, 45% B; T = 12, 53% B; T = 16, 100% B, T = 20, 100% B; A: water + 0.1% formic acid, B: acetonitrile + 0.1% formic acid; and 0.8 mL/min. The diode array detector was set to measure absorbance at 320 nm. The mass spectrometer was set to precursor ion scan mode with the precursor ions m/z: 450 to 550, collision energy = 30 V, fragmentor = 70 V, and daughter ions m/z: 139.2 or 154.2. The resulting data were analyzed offline with Agilent MassHunter Qualitative Analysis software. Chromatograms were extracted for each parent–daughter ion mass transition, and the integrated areas for the major PTM congeners (SI Appendix, Fig. S28) were used to compare PTM production. One PTM peak had identical retention times and fragmentation spectra as an authentic standard of maltophilin (sourced from EMC Microcollections, GmbH; SI Appendix, Fig. S29), a stereoisomer of 10-epi-maltophilin produced by Streptomyces sp. strain SCSIO 40010, which has a similar PTM BGC to strain JV180 (66). The sums of PTM peak areas were normalized by total protein, and the relative PTM production was calculated relative to the appropriate control strain, typically JV180 rpsL. PTM production experiments were generally carried out in triplicates, unless specified. The statistical significance in the differences observed was calculated by two-tailed Student’s t test, typically relative to JV180 or its rpsL mutant, JV307, or otherwise indicated. *P < 0.05, **P < 0.01, and ***P < 0.001. Error bars represent SD.
Generally, the PTM production data reported were based on liquid media cultures that were used to collect the corresponding qRT-PCR data. Relative PTM production trends were consistent between solid and liquid media. Some figures in the SI Appendix show relative PTM production on solid media, where flask culture/qRT-PCR data were not collected.
qRT-PCR.
Strains were cultivated in flasks, as described in PTM Production and Analysis. After 4 d, 1 mL culture added to 2 mL RNAlater and vortexed to stabilize RNA. The mixture was centrifuged at 3,214× g for 10 min, and the supernatant was discarded. The RNA-stabilized pellet was resuspended in 250 µL 10-mg/mL lysozyme (Sigma-Aldrich) and incubated at 37 °C for 30 min. To the lysate, 750 µL TRIzol reagent (Thermo Fisher Scientific) was added, and protein and RNA were extracted following the manufacturer’s protocol from this point. Protein concentration was measured by Bradford assay. The RNA was resuspended in 84 µL nuclease-free water. DNase treatment was carried out by adding 10 µL 10× Turbo DNase buffer, 4 µL Turbo DNase with 2 µL RNAsin (Promega), for ∼6 h at 37 °C. Removal of leftover DNA was confirmed by PCR and gel electrophoresis before the DNase inactivation reagent from the Turbo DNase kit (Thermo Fisher Scientific) was added. RNA concentration was measured with a NanoDrop, and 5 µg RNA was used for reverse transcription with SuperScript II (Thermo Fisher Scientific) following the manufacturer’s protocol.
Primers for qPCR were designed using the Integrated DNA Technologies (IDT) PrimerQuest Tool. Real-time PCR was performed on a CFX Connect Real-Time PCR Detection System (BioRad) with the following program: 1 cycle at 95 °C for 3 min, 40 cycles of 95 °C for 10 s, and 55 °C for 30 s. Each reaction contained 5 µL iTaq Universal SYBR Green Supermix (BioRad), 2 µL nuclease-free water, 1 µL 10-µM forward primer, 1 µL 10-µM reverse primer, and 1 µL template cDNA. The relative transcript abundance was calculated using the ΔΔCT method, and hrdB was used as the housekeeping gene (67). The primer efficiency was determined as described by the qPCR instrument manufacturer for several pairs of PTM BGC probes, and the ftdB primers YQ376-180ftdB1153 and YQ377-180ftdB1278 were chosen for subsequent experiments, as they had the highest efficiency and produced the most consistent results. Data shown represent at three technical replicates, each for at least two biological replicates. The statistical significance in the differences observed was calculated by two-tailed Student’s t test, typically relative to JV180 or its rpsL mutant, JV307, or otherwise indicated. *P < 0.05, **P < 0.01, and ***P < 0.001. Error bars represent SD.
Expression and Purification of His-Tagged AdpA.
