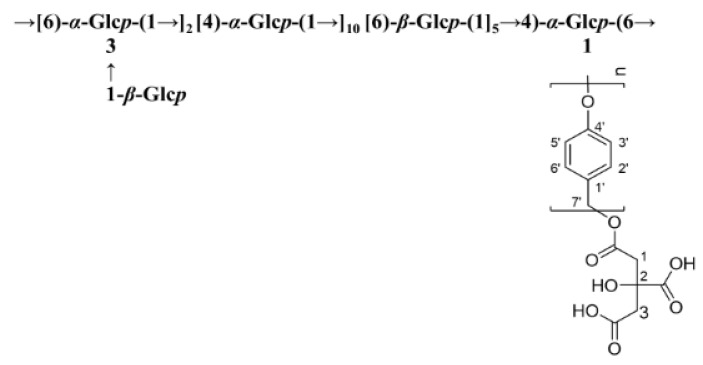Abstract
A novel homogeneous polysaccharide named GEP-1 was isolated and purified from Gastrodia elata (G. elata) by hot-water extraction, ethanol precipitation, and membrane separator. GEP-1, which has a molecular weight of 20.1 kDa, contains a polysaccharide framework comprised of only glucose. Methylation and NMR analysis showed that GEP-1 contained 1,3,6-linked-α-Glcp, 1,4-linked-α-Glcp, 1,4-linked-β-Glcp and 1,4,6-linked-α-Glcp. Interestingly, GEP-1 contained citric acid and repeating p-hydroxybenzyl alcohol as one branch. Furthermore, a bioactivity test showed that GEP-1 could significantly promote the growth of Akkermansia muciniphila (A. muciniphila) and Lacticaseibacillus paracasei (L. paracasei) strains. These results implied that GEP-1 might be useful for human by modulating gut microbiota.
Keywords: Gastrodia elata, structural characterization, polysaccharide, gut microbiota
1. Introduction
Gastrodia elata (G. elata), from the family Ochidaceae, is a traditional Chinese medicine, which is widely used in the clinic. Modern pharmacological research shows that G. elata has a variety of biological activities, such as anti-convulsion [1,2], improving memory [3], preventing senescence [4], neuroprotection [5], etc. [6,7,8,9]. In the past, more attention was paid to the small molecular compounds in G. elata, including gastrodin, parishin, phenolic compounds, 4-hydroxybenzyl alcohol, β-sitosterol and so on [10,11,12,13,14]. In recent years, with the progress of separation, purification and detection technology, polysaccharide macromolecules in G. elata are receiving increasing attention. G. elata contains more than 10% polysaccharide components, which also show a variety of biological activities [15,16,17,18,19]. For example, Xie et al. reported that polysaccharides of G. elata can improve the learning and memory ability of D-galactose–induced aging mice to improve the activity of enzymes related to oxidative metabolism in the body [20]. In addition, polysaccharides of G. elata are shown to increase the number of brain-derived neurotrophic factor (BDNF)-positive cells and stem cell factor (SCF)-positive cells and decrease the average gray value, suggesting that polysaccharides from G. elata (PGB) have neuroprotective effects by up-regulating BDNF and SCF expression in brain tissues around ischemic lesions [21]. Therefore, it is of great significance to study the isolation, purification, and biological activities of polysaccharides from G. elata.
Since oral polysaccharides are hardly absorbed, they may not directly act on the target organs in vivo to produce biological effects. Although polysaccharides cannot be directly absorbed by the human body, they can be decomposed and utilized by gut microbiota to produce a series of bioactive metabolites [22]. In recent years, with the deepening of research, more and more researchers believe that polysaccharides can affect the structure and function of gut microbiota and play a regulatory role in human health by remodeling intestinal microecology or intestinal homeostasis [23,24,25]. For example, Lycium barbarum polysaccharides can promote the production of short-chain fatty acids and modulate the composition of the gut microbiota, increasing the relative abundances of Bacteroidaceae, Lactobacillaceae, Prevotellaceae and Verrucomicrobiaceae, which are positively associated with immunomodulatory traits [26]. In 2013, Zhang and his co-workers reported that polysaccharide from the fungus Flammuliana velutipes improves colitis via regulation of colonic microbial dysbiosis and inflammatory responses [27]. In conclusion, gut microbiota have become an important bridge for polysaccharides to exert their biological activities in vivo.
Based on the above literature reports, we carried out related research on the ability of polysaccharide of G. elata to promote the growth of probiotics. First, the polysaccharides from G. elata were extracted and separated, and a homogeneous polysaccharide named GEP-1 was obtained. The structure of GEP-1 was characterized by HPGPC, PMP-HPLC (1-phenyl-3-methyl-5-pyrazolone pre-column derivatization prior to HPLC), LC/MS, FT-IR, NMR, and SEM methods. Then, two gut microbiota, Akkermansia muciniphila (A. muciniphila) and Lacticaseibacillus paracasei (L. paracasei), were selected to evaluate the activity of GEP-1. GEP-1 demonstrated promoting effects on the growth of the two selected gut microbiota. Therefore, in this paper, we will report the isolation, purification and structural characterization of GEP-1, as well as its growth promoting effect on gut microbiota.
