Abstract
Spinal muscular atrophy (SMA) is a leading genetic cause of infant death worldwide that is characterized by loss of spinal motor neurons leading to muscle weakness and atrophy. SMA results from the loss of survival motor neuron 1 (SMN1) gene but retention of its paralog SMN2. The copy numbers of SMN1 and SMN2 are variable within the human population with SMN2 copy number inversely correlating with SMA severity. Current therapeutic options for SMA focus on increasing SMN2 expression and alternative splicing so as to increase the amount of SMN protein. Recent work has demonstrated that not all SMN2, or SMN1, genes are equivalent and there is a high degree of genomic heterogeneity with respect to the SMN genes. Because SMA is now an actionable disease with SMN2 being the primary target, it is imperative to have a comprehensive understanding of this genomic heterogeneity with respect to hybrid SMN1–SMN2 genes generated by gene conversion events as well as partial deletions of the SMN genes. This review will describe this genetic heterogeneity in SMA and its impact on disease phenotype as well as therapeutic efficacy.
Keywords: spinal muscular atrophy, copy number variation, SMN1, SMN2, modifier gene, precision medicine, therapeutics, gene conversion, hybrid gene
1. Introduction
Proximal spinal muscular atrophy (SMA) is a leading genetic cause of infant death worldwide. SMA is an early-onset disease that is characterized by the loss of α-motor neurons in the anterior horn of the spinal cord, i.e., lower motor neurons [1,2]. The incidence of SMA is 1 in 6000–10,000 live births [3,4,5]. SMA has a carrier frequency of 1:25–50 in most populations [5,6,7,8], although it is lower for some ethnic groups [9,10,11,12]. SMA results from the loss of α-motor neurons in the ventral spinal cord, leading to denervation and muscle weakness, with proximally innervated muscles being preferentially targeted. Following onset of symptoms, the denervation is progressive over time, as shown in SMA patients using motor unit number estimation (MUNE) and maximum compound muscle action potential amplitude (CMAP) analysis [13].
Based on age of onset and severity of the disease, SMA can be classified into five distinct phenotypes [14,15]. Type 0 SMA infants present with very severe hypotonia and require respiratory support from birth. These SMA infants usually do not survive beyond 6 months. Type I SMA (Online Inheritance in Man (OMIM) database #253300) patients have an age of onset before 6 months and they present with limb weakness due to hypotonia and the inability to sit independently. Abnormal respiratory patterns have been observed in type I SMA infants due to weakness in the intercostal muscles but not the diaphragm. These patients typically have shortened lifespans. Type II SMA (OMIM #253500) patients have an age of onset before 18 months. They are poor crawlers and weak sitters; most of these patients can rarely stand and only with support. Their legs are generally weaker than their arms. These patients generally have a life expectancy into adulthood due to improvements in the standards of care. Type III SMA (OMIM #253400) patients have an age of onset greater than 18 months. These patients are able to walk with difficulty (waddling gait) and the legs are weaker than the arms. Type III SMA individuals generally live for a normal lifespan but some of them may require mobility support as the disease progress. Adult-onset (type IV) SMA (OMIM #271150) patients typically exhibit a slowly progressive limb weakness but the disease course is fairly benign.
While spinal motor neurons are the primary cell type affected in SMA, other types of cells aside from the motor neurons may also be affected by SMA [16,17]. For example, there are immature myoblasts present within muscles of SMA patients [18] and type I SMA patients tend to have smaller myotubes [19]. In addition to motor neuron degeneration, axonal degeneration of sensory neurons has also been observed in patients with severe SMA but not in milder forms of the disease [20,21]. Imaging and electrophysiology studies have shown degeneration of the thalamic nuclei within the cerebrum of type I SMA patients [22,23]. Type I SMA patients also manifest cardiac abnormalities including bradycardia and septal defects [24], while heart abnormalities are not observed in milder, types II and III SMA patients [25]. Distal digital necrosis of the blood vesssels occurs in type I SMA patients [24,26]. Type I SMA patients also show an abnormal increase in pancreatic islet α cells, leading to abnormal glucose levels in some patients [27]. Other metabolic manifestations observed in SMA include abnormalities in fatty acid metabolism in SMA patients [28,29,30] and elevated serum leptin levels [31]. The multisystem nature of SMA tends to be more clinically prominent in more severe forms of the disease. It is unclear at present if these systemic clinical manifestations are a consequence of motor neuron dysfunction but it is important, nevertheless, to consider these systemic clinical manifestations in the care for and treatment of SMA patients.
