Summary
Characterizing mRNA and protein expression with temporal and spatial resolution is a valuable component of nearly every developmental study. Here, we describe a protocol that combines in situ hybridization chain reaction (HCR) and immunofluorescence, allowing for the detection of mRNAs and proteins simultaneously, in zebrafish embryos and larvae. This protocol expands the flexibility of multiplexed HCR by coupling it with traditional immunofluorescence detection.
For complete details on the use and execution of this protocol, please refer to Choi et al. (2010, 2016, 2018) and Howard et al. (2021).
Subject areas: Developmental biology, Microscopy, Model Organisms, Molecular Biology, Antibody, In Situ Hybridization
Graphical abstract
Highlights
-
•
Combined in situ HCR and immunofluorescence for a modular and flexible procedure
-
•
Simultaneous detection of mRNA and protein in zebrafish embryos and larvae
-
•
Highly customizable protocol with a broad variety of targets and applications
-
•
Use of commercially available antibodies in a simple and streamlined protocol
Characterizing mRNA and protein expression with temporal and spatial resolution is a valuable component of nearly every developmental study. Here, we describe a protocol that combines in situ hybridization chain reaction (HCR) and immunofluorescence, allowing for the detection of mRNAs and proteins simultaneously, in zebrafish embryos and larvae. This protocol expands the flexibility of multiplexed HCR by coupling it with traditional immunofluorescence detection.
Before you begin
This protocol describes steps carried out for the detection of mRNA and protein expression in zebrafish embryos/larvae using previously confirmed Hybridization Chain Reaction (HCR) probe sets and antibodies. We have used this protocol to perform a 3-marker Whole-mount Immuno-Coupled Hybridization Chain Reaction (WICHCR) and a 5-marker WICHCR, by selecting appropriate fluorophores and spectrally deconvolving the correct wavelengths (for the 5-marker experiment), to produce well defined and distinct fluorescence channels.
Experimental design
-
1.Due to the potential to analyze complex experimental conditions involving numerous probes, the user should minimize the overlap between emission spectra of fluorophores when designing experiments relying on multichannel images.
-
a.When choosing markers that will be used for HCR analysis ensure a unique fluorophore pairs with a specific marker by matching a specific B isoform with the corresponding probe set.Note: The B isoform designates a specific and unique sequence among the HCR probes which will initiate amplifier polymerization. For further details on this process please refer to the seminal literature on the HCR protocol (Choi et al., 2010, 2016, 2018).
-
b.Apply a similar thought process when choosing the optimal primary and secondary antibodies, by selecting correct antibody serotypes.
-
a.
Note: We recommend use of FPbase.org when planning each experiment (Lambert, 2019) for comparing fluorophore emission spectra and choosing the best combinations to suit your experimental design.
-
2.
Validate HCR probe sets and antibodies separately before the start of the WICHCR protocol to ensure efficient detection of the individual markers and to determine ideal concentrations. Some HCR probe sets and antibodies have been found to work better at higher concentrations.
-
3.Protect samples from direct light to prevent fluorophore quenching after amplifiers have been added. When manipulating the samples try to do it as fast as possible, but with care, to prevent prolonged photo-exposure.
-
a.During incubation steps, make sure to cover the samples with foil paper and keep them out of direct light, such as a desk drawer or opaque refrigerator.
-
a.
-
4.
WICHCR samples can be stored at 4°C for a week with minimal loss of signal; however, it is strongly recommended that samples be imaged within that period.
HCR probe sets and amplifier generation
Timing: 1–2 weeks
Note: HCR probe sets and amplifiers are ordered from Molecular Instruments, Inc. (https://www.molecularinstruments.com/) and usually take ∼2 weeks for production and shipping.
-
5.Identify the target RNA sequence and NCBI Accession number of your gene of interest for HCR probe set design.
-
a.Carefully select the B isoform and probe set size for each HCR probe set. A probe set consisting of 20 probe pairs is ideal for the WICHCR protocol, though fewer probes per set may still yield sufficient detection.
-
a.
Note: Some of the HCR probe sets used in this paper are smaller than 20, however they still provide excellent detection of the target mRNA.
CRITICAL: The B isoform for each particular HCR probe set dictates the probe sets you can multiplex in the same WICHCR experiment. Choose carefully when designing the experiments which markers you wish to image in the same experiment. For example, you would want to ensure a unique fluorophore identifies each target, similar to how we demonstrate in the expected results that Alexa Fluor 488 denotes elavl3 expression and Alexa Fluor 546 marks phox2bb mRNA (Figures 1C and 1E). Specific amplifiers and probes share a unique B isoform, which confers the specificity of each pair. For further details on topic of multiplexing probes, please refer to the seminal papers on HCR methodology (Choi et al., 2010, 2016, 2018).
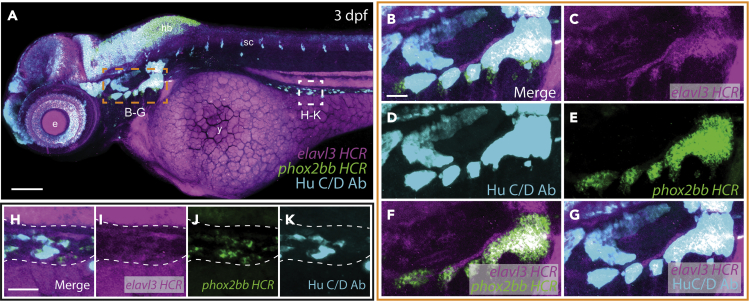
Figure 1.
WICHCR allows detection of mRNA and protein with high resolution in zebrafish
(A) WICHCR against elavl3, phox2bb and Hu C/D in 3 dpf WT zebrafish larvae, orange boxed region depicts the developing cranial ganglia (B–G) and white box marks a region of the developing gut (H–K, dotted line). Scale bar = 100 μm for A and 25 μm for B–K. Anterior to the right and posterior to the left for all the panels. y = yolk sac, e = developing eye, hb = developing hindbrain, sc = developing spinal cord.
-
6.