2. Results and Discussion
2.1. Extraction, Isolation and Purification of GEP-1
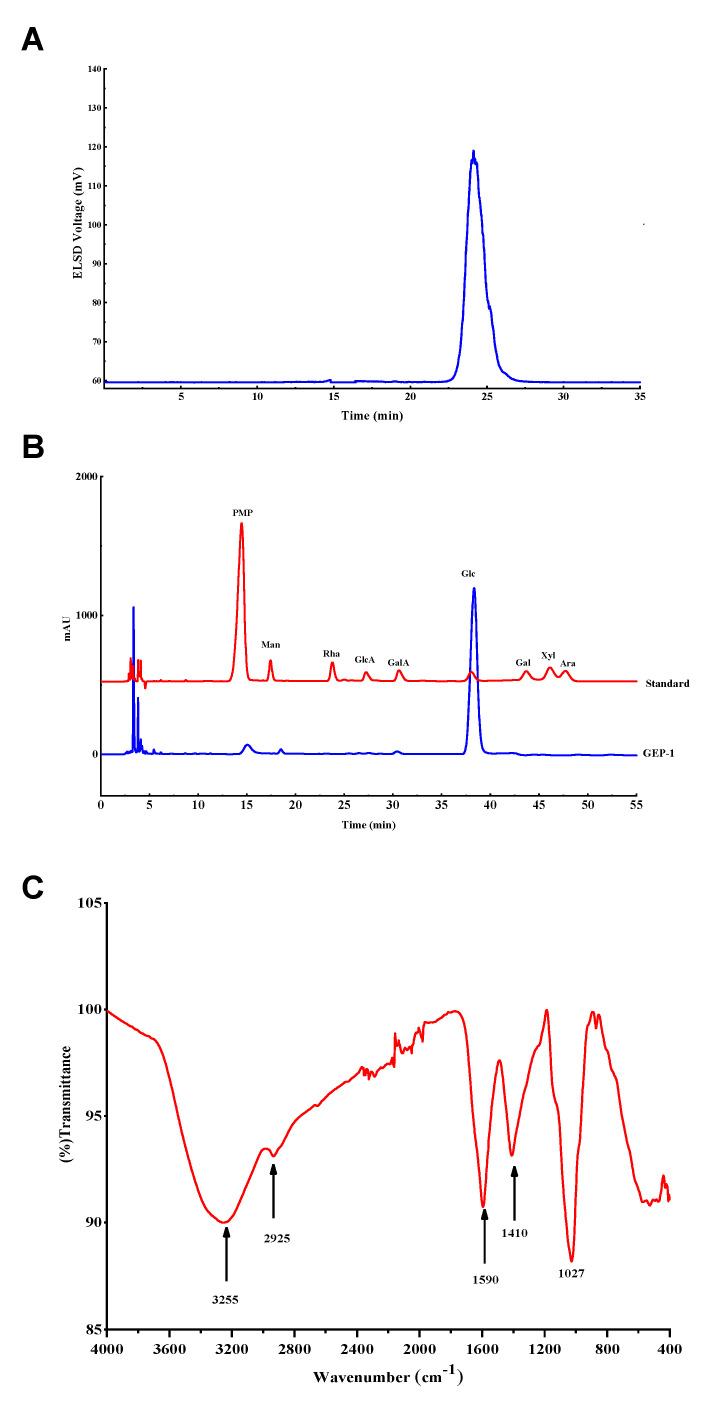
Crude polysaccharides GEP50 (200 g, yield: 5.56%) were extracted from the rhizome of G. elata (3.6 kg) by water extraction and alcohol precipitation. Then GEP50 (180 g) was dissolved in water (2.5 L), separated by the membrane separation equipment with 10 kDa membrane and eluted with H2O to obtain a homogeneous polysaccharide, GEP-1 (20 g, yield: 11.11%). The elution curve of is displayed in Figure S1. As shown in Figure 1A, GEP-1 showed a symmetrical peak, indicating it was a homogeneous polysaccharide and the molecular weight was 20.15 kDa according to the standard curve. Furthermore, the protein content of GEP-1 was 0.78 ± 0.03% tested using a BCA reagent kit.
Figure 1.
(A): HPGPC of GEP-1; (B): HPLC chromatograms of PMP derivatives; (C): FT-IR spectrum of GEP-1.
2.2. Monosaccharide Composition, Linkage Pattern and CA and Repeated HA Analysis
The monosaccharide composition of GEP-1 was detected by PMP pre-column derivative method. The HPLC result (Figure 1B) showed that GEP-1 was composed of Glc compared with standards. The linkage patterns of glycosidic residues in GEP-1 were determined by methylation method and GC-MS analysis.
The result (Table 1) demonstrated that fully O-methylated GEP-1 was comprised of 2,3,4,6-Me4-Glcp, 2,3,6-Me3-Glcp, 2,3,4-Me3-Glcp, 2,3-Me2-Glcp and 2,6-Me2-Glcp with the molar ratio of 2.23:9.69:4.93:1.08:2.07. Interestingly, the signals around 7.0 ppm and 2.0 ppm in 1H-NMR and the signals between 110 ppm and 180 ppm in the 13C-NMR spectra of GEP-1 indicated that non-polysaccharides constituents might be linked to polysaccharide part in GEP-1. Also, non-polysaccharide constituents might be p-HA and CA based on the NMR signals and reported structures of constituents in G. elata [28,29]. In order to understand the p-HACA in GEP-1, GEP-1 (3 mg) was hydrolyzed with 2 M TFA to obtain the full hydrolysates. The LC-MS results (Figure S2 and Table 2) showed that m/z 193.0343 [CA + H]+, m/z 107.0489 [p-HA − H2O + H]+, and m/z 163.0597 [Glc − H2O + H]+ confirmed the existence of CA and p-HA. Interestingly, consecutive neutral losses of 106.04 Da between m/z 107.0489 [HA − H2O + H]+, m/z 213.0913 [2HA − H2O + H]+, m/z 319.1326 [3HA − H2O + H]+, m/z 425.1745 [4HA − H2O + H]+, m/z 531.2161 [5HA − H2O + H]+, m/z 637.2576 [6HA − H2O + H]+, and m/z 743.3001 [7HA − H2O + H]+ in positive mode, and also among m/z 105.0349 [HA − H2O − H]−, m/z 211.0766 [2HA − H2O − H]−, m/z 317.1182 [3HA − H2O − H]−, m/z 423.1605 [4HA − H2O − H]−, m/z 529.2029 [5HA − H2O − H]−, m/z 635.2458 [6HA − H2O − H]−, m/z 741.2882 [7HA − H2O − H]−, m/z 847.3228 [8HA − H2O −H]−, m/z 953.3691 [9HA − H2O − H]− and m/z 1059.4114 [10HA − H2O − H]− in negative mode suggested the repeating p-HA unit in GEP-1. Furthermore, GEP-1 was hydrolyzed using 4 mol/L of NaOH in 50% methanol and acidized by HCl to determine the content of CA. Finally, the content of CA in GEP-1 was about 3.93% by comparing the area of its full alkaline hydrolysates with the CA standard.
