Significance
Antimicrobial resistance (AMR) is typically caused by genes present on plasmids, genetic parasites that can rapidly spread between bacterial cells. We demonstrate that plasmids can evolve increased transmission when hosts are abundant by increasing the number of plasmid molecules present within cells. A side effect of having more plasmids in a cell is a greater degree of cellular AMR. Selection for higher cellular levels of AMR in turn resulted in increased transmission via the same mechanism. Opportunities for plasmid transmission thus promote the evolution of plasmids that are highly infectious and confer high levels of AMR on their hosts.
Keywords: plasmid copy number, conjugative plasmids, antibiotic resistance, plasmid transfer
Abstract
Conjugative plasmids are mobile elements that spread horizontally between bacterial hosts and often confer adaptive phenotypes, including antimicrobial resistance (AMR). Theory suggests that opportunities for horizontal transmission favor plasmids with higher transfer rates, whereas selection for plasmid carriage favors less-mobile plasmids. However, little is known about the mechanisms leading to variation in transmission rates in natural plasmids or the resultant effects on their bacterial host. We investigated the evolution of AMR plasmids confronted with different immigration rates of susceptible hosts. Plasmid RP4 did not evolve in response to the manipulations, but plasmid R1 rapidly evolved up to 1,000-fold increased transfer rates in the presence of susceptible hosts. Most evolved plasmids also conferred on their hosts the ability to grow at high concentrations of antibiotics. This was because plasmids evolved greater copy numbers as a function of mutations in the copA gene controlling plasmid replication, causing both higher transfer rates and AMR. Reciprocally, plasmids with increased conjugation rates also evolved when selecting for high levels of AMR, despite the absence of susceptible hosts. Such correlated selection between plasmid transfer and AMR could increase the spread of AMR within populations and communities.
Conjugative plasmids are mobile genetic elements that transmit horizontally within and between species of bacteria (1). Plasmids often confer adaptive traits such as new metabolic abilities, virulence, and antimicrobial resistance (AMR); therefore, an understanding of variation in plasmid transmission rate (transfer) (2) is critical (3, 4). While greater transfer is expected to directly increase the frequency of plasmids in a population, it might also affect the phenotypes conferred by plasmids in other ways. Greater transfer may confer a cost to the host (5) (limiting growth and reducing the frequency of plasmid-carried traits), directly or indirectly modifying the expression of other plasmid-encoded traits (6) or promoting loss of accessory genes (7). Finally, plasmids that are at high frequencies within communities may rapidly adapt to ameliorate their costs (8), further increasing their frequency (9–11). Here, we experimentally investigate the consequences of selection for increased plasmid transfer rates on plasmid-encoded AMR.
Increased plasmid transfer (or parasite transmission in general) is predicted to be favored only when susceptible hosts are present in abundance (12). This is because the cost to host bacteria limits plasmid vertical transmission, leading to a trade-off between horizontal and vertical transmission (5). Indeed, evolution experiments in conditions with low opportunity for transfer observe decreased transfer rates and carriage costs (10, 13–15), but horizontal transmission can increase when susceptible hosts are available (5, 16–18). Here, we evolved conjugative plasmids under variable host availability and followed the evolution of transfer rates and AMR. We used two plasmids, R1 and RP4, which have variable transfer rates in closely related host strains (19), suggesting the potential for rapid evolution. R1 and RP4 are both conjugative multidrug-resistant plasmids, but differ in replication mechanisms, host range, and transfer regulation (20, 21).
Results
R1 Plasmid Evolves High Transfer Rates in the Presence of Susceptible Hosts.
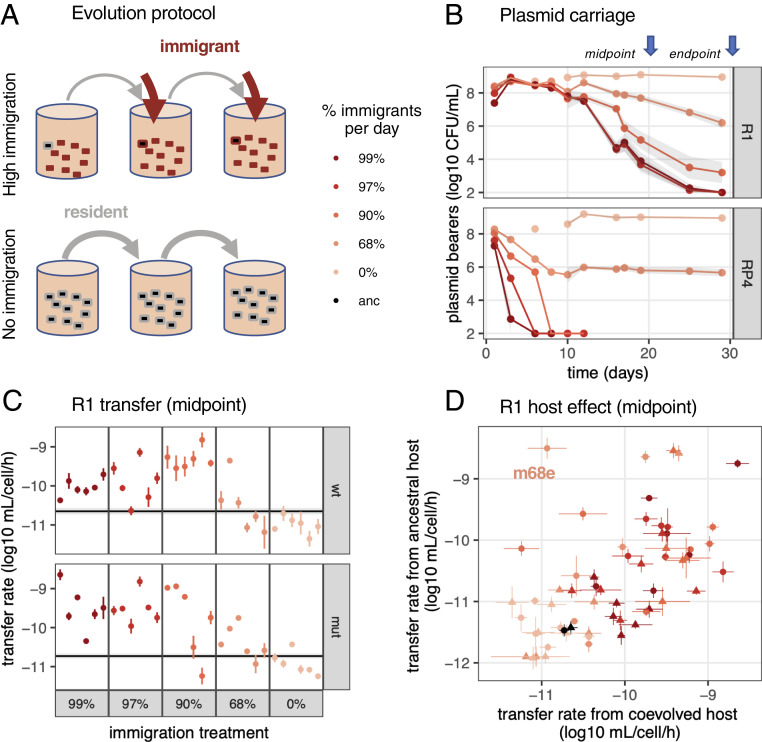
To vary the importance of horizontal transmission in plasmid life cycle without enforcing direct selection for plasmid-bearing cells, we passaged an initial culture of plasmid-bearing cells (either plasmid RP4 or R1) with regular influx of various proportions of immigrant plasmid-free cells and in the absence of any antibiotic selection (Fig. 1A). We used both wild-type (wt) and mutator (mut) Escherichia coli hosts, the latter to increase genetic variation available for selection. Plasmid maintenance in immigration treatments required horizontal transmission, or they would be rapidly diluted out, with plasmids in high immigration treatments experiencing higher selection for horizontal transmission (SI Appendix, Fig. S1). RP4 plasmid was rapidly lost from all treatments with ≥90% immigration per day, but stably maintained over 30 d, after an initial decline, under 68% immigration. R1 plasmid was maintained for longer than RP4, but R1-bearing cell density decreased steadily for all immigration treatments, with a faster decrease under higher immigration (Fig. 1B).