Carefully select an HCR amplifier for the respective B isoform with desired fluorescent label, for detection of the correct target at the preferred wavelength, bearing in mind the desired detection wavelengths available to label the antibodies.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-zebrafish Sox10 | GeneTex | Catalog # GTX128374; RRID: AB_2885766 |
| Mouse anti-Hu C/D | Thermo Fisher Scientific | Catalog # A-21271; RRID: AB_221448 |
| Mouse anti-Histone H3 (phospho S10) | Abcam | Catalog # ab14955; RRID: AB_443110 |
| Alexa Fluor 488 goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | Catalog # A-11008, RRID: AB_143165 |
| Alexa Fluor 594 goat anti-mouse IgG1 | Thermo Fisher Scientific | Catalog # A-21125, RRID: AB_2535767 |
| Alexa Fluor 647 goat anti-mouse IgG2b | Thermo Fisher Scientific | Catalog # A-21242, RRID: AB_2535811 |
| Chemicals, peptides, and recombinant proteins | ||
| PBS (10×), pH 7.4 (no calcium, no magnesium) | Gibco | Catalog # 70011044 |
| Tween™ 20 | Fisher Scientific | Catalog # BP337 |
| Acetone (certified ACS) | Fisher Scientific | Catalog # A18 |
| 20× SSC | Thermo Fisher | Catalog # 15557036 |
| Glycerol (certified ACS) | Fisher Scientific | Catalog # G33 |
| Goat serum | LAMPIRE Biological Laboratories | Catalog # 7332500 |
| Paraformaldehyde | Sigma-Aldrich | Catalog # P6148 |
| Methylene Blue | Fisher Chemical | Catalog # M291-25 |
| Methanol (100%, HPLC grade) | Millipore | Catalog # MX0475 |
| Pronase | Millipore | Catalog # 10165921001 |
| 1-Phenyl 2-thiourea (PTU) | Sigma-Aldrich | Catalog # P7629 |
| Sodium Chloride (NaCl) | Fisher Scientific | Catalog # S271 |
| Calcium chloride, dihydrate (CaCl2 ⋅ 2H2O) | Millipore | Catalog # 208291 |
| Potassium chloride (KCl) | Fisher Scientific | Catalog # P217 |
| Magnesium sulfate, 7-hydrate (MgSO4 ⋅ 7H2O) | J.T. Baker | Catalog # 250001 |
| Nuclease-Free Water (not DEPC-Treated) | Thermo Fisher | Catalog # AM9932 |
| Proteinase K, Lyophilized | Millipore | Catalog # 70663 |
| Critical commercial assays | ||
| HCR Probe Hybridization Buffer | Molecular Instruments, Inc. | n/a |
| HCR Probe Wash Buffer | Molecular Instruments, Inc. | n/a |
| HCR Amplification Buffer | Molecular Instruments, Inc. | n/a |
| Experimental models: organisms/strains | ||
| AB wildtype zebrafish line | ZIRC | Catalog # ZL1, RRID:ZIRC_ZL1 |
| Oligonucleotides | ||
| Zebrafish elavl3 HCR v3.0 probe set (B2) set size: 20 | Molecular Instruments, Inc. | Genbank: NM_131449 |
| Zebrafish phox2bb HCR v3.0 probe set (B1) set size: 20 | Molecular Instruments, Inc. | GenBank: NM_001014818.1 |
| Zebrafish barx1 HCR v3.0 probe set (B3) set size: 18 | Molecular Instruments, Inc. | GenBank: NM_001024949.1 |
| Zebrafish mitfa HCR v3.0 probe set (B1) set size: 12 | Molecular Instruments, Inc. | GenBank: NM_130923 |
| Zebrafish sox10 HCR v3.0 probe set (B5) set size: 20 | Molecular Instruments, Inc. | GenBank: NM_131875 |
| B1 546 HCR Amplifier | Molecular Instruments, Inc. | n/a |
| B2 488 HCR Amplifier | Molecular Instruments, Inc. | n/a |
| B3 514 HCR Amplifier | Molecular Instruments, Inc. | n/a |
| B5 546 HCR Amplifier | Molecular Instruments, Inc. | n/a |
| B1 647 HCR Amplifier | Molecular Instruments, Inc. | n/a |
| Software and algorithms | ||
| Fiji | Fiji | RRID:SCR_002285 |
| Confocal microscope software | Olympus | FV31S-SW acquisition software |
| Olympus cellSens Dimensions Software V2.1 | Olympus | RRID:SCR_014551 |
| Imaris | Bitplane | RRID:SCR_007370 |
| Other | ||
| Olympus MVX10 Macroview Stereo Fluorescence Microscope (or similar, for analysis of signal detection) | Olympus | RRID:SCR_018612 |
| Olympus Confocal Laser Scanning Microscope Fluoview FV3000 (or similar, for high quality imaging) | Olympus | RRID:SCR_017015 |
Note: This protocol indicates antibodies and HCR probe sets used for the particular experiments carried out for this manuscript. Antibodies and HCR probes need to be adjusted for the targets the user wants to detect.
Note: All Molecular Instruments, Inc. products (HCR probe sets, amplifiers, and buffers) can be found in the following website: https://www.molecularinstruments.com/
Materials and equipment
1× PBS
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× PBS | 1× | 100 mL |
| MilliQ H2O | n/a | Add to 1 L |
| Total | n/a | 1 L |
Note: Mix thoroughly and store at 20°C–25°C for long term storage.
CRITICAL: avoid using calcium chloride and magnesium chloride in PBS as this leads to increased autofluorescence in the embryos/larvae.
1× PBST
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× PBS | 1× | 100 mL |
| Tween-20 | 0.1% | 1 mL |
| MilliQ H2O | n/a | Add to 1 L |
| Total | n/a | 1 L |
Note: Mix thoroughly and store at 20°C–25°C for long term storage for up to a year. Discard if precipitates form.
4% Paraformaldehyde (PFA)
| Reagent | Final concentration | Amount |
|---|---|---|
| Paraformaldehyde | 4% | 20 g |
| 1× PBS | 1× | Add to 500 mL |
| Total | n/a | 500 mL |
CRITICAL: PFA is a hazardous material, it should be prepared with extreme care under a fume hood and using appropriate personal protective equipment (PPE).