2. Genetics and Biology of SMA
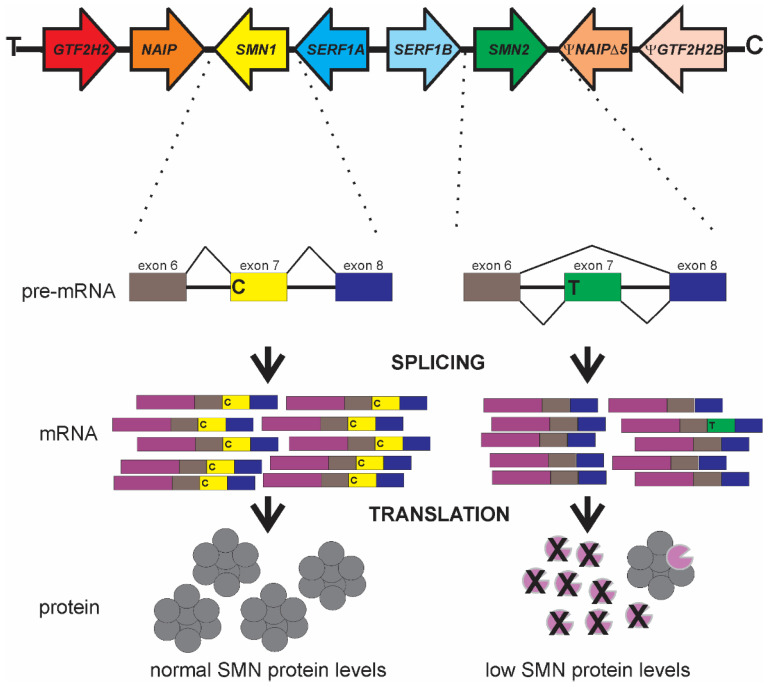
The SMA gene locus [32] maps to the 5q13 region of chromosome 5 (reviewed in [33]). Within this region there lies a 500 kilobase (kb) inverted segmental duplication that is unique to human lineages [34,35,36]. There are four genes within this segmental duplication region (Figure 1): SMN (survival motor neuron; [37]), NAIP (neuronal apoptosis inhibitor protein; [38]), GTF2H2A (general transcription factor IIH, p44; [39,40]) and SERF1A (small EDRK-rich factor 1A, H4F5A; [41]). These duplicated genes are either identical to their partner gene (SERF1B), differ by a small number of nucleotides but still produce functional genes (SMN2) or are pseudogenes (ΨGTF2H2B and ΨNAIPΔ5).
Figure 1.
Genomic organization of the SMA-associated segmental duplication at chromosome 5q13 and the functional differences between SMN1 and SMN2 with respect to SMN gene regulation. Adapted from [42,43].
In more than 95% of cases, proximal SMA results from the loss of SMN1 but retention of SMN2, regardless of clinical severity [37]. In more severe types of SMA, other genes within this region of segmental duplication (such as NAIP, GTF2H2A and SERF1A) may also be lost, but not always [39,40,44,45,46,47]. Intragenic mutations in SMN1 [37,48] account for the remaining 5% of SMA cases (see Section 8) providing additional evidence to support SMN1 as the gene responsible for SMA. In SMA patient-derived cell lines as well as in patient tissues, SMN protein levels are inversely correlated with disease severity [49,50,51,52,53,54,55,56]. SMN is a ubiquitously expressed protein that is required for the assembly of diverse ribonucleoprotein (RNP) complexes, including small nuclear RNPs (snRNPs) required for spliceosome assembly and messenger RNPs (mRNPs) needed for transport of mRNAs along axons [48,57,58]. While it is well established that SMN is required for RNP assembly, it still remains to be resolved which types of RNPs are affected in SMA and how motor neurons are preferentially affected.