Fig. 1.
Evolution of plasmid transfer rate with varying host availability. (A) Experimental evolution regime. Plasmid-bearing cells (black) were first mixed with plasmid-free cells, then evolving populations were diluted every day to fresh medium (gray arrows) mixed in varying frequencies to plasmid-free cells (red arrows). Each treatment was performed in six replicates with either WT or mut E. coli hosts. (B) Plasmid population dynamics. Initially one of six replicates was measured until day 12; the treatment average is shown ± SEM (shaded area). (C) Midpoint R1 conjugation rates. The black line and shaded area are the geometric mean and SE, respectively, of ancestral plasmid transfer rate; each colored dot and line indicates, respectively, the geometric mean and SE of evolved clones (n = 3). WT clones are shown on the Top, and mut clones on the Bottom. (D) Effect of host evolution on R1 conjugation. The x axis shows transfer rates measured from plasmid-bearing coevolved hosts (same as in C); the y axis shows rates from ancestral hosts carrying evolved plasmids. Dots and lines indicate, respectively, the geometric mean and SE (n = 3). Circles show the WT background and triangles, the mut background.
We first measured transfer rates from coevolved plasmid–host clones to ancestral recipients. We detected no significant changes in endpoint (29 d) conjugation rates for RP4 but increased transfer for R1 with 68% immigration (SI Appendix, Fig. S2, R1 transfer rate ∼ immigration, F2,86 = 5.2, P = 0.007). For R1, we next focused on midpoint clones, before plasmid lineages under high immigration went extinct. We observed significant effects of immigration treatment on evolved transfer rates (Fig. 1C, transfer rate ∼ immigration, F5,198 = 43.1, P < 2.10−16). Specifically, treatments with ≥90% immigration had significantly increased transfer rates compared to the ancestor and ≤68% immigration treatments (Tukey’s test, P < 2.10−5) but were not significantly different from each other (see SI Appendix, Supplementary Text for detailed statistics and host effect). Thus, high transfer rates evolved in the presence of abundant plasmid-free recipients.
To tease out effects of host evolution on evolved transfer rates, we also measured transfer rates of midpoint evolved plasmids from the ancestral host. Transfer rates were still greater in immigration treatments (SI Appendix, Fig. S3A). Transfer rates from coevolved hosts correlated with transfer rates from ancestral hosts (Fig. 1D, bivariate model Cov(coevolved, ancestral) = 0.39 ± 0.10, clone covariance effect χ = 23, P = 1.5.10−6; correlation coefficient = 0.60 ± 0.09). Thus, increased transfer rates are mostly due to genetic changes in plasmids rather than hosts. However, a few clones diverged from this pattern, most significantly clone mut68e with a 1,000-fold increase in transfer rate when the plasmid was present in an ancestral host, but not in its coevolved host (Fig. 1D). This suggests that some hosts evolved repression of plasmid transfer, potentially because of transfer costs to the host.
R1 Evolves Higher Costs.
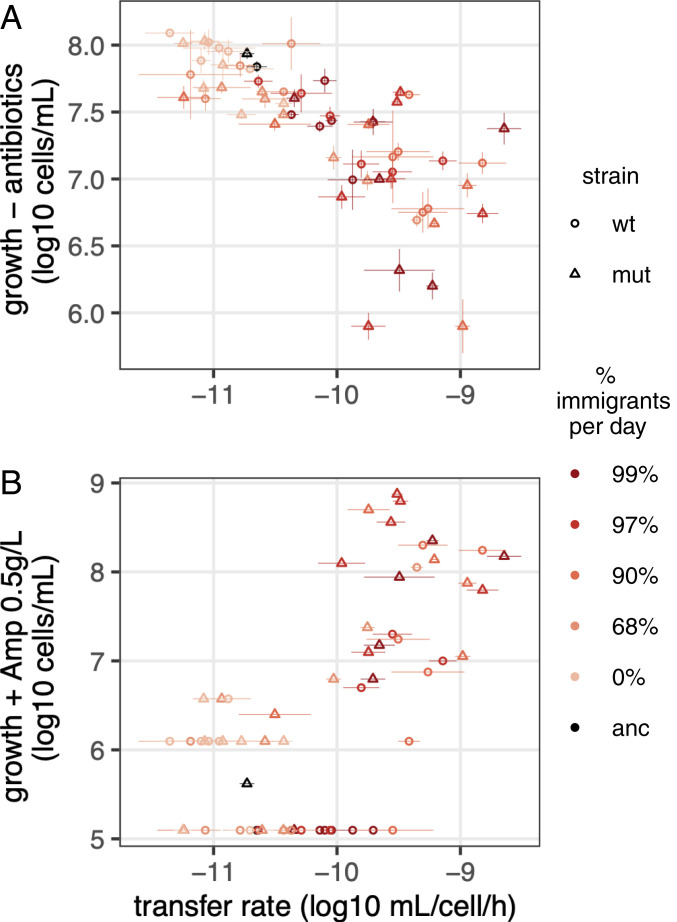
As a proxy for plasmid cost, we measured the densities reached by donor hosts during conjugation assays. Donor density was negatively correlated with transfer rates (Fig. 2A, donor density ∼ transfer rate, estimate = −0.53 ± 0.03, r2 = 0.56, P < 2.2 10−16). Host density still correlated with transfer rates when plasmids were carried by the same, ancestral host (SI Appendix, Fig. S3B), confirming that some of the reduction in cell growth is due to plasmid evolution. This trade-off likely limits the fitness benefit of increased transfer rate, by limiting plasmid vertical transmission.