Note: Preheat 300 mL of 1× PBS at 60°C, add PFA and stir continuously until the solution is clear. Allow solution to cool and complete to 500 mL with 1× PBS. The solution should be aliquoted and stored at −20°C for long term storage. Freshly thawed 4% PFA should be allowed to cool to 4°C before using, thawed 4% PFA can be stored at 4°C for up to a week. Do not use thawed 4% PFA older than recommended.
Proteinase K (2 mg/mL) stock
| Reagent | Final concentration | Amount |
|---|---|---|
| Proteinase K | 2 mg/mL | 2 mg |
| Nuclease Free Water | n/a | 1 mL |
| Total | n/a | 1 mL |
Note: Make 20 μL aliquots and store at −20°C for long term storage.
CRITICAL: PK stock aliquots can be cyclically frozen and thawed a maximum of 4 times before diminished efficacy. After using an aliquot, mark how many times it has been used and store at −20°C or discard accordingly.
60× E3 stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Chloride | 300 mM | 140 g |
| Calcium Chloride dihydrate | 200 mM | 23.4 g |
| Potassium Chloride (anhydrous) | 100 mM | 6.1 g |
| Magnesium sulfate heptahydrate | 2 mM | 3.9 g |
| MilliQ H2O | n/a | Bring up to 8 L |
| Total | n/a | 8 L |
Note: Store at 20°C–25°C for up to 12 months.
20× Methylene Blue stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Methylene Blue | 2.6 mM | 1 g |
| MilliQ H2O | n/a | 1000 mL |
| Total | n/a | 1000 mL |
Note: Store at 20°C–25°C for up to 24 months.
1× E3 culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| 60× E3 stock solution | 1× | 166 mL |
| 20× Methylene Blue stock solution | 1× | 500 μL |
| MilliQ H2O | n/a | Bring up to 10 L |
| Total | n/a | 10 L |
Note: Store at 20°C–25°C for up to 12 months. Store at 28°C for up to 6 months. Discard if microbial growth begins.
25× PTU stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 1-phenyl-2-thiourea | 5 mM | .75 g |
| MilliQ H2O | n/a | 1000 mL |
| Total | n/a | 1000 mL |
Note: Store at 4°C for 6 months, protected from ambient light exposure. Smaller aliquots may be prepared.
1.5× PTU/E3 Working stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 25× PTU stock solution | 0.3 mM | 60 mL |
| 60× E3 | 1× | 16.67 mL |
| 20× Methylene Blue stock solution | 1× | 50 μL |
| MilliQ H2O | n/a | 923.33 mL |
| Total | n/a | 1000 mL |
Note: Store between 25°C–28°C for 4 months, protected from ambient light exposure.
Pronase (30 mg/mL) stock
| Reagent | Final concentration | Amount |
|---|---|---|
| Pronase | 30 mg/mL | 1.5 g |
| MilliQ H2O | n/a | 50 mL |
| Total | n/a | 50 mL |
Note: Make 1 mL aliquots and store at −20°C for long term storage.
5% goat block solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Goat serum | 5% | 500 μL |
| 1× PBST | 1× | Add to 10 mL |
| Total | n/a | 10 mL |
Note: Thaw a fresh aliquot of goat serum, add to the corresponding volume of 1× PBST, and vortex vigorously. It is recommended to prepare a fresh batch of 5% block solution at the beginning of each WICHCR protocol not performed in parallel. The 5% block solution can be stored at 4°C for up to a week or at −20°C for 3 months.
Alternatives: Animal serum should be selected according to the antibody combination that will be used. We have successfully carried out WICHCR protocol with goat and donkey serums. While we recommend using diluted animal serum for optimal results, BSA solutions may serve as a suitable alternative for select applications, however the use of BSA as a blocking agent was not tested in the development of this protocol.
5× SSCT
| Reagent | Final concentration | Amount |
|---|---|---|
| 20× SSC | 5× | 10 mL |
| Tween-20 | 0.1% | 40 μL |
| MilliQ H2O | n/a | Add to 40 mL |
| Total | n/a | 40 mL |
Note: Mix thoroughly and store at 20°C–25°C for long term storage for up a year. Discard if precipitates form.
Step-by-step method details
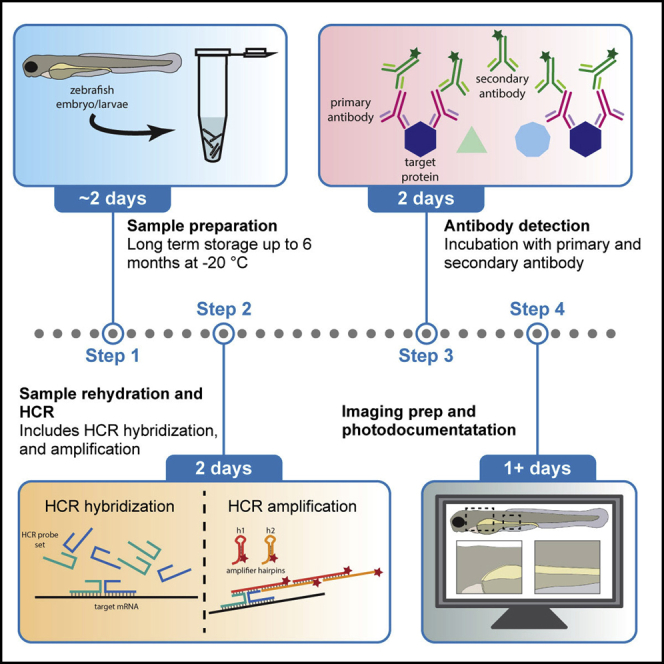
Sample preparation
Timing: 2 days
In this step, zebrafish embryos/larvae are sorted and collected at the time points required for the analysis the researcher wishes to carry out. In this protocol, zebrafish larvae were collected at 3 days post fertilization (dpf).
Note: All incubations in this section are carried out at 20°C–25°C without agitation, unless otherwise described in the protocol.
-
1.