Table 1.
Linkage patterns analysis of GEP-1.
| Methylated Sugars | Linkages | Major Mass Fragments (m/z) | Molar Ratio |
|---|---|---|---|
| 2,3,4,6-Me4-Glc | 1-linked-Glc | 43, 71, 87, 102, 118, 129, 145, 161, 162, 205 | 2.23 |
| 2,3,6-Me3-Glc | 1,4-linked-Glc | 43, 59, 71, 87, 102, 118, 129, 162, 233 | 9.69 |
| 2,3,4-Me3-Glc | 1,6-linked-Glc | 43, 59, 87, 99, 101, 118, 129, 162, 189, 233 | 4.93 |
| 2,3-Me2-Glc | 1,4,6-linked-Glc | 43, 59, 85, 102, 118, 127, 162, 201, 161 | 1.08 |
| 2,6-Me2-Glc | 1,3,4-linked-Glc | 43, 59, 87, 118, 129, 160, 185, 305 | 2.07 |
Table 2.
LC-MS results of non-polysaccharide constituents in GEP-1 hydrolysate.
| Name | Molecular Formula | Theoric Mass | tR (min) |
m/z Experimental |
pos/neg | Error (ppm) |
|---|---|---|---|---|---|---|
| CA | C6H8O7 | 192.0265 | 2.72 | 193.0343 | [M + H]+ | −0.047 |
| 1HA | C7H8O2 | 124.0519 | 1.12 | 107.0489 | [M − H2O + H]+ | −2.535 |
| 2HA | C14H14O3 | 230.0937 | 8.44 | 213.0913 | [M − H2O + H]+ | 1.473 |
| 3HA | C21H20O4 | 336.1356 | 8.86 | 319.1326 | [M − H2O + H]+ | -0.894 |
| 4HA | C28H26O5 | 442.1775 | 9.16 | 425.1745 | [M − H2O + H]+ | −0.554 |
| 5HA | C35H32O6 | 548.2193 | 9.40 | 531.2161 | [M − H2O + H]+ | −1.018 |
| 6HA | C42H38O7 | 654.2612 | 9.52 | 637.2576 | [M − H2O + H]+ | −1.311 |
| 7HA | C49H44O8 | 760.3031 | 9.58 | 743.3001 | [M − H2O + H]+ | −0.377 |
| Glc | C6H12O6 | 180.0628 | 1.02 | 163.0597 | [M − H2O + H]+ | −2.207 |
| CA | C6H8O7 | 192.0265 | 2.75 | 191.0196 | [M − H]− | 5.293 |
| Glc | C6H12O6 | 180.0628 | 1.02 | 179.0561 | [M − H]− | 0.160 |
| 1HA | C7H8O2 | 124.0519 | 1.12 | 105.0349 | [M − H2O + H]− | 2.493 |
| 2HA | C14H14O3 | 230.0937 | 8.44 | 211.0766 | [M − H2O − H]− | 0.839 |
| 3HA | C21H20O4 | 336.1356 | 8.92 | 317.1182 | [M − H2O − H]− | −0.403 |
| 4HA | C28H26O5 | 442.1775 | 9.16 | 423.1605 | [M − H2O − H]− | 0.656 |
| 4HA | C28H26O5 | 442.1775 | 9.16 | 411.1606 | [M − CH2O − H]− | 1.113 |
| 5HA | C35H32O6 | 548.2193 | 9.13 | 529.2029 | [M − H2O − H]− | 1.681 |
| 6HA | C42H38O7 | 654.2612 | 9.38 | 635.2458 | [M − H2O − H]− | 2.956 |
| 7HA | C49H44O8 | 760.3031 | 9.49 | 741.2882 | [M − H2O − H]−- | 3.201 |
| 8HA | C56H50O9 | 866.3449 | 9.73 | 847.3228 | [M − H2O − H]− | 0.140 |
| 9HA | C63H56O10 | 972.2868 | 9.76 | 953.3691 | [M − H2O − H]− | −0.447 |
| 10HA | C70H62O11 | 1078.4287 | 9.85 | 1047.4118 | [M − CH2O − H]− | 0.476 |
| 10HA | C70H62O11 | 1078.4287 | 9.85 | 1059.4114 | [M − H2O − H]− | 0.008 |
2.3. FT-IR Analysis
The FT-IR spectrum of GEP-1 illustrated in Figure 1C showed strong absorption bands at 3300 and 2900 cm−1. These bands were attributed to the stretching vibrations of OH and CH groups, respectively. Meanwhile, the intense peaks at 1600 and 1400 cm−1 correspond to the asymmetric and symmetric C=O or C=C stretching vibrations. The absorption bands observed at 1030 and 600 cm−1 suggested that GEP-1 comprised a pyranose structure. In general, all the spectroscopic bands identified were characteristics of polysaccharides, p-HA and CA.