Fig. 2.
Breakdown of the trade-off between transfer and host fitness under antibiotic selection. Host growth in the absence of antibiotics (A, donor cell density during conjugation assays) and in the presence of high levels of antibiotics (B, density of cells able to grow on Amp 0.5 g/L) is shown for midpoint coevolved clones as a function of their transfer rate. Dots and lines indicate, respectively, the geometric average and SE (n = 3).
R1 Also Evolves Increased AMR.
To evaluate whether the benefits conferred by plasmids in the presence of antibiotics also evolved, we measured cell growth in the presence of high levels of antibiotics (Fig. 2B). Many evolved clones conferred on their hosts an ability to grow in conditions in which the ancestor could not (Fig. 2B). Most of these also had increased transfer rate (Spearman’s rank correlation rs = 0.62, P = 10−7), suggesting that increased AMR is a side effect of selection for transfer.
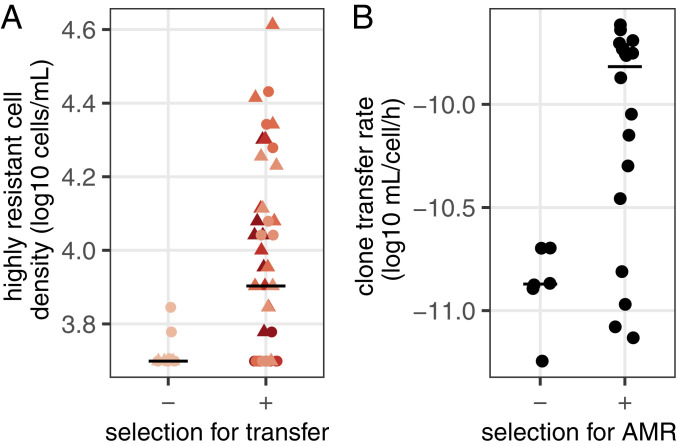
This apparent coupling between transmission and AMR prompted us to address whether selection for high transmission coselects increased AMR and vice versa. First, we plated early populations from the evolution experiment directly on ampicillin (Amp) 0.5 g/L (Fig. 3A). Highly resistant cells were more common in immigration treatments, especially in the mutator host (log10 cell density ∼ immigration * host, F1,56 = 12.8, P = 0.0007). In some lineages, nearly all plasmids conferred upon their hosts ability to grow at high antibiotic concentrations and showed increased transfer compared to the ancestor plasmid R1WT (SI Appendix, Fig. S4). Thus, immigration treatments selecting for increased transfer collaterally promote the appearance of high AMR.
Fig. 3.
Reciprocal selection for high transfer rate and AMR. (A) Host availability promotes the evolution of highly antibiotic-resistant plasmids. The density of cells able to grow in the presence of 0.5 g/L Amp is shown for populations evolved for 9 d with or without immigration. (B) Selecting for high level antibiotic resistance promotes evolution of highly transmissible plasmids. Six independent populations of an R1WT clone were plated with or without high concentrations of antibiotics, and the transfer rate of three mutants per population was measured (geometric average of n = 3 replicates per mutant). Horizontal lines show the median value for each treatment.
Next, we determined whether selection for elevated AMR also increases transfer rates in the absence of direct selection for transmission. We plated independent cultures of the ancestral host carrying R1WT on high levels of antibiotics, and measured plasmid transfer rate for highly resistant mutants (Fig. 3B). Selection for AMR increased transfer rate overall (log10 transfer rate ∼ selection, F1,70 = 30.6, P = 5.10−7), and 13 of 18 mutants displayed increased transfer compared to R1WT, with ∼10-fold higher transfer rates (Fig. 3B).
The Presence of Susceptible Hosts Favors R1 Copy Number Variants.