Recently fertilized zebrafish embryos are collected and cultured in 1× E3 culture media and switched to 0.0045% 1-phenyl 2-thiourea (PTU)/E3 solution after 22 hpf to maintain transparency, which allows better imaging of older embryos/larvae. PTU has been shown at this concentration to not exhibit developmental effects (Karlsson et al., 2001).
-
2.
Embryos can be dechorionated as early as 16 hpf by adding 100 μL of Pronase stock and incubating for 30 min. Swirl plates occasionally to facilitate chorion removal, once chorions are removed rinse embryos with 1× E3 culture media 3 times, to remove any residual Pronase and chorion debris.
Alternatives: embryos can be manually dechorionated by using a pair of fine tweezers.
-
3.
At the desired developmental stage, sort and collect previously dechorionated zebrafish embryos/larvae into 1.5 mL microcentrifuge tubes, between 15 and 20 specimens per tube are recommended. Sex characteristics are indistinguishable in embryonic and larval zebrafish. Both sexes are typically represented in a clutch.
-
4.
Add 1 mL of freshly thawed 4% PFA per tube, to fix samples for ∼16 h at 4°C.
Note: The fixation in 4% PFA should be performed for at least 16 hours to a maximum of two days at 4°C. We have seen that over fixation leads to higher background, especially in the fluorescent range similar to the GFP emission spectra.
Alternatives: Samples can also be fixed at 20°C–25°C for 4 hours without affecting WICHCR signal.
-
5.
Wash samples with 1 mL of 1× PBS 3 times for 5 min per wash.
CRITICAL: Discard the liquid waste according to the PFA handling specifications.
-
6.Dehydrate and permeabilize the samples with a series of methanol (MeOH) washes (1 mL each):
-
a.100 % MeOH for 10 min
-
b.100% MeOH for 10 min
-
a.
-
7.
Store samples at −20°C for ∼16 h before use.
Pause point: Samples can be stored for up to six months at −20°C after this step. If relying on endogenous fluorescence for the detection of one marker, the embryos should be as freshly prepared as possible due to loss of endogenous fluorescence.
Sample rehydration and permeabilization
Timing: ∼4 h (depending on the embryo/larvae stage)
After samples have been dehydrated and chilled in methanol, in this section the tissue is rehydrated and permeabilized for easy access of the HCR probes and antibodies inside of the embryo/larvae.
Note: All incubations in this section are carried out at 20°C–25°C without agitation, unless otherwise described. The volume added in each step is 500 μL, except where specified volumes are used.
-
8.Rehydrate with a series of graded MeOH/PBST for 5 min each:
-
a.75% MeOH/ 25% 1× PBST
-
b.50% MeOH/ 50% 1× PBST
-
c.25% MeOH/ 75% 1× PBST
-
d.2 times in 100% 1× PBST
-
a.
-
9.
Incubate samples for 12 min at −20°C in 100% acetone to further permeabilize embryos.
Note: The acetone permeabilization is performed, irrespective of the developmental stage of the samples collected, to improve permeability. Acetone must be pre-chilled at −20°C.
-
10.
Wash 3 times for 5 min each in 1× PBST.
-
11.
Incubate with 10 μg/mL Proteinase K diluted in 1× PBST. The incubation time depends on the embryo/larvae age, measured in either hours post fertilization (hpf) or days post fertilization (dpf). Optimal times for Proteinase K digestion per stage are depicted in Table 1.
CRITICAL: PK stock aliquots should be thawed a maximum of 4 times before discarding. We have found that many cycles of freeze/thaw decrease the enzyme efficiency, thus affecting embryo permeability.
-
12.
Wash samples 3 times with PBST without incubation.
-
13.
Fix samples with 4% PFA for 20 min.
-
14.
Wash samples 5 times for 5 min each with 1 mL 1× of PBST.
Note: If you are validating a new antibody for the first time in this protocol, we recommend that you do so first in a pilot assay in wild-type tissue staged similarly to your planned experimental assay without completing the HCR step. Please proceed directly to step 28 and continue the protocol as written.
Pause point: If the protocol cannot be immediately continued, samples can be left in PBST at 4°C before moving on to next step for up to 4 days. However, the HCR hybridization step is recommended to be started the same day the permeabilization is performed.
Table 1.
Recommended Proteinase K digestion times per zebrafish stage
| Embryonic/Larval age | Proteinase K digestion time (minutes) |
|---|---|
| 24 hpf | 12 |
| 26 hpf | 16 |
| 48 hpf | 20 |
| 52 hpf | 23 |
| 72 hpf | 30 |
| 4 dpf | 40 |
| 5 dpf | 50 |
HCR hybridization
Timing: 1 h + 12–16 h hybridization
In this step, zebrafish embryos/larvae are equilibrated in probe hybridization buffer and then incubated with HCR probe sets for hybridization with its target mRNA. Here, 3 dpf zebrafish larvae were hybridized with probe sets against elavl3 and phox2bb (for the 3-marker experiment), or with sox10, barx1, and mitfa (for the 5-marker spectrally deconvolved experiment), for more specifics on marker combinations please see the expected outcomes section. Details on probe hybridization buffer are listed in the key resources table.
-
15.
Pre-warm probe hybridization buffer to 37°C.
CRITICAL: Probe hybridization buffer contains formamide, careful handling is recommended.
-
16.
Pre-hybridize samples with 250 μL of probe hybridization buffer for 30 min at 37°C.
-
17.Thaw all the HCR probe sets that will be used for the experiment while the samples are pre-hybridizing.
-
a.Prepare HCR probe mixture solution by adding 2–3 pmol of each probe set (2–3 μL of each 1 μM probe set stock) into 250 μL of probe hybridization buffer. Mix gently by pipetting up and down.Note: Larger volumes of probe mixture solution may be prepared if analyzing multiple tissue conditions (multiple tubes of embryos/larvae) with identical probe set combinations. For example, if analyzing three different experimental conditions of embryos with the same markers, prepare 6–9 μL of each probe set into 750 μL of probe hybridization buffer, allotting 250 μL of probe solution per condition.Note: Some HCR probe sets, especially the ones with smaller probe set sizes, have been found to work better at higher concentrations. Using 4–5 μL with such probes has given us great results.