The full-length adpA gene of Streptomyces sp. strain JV180 was amplified by PCR using primers pET11a-AdpA-F and pET11a-AdpA-R. The PCR product was cloned into the expression plasmid pET11a. The expression recombinant plasmid, pKN052, contained the sequence adpA-CTC-GAG-(CAC)6-TGA under the control of the T7 promoter, similar to the construct reported by Yamazaki et al. (30). E. coli BL21 (DE3) Rosetta-harboring pKN052 was cultured in lysogeny broth (LB) medium with 100 µg/mL ampicillin at 37 °C overnight. A total of 1 mL seed culture was transferred to 150 mL LB medium with 100 µg/mL ampicillin and incubated by shaking at 37 °C at 250 rpm. When OD600 reached 0.6 to 0.8, the cells were chilled on ice for 1 h. After adding IPTG to 1 mM, the culture was continued for shaking at 250 rpm for 22 h at 18 °C. The cells were harvested by centrifugation at 5,000 rpm for 30 min, resuspended in Tris⋅HCl buffer (20 mM Tris⋅HCl, 200 mM NaCl, and 10% glycerol: pH:8.0), and stored at −80 °C. To purify the protein, cells were disrupted by sonication (3 min: 10 s on/10 s off at 20% amplitude), and His-tagged AdpA was purified with nickel-nitrilotriacetic acid (Qiagen) resin by eluting with 250 mM imidazole. Protein expression was verified with SDS–polyacrylamide gel electrophoresis before downstream experiments.
Electrophoretic Mobility Shift Assay.
Double-stranded DNA probes containing the 20 bp putative PftdA AdpA binding site plus 15 bp additional flanking sequences were synthesized by IDT. The DNA was resuspended in TEN buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl, and pH 8.0). For each probe, 50 nM probes were prepared using the Dig Gel Shift Kit second generation (version 10, Roche) with the following conditions: 20 µL DNA (from 100 nM stock), 8 µL 5× labeling buffer (1 M potassium cacodylate; 125 mM Tris⋅HCl; 1.25 mg/mL bovine serum albumin; and pH 6.6), 8 µL CoCl2 (25 mM), 1.5 µL digoxigenin (DIG)-11-DDUTP (1 mM), 1.5 µL terminal transferase, and water up to 40 µL. After brief mixing and centrifugation, reactions were incubated at 37 °C for 30 min and then chilled on ice. The reaction was quenched by adding 2 µL 200-mM EDTA (pH 8.0).
The protein–DNA interaction assay protocol was based on that described by Ming et al. (34). Purified AdpA-His was serially diluted in protein buffer (10 mM Tris⋅HCl, pH7.5; 100 mM NaCl; 2.5% [weight/volume] glycerol; and 0.25 mM DTT). The reaction mixtures contained 2 µL binding buffer (200 mM Tris⋅HCl, pH 7.5; 1 M KCl; 2.5 mg/mL bovine serum albumin; and 1% nonidet P-40), 1.2 µL 1-ng/µL poly[d(I-C)], 1.8 µL DNA probe (4.5 nM), the desired volume of purified protein, and water up to 20 µL. Samples were incubated for 1 h on ice. Gel electrophoresis was performed with a 5% native acrylamide gel (Bio-RAD, mini protein Tris-borate-EDTA [TBE] precast gel) in 0.5× TBE buffer (10× concentration: 890 mM Tris; 80 mM boric acid; and 20 mM EDTA, pH 8.3) at 85 V for 145 min. Electroblotting was performed using a BioRad Turbo Transfer System (Trans-Blot TurboTM System) on positively charged nylon membrane (Sigma-Aldrich). Cross-linking was performed by baking the nylon membrane at 120 °C for 30 min. Subsequently, the chemiluminescent detection was done as mentioned in the DIG kit protocol with very slight modification (overnight blocking, 2 h anti-DIG–AP treatment, and washing for 30 min each time), and the imaging was done with a LI-COR (Odyssey Fc) imaging system (model No. 2800).
Supplementary Material
Acknowledgments
We thank Jahdiel Berrios, Mayra Banuelos, and Adam Robinson for their bioinformatics assistance; Brandon Chia and John D’Alessandro for cloning assistance; and Prof. Doug Chalker for technical advice. We also thank Prof. Arpita Bose and members of the Bose Laboratory for useful discussions and proofreading assistance. This work was supported by Washington University in St. Louis new faculty start-up funds and the NSF award under NSF-CAREER 1846005 to J.A.V.B.