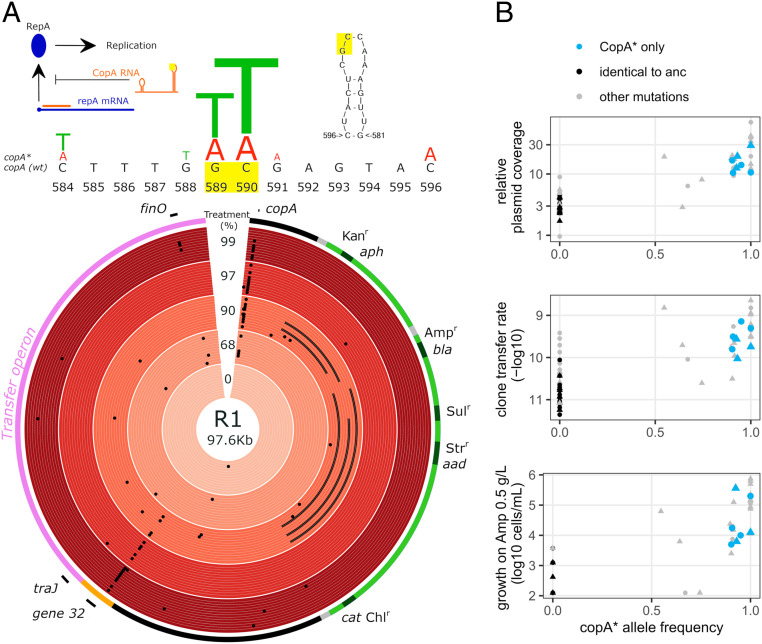
To understand the genetic basis of the evolved phenotypic changes, we sequenced midpoint evolved clones. The summary of mutations is shown in Dataset S1. Plasmid mutations were found almost exclusively in clones from immigration treatments (Fig. 4A). We focused next on loci with parallel mutations in independently evolved lineages, a sign of convergent molecular evolution (but see SI Appendix, Supplementary Text for other mutations). First, four evolved plasmids from the 90% immigration treatment contain large deletions (∼10 to 20 kb), which include one or several antibiotic resistance determinants (SI Appendix, Fig. S5). Rapid loss of resistance genes by R1 was observed previously (7, 14), and is likely underestimated here as we used Amp resistance as a proxy for plasmid carriage. Next, four evolved plasmids carried the same 1-bp insertion in the coding region of finO, the main repressor of the transfer operon (20), and another carried a 7-bp insertion, causing a frameshift. These mutations are known to cause loss of function and derepression of the transfer operon (22). Seventeen variants had a G deletion and one a GG deletion in a polyG tract ∼50 bp before the coding region of gene32, part of the leading region first transferred to recipient cells. gene32 function is unknown but it is conserved and likely important for conjugative transfer (23). Finally, the most common mutations were in the copA locus. The untranslated antisense CopA RNA plays a central role in R1 replication and copy number control by inhibiting translation of the initiator for plasmid replication RepA (24). We observed overall 27 point mutation variants contained in a 13-bp region within copA: nine different types of mutations were observed, but all from C or G nucleotides toward A or T, denoted as copA* below (Fig. 4A). These mutations structurally alter the head of CopA RNA’s loop II, which is involved in binding to RepA mRNA (24). Identical or similar mutations were shown to increase R1 plasmid copy number (PCN) by decreasing CopA binding to RepA mRNA (25). Accordingly, we observed increased sequencing coverage for copA* plasmids consistent with increased PCN (Fig. 4 B, Top, detail in SI Appendix, Fig. S5).
Fig. 4.
Molecular evolution and mechanistic basis for increased R1 transfer and AMR. (A) Variants detected in evolved plasmids. The outermost ring of the circos plot shows a simplified plasmid map. Evolved plasmids are then shown by treatment, from outside in wt clones a to f then mut clones a to f. Black dots indicate small polymorphisms; lines show large deletions. Inset diagrams above show the secondary structure of CopA antisense RNA and its mode of action. Significant 13 bp of evolved variants are summarized with letter size proportional to the relative abundance of each mutation. The position of the most common mutations in the second loop of CopA RNA is highlighted in yellow (modified from ref. 24). (B) Phenotypic effects of copA* variants. Evolved plasmid coverage, transfer rate, and growth on Amp 0.5 g/L are shown as a function of copA* mutation frequency in the evolved clone (as not all plasmid copies necessarily have copA* mutation). Blue dots indicate clones for which no other mutation than copA* was present on the plasmid, and black dots indicate clones for which no mutation was detected on the plasmid.
An Increase in R1 Copy Number Couples Transfer and AMR.
Across all clones, only copA* and gene32 mutations were significantly associated with high transfer rates, copA* having the strongest effect (log10 transfer rate ∼ copA* + gene32 + finO, copA* effect 1.01 ± 0.15, F1,200 = 236, P < 2.10−16, gene32 effect 0.30 ± 0.16, F1,200 = 12.0, P = 0.0007). finO inactivation can strongly increase conjugation rate (Fig. 5 A), however no evolved plasmid carried finO mutations alone here, and its effect on transfer was nonsignificant (effect 0.22 ± 0.27, F = 2.4, P = 0.12). Strikingly, seven evolved plasmids carried only a copA* mutation and were otherwise identical to R1WT (blue dots in Fig. 4B); these had on average a 17-fold increased transfer rate (copA* effect, F1,97 = 170, P < 2.10−16) in coevolved hosts, and a 19-fold increased transfer rate when in the ancestral host (SI Appendix, Fig. S6, F1,109 = 160, P < 2.10−16). copA* variants also conferred a 55-fold increase in AMR on their hosts (F1,25 = 45, P = 5.10−7), which is consistent with previous results on PCN effect on AMR (26–28). Thus, copA* mutations are sufficient to increase R1 transfer rate 17-fold and explain (via increased PCN) the increase in AMR. At high doses of other antibiotics to which R1 confers resistance (ampicillin, chloramphenicol, and streptomycin), R1copA* also increased cell survival compared to R1WT and R1finO (SI Appendix, Fig. S7). Thus, increased gene dosage affects both the transfer region and other phenotypes encoded by R1.
Fig. 5.