-
b.Pre-warm the probe mixture solution at 37°C for 15 min, before adding to the samples. This can be done in parallel with sample pre-hybridization.
-
a.
-
18.
Replace pre-hybridization solution with probe mixture solution and incubate the samples for 12–16 h at 37°C.
HCR probe washes and HCR amplification
Timing: 2 h + 12–16 h HCR amplification
The goal of this step is to remove unbound HCR probes and initiate HCR signal amplification by choosing the correct amplifier for the wavelength desired for detection. For more details on amplifiers used for the 3-marker experiment and 5-marker experiment performed for this protocol, go to expected outcomes. Details on probe wash buffer and amplification buffer are listed in the key resource table.
Note: All incubations in this section are carried out at 20°C–25°C without agitation, unless otherwise described.
Note: Preheat probe wash buffer to 37°C before use. Additionally, let amplification buffer equilibrate at 20°C–25°C while washing samples.
-
19.
Remove probe mixture solution and wash excess probe 4 times for 15 min each wash with 250 μL of pre-warmed probe wash buffer. Maintain samples at 37°C during washes.
Note: Probe mixture solution can be saved at −20°C for reuse, however it is possible to see reduced signal with repeated use. Label tube containing probe mixture solution with probe sets used and number of times used.
-
20.
Wash larvae 2 times for 5 min each with 500 μL 5× SSCT.
Note: Make sure the amplification buffer has warmed up to 20°C–25°C before proceeding.
-
21.
Pre-amplify larvae with 250 μL of 20°C–25°C probe amplification buffer for 30 min.
-
22.Thaw all the amplifier hairpins that will be used for the experiment while the samples are pre-amplifying.
-
a.Add 3–5 μL of the first amplifier hairpin h1 (3 μM stocks), per tissue condition, in a sterile tube. In a separate tube add an equal volume of amplifier hairpin h2. Repeat this step for each different B isoform that was used during probe hybridization, taking care such that each amplifier corresponds to only one B isoform. Carefully label each tube for the corresponding hairpin number and B isoform, as well as detection wavelength.
 CRITICAL: Make sure to select both hairpins h1 and h2 for the correct B isoform and wavelength desired.
CRITICAL: Make sure to select both hairpins h1 and h2 for the correct B isoform and wavelength desired. CRITICAL: Amplifier hairpins should be protected from light as much as possible.Note: If analyzing multiple tissue conditions (multiple tubes of embryos/larvae) with identical probe set combinations, the amount of amplifier hairpins should be scaled accordingly. For example, if analyzing three different experimental conditions with the same markers, prepare 9–15 μL of amplifier hairpin h1 and an equal volume of amplifier hairpin h2 in separate tubes for each B isoform used in the experiment.
CRITICAL: Amplifier hairpins should be protected from light as much as possible.Note: If analyzing multiple tissue conditions (multiple tubes of embryos/larvae) with identical probe set combinations, the amount of amplifier hairpins should be scaled accordingly. For example, if analyzing three different experimental conditions with the same markers, prepare 9–15 μL of amplifier hairpin h1 and an equal volume of amplifier hairpin h2 in separate tubes for each B isoform used in the experiment. -
b.Snap cool the amplifier hairpins by heating all amplifier hairpin tubes at 95°C for 90 s and then let them cool to 20°C–25°C in a dark drawer for 30 min.
-
a.
-
23.
Prepare amplifier hairpin mixture solution by adding the snap cooled h1 and h2 hairpins to 250 μL of amplification buffer per tissue condition.
-
24.
Replace pre-amplification buffer with amplifier hairpin mixture solution, adding 250 μL of solution per tube. Incubate all samples for 12–16 h at 20°C–25°C protected from light.
CRITICAL: From now on samples should be covered with an opaque cover, such as aluminum foil, and protected from light at all times. We have found desk drawers work well for incubations. Carry out the sample manipulations very carefully but as fast as possible to minimize light exposure.
HCR amplification hairpin washes and antibody detection
Timing: 3 days
In this step, we finish the HCR component of the WICHCR protocol by removing excess amplifier hairpins and prepare the samples for primary antibody detection. For more details on antibodies and their respective concentrations used for the 3-marker experiment and 5-marker experiment performed for this protocol, please review the expected outcomes section.
Note: All incubations in this section are carried out at 20°C–25°C without agitation, unless otherwise described. The volume added in each step is 500 μL, except where specified volumes are used.
CRITICAL: Continue to keep samples protected from exposure to light, such as with aluminum foil covers and storage in a dark drawer during incubations. Additionally, we recommend working quickly to minimize prolonged light exposure when manipulating samples.
-
25.Wash excess amplifier hairpins by washing with successive 5× SSCT as follows:
-
a.2 washes for 5 min each
-
b.2 washes for 30 min each
-
c.1 final wash for 5 min
-
a.
-
26.
Remove 5× SSCT to wash tissue with 1× PBST 3 times for 5 min per wash.
-
27.
Check fluorescence under a fluorescence macroscope (Olympus MVX10 Macroview Stereo Fluorescence Microscope or similar) to confirm correct HCR pattern.
CRITICAL: Visually confirm that HCR signal is detectable every time you reach this step before moving on to the following steps. HCR signal may be faint for some fluorophore combinations, requiring close examination. Minimize exposure of tissue to high intensity lasers to reduce photobleaching. If no signal is detected please see the troubleshooting section.
Note: If you are validating a new HCR probe for the first time in this protocol, we recommend that you do so first in a pilot assay in wild-type tissue staged similarly to your planned experimental assay without completing the antibody detection step. Please proceed directly to step 36 and continue the protocol as written.
Pause point: Samples can be left in 1× PBST for a few hours up to 12–16 hours, if needed, before moving on to next step.
-
28.
Dispense 5% goat block solution to samples for 1–2 h at 20°C–25°C or for 12–16 h at 4°C.
Pause point: Samples can also be blocked at 4°C for 12–16 hours, due to experimental time constraints. We have not observed any notable benefit to 12–16 hours blocking with the antibodies tested in our lab.