2.4. NMR Analysis
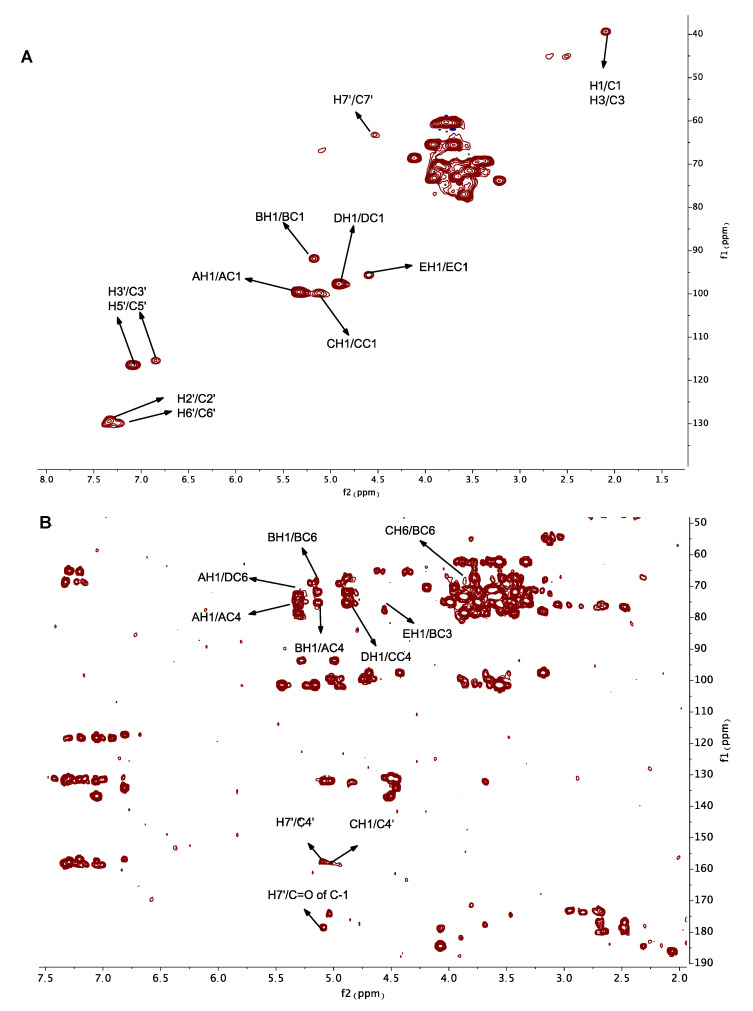
NMR plays an essential role in the structural characterization of polysaccharides. In this study, 1H, 13C, HSQC and HMBC spectra (Figure 2) of GEP-1 were recorded. Due to the existence of repeating p-HA and CA in GEP-1, the first step in analyzing the NMR spectra was the identification of repeating p-HA and CA. Based on the literature [30], peaks at 2.07/39.50 ppm, 7.29/129.38 ppm or 7.23/130.15 ppm, 6.85/115.56 ppm or 7.09/116.56 ppm, and 5.14/67.94 ppm in HSQC were attributed to H1/C1 (H3/C3), H2′/C2′ (H6′/C6′), H3′/C3′(H5′/C5′), and H7′/C7′, and signals at 68.35 ppm, 129.55 ppm, 157.55 ppm, 179.56 ppm, 182.52 ppm and 181.65 ppm in the 13C-NMR spectra were attributed to C2, C1′, C4′, C=O of C-1, C=O of C-2 and C=O of C-3 of p-HACA and repeated p-HA in GEP-1. And the signals at 5.14/179.56 ppm (H7′/C=O of C-1) and 5.14/157.55 ppm (H7′/C4′) in HMBC spectrum indicated C=O of C-1 of CA was linked to H-7′ of HA, H-7′ of HA linked to C-4′ of HA, confirming the existences of p-HA CA and repeated p-HA.
Figure 2.
HSQC (A) and HMBC (B) spectra of GEP-1.
Secondly, the identification of anomeric carbon and anomeric proton signals were analyzed. Due to the fact 1,4-Glcp was the most abundant glycosidic residue according to the methylation result the most intense anomeric proton signal at 5.33 ppm in the 1H-NMR spectrum was assigned to H1 of 1,4-Glcp, and the signal at δH 5.33 ppm was corresponding to δC 99.67 ppm in the HSQC spectrum, so the peak at 5.33/99.67 ppm was assigned to H1/C1 of 1,4-Glcp. Therefore, based on the results of methylation analysis, the NMR spectra and the literature, the anomeric signals at 5.33/99.67 ppm, 5.18/91.93 ppm, 5.12/100.00 ppm, 4.91/97.90 ppm and 4.60/95.73 ppm were distributed to⟶4)-α-Glcp-(1⟶, ⟶3,6)-α-Glcp-(1⟶, ⟶4,6)-α-Glcp-(1⟶, ⟶6)-β-Glcp-(1⟶ and β-Glcp-(1⟶, and named as residue A, B, C, D and E, respectively. Based on the literature and 2D NMR spectra, the signals at 3.56/73.15 ppm, 3.89/74.93 ppm, 3.89/73.37 ppm, 3.89/74.93 ppm and 3.70/61.87 ppm were assigned to H2/C2, H3/C3, H4/C4, H5/C5 and H6/C6 of 1,4-Glcp.
The detailed chemical shifts of GEP-1 are shown in Table 3. The linkage sequences of residues in GEP-1 were speculated by signals in HMBC spectrum. The peaks at 3.90/67.21 ppm (CH6/BC6), 4.60/76.34 ppm (EH1/BC3), 5.33/68.76 ppm (AH1/DC6), 5.18/73.37 ppm (BH1/AC4), 4.91/74.33 ppm (DH1/CC4) indicated that O-6 of residue C was linked to C-6 of residue B, O-1 of residue E linked to C-3 of residue B, O-1 of residue A linked to linked to C-6 of residue D, O-1 of residue B linked to C-4 of residue A, O-1 of residue D linked to C-4 of residue C. Also, the peaks at 5.33/73.37 ppm (AH1/AC4), 5.18/67.21 ppm (BH1/BC6) and 4.91/68.76 ppm (DH1/DC6) illustrated the linkage between O-1 of residue A and C-4 of residue A, the linkage between O-1 of residue B and C-6 of residue B and the linkage between O-1 of residue D and C-6 of residue D, indicating that residue A, residue B and residue D were repeated in the structure of GEP-1. Further, the signal at 5.10/157.55 ppm (CH1/C4’) revealed that O-1 of residue C was linked to C4’ of repeated p-HA. Together, the possible repeating unit of GEP-1 is shown in Figure 3.
Table 3.