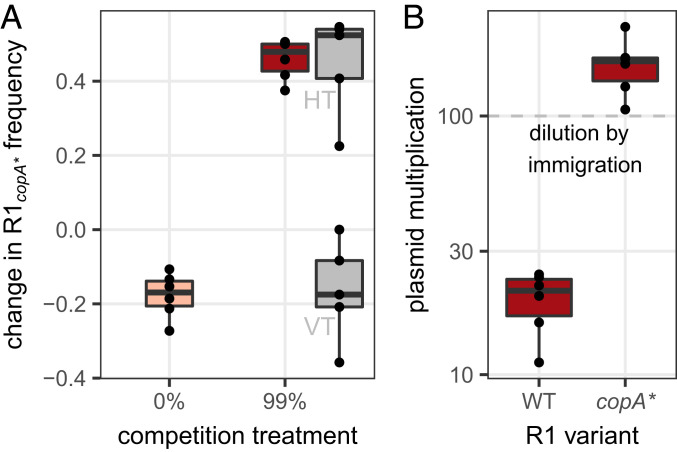
copA* variants trade-off vertical against horizontal transmission. (A) Change in frequency of R1copA* in competition with R1WT, in the absence or presence of immigration. In the presence of immigration (99% treatment), the change in frequency within initial donors (VT) and in transconjugants (HT) is shown in gray. The center value of the boxplots shows the median, boxes show the first and third quartile, and whiskers represent 1.5 times the interquartile range; individual data points are shown as dots (n = 6). (B) Detail of the change in plasmid-carrying cell density for each variant in the presence of immigration. The center value of the boxplots shows the median, boxes show the first and third quartile, and whiskers represent 1.5 times the interquartile range; individual data points are shown as dots.
To understand the mechanism by which transfer increases, we measured mobilization of pMOB, a nonconjugative construct carrying R1 oriT (origin of transfer). pMOB can be mobilized by R1 transfer machinery, but its PCN is independently regulated; thus increases in pMOB transfer can only result from increased availability of R1 transfer machinery. A finO variant, which has derepressed expression of the transfer machinery (22) but no change in PCN, increased transfer of both itself and pMOB by around 1,000-fold (SI Appendix, Fig. S8). A copA* variant also increased transfer for both plasmids but mobilized pMOB ∼4-fold less than itself (R1copA* 26-fold, pMOB 6.5-fold, paired two-tailed t test, P = 0.0003). Thus, increased transfer of copA* variants is due to the combination of increased transfer machinery expression (6.5-fold increased pMOB mobilization), and increased number of copies of oriT (4-fold R1-specific increase in mobilization). Both effects are approximately of the same size as the 5-fold increase in PCN, suggesting they are due directly to increased gene dosage.
CopA* Variants Spread Better in the Presence of Immigration, Despite Their Cost.
Finally, we characterized the fitness effects of a representative copA* variant (w90d, carrying the unique mutation G589T) in competition with the ancestral plasmid (Fig. 5A). In the absence of plasmid-free cells, R1copA* conferred a fitness cost to its host when competing with R1WT. In the presence of 99% plasmid-free recipients, R1copA* instead had a selective advantage against R1WT. This was due to horizontal transmission, as donor hosts containing R1copA* still decreased in frequency (Fig. 5A, gray). Increased horizontal transmission also allowed copA* variants to overcome the dilution caused by immigration, which R1WT was unable to do (Fig. 5B). Thus, copA* variants trade off increased horizontal transmission against decreased vertical transmission, which is beneficial in conditions of high immigration. finO variants imposed a much higher cost to their hosts (SI Appendix, Fig. S9), which presumably limits the spread of finO variants relative to increased PCN.
Discussion
In the presence of susceptible hosts, we observed rapid evolution of increased horizontal transmission for R1, but not for RP4. Mutations able to increase transfer rate in RP4 might be constrained or too costly: regulation of transfer in RP4 makes increases in transfer expression hard to achieve (21), and increasing PCN, while possible, can be extremely costly (29). For R1, despite a large increase in horizontal transmission with full derepression of the transfer operon, few clones evolved finO inactivation, likely because of its high cost to vertical transmission (30). Instead, the most common mechanism for increased transfer was the evolution of increased PCN due to parallel mutations in copA, which imposed some, but much lower, fitness costs. Similar PCN evolution could be common in other plasmids, as point mutations are frequently sufficient to change PCN, via changes in regulatory RNAs (25, 26, 31), or in Rep proteins (32, 33). Such changes likely always increase mobilization in both conjugative and mobilizable plasmids, because of a direct increase in oriT dosage. For conjugative plasmids, increased gene dosage of the transfer operon might further increase transfer machinery activity, similarly to R1. Note however that, in contrast to our findings, high copy mutations in a conjugative IncI plasmid conferring colistin resistance were shown recently to negatively affect plasmid fitness despite an increase in horizontal transmission (34).
In addition to increased horizontal transmission, copA* mutations also increased AMR through increased gene dosage. High levels of AMR correlated with high PCN have been observed previously (24, 26–28); we show here this can be driven by selection for transfer. Interestingly, specific coupling between horizontal transmission and AMR has been observed in other mobile elements (6, 35), in which increased resistance gene expression also caused increased expression of transfer genes located immediately downstream. The effect we uncover here is more general, as all genes carried on the plasmid experience higher dosage. In R1, this translates into increased resistance at antibiotics concentrations not likely to be encountered naturally even under antibiotic treatment. However, similar increases in PCN might lead to clinically and environmentally relevant fitness benefits depending on plasmid accessory gene content. High PCN has been linked to increased resistance to low doses of antibiotics against which initial resistance is minimal (33, 35–37). Virulence and metabolic genes are also frequently carried on conjugative plasmids, and increased PCN has been linked to increased virulence in animal (38) and plant (39) pathogens and to increased ability to grow on limiting nutrients (40).