Footnotes
Competing interest statement: J.A.V.B. was a former employee, consultant, and advisory member of Warp Drive Bio, Inc. Warp Drive Bio is now a wholly owned subsidiary of Revolution Medicines where J.A.V.B. is a minority shareholder.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103515118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Change History
October 8, 2021: Figure 3 has been updated.
References
- 1.Bérdy J., Bioactive microbial metabolites. J. Antibiot. (Tokyo) 58, 1–26 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Miyadoh S., Research on antibiotic screening in Japan over the last decade: A producing microorganism approach. Actinomycetologica 7, 100–106 (1993). [Google Scholar]
- 3.Laxminarayan R., et al., Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013). Correction in: Lancet Infect. Dis. 14, 11, 182 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Nett M., Ikeda H., Moore B. S., Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26, 1362–1384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherlach K., Hertweck C., Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Hoskisson P. A., Seipke R. F., Cryptic or silent? The known unknowns, unknown knowns, and unknown unknowns of secondary metabolism. MBio 11, e02642-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutledge P. J., Challis G. L., Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gramajo H. C., Takano E., Bibb M. J., Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7, 837–845 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Bibb M., 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology (Reading) 142, 1335–1344 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Rigali S., Anderssen S., Naômé A., van Wezel G. P., Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem. Pharmacol. 153, 24–34 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Antosch J., Schaefers F., Gulder T. A. M., Heterologous reconstitution of ikarugamycin biosynthesis in E. coli. Angew. Chem. Int. Ed. Engl. 53, 3011–3014 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Blodgett J. A. V., et al., Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 11692–11697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang G., Zhang W., Saha S., Zhang C., Recent advances in discovery, biosynthesis and genome mining of medicinally relevant polycyclic tetramate macrolactams. Curr. Top. Med. Chem. 16, 1727–1739 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Guo Y., Zheng W., Rong X., Huang Y., A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: Use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58, 149–159 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Luo Y., et al., Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 4, 2894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohnishi Y., Yamazaki H., Kato J.-Y., Tomono A., Horinouchi S., AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Qi Y., et al., Draft genome sequences of two polycyclic tetramate macrolactam producers, Streptomyces sp. Strains JV180 and SP18CM02. Microbiol. Resour. Announc. 9, e01066-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y., Zhang L., Barton K. W., Zhao H., Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth. Biol. 4, 1001–1010 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Ju K.-S., et al., Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 112, 12175–12180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y., Ding E., Blodgett J. A. V., Native and engineered clifednamide biosynthesis in multiple Streptomyces spp. ACS Synth. Biol. 7, 357–362 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Chiang Y.-M., Chang S.-L., Oakley B. R., Wang C. C. C., Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr. Opin. Chem. Biol. 15, 137–143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ingram C., Brawner M., Youngman P., Westpheling J., xylE functions as an efficient reporter gene in Streptomyces spp.: Use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 171, 6617–6624 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Maclntyre L. W., Brady S. F., Refactoring biosynthetic gene clusters for heterologous production of microbial natural products. Curr. Opin. Biotechnol. 69, 145–152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed Y., Rebets Y., Tokovenko B., Brötz E., Luzhetskyy A., Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci. Rep. 7, 9784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bu X.-L., Weng J.-Y., Yu He-L., Xu M.-J., Xu J., Three transcriptional regulators positively regulate the biosynthesis of polycyclic tetramate macrolactams in Streptomyces xiamenensis 318. Appl. Microbiol. Biotechnol. 104, 701–711 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Jeong Y., et al., The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2). Nat. Commun. 7, 11605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y., et al., The transcription unit architecture of Streptomyces lividans TK24. Front. Microbiol. 10, 2074 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bibb M. J., White J., Ward J. M., Janssen G. R., The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol. Microbiol. 14, 533–545 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Siegl T., Tokovenko B., Myronovskyi M., Luzhetskyy A., Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 19, 98–106 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki H., Ohnishi Y., Horinouchi S., An A-factor-dependent extracytoplasmic function ς factor (ς(AdsA)) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182, 4596–4605 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borovok I., Gorovitz B., Schreiber R., Aharonowitz Y., Cohen G., Coenzyme B12 controls transcription of the Streptomyces class Ia ribonucleotide reductase nrdABS operon via a riboswitch mechanism. J. Bacteriol. 