The major functional difference between these two SMN genes is a C-to-T transition in exon 7 (SMN2 c.850C>T) [59,60]. While translationally silent, this position on exon 7 is in the middle of an exonic splicing enhancer (ESE) sequence important for the inclusion of exon 7 in SMN transcripts (Figure 1). This ESE is disrupted in SMN2, thereby causing the exclusion of exon 7 (SMNΔ7) from the majority (~90%) of SMN2-derived mRNAs. The resultant SMNΔ7 protein is unstable and is unable to associate with itself [61,62,63]. Some SMN2 mRNAs contain exon 7, depending on cell type, and can produce some full-length, functional SMN proteins. The SMNΔ7 protein is still partially functional since transgenic overexpression of SMNΔ7 in severe SMA mice partially ameliorates their phenotype [64].
3. SMN2 as a Disease Modifier for SMA
The number of SMN2 copies in the human genome varies between 0 and 8. Numerous studies have demonstrated an inverse relationship between SMN2 copy number and disease severity in SMA [13,37,49,50,54,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94]. Patients with milder forms of SMA have higher SMN2 copy numbers than severe SMA patients. SMN2 copy number is being used as a prognostic tool to guide therapeutic strategies and care plans for SMA patients across the spectrum of phenotype severity [95,96]. The variability in SMN2 copy number within the SMA patient population and its relationship to disease severity makes it an ideal target for therapeutics development.
Animal models such as zebrafish, fruit flies and mice have a single Smn gene which is orthologous to SMN1 [97,98]. Loss of Smn in mice (mSmn) leads to embryonic lethality or cell type-specific death, if using conditional gene knockout approaches [99,100,101,102]. Transgenic insertion of SMN2 rescues the embryonic lethality observed in mSmn nullizygous mice [103,104,105]. While two copies of SMN2 rescues embryonic lethality in mSmn-deficient mice, these mice develop a very severe SMA phenotype and die within 8 days after birth [103,104]. Those mSmn-deficient mice with 3–4 SMN2 copies exhibit a milder SMA phenotype than the two-copy SMN2 SMA mice [104,105]. If the SMN2 copy number is high (i.e., eight), then the resultant mSmn-deficient mice exhibit no signs of SMA and are phenotypically normal [103]. Introduction of SMN2 onto a Smn nullizygous background in zebrafish also rescues embryonic lethality in this animal model [106,107]. SMN2 CNV, therefore, is a major modifier of disease severity in SMA.
4. Measurement of SMN1 and SMN2 CNV
Because SMN2 copy number influences disease severity in SMA, there is prognostic value in accurate measurement of SMN2 copy number from patients being evaluated for SMA. Molecular diagnosis of SMA—i.e., loss of SMN1—has historically been made using a polymerase chain reaction (PCR)-based assay followed by digestion of the PCR product with specific restriction endonucleases (PCR-RFLP) [37,75]. Different types of genotyping assays—including radioactive PCR [49,65], fluorescent PCR [79], quantitative (real-time) PCR (qPCR) [76,77,78], competitive PCR/primer extension [80], denaturing high-performance liquid chromatography [81], multiplex ligation-dependent probe amplification (MLPA) [82,83,84,85,86,108], quantitative capillary electrophoresis fragment analysis [87], short-amplicon melt profiling [88], fluorescent multiplex PCR/capillary electrophoresis [89,90] and universal fluorescent triprobe ligation [91]—have since been developed to quantify SMN2 copy number in DNA samples from SMA patients. An important limitation of these established PCR-based copy number assays is the requirement for a parallel-run calibration curve to assign a necessary breakpoint that identifies placement of an ordinal SMN2 value. Additionally, these techniques cannot easily distinguish unit differences in SMN1 or SMN2 when the copy number is greater than three [78,85,109]; however, recent refinements to MLPA assays can accurately measure four or five copies of SMN1 or SMN2 [110]. Digital PCR (dPCR) can accurately measure SMN1 and SMN2 over a large range of unit copies (0–6) without the need for an external calibration curve [70,93,111,112,113,114,115]. Next-generation sequencing approaches have recently been shown to be useful for SMA carrier detection [116,117,118,119,120] as well as for SMN2 copy number measurements [120].