Increased PCN could promote further evolution because multicopy plasmids have increased mutation supply (27). Indeed, copA* variants have significantly more additional single nucleotide polymorphisms (SNPs) than other evolved variants (Wilcoxon rank sum test, W = 238, P = 0.0009), suggesting that evolution of PCN is sufficient to rapidly increase evolvability. However, high PCN can also limit evolvability because any new mutation will be present on only one plasmid copy, affecting both genetic drift and selection (41). Overall, PCN is a crucial component of plasmid life history and modifying it can have far-reaching consequences through both gene dosage and evolvability (42).
Direct selection for cells containing conjugative plasmids usually removes plasmid-free hosts available for transfer, thus decreasing horizontal transmission overall (43). We show here that selection for high gene dosage of plasmid-carried genes can, by contrast, favor highly transmissible plasmids, because of the pleiotropic effects of increased PCN. Another factor to consider in order to understand longer-term coevolution is the cost plasmid transmission imposes on hosts and the resultant selection pressure. We observed here a few clones which evolved repression of plasmid transfer. Alternatively, selection for plasmid genes might favor alleles promoting transfer if plasmids are transferred to kin (44). Increased horizontal transmission via high PCN creates a positive relationship between two traits central to plasmid life history, coupling vertical and horizontal transmission components in ways that might complicate AMR management.
Methods
Strains, Plasmids, and Growth Conditions.
The ancestral plasmid-bearing strains were E. coli MG1655 (wt) and MG1655 mutL::KnR (mut) obtained by transduction from the Keio collection (45). Conjugative plasmids were R1 and RP4: R1 is an narrow-host-range IncFII plasmid, conferring resistance to Amp, kanamycin, sulphonamides, streptomycin, and chloramphenicol. RP4 is a broad-host-range IncP plasmid conferring resistance to Amp, kanamycin, and tetracycline. Plasmids were conjugated into ancestral strains from the MFDpir donor strain (19). Plasmid-free immigrants were variants of wt and mut strains marked with td-Cherry (44) for transfers 1 to 15 and with a Δlac deletion (46) for transfers 16 to 29; this allowed us to determine whether plasmid-bearing clones were the original hosts, early or recent transconjugants, by plating on 5-bromo-4-chloro-3-indolyl- b-D-galactopyranoside + isopropyl-b-D-thiogalactopyranoside and checking for red fluorescence. Cells were grown in lysogeny broth (LB), and plasmid-bearing clones were selected with Amp 100 mg/L. Spontaneous rifampicin-resistant (RifR) and nalidixic acid-resistant (NalR) mutants of MG1655 (19) were used, respectively, as a standard donor host for measuring plasmid transfer from an ancestral genotype, and as a standard recipient in conjugation assays, using Rif 100 mg/L and Nal 30 mg/L for selection. Plasmids were transferred to plasmid-free backgrounds by conjugation, then transconjugants were selected on Amp + appropriate selection for the recipient phenotype. High-level evolved resistance to Amp was measured by plating on LB–agar + Amp 0.5 g/L; when no colonies were detected, a threshold value was calculated assuming one colony at the lowest dilution plated. Spontaneous mutants with increased antibiotic resistance using R1-carrying cells were obtained by plating 100 µL overnight cultures on Amp 500 mg/L + streptomycin (Str) 200 mg/L (as Amp alone was not sufficient to kill R1WT-carrying cells at high cell density).
Evolution Experiment and Evolved Clones.
Evolving populations were grown in 200 µL LB medium in 96-well plates, at 37 °C with 180 rpm shaking and 100-fold overall dilution every 24 h, in the absence of any antibiotic. For immigration treatments, plasmid-free immigrants were grown fresh from glycerol stock and mixed with the evolving resident cultures in 99:1, 97:3, 90:10, 68:32, and 0:100 ratios at each passage. Plasmid-free td-Cherry strains were also passaged with 100-fold daily dilution. Six replicate lineages were evolved per treatment × host strain (wt or mut) combination. One individual clone per lineage was picked for characterization at 19 d (midpoint) and 29 d (endpoint) (SI Appendix, Table S1). Plating and sequencing revealed a few instances of contamination between wells, detailed in SI Appendix, Supplementary Text.
Conjugation Assays.
Plasmids-carrying donors and MG1655 NalR were grown overnight without antibiotic selection. Donor strains were first diluted 5-fold into LB medium and grown at 37 °C for 1 h, then 20 µL diluted donor cultures were mixed with 20 µL recipients and 160 µL prewarmed LB. After a 1-h mating, serial dilutions were plated on Amp, Nal, and Amp + Nal to estimate densities of donors + transconjugants, recipients, and transconjugants, respectively. Conjugation rates were computed using the endpoint method (47), using 0.8 h−1 as the exponential growth rate (estimated independently from growth curves made with 100-fold dilution in LB). When no transconjugant was detected, a threshold conjugation rate was calculated by assuming that one single transconjugant colony was observed. The short 1-h mating time was chosen in order to limit the effect of potential differences in growth rate or transfer rate between donors, recipients, and transconjugants (48). Replicates from ancestral host assays (Fig. 1D, y axis and SI Appendix, Fig. S3) with donor densities <4.108 cells/mL, and no transconjugants were discarded as they were found to correspond to a few wells in which strong evaporation occurred in the overnight donor culture prior to the conjugation assay.
Sequencing and Bioinformatics.
Clones selected for sequencing (SI Appendix, Table S1) were grown overnight, with Amp selection for plasmid-carrying clones, and DNA was extracted using CTAB extraction. Illumina whole-genome sequencing was provided by MicrobesNG (https://microbesng.com/). Sequencing data were mapped to a reference genome combining MG151655 (GenBank accession NC_000913) and R1 plasmid (GenBank accession KY749247), using breseq 0.35.1 (49), run in polymorphism mode to account for the coexistence of several plasmid alleles in high PCN clones. Variants not present at frequency >20% in at least one clone, and variants also detected in ancestral clones were discarded. Large deletions were called manually using coverage data, generated with breseq command bam2cov with default settings. Circos plots were made with R package OmicCircos (50).