Note: Animal serum used for the block solution should be carefully selected based on the antibody combinations that will be used. Sterile technique is encouraged when working with animal serums to avoid bacterial contamination of tissue samples. We have successfully carried out WICHCR protocol with goat and donkey serums.
-
29.
Prepare primary antibody solution by diluting the appropriate antibodies with the previously confirmed working concentration in block solution.
-
30.
Remove blocking solution and replace with diluted primary antibody solution. Incubate tissue with primary antibody solution for ∼16 h at 4°C.
Note: Some antibodies may demonstrate higher specificity with longer primary incubation times. We highly recommend performing optimizations for concentration, incubation time, and secondary antibody compatibility prior to starting a WICHCR protocol. Volume of antibody solution added may vary based on dilution constraints. The volume should always be enough to cover the embryos, which is typically between 100–500 μL per tissue condition.
-
31.
Wash excess primary antibody 6 times for 30 min with 1× PBST.
-
32.
Prepare secondary antibody solution by diluting the corresponding secondary antibodies with the previously confirmed working concentration in block solution.
-
33.
Incubate samples in secondary antibody solution for ∼16 h at 4°C.
Note: Some antibodies may demonstrate improved signal with longer secondary incubation times. We highly recommend performing optimizations for concentration, incubation time, and primary antibody compatibility prior to starting a WICHCR protocol. Volume of antibody solution added may vary based on dilution constraints. The volume should always be enough to cover the embryos, which is typically between 100–500 μL per tissue condition.
-
34.
Wash out excess secondary antibody 6 x 30 min with 1× PBST.
-
35.
Check antibody and HCR signal under a fluorescence macroscope (Olympus MVX10 Macroview Stereo Fluorescence Microscope or similar) to confirm retained HCR signal and correct antibody staining.
CRITICAL: Always confirm antibody signal is detectable before moving on.
Imaging preparation and photodocumentation
Timing: 1 day + Imaging time
In this stage, finished WICHCR samples with confirmed signal are prepared for imaging by glycerol clearing and then mounting them for high resolution imaging.
Alternatives: This imaging prep protocol can be adapted or substituted according to each lab’s imaging preparations.
CRITICAL: Continue protecting samples from ambient light by covering with foil and storing in a dark drawer during incubations to reduce photobleaching.
-
36.Clear embryos in serial glycerol solutions at 20°C–25°C by sequentially adding 500 μL of:
-
a.25% glycerol/ 75% 1× PBST
-
b.50% glycerol/ 50% 1× PBST
-
c.75% glycerol/ 25% 1× PBST
-
a.
Note: Embryos/larvae should fall to the bottom of the tube before moving on to the next glycerol concentration.
Note: Samples can be stored in 75% glycerol/ 25% 1× PBST, covered from light for up to one week at 4°C. However, it is highly recommended to image them as soon as possible.
-
37.
Mount embryos in 75% glycerol/ 25% 1× PBST for imaging in confocal microscope, to achieve a high resolution image. Image using the settings required to capture the signal in the regions of interest.
-
38.
After imaging, process and analyze the data according to the experimental outline and using the image analysis software of preference.
Expected outcomes
WICHCR is designed with complex imaging schemes in mind, allowing for combined detection of protein and mRNA in the same sample. Here we show two experimental conditions, that show the potential of the WICHCR protocol for the analysis of both downstream gene products, where we have combined 3 markers (Figure 1) and 5 markers (Figure 2) for both mRNA and protein. The details for each of the two experiments are depicted in Table 2.
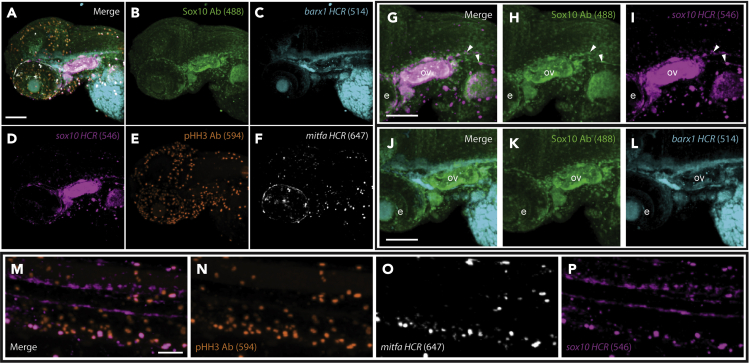
Figure 2.
WICHCR supports complex fluorophore combinations for advanced experimental design
(A) WICHCR against Sox10, barx1, sox10, pHH3 and mitfa in 3 dpf WT zebrafish larvae.
(B–F) Individual panels for each channel after spectral deconvolution. Channels were initially imaged as the following sets: 488/514 (Sox10 AB/barx1), 546/594 (sox10 HCR/pHH3), and 647 (mitfa).
(G–I) HCR probe against sox10 and antibody against Sox10 show expected overlapping expression of mRNA and protein.
(J–L) Channel deconvolution efficiently separates an immunologically and HCR labeled marker with nearby excitation/emission wavelengths. Sox10 antibody (J and K) and barx1 mRNA (J and L) are shown. M-P. WICHCR permits the detection of post-translational modifications and cell identity. Scale bar = 100 μm for A–L and 50 μm for M–P. Anterior to the right and posterior to the left for all the panels. ov = otic vesicle, e = developing eye.
Table 2.