Chemical shifts of main residues of GEP-1.
| Code | Residues | Chemical Shifts (ppm) | |||||
|---|---|---|---|---|---|---|---|
| H1/C1 | H2/C2 | H3/C3 | H4/C4 | H5/C5 | H6/C6 | ||
| A | ⟶4)-α-Glcp-(1⟶ | 5.33/99.67 | 3.56/73.15 | 3.89/74.93 | 3.89/73.37 | 3.89/74.93 | 3.70/61.87 |
| B | ⟶3,6)-α-Glcp-(1⟶ | 5.18/91.93 | 3.64/73.0 | 3.76/76.34 | 3.39/71.67 | 3.81/71.91 | 3.63/67.21 |
| C | ⟶4,6)-α-Glcp-(1⟶ | 5.10/100.00 | 3.58/73.42 | 3.90/74.93 | 3.67/74.33 | 3.83/71.78 | 3.90/66.07 |
| D | ⟶6)-β-Glcp-(1⟶ | 4.91/97.90 | 3.38/76.4 | 3.63/77.4 | 3.45/72.6 | 3.63/77.7 | 3.74/68.76 |
| E | β-Glcp-(1⟶ | 4.60/95.73 | 3.19/73.96 | 3.72/72.89 | 3.34/69.37 | 3.55/74.57 | 3.85/60.52 |
Figure 3.
The putative repeating unit of GEP-1.
2.5. SEM Analysis
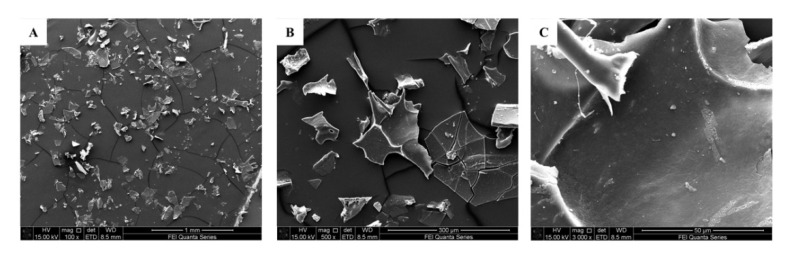
The surface morphologies of GEP-1 were observed through SEM at 100 ×, 500 × and 3000 × magnifications. The SEM images of GEP-1 were displayed in Figure 4, GEP-1 exhibited an irregular, laminated, wrinkled, sheet-like morphology with a little rough surface, which was accordance with its branched structure.
Figure 4.
SEM images of GEP-1 at different magnifications (A: 100 ×; B: 500 ×; C: 3000 ×).
2.6. GEP-1 Induced the Growth A. muciniphila and L. paracasei
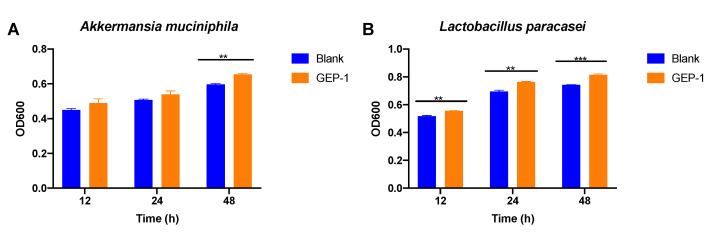
To test whether GEP-1 could have effect on A. muciniphila and L. paracasei strains, the two strains were cultured supplemented with 1 mg/mL of GEP-1. As shown in Figure 5A, it was showed that the growth of A. muciniphila was promoted by polysaccharide GEP-1, and significant promotion was detected at 48 h. In Figure 5B, GEP-1 significantly induced the growth of L. paracasei at 12 h, 24 h and 48 h. These results indicated that GEP-1 had bioactivity on human gut probiotic A. muciniphila and L. paracasei in vitro.
Figure 5.
(A): The growth of A. muciniphila in the medium containing 1 mg/mL GEP-1 for 12, 24 and 48 h; (B): The growth of L. paracasei in the medium containing 1 mg/mL GEP-1 for 12, 24h and 48 h. (n = 3, by student t test, ** p < 0.01, *** p < 0.001.).
Previous studies suggested that most homogeneous polysaccharides from G. elata consisted only of glucose and have a main chain of 1-4 linked glycosidic bonds. GEP-1 also had a backbone of 1,4-linked glucose and 1,6-linked glucose. But what was special was that GEP-1 contained a branch of CA and repeating p-HA. Most of the isolated glucans exhibited anti-cancer, antioxidant and immunological activities [31]. In this experiment, GEP-1 obviously induced the growth of A. muciniphila and L. Paracasei, and the regulatory effects of polysaccharides from G. elata on two bacteria from human intestinal flora were demonstrated.
3. Materials and Method
3.1. Materials
The dried rhizomes of G. elata were obtained from Zhaotong (Yunnan Province, China) and authenticated by Menghua Tian from Zhaotong Tama Research Institute of Yunnan Province (China). Monosaccharide standards, glucose (Glc), mannose (Man), rhamnose (Rha), glucuronic acid (GlcA), galacuronic acid (GalA), galactose (Gal), xylose (Xyl) and arabinose (Ara), and citric acid (CA) were purchased from Sigma (St. Louis, MO, USA). DEAE-52 were purchased from Whatman (Kent, UK). BCA reagent kit and MRS medium was purchased from Dalian Meilun Biotech Co., Ltd. (Dalian, China). Defined medium was in-house prepared. A. muciniphila strain ATCC BAA-835 and L. paracasei were obtained from BeiNa Chuanglian Biotechnology Co., Ltd. (Beijing, China).
3.2. Extraction, Isolation and Purification of GEP-1
The dried rhizomes of G. elata (3.6 kg) were first crushed to fine particles and extracted with 30 volumes of distilled water at 70 °C for 4 h for three times. The concentrated water extract was precipitated with 95% ethanol to a final concentration of 50% and deproteinized with Sevage method [32] to obtain crude polysaccharide (GEP50). GEP50 was applied on the membrane separation equipment (Shanghai Mosu Science Equipment Co., Ltd., Shanghai, China) equipped with a 10 kDa membrane and eluted with H2O. Finally, a homogeneous polysaccharide GEP-1 was obtained.