188, 2512–2520 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emory S. A., Bouvet P., Belasco J. G., A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 6, 135–148 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao M. D., et al., Complex structure of the DNA-binding domain of AdpA, the global transcription factor in Streptomyces griseus, and a target duplex DNA reveals the structural basis of its tolerant DNA sequence specificity. J. Biol. Chem. 288, 31019–31029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higo A., Hara H., Horinouchi S., Ohnishi Y., Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19, 259–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato J.-y., Miyahisa I., Mashiko M., Ohnishi Y., Horinouchi S., A single target is sufficient to account for the biological effects of the A-factor receptor protein of Streptomyces griseus. J. Bacteriol. 186, 2206–2211 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller-Hill B., Some repressors of bacterial transcription. Curr. Opin. Microbiol. 1, 145–151 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Browning D. F., Busby S. J., The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 57–65 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Seipke R. F., Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS One 10, e0116457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnishi Y., et al., Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li A., Piel J., A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem. Biol. 9, 1017–1026 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Atkinson D. J., Brimble M. A., Isolation, biological activity, biosynthesis and synthetic studies towards the rubromycin family of natural products. Nat. Prod. Rep. 32, 811–840 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Pablo Gomez-Escribano J., et al., Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 3, 2716–2720 (2012). [Google Scholar]
- 44.Wang L., McVey J., Vining L. C., Cloning and functional analysis of a phosphopantetheinyl transferase superfamily gene associated with jadomycin biosynthesis in Streptomyces venezuelae ISP5230. Microbiology (Reading) 147, 1535–1545 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Diacovich L., et al., Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem. 277, 31228–31236 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Schujman G. E., Paoletti L., Grossman A. D., de Mendoza D., FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell 4, 663–672 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Wang W., et al., Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat. Biotechnol. 38, 76–83 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Li S., Li Z., Pang S., Xiang W., Wang W., Coordinating precursor supply for pharmaceutical polyketide production in Streptomyces. Curr. Opin. Biotechnol. 69, 26–34 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Amos G. C. A., et al., Comparative transcriptomics as a guide to natural product discovery and biosynthetic gene cluster functionality. Proc. Natl. Acad. Sci. U.S.A. 114, E11121–E11130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Wezel G. P., McDowall K. J., The regulation of the secondary metabolism of Streptomyces: New links and experimental advances. Nat. Prod. Rep. 28, 1311–1333 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Okada B. K., Seyedsayamdost M. R., Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 41, 19–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henderson I. R., Owen P., Nataro J. P., Molecular switches—The ON and OFF of bacterial phase variation. Mol. Microbiol. 33, 919–932 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Bowman B. R., et al., “Compositions and methods for the production of compounds.” US Patent US10907188B2 (2019).
- 54.Shigdel U. K., et al., Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface. Proc. Natl. Acad. Sci. U.S.A. 117, 17195–17203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Myronovskyi M., Luzhetskyy A., Heterologous production of small molecules in the optimized Streptomyces hosts. Nat. Prod. Rep. 36, 1281–1294 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Culp E. J., et al., Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 37, 1149–1154 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Liu G., Chater K. F., Chandra G., Niu G., Tan H., Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bednarz B., Kotowska M., Pawlik K. J., Multi-level regulation of coelimycin synthesis in Streptomyces coelicolor A3(2). Appl. Microbiol. Biotechnol. 103, 6423–6434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Dessel W., Van Mellaert L., Geukens N., Anné J., Improved PCR-based method for the direct screening of Streptomyces transformants. J. Microbiol. Methods 53, 401–403 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Sambrook J., Fritsch E. F., Maniatis T., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, ed. 2, 1989). [Google Scholar]
- 61.Ko B., et al., Construction of a new integrating vector from actinophage φOZJ and its use in multiplex Streptomyces transformation. J. Ind. Microbiol. Biotechnol. 47, 73–81 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A., Practical Streptomyces Genetics (John Innes Foundation, 2000). [Google Scholar]
- 63.Hirsch C. F., Ensign J. C., Nutritionally defined conditions for germination of Streptomyces viridochromogenes spores. J. Bacteriol. 126, 13–23 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blodgett J. A. V., et al., Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat. Chem. Biol. 3, 480–485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bose A., Newman D. K., Regulation of the phototrophic iron oxidation (pio) genes in Rhodopseudomonas palustris TIE-1 is mediated by the global regulator, FixK. Mol. Microbiol. 79, 63–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu W., et al., Genome mining of marine-derived Streptomyces sp. SCSIO 40010 leads to cytotoxic new polycyclic tetramate macrolactams. Mar. Drugs 17, 663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Claesen J., Bibb M. J., Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J. Bacteriol. 193, 2510–2516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.