Mobilization Experiment.
R1 oriT was amplified using primers 5′-CACGAAGCT-TGCCTGCACTTTCGCCATATG-3′ and 5′-CACCGAATTCAATCAGTGGCCTGGCAG-ATC-3′, then cloned into pHERD30T plasmid (51), using restriction enzymes HindIII and EcoRI. The resulting plasmid pMOB was transformed into MG1655. pMOB-bearing cells were selected using 50 mg/L gentamycin (Gen), a dose necessary to avoid low-level resistance conferred by R1 variants. Clones containing both pMOB and an R1 variant were obtained by conjugation of R1 into MG1655 + pMOB using Amp + Gen selection. Mating was conducted as described above, but donor clone overnight cultures contained Amp + Gen, and mating time was reduced to 30 min to limit secondary transfer; cells were also plated on Gen and Gen + Nal.
Plasmid Fitness Assays.
After overnight growth without antibiotics, strains were mixed with equal ratios of plasmid-bearing competitors, and grown for 24 h after 100-fold overall dilution in 96-well plates. Densities were estimated immediately after mixing and after 24 h, by plating serial dilutions on selective medium. Competition assays in the absence of immigration were run using two-way competitions with td-Cherry– and Δlac-marked MG1655 hosts, and plasmid variants were identified by the host background phenotype, as no transfer is taking place (n = 6 for each combination, results with opposite host markers were pooled). Competition assays with immigration were run mixing 1% plasmid-carrying hosts with 99% MG1655 NalR. In this case, R1copA* variants were distinguished from R1WT by streaking individual colonies on plates containing Amp 800 mg/L and Strep 200 mg/L (on which only R1CopA* clones are able to grow).
Antibiotics Effects.
Plasmid effect on survival in the presence of antibiotics was measured with plasmids carried by MG1655Δlac. Each strain was grown overnight in eight independent replicates, without antibiotics, then dilution series were plated on agar with various concentrations of antibiotics. Survival was defined as colony-forming units (CFUs) on antibiotic medium/CFUs on LB agar without antibiotics. To survey growth in the presence of antibiotics for all evolved midpoint clones, cultures of evolved clones were first grown overnight in LB + Amp 100 mg/L, then plated on agar with Amp 0.5 g/L.
Data Analysis.
Statistical analysis was performed using R version 3.4.1 (52). Transfer rate and cell density values were log transformed before analysis. To analyze the effect of host background on clone transfer rate (Fig. 2A), where data from each background were obtained in separate experiments, a bivariate model was run using the ASReml package with clone as a random effect (53).
Supplementary Material
Acknowledgments
We thank Alastair Wilson for help with statistical analysis. The work was funded by Natural Environment Research Council Grant NE/S000771/1 (to A.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107818118/-/DCSupplemental.
Data Availability
The datasets supporting this publication are openly available from the University of Exeter's institutional repository at: https://ore.exeter.ac.uk/repository/handle/10871/126437 (54). Short read sequencing data have been deposited in the European Nucleotide Archive (PRJEB40537) (55).
References
- 1.Frost L. S., Leplae R., Summers A. O., Toussaint A., Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Lopatkin A. J., et al., Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 8, 1689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Hal S. J., Wiklendt A., Espedido B., Ginn A., Iredell J. R., Immediate appearance of plasmid-mediated resistance to multiple antibiotics upon antibiotic selection: An argument for systematic resistance epidemiology. J. Clin. Microbiol. 47, 2325–2327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker K. S., et al., Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat. Commun. 9, 1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner P. E., Cooper V. S., Lenski R. E., Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution 52, 315–329 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Turner P. E., et al., Antibiotic resistance correlates with transmission in plasmid evolution. Evolution 68, 3368–3380 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Smith J., Superinfection drives virulence evolution in experimental populations of bacteria and plasmids. Evolution 65, 831–841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.San Millan A., MacLean R. C., Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 5, 65–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Millan A., et al., Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 5, 5208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison E., Guymer D., Spiers A. J., Paterson S., Brockhurst M. A., Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 25, 2034–2039 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Loftie-Eaton W., et al., Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 1, 1354–1363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson R. M., May R. M., Coevolution of hosts and parasites. Parasitology 85, 411–426 (1982). [DOI] [PubMed] [Google Scholar]
- 13.Porse A., Schønning K., Munck C., Sommer M. O., Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlberg C., Chao L., Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165, 1641–1649 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dionisio F., Conceição I. C., Marques A. C. R., Fernandes L., Gordo I., The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 1, 250–252 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Gelder L., Williams J. J., Ponciano J. M., Sota M., Top E. M., Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178, 2179–2190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kottara A., Hall J. P. J., Harrison E., Brockhurst M. A., Multi-host environments select for host-generalist conjugative plasmids. BMC Evol. Biol. 16, 70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messenger S. L., Molineux I. J., Bull J. J., Virulence evolution in a virus obeys a trade-off. Proc. Biol. Sci. 266, 397–404 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitriu T., Marchant L., Buckling A., Raymond B., Bacteria from natural populations transfer plasmids mostly towards their kin. Proc. Biol. Sci. 286, 20191110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost L. S., Koraimann G., Regulation of bacterial conjugation: Balancing opportunity with adversity. Future Microbiol. 5, 1057–1071 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Zatyka M., Thomas C. M., Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 21, 291–319 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Yoshioka Y., Ohtsubo H., Ohtsubo E., Repressor gene finO in plasmids R100 and F: Constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J. Bacteriol. 