WICHCR markers and detection wavelengths for an example of 3 and 5 marker experiment
|
Figure 1 (3-marker WICHCR) |
Figure 2 (5-marker WICHCR) |
|||||
|---|---|---|---|---|---|---|
| Marker | Type and concentration | Detection wavelength | Marker | Type and concentration | Detection wavelength | |
| 1 | elavl3 | HCR (B2) | 488 | Sox10 | Ab (rabbit) (1:250) |
488 |
| 2 | phox2bb | HCR (B1) | 546 | barx1 | HCR (B3) | 514 |
| 3 | Hu C/D | Ab (mouse IgG2b) (1:250) |
647 | sox10 | HCR (B5) | 546 |
| 4 | n/a | n/a | n/a | pHH3 | Ab (mouse IgG1) (1:1000) |
594 |
| 5 | n/a | n/a | n/a | mitfa | HCR (B1) | 647 |
As a proof of concept, we first performed a 3-marker WICHCR on 3 dpf WT zebrafish embryos, using HCR probe sets against the neural markers elavl3 , an ortholog of HuC (Kim et al., 1996), and phox2bb (Elworthy et al., 2005) and coupling it with an antibody against Hu C/D (Olden et al., 2008). WICHCR protocol permitted us to detect these markers with high resolution (Figure 1). The cervical ganglia served as a useful tissue to validate the expression of our target gene products (Figures 1B–1G) due to its relatively large size, reactivity to all probes used, and distinct shape at this developmental stage. Both HCR markers against elavl3 (Figure 1C) and phox2bb (Figure 1E) reliably labeled the ganglia, with nested adjacent expression domains (Figure 1F). Additionally, both the HCR and antibody detection of Hu+ cells (Figure 1D) were largely coincident (Figure1G) as expected. It was also possible to detect the expression of these markers in the nascent enteric neural cell population within the developing gut (Figures 1H–1K). These results show that WICHCR works for the detection of mRNA and protein in superficial and internal structures in the embryo.
To further characterize the power and flexibility of the WICHCR protocol, we went on to analyze 5 markers simultaneously. Relying on our expertise in neural crest stem cell biology to guide which probes to examine, we have employed antibodies against Sox10, which labels migrating neural crest cells (NCC) (Carney et al., 2006), and phosphorylated Histone H3 (pHH3) , a canonical marker for cell division (Kim et al., 2017), as well as HCR probe sets against barx1, mitfa, and sox10, labeling mesenchyme, melanophore/pigment progenitors, and NCC respectively (Howard et al., 2021) (Figure 2). Following the WICHCR protocol, confocal images were spectrally deconvolved, distinguishing the signals in the corresponding wavelength for each of the fluorescent markers used (Figures 2A–2F). As expected, we readily detected Sox10 protein (Figure 2H) and sox10 mRNA (Figure 2I) in overlapping domains throughout the zebrafish larvae, in the otic vesicle and other regions (Figures 2G–2I, white arrowheads) by 3 dpf. Spectrally deconvolved channels for the Sox10 protein and an HCR marker barx1 (Figures 2J–2L) highlighted the precision provided by the protocol, revealing distinct regions of expression for each marker. More importantly, we are able to combine detection of phosphorylated Histone H3, a type of post translationally modified histone, along with mRNA labeling to reveal both nascent cell identity and ongoing cellular processes (Figure 2M–2P). The ability to detect mRNA expression and proteins with post translational modifications simultaneously, builds on the historical context of mRNA probes to distinguish tissue identity to include powerful information about physiological processes within those cells, which would otherwise remain undetectable by mRNA probes alone (Figure 2M–2P). Considering these examples together, we aim to show the interrogative power of the WICHCR protocol. By taking advantage of both the remarkable flexibility of HCR and robust detection of traditional immunohistological assays, we offer to the community a simple, sensitive, and modular assay equipped to simultaneously discern both protein and mRNA detection within their endogenous tissue contexts.
Limitations
The WICHCR protocol can be applied to perform complex experiments to detect mRNAs and protein in the same sample, this is of particular importance when no commercial antibodies are available. The current version of the protocol has not been tested in older larvae or adult samples, and tissue permeability might be problematic in older specimens and could lead to poor detection of the markers. In this case, the researcher should use a different permeabilization method designed for older specimens, such as treatment with collagenase, and confirm the markers can be detected individually at the desired age before attempting WICHCR. Additionally, it is possible that this protocol may be performed in tissue sections to increase permeability, which is an objective for further optimization of this protocol.
Troubleshooting
Problem 1
HCR signal not detected or is very poor (step 27)
Potential solution
Sample might be poorly permeabilized. Use a fresh PK stock when permeabilizing embryos/larvae and ensure the correct amount of time has lapsed for the developmental stage being assayed (step 11).
HCR probe set was not previously validated in wild-type whole mount tissue. Prior validation is a crucial step to ensure reproducibility in the WICHCR protocol. Validate HCR probe set efficiency by using wild-type embryos/larvae at similar stages to those planed for future experiments to determine both the efficacy of the probe set as well as the regions in which the gene of interest is expressed.
Make sure that the correct HCR amplifiers were selected for the markers wished to detect and are visualized at the correct wavelength (steps 22–24).
If the HCR signal is weak or highly variable between embryos/larvae, HCR probe set concentration used may not be optimal. In this case, increase HCR probe set concentration. We have found some probe sets work at higher concentrations (step 17).
Use a positive control probe set for confirmed detection at the developmental stage wished to be analyzed. For example, we have found elavl3 is a great positive control in 3 dpf zebrafish larvae.
Problem 2
High background from HCR labeling (step 27)
Potential solution
-
•
Ensure that all wash and incubation steps are performed at the correct temperature specified in the protocol. Improper temperature regulation in washes/incubations may lead to non-specific amplifier trapping in tissue (steps 16–26).
-
•
Ensure that each amplifier has been snap-cooled in a separate tube to form proper hairpin looping. Snap-cooling hairpins in a single vial may yield uninterpretable and inconsistent results (step 22).
-
•
Duration of washes was insufficient. Increase length of time for each wash or add an additional wash step (steps 16–26).
-
•
HCR probe set was not previously validated in wild-type whole mount tissue. Prior validation is a crucial step to ensure reproducibility in the WICHCR protocol. Validate HCR probe set efficiency by using wild-type embryos/larvae at similar stages to those planed for future experiments to determine both the efficacy of the probe set as well as the regions in which the gene of interest is expressed.
Problem 3
No antibody fluorescence signal is detected in the samples (step 35)
Potential solution
-
•
Validate correct antibody detection by using wild type-embryos/larvae at similar stages to those planed for future experiments to determine both the efficacy of the antibody, as well as the tissues in which the gene of interest is expressed.