3.3. Estimation of Homogeneity and Molecular Weight
The homogeneity and molecular weight of GEP-1 was estimated by HPGPC using an Agilent Technologies system (Santa Clara, CA, USA) equipped with an ELSD. The sample (2 mg/mL and 20 µL) was applied to series TSK G5000 and G3000 columns and eluted with H2O at 0.8 mL/min. Commercially available dextrans (Mw: 2.70, 5.25, 9.75, 13.05, 64.65, 135.30, 300.60 kDa) were used as standard molecular markers for the construction of the calibration curve. The molecular weight was estimated based on the calibration curve.
3.4. Analysis of Composition
The carbohydrate content was determined by phenol-sulfuric acid colorimetric method using glucose as standard. The protein content was measured using BCA reagent kit. Based on the previous studies that investigated small molecules, many of which contained p-hydroxylbenzyl alcohol (p-HA) and/or CA [10], CA and repeated p-HA might be present in the polysaccharides from G. elata. Therefore, the sample was fully hydrolyzed with 2 M TFA at 110 °C for 5 h and then dissolved in 50% methanol and components in the hydrolysate were identified by LTQ-Orbitrap Velos Pro hybrid mass spectrometer (Thermo Fisher, Scientific, Carlsbad, CA, USA). The chromatographic separation was carried out on an HSS T3 column (2.1 × 150 mm i.d., particle size 1.8 µm, Waters, Milford, CT, USA) operated at 30 °C. The mobile phase consisted of 0.1% formic acid (FA) in water (A) and acetonitrile (B) with a flow rate of 0.3 mL/min. The gradient elution was as follows: 0–12 min, 0–100% B; 12-15 min, 100% B; 15–20 min, 100%–0% B. Later, GEP-1 was hydrolysed using 4 M NaOH in 50% methanol (2 mL) for 3 h at room temperature, neutralized by HCl, and then the content of citric acid in GEP-1 was determined by AQUITY UPLC system (Waters) equipped with a C18 column using CA as the standard solution. The binary mobile phase consisted of 0.1% FA in 90% water (A) and 0.1% FA in 90% acetonitrile (B). The elution gradient was set as follows: 0–6 min, 0 %–100%B.
3.5. Monosaccharide Composition
The monosaccharide composition of the polysaccharide was determined by PMP-derivative method [33]. 3 mg of GEP-1 was hydrolyzed in 2 mL of 2 mol/L trifluoracetic acid (TFA) solution at 110 °C for 5 h. After TFA fully being eliminated by nitrogen, the hydrolysate was dissolved in 500 µL ammonium solution (25%) and mixed with 500 µL of 0.5 mol/L PMP-methanol solution, and the reaction was conducted at 70 °C for 2 h. After the elimination of ammonia by nitrogen, the reaction product was dissolved in 500 µL water and extracted with chloroform. Finally, the supernatant was tested on Agilent 1260 HPLC equipped with an Agilent C18 column (4.6 × 200 mm, 5 µm). The sample was eluted with 20 mM ammonium acetate in water (82.5%) and acetonitrile (17.5%) at 30 °C with 1 mL/min flow rate.
3.6. Fourier Transform Infrared (FT-IR) Spectroscopy
2 mg sample dissolved in chloroform was added on the KBr window and dried under infrared light, and then measured by Thermo Nicolet iS5 FTIR spectrometer in the middle infrared range of 4000—400 cm−1.
3.7. Methylation Analysis
The methylation analysis of GEP-1 was referred to the Hakomori method based on the previous literature [34]. The sample (20 mg) was per-O-methylated by repeated reaction with NaH-DMSO and methyl iodide until the methylation was completed. Then, the residue was hydrolyzed by 2 M TFA at 110 °C for 5 h. After that, the sample was reduced with NaBD4 (25 mg) and acetylated with acetic anhydride/pyridine. Finally, the partially methylated alditol acetates (PMAAs) were analyzed by GC-MS (Agilent 7890A).
3.8. NMR Analysis
GEP-1 (60 mg) was dissolved in deuterium oxide (D2O, 500 µL), and exchanged hydrogen with deuterium for three times by repeated freeze-drying. After lyophilization, the sample was dissolved in D2O (500 µL), centrifuged and transferred into an NMR tube. The NMR spectra (1H, 13C, HSQC and HMBC) were obtained by a 600 MHz Bruker spectrometer (Bruker, Ettlingen, Germany).
3.9. Scanning Electron Microscopy (SEM) Analysis
The microscopic features of GEP-1 were observed by a scanning electron microscope (SEM) according to a previously reported method [35]. In brief, GEP-1 was ground into powder and distributed evenly on a silicon wafer using double-sided adhesive tape (1 cm × 1 cm). Subsequently, the wafer was sprayed with gold powder using a sputter coater. Finally, SEM images of GEP-1 at different magnifications were recorded by SEM (Quanta 250, FEI, Hillsboro, OR, USA).
3.10. Strains
A. muciniphila and L. paracasei were cultured in MRS medium or defined medium. Bacteria were seeded (OD600 = 0.1) in the medium containing 1 mg/mL of GEP-1. The culture medium was then collected for the determination of OD600 through microplate reader.
4. Conclusions
A homogeneous polysaccharide with molecular weight 20.15 kDa and named GEP-1 was isolated and purified from G. elata. The monosaccharide composition of GEP-1 contained only glucose. The backbone of GEP-1 was composed of →[6)-α-Glcp-(1→]2 [4)-α-Glcp-(1→]10 [6)-β-Glcp-(1]5→4)-α-Glcp-(6→, with three branches of β-Glcp and CA-repeating p-HA attached to the backbone chain at O-3 position of 1,3,6-linked α-Glcp and O-1 position of 1,4,6-linked α-Glcp. Bioactivity tests showed that GEP-1 could promote the growth of A. muciniphila and L. paracasei strains. These results imply that GEP-1 could be employed to modulate human gut microbiota for the treatment of diseases.
Supplementary Materials
The following are available online, Figure S1: The BPI chromatogram (A) and MS/MS spectra of 10HA in GEP-1 hydrolysate in negative mode (B), Figure S2: The mass spectra of PMAAs derived from GEP-1, Figure S3: 1H- and 13C-NMR spectra of GEP-1.