5. SMN1 to SMN2 Gene Conversions and Partial Deletions
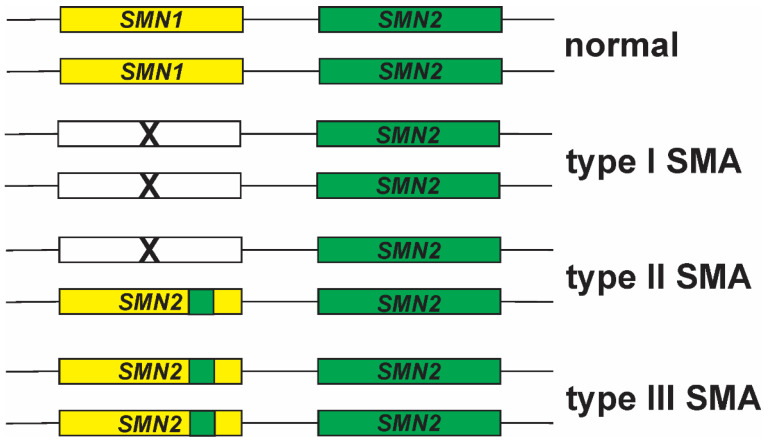
Gene conversion is one mechanism to account for increased SMN2 copy number in the absence of SMN1 in SMA [121]. In this scenario, the SMN1 gene actually contains part of SMN2, in particular within exon 7 [46,122,123,124,125,126]. Gene conversion events between SMN1 and SMN2 have been observed by multiple groups using different approaches [89,93,122,123,124,125,127,128,129,130,131,132,133]. Gene conversion events may account for the inverse relationship between SMN2 copy number and disease severity in SMA (Figure 2). Deletion of SMN1 on both chromosomes is hypothesized to cause the more severe type I SMA. Milder forms of SMA result from conversion of SMN1 to SMN2 on one or both chromosomes (reviewed in [121]). Gene conversion events lead to the generation of hybrid SMN genes, i.e. some portions are SMN1 while other sections of the gene are SMN2.
Figure 2.
Relationship between SMN1–SMN2 gene conversion and disease severity in SMA. Adapted from [134].
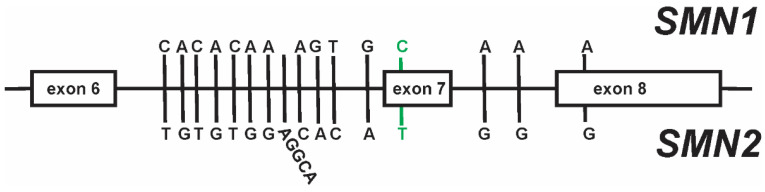
Most gene conversion events occur at the canonical c.840C>T nucleotide difference at exon 7 [59,60]. There are, however, at least 15 other paralogous structural variants (PSVs) between SMN1 and SMN2 (Figure 3; [60,108,120,135,136]). Gene conversion events at exon 8 (SMN2c.1155G>A) as well as those within intron 6 (SMN2c.835-44G>A) and intron 7 (SMN2c.888+100A>G and SMN2c.888+215A>G) have been observed in SMA, as well as in control populations [127,129,133]. Some of these PSVs, such as c.835-44G>A and c.888+100A>G, can affect exon 7 inclusion in spliced SMN mRNAs [137,138]. Some hybrid SMN2 genes produce greater amounts of SMN protein than expected and SMA patients harboring these hybrid genes have milder than expected clinical phenotypes. Further characterization of these gene conversion events will aid in the understanding of the functional consequence of these hybrid genes on SMN expression.
Figure 3.
Paralogous sequence variants (PSVs) between SMN1 and SMN2. The canonical PSV at exon 7 that functionally distinguishes SMN1 from SMN2 is highlighted in green.
Even though the relationship between SMN2 CN and disease severity is strong, there are exceptions to this inverse relationship. Some SMA patients displaying a type II or III clinical presentation only have two copies of SMN2 as opposed to the predicted SMN2 copy number for milder forms of SMA [139,140,141]. SMN2 sequencing identified the presence of a rare single-nucleotide variant (SMN2 c.859G>C) in exon 7 [139,140,141]. This variant regulates the splicing of SMN2 pre-mRNAs so that a greater proportion of SMN2 transcripts contain exon 7.
While most cases of SMA result from a complete loss of SMN1, partial deletions in SMN1 have been identified in some samples from SMA patients—as well as in healthy controls—using PCR [142,143], microsatellite analysis [85,143], MLPA [83,85,108,110,127,142], whole-genome sequencing [120,135], long-range PCR [129] and dPCR [93]. Additionally, partial deletions have been observed in SMN2. The most common partial deletion of SMN1 or SMN2 encompasses exons 7 and 8 (SMN1/2Δ78) and is roughly 6.3 kb in length [120,135]. This partial deletion spans 6.3 kb of DNA, although it is possible that the sizes of these SMN1/2Δ78 partial deletions may be variable. Whole-genome sequencing revealed the presence of a 1.9 kb partial deletion in a single sample that spans exon 7 and part of the flanking intronic regions [120]. In addition to SMN1/2Δ78, partial deletions within the SMN genes have been observed in other regions of the SMN genes, including losses of exons 5 and 6 (SMN1Δ56) [144], of exons 2a through 5 (SMN1Δ2a5) [145] and of exons 1 through 6 (SMNΔ16) [85,143]. We and others [93,142] have detected partial deletions of exon 8 in SMN1 (SMN1Δ8). Even though this exon is downstream of the protein-encoding region of SMN1 mRNA, it may affect SMN1 mRNA stability as well as post-transcriptional gene regulation.
The reason for specific breakpoints to be favored in partial deletion of SMN1 is currently not known. There are numerous intrachromosomal repeats within human chromosome 5 [34], including within the SMA gene locus. Alu repeat elements are primate-specific, 300-base segments of repetitive DNA found throughout the human genome [146]. Within the SMN genes, there are numerous Alu repeat elements of different types [147]. Some of these Alu repeat elements may cause partial deletions of SMN1 (or SMN2) by nonallelic homologous recombination [148]. The most common partial deletion (6.3 kb) of SMN1/2, SMN1/2Δ78, is flanked by Alu repeat elements [135]. Other partial deletions of SMN1—such as SMN1Δ56 [144], SMN1Δ2a5 [145] and SMN1Δ16 [85,143]—are also flanked by Alu repeat elements. Alu/Alu-mediated rearrangements, therefore, may account for these partial deletions within SMN1.
6. SMN2 Copy Number and Therapeutic Efficacy
The Food and Drug Administration has approved three therapeutic agents for SMA patients: nusinersen (SpinrazaTM, Ionis Pharmaceuticals (Carlsbad, CA, USA) and Biogen (Cambridge, MA, USA) [149,150]), onasemnogene abeparvovec (ZolgensmaTM, AveXis (Bannockburn, IL, USA) and Novartis (Basel, Switzerland) [151]) and risdiplam (EvrysdiTM, Genentech (South San