-
•
Increase primary antibody concentration and increase incubation time. Additionally, secondary antibody concentration can be increased, however secondary antibody incubation time should remain the same (steps 30 and 32).
-
•
Use a positive control antibody for confirm correct antibody detection at the developmental stage to be analyzed (step 30).
-
•
Sample might be poorly permeabilized. Use a fresh PK stock when permeabilizing embryos (step 11). Additionally, skipping the acetone permeabilization will also cause some antibodies to fail (step 9).
-
•
Investigate the similarity of the immunogen used to validate the antibody to that of the zebrafish target. Immunogens from other species with highly dissimilar sequences may not be suitable for use in this protocol. In this case we recommend finding an alternative antibody provider to test or having an antibody generated for your specific application.
Problem 4
High background from antibody label (steps 35 and 37)
Potential solution
-
•
This could be because of non-specific binding of the antibody, increase blocking time or concentration of the blocking solution (step 28).
-
•
Washes were not sufficient, increase the number of washes to remove non-specific staining (steps 31 and 34).
-
•
Use a negative control where, using the same conditions as the experimental samples, you incubate the samples with only secondary antibody to rule out any unspecific binding of the secondary antibody (step 33).
Problem 5
Poor segregation of fluorescent signals after image acquisition (step 38)
Potential solution
-
•
Fluorophores used present overlapping emission spectra. Address this issue by performing spectral deconvolution of the fluorescent signals or selecting different non-overlapping fluorophores for detection of targets (steps 22–24, 32, 33, and 38).
-
•
Multiplexed targets are indistinguishable after spectral deconvolution. We recommend first using alternative fluorophores to label the problematic targets. If this is not possible, ensure that unique fluorophores are used to label each target (steps 22–24, 32, and 33).
-
•
Microscope settings for imaging of spectrally overlapping fluorophores are suboptimal. Utilizing wild-type tissue with previously validated labeling methods or fluorescent beads, optimize microscopic settings to distinguish each fluorophore. The most common changes in imaging parameters to address this problem are increasing the wavelength over which each fluorophore spectrum is acquired, decreasing step sizes between individual wavelength scans of the tissue, and optimizing settings to decrease photobleaching of the fluorophores. We strongly encourage that every user annotate the precise emission and excitation spectra for each fluorophore as they optimize their settings using FPbase.org (Lambert, 2019) (step 37).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Rosa A. Uribe (rosa.uribe@rice.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
Funding was provided by the Cancer Prevention and Research Institute of Texas (CPRIT) Recruitment of First-Time Tenure Track Faculty Members (CPRIT-RR170062) and the NSF CAREER Award (1942019). Imaris image analysis was performed using Rice University’s Shared Equipment Authority (SEA) IMARIS workstation. The authors would like to thank Dr. Dan S. Wagner for discussions and his shared laboratory and animal resources. We also would like to thank Dr. Eric Bridenbaugh from Olympus for his deep technical insight, training, and continued help in optimizing our multispectral imaging workflow. Lastly, we thank Tom Chaplin and Bleta Rexha for technical assistance.
Author contributions
Conceptualization, R.I.-G.-P., A.G.A.H., and R.A.U.; formal analysis, R.I.-G.-P. and A.G.A.H.; validation, R.I.-G.-P., A.G.A.H., and E.W.S.; investigation, R.I.-G.-P., A.G.A.H., and E.W.S.; visualization, R.I.-G.-P., A.G.A.H., and R.A.U.; methodology, R.I.-G.-P. and A.G.A.H.; writing – original draft, R.I.-G.-P., A.G.A.H., and R.A.U.; writing – reviewing and editing, E.W.S. and R.A.U.; resources, R.A.U.; supervision, R.A.U.; funding acquisition, R.A.U.; project administration, R.A.U.
Declaration of interests
The authors declare no competing interests.
References
- Carney T.J., Dutton K.A., Greenhill E., Delfino-Machin M., Dufourcq P., Blader P., Kelsh R.N. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;113:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Choi H.M.T., Chang J.Y., Trinh L.A., Padilla J.E., Fraser S.E., Pierce N.A. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.M.T., Calvert C.R., Husain N., Huss D., Barsi J.C., Deverman B.E., Hunter R.C., Kato M., Lee S.M., Abelin A.C.T. Mapping a multiplexed zoo of mRNA expression. Development. 2016;143:3632–3637. doi: 10.1242/dev.140137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.M.T., Schwarzkopf M., Fornace M.E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N.A. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 2018;145:dev165753. doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S., Pinto J.P., Pettifer A., Cancela M.L., Kelsh R.N. Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mech. Dev. 2005;122:659–669. doi: 10.1016/j.mod.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Howard A.G.A., Baker P.A., Ibarra-García-Padilla R., Moore J.A., Rivas L.J., Tallman J.J., Singleton E.W., Westheimer J.L., Corteguera J.A., Uribe R.A. An atlas of neural crest lineages along the posterior developing zebrafish at single-cell resolution. Elife. 2021;10:e60005. doi: 10.7554/eLife.60005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J., Von Hofsten J., Olsson P.E. Generating transparent zebrafish: A refined method to improve detection of gene expression during embryonic development. Mar. Biotechnol. 2001;3:522–527. doi: 10.1007/s1012601-0053-4. [DOI] [PubMed] [Google Scholar]
- Kim C.H., Ueshima E., Muraoka O., Tanaka H., Yeo S.Y., Huh T.L., Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Kim J.-Y., Jeong H.S., Chung T., Kim M., Lee J.H., Jung W.H., Koo J.S. The value of phosphohistone H3 as a proliferation marker for evaluating invasive breast cancers: A comparative study with Ki67. Oncotarget. 2017;8:65064–65076. doi: 10.18632/oncotarget.17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert T.J. FPbase: a community-editable fluorescent protein database. Nat. Methods. 2019;16:277–278. doi: 10.1038/s41592-019-0352-8. [DOI] [PubMed] [Google Scholar]
- Olden T., Akhtar T., Beckman S.A., Wallace K.N. Differentiation of the zebrafish enteric nervous system and intestinal smooth muscle. Genesis. 2008;46:484–498. doi: 10.1002/dvg.20429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.