Author Contributions
J.H. (Jiangyan Huo), F.L., J.H. (Jinjun Hou), Z.Z. and H.L. performed in the initial task of isolation and purification of polysaccharides and wrote an initial draft for her thesis. M.L. interpretation of the research work and wrote the manuscript. X.Z., Y.L. and C.X. performed the bioassays. W.W. acquired the funding and supervised of the research work. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Special Fund for National Key R & D Program of China [2018YFC1707001, 2018YFC1707900, 2018YFC1707903, 2018YFC1707905], Research Project of Science and Technology Commission of Shanghai Municipality [18DZ2200800].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be provided upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the GEP-1 is available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shin E.J., Bach J.-H., Nguyen T.T.L., Jung B.-D., Oh K.W., Kim M.J., Jang C.G., Ali S.F., Ko S.K., Yang C.H., et al. Gastrodia elata Bl attenuates cocaine-induced conditioned place preference and convulsion, but not behavioral sensitization in mice: Importance of GABA(A) receptors. Curr. Neuropharmacol. 2010;9:26–29. doi: 10.2174/157015911795017326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh C.-L., Chiang S.-Y., Cheng K.-S., Lin Y.-H., Tang N.-Y., Lee C.-J., Pon C.-Z., Hsieh C.-T. Anticonvulsive and free radical scavenging activities of Gastrodia elata Bl. in kainic acid-treated rats. Am. J. Chin. Med. 2001;29:331–341. doi: 10.1142/S0192415X01000356. [DOI] [PubMed] [Google Scholar]
- 3.Wu C.R., Hsieh M.T., Huang S.C., Peng W.H., Chang Y.S., Chen C.F. Effects of Gastrodia elata and its active constituents on scopolamine-induced amnesia in rats. Planta Med. 1996;62:17–321. doi: 10.1055/s-2006-957892. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z.-W., Li Y., Liu D.-H., Mu Y., Dong H.-J., Zhou H.-L., Guo L.-P., Wang X. Four new phenolic constituents from the rhizomes of Gastrodia elata Blume. Nat. Prod. Res. 2018;33:1140–1146. doi: 10.1080/14786419.2018.1460836. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z.-K., Ng C.-F., Shiu H.-T., Wong H.-L., Chin W.-C., Zhang J.-F., Lam P.-K., Poon W.-S., Lau C.B.-S., Leung P.-C., et al. Neuroprotective effect of Da Chuanxiong Formula against cognitive and motor deficits in a rat controlled cortical impact model of traumatic brain injury. J. Ethnopharmacol. 2018;217:11–22. doi: 10.1016/j.jep.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Liu J., Mori A. Antioxidant and free radical scavenging activities of Gastrodia elata Bl. and Uncaria rhynchophylla (Miq.) jacks. Neuropharmacology. 1992;31:1287–1298. doi: 10.1016/0028-3908(92)90058-W. [DOI] [PubMed] [Google Scholar]
- 7.Ha J.H., Shin S.M., Lee S.K., Kim J.-S., Shin U.-S., Huh K., Kim J.-A., Yong C.-S., Lee N.-J., Lee D.-U. In vitro effects of hydroxybenzaldehydes from Gastrodia elata and their analogues on GABAergic neurotransmission, and a structureactivity correlation. Planta Med. 2001;67:877–880. doi: 10.1055/s-2001-18844. [DOI] [PubMed] [Google Scholar]
- 8.Ahn E.K., Jeon H.J., Lim E.J., Jung H.J., Park E.H. Anti-inflammatory and anti-angiogenic activities of Gastrodia elata Blume. J. Ethnopharmacol. 2007;110:476–482. doi: 10.1016/j.jep.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Hu K., Jeong J.H. A convergent synthetic study of biologically active benzofuran derivatives. Arch. Pharm. Res. 2006;29:476–478. doi: 10.1007/BF02969419. [DOI] [PubMed] [Google Scholar]
- 10.Choi J.H., Lee D.U. A new citryl glycoside from Gastrodia elata and its inhibitory activity on GABA transaminase. Chem. Pharm. Bull. 2006;54:1720–1721. doi: 10.1248/cpb.54.1720. [DOI] [PubMed] [Google Scholar]
- 11.Matias M., Silvestre S., Falcão A., Alves G. Gastrodia elata and epilepsy: Rationale and therapeutic potential. Phytomedicine. 2016;23:1511–1526. doi: 10.1016/j.phymed.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh C.L., Lin J.J., Chiang S.Y., Su S.Y., Tang N.Y., Lin G.G., Lin I.H., Liu C.H., Hsiang C.Y., Chen J.C., et al. Gastrodia elata modulated activator protein 1 via c-Jun N-terminal kinase signaling pathway in kainic acid-induced epilepsy in rats. J. Ethnopharmacol. 2007;109:241–247. doi: 10.1016/j.jep.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y., Gong X.J., Zhou X., Kang Z.J. Relative bioavailability of gastrodin and parishin from extract and powder of Gastrodiae rhizoma in rat. J. Pharm. Biomed. Anal. 2014;100:309–315. doi: 10.1016/j.jpba.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Shu C.H., Qiao N., Piye N., Ming W.H., Xiao S.S., Feng S., Sheng W., Opler M. Protective effects of Gastrodia elata on aluminium-chloride-induced learning impairments and alterations of amino acid neurotransmitter release in adult rats. Restor. Neurol. Neurosci. 2008;26:467–473. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X.H., Guo X.N., Zhan J.P., Xie Z.L., Wang J.M., Zhang Y.T., Chen Y.L., Li X.B. The effects of polysaccharide from Gastrodia elata B1 on cell cycle and caspase proteins activity in H22 tumor bearing mice. Chin. J. Gerontol. 2015;20:5681–5682. [Google Scholar]
- 16.Glisic S., Ivanovic J., Ristic M., Skala D. Extraction of sage (Salvia officinalis L.) by supercritical CO2: Kinetic data, chemical composition and selectivity of diterpenes. J. Supercrit. Fluids. 2010;52:62–70. doi: 10.1016/j.supflu.2009.11.009. [DOI] [Google Scholar]