169, 619–623 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox K. E. L., Schildbach J. F., Sequence of the R1 plasmid and comparison to F and R100. Plasmid 91, 53–60 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Nordström K., Plasmid R1—Replication and its control. Plasmid 55, 1–26 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Persson C., Wagner E. G., Nordström K., Control of replication of plasmid R1: Structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 9, 3767–3775 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Lopez A., et al., A naturally occurring SNP in plasmid pB1000 produces a reversible increase in antibiotic resistance. Antimicrob. Agents Chemother. 61, e01735-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.San Millan A., Escudero J. A., Gifford D. R., Mazel D., MacLean R. C., Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat. Ecol. Evol. 1, 0010 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Uhlin B. E., Nordström K., R plasmid gene dosage effects in Escherichia coli K-12: Copy mutants of the R plasmic R1drd-19. Plasmid 1, 1–7 (1977). [DOI] [PubMed] [Google Scholar]
- 29.Haugan K., Karunakaran P., Tøndervik A., Valla S., The host range of RK2 minimal replicon copy-up mutants is limited by species-specific differences in the maximum tolerable copy number. Plasmid 33, 27–39 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Haft R. J. F., Mittler J. E., Traxler B., Competition favours reduced cost of plasmids to host bacteria. ISME J. 3, 761–769 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Muesing M., Tamm J., Shepard H. M., Polisky B., A single base-pair alteration is responsible for the DNA overproduction phenotype of a plasmid copy-number mutant. Cell 24, 235–242 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Thompson M. G., et al., Isolation and characterization of novel mutations in the pSC101 origin that increase copy number. Sci. Rep. 8, 1590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen R. C., Brown S. P., Modified antibiotic adjuvant ratios can slow and steer the evolution of resistance: Co-amoxiclav as a case study. mBio 10, e01831-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., et al., A ProQ/FinO family protein involved in plasmid copy number control favours fitness of bacteria carrying mcr-1-bearing IncI2 plasmids. Nucleic Acids Res. 49, 3981–3996 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beabout K., et al., Rampant parasexuality evolves in a hospital pathogen during antibiotic selection. Mol. Biol. Evol. 32, 2585–2597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Z., et al., Increased plasmid copy number contributes to the elevated carbapenem resistance in OXA-232-producing Klebsiella pneumoniae. Microb. Drug Resist. 26, 561–568 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Beltran J., et al., Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat. Ecol. Evol. 2, 873–881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., et al., Increased plasmid copy number is essential for Yersinia T3SS function and virulence. Science 353, 492–495 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Cho H., Winans S. C., VirA and VirG activate the Ti plasmid repABC operon, elevating plasmid copy number in response to wound-released chemical signals. Proc. Natl. Acad. Sci. U.S.A. 102, 14843–14848 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maisnier-Patin S., Roth J. R., Selection and plasmid transfer underlie adaptive mutation in Escherichia coli. Genetics 210, 821–841 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilhan J., et al., Segregational drift and the interplay between plasmid copy number and evolvability. Mol. Biol. Evol. 36, 472–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Beltrán J., DelaFuente J., León-Sampedro R., MacLean R. C., San Millán Á., Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 19, 347–359 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Hall J. P. J., Williams D., Paterson S., Harrison E., Brockhurst M. A., Positive selection inhibits gene mobilisation and transfer in soil bacterial communities. Nat. Ecol. Evol. 1, 1348–1353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimitriu T., et al., Indirect fitness benefits enable the spread of host genes promoting costly transfer of beneficial plasmids. PLoS Biol. 14, e1002478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T., et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dimitriu T., et al., Negative frequency dependent selection on plasmid carriage and low fitness costs maintain extended spectrum β-lactamases in Escherichia coli. Sci. Rep. 9, 17211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonsen L., Gordon D. M., Stewart F. M., Levin B. R., Estimating the rate of plasmid transfer: An end-point method. J. Gen. Microbiol. 136, 2319–2325 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Huisman J. S., et al., Estimating the rate of plasmid transfer in liquid mating cultures. bioRxiv [Preprint] (2020). 10.1101/2020.03.09.980862 (Accessed 15 July 2021). [DOI]
- 49.Deatherage D. E., Barrick J. E., “Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq” in Engineering and Analyzing Multicellular Systems, Sun L., Shou W., Eds. (Springer, New York, 2014), vol. 1151, pp. 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y., et al., OmicCircos: A simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 13, 13–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D., PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74, 7422–7426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Development Core Team , R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0 (2008). [Google Scholar]
- 53.Houslay T. M., Wilson A. J., Avoiding the misuse of BLUP in behavioural ecology. Behav. Ecol. 28, 948–952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dimitriu T., Matthews A. C., Buckling A., Increased copy number couples the evolution of plasmid horizontal transmission and plasmid-encoded antibiotic resistance (dataset). University of Exeter's institutional repository. https://ore.exeter.ac.uk/repository/handle/10871/126437. Deposited 15 July 2021. [DOI] [PMC free article] [PubMed]
- 55.University of Exeter, Increased copy number couples the evolution of plasmid horizontal transmission and antibiotic resistance. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB40537. Deposited 28 September 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this publication are openly available from the University of Exeter's institutional repository at: https://ore.exeter.ac.uk/repository/handle/10871/126437 (54). Short read sequencing data have been deposited in the European Nucleotide Archive (PRJEB40537) (55).