- 17.Chen J.C., Tian S., Shu X.Y., Du H.T., Li N., Wang J.R. Extraction, characterization and immunological activity of polysaccharides from Rhizoma gastrodiae. Int. J. Mol. Sci. 2016;17:1011. doi: 10.3390/ijms17071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B.H., Tan J., Zhang C., Wu Y. Neuroprotective effect of polysaccharides from Gastrodia elataume against corticosterone-induced apoptosis in PC12 cells via inhibition of the endoplasmic reticulum stress-mediated pathway. Mol. Med. Rep. 2018;17:1182–1190. doi: 10.3892/mmr.2017.7948. [DOI] [PubMed] [Google Scholar]
- 19.Kim K.J., Lee O.H., Han C.K., Kim Y.C., Hong H.D. Acidic Polysaccharide extracts from Gastrodia rhizomes suppress the atherosclerosis risk index through inhibition of the serum cholesterol composition in sprague dawley rats fed a high-fat diet. Int. J. Mol. Sci. 2012;13:1620–1631. doi: 10.3390/ijms13021620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X.Y., Chao Y.M., Du Z., Zhang Y. Effects of polysaccharides from Gastrodia elata on anti-aging of ageing Mice. Pharm. J. Chin. PLA. 2010;26:206–209. [Google Scholar]
- 21.Li H.B., Wu F., Miao H.C., Xiong K.R. Effects of polysaccharide of Gastrodia elataume and electro-acupuncture on expressions of brain-derived neurotrophic factor and stem cell factor protein in caudate putamen of focal cerebral ischemia rats. Med. Sci. Monit. Basic. Res. 2016;22:175–180. doi: 10.12659/MSMBR.901524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H., Zhang M., Ma Q., Tian B., Nie C., Chen Z., Li J. Health beneficial effects of resistant starch on diabetes and obesityviaregulation of gut microbiota: A review. Food Funct. 2020;11:5749–5767. doi: 10.1039/D0FO00855A. [DOI] [PubMed] [Google Scholar]
- 23.Koropatkin N.M., Cameron E.A., Martens E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Yang X., Wu Z., Wang H., Li Q., Mei H., You R., Zhang Y. Dendrobium officinale polysaccharide protected CCl4-induced liver fibrosis through intestinal homeostasis and the LPS-TLR4-NF-kappa B signaling pathway. Front. Pharmacol. 2020;11:240. doi: 10.3389/fphar.2020.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C.-J., Lin C.-S., Lu C.-C., Martel J., Ko Y.-F., Djcius D.M., Tseng S.-F., Wu T.-R., Chen Y.-Y.M., Young J.D., et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 2015;6:7489. doi: 10.1038/ncomms8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Y., Yan Y.M., Chen D., Ran L., Mi J., Lu L., Jing B., Li X., Zeng X., Cao Y. Modulating effects of polysaccharides from the fruits of Lycium barbarum on the immune response and gut microbiota in cyclophosphamide-treated mice. Food Funct. 2019;10:3671–3683. doi: 10.1039/C9FO00638A. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R., Yuan S., Ye J., Wang X., Zhang X., Shen J., Yuan M., Liao M. Polysaccharide from flammuliana velutipes improves colitis via regulation of colonic microbial dysbiosis and inflammatory responses. Int. J. Biol. Macromol. 2005;149:1252–1261. doi: 10.1016/j.ijbiomac.2020.02.044. [DOI] [Google Scholar]
- 28.Lai C.-J.-S., Yuan Y., Liu D.-H., Kang C.-Z., Zhang Y., Zha L., Kang L., Chen T., Nan T.-G., Hao Q.-X., et al. Untargeted metabolite analysis-based UHPLC-Q-TOF-MS reveals significant enrichment of p-hydroxybenzyl dimers of citric acids in fresh beige-scape Gastrodia elata (Wutianma) J. Pharm. Biomed. 2017;140:287–294. doi: 10.1016/j.jpba.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 29.Zhan H.-D., Zhou H.-Y., Sui Y.-P., Du X.-L., Wang W., Dai L., Sui F., Huo H.-R., Jiang T.-L. The rhizome of Gastrodia elata Blume—An ethnopharmacological review. J. Ethnopharmacol. 2016;189:361–385. doi: 10.1016/j.jep.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y.W., Li Z.F., He M.Z., Feng Y.L., Wang Q., Li X., Yang S.L. Chemical constituents of Gastrodia elata. Chin. Tradit. Herbal Drugs. 2013;44:2974–2976. [Google Scholar]
- 31.Zhu H., Liu C., Hou J., Long H., Wang B., Guo D., Lei M., Wu W. Gastrodia elata Blume polysaccharides: A review of their acquisition, analysis, modification, and pharmacological activities. Molecules. 2019;24:2436. doi: 10.3390/molecules24132436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Y., Lu W., Chu Y., Yan J.K., Wang S.J., Xu H.X., Ma H.L. Extraction of polysaccharide from maca: Characterization and immunoregulatory effects on CD4+ T cell. Int. J. Biol. Macromol. 2020;154:477–485. doi: 10.1016/j.ijbiomac.2020.03.098. [DOI] [PubMed] [Google Scholar]
- 33.Huo J.Y., Lu Y., Jiao Y.K., Chen D.F. Structural characterization and anticomplement activity of an acidic polysaccharide from Hedyotis diffusa. Int. J. Biol. Macromol. 2020;155:1553–1560. doi: 10.1016/j.ijbiomac.2019.11.132. [DOI] [PubMed] [Google Scholar]
- 34.Hakomori S. A rapid premethylation of glycolipid and polysaccharide catalyzed methysufinyl carbanion in dimethyl sulfoxide. J. Biochem. 1964;55:205–208. [PubMed] [Google Scholar]
- 35.Lin Z., Li T., Yu Q., Chen H., Zhou D., Li N., Chunyan Yan C. Structural characterization and in vitro osteogenic activity of ABPB-4, a heteropolysaccharide from the rhizome of Achyranthes bidentata. Carbohyd. Polym. 2021;259:117553. doi: 10.1016/j.carbpol.2020.117553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be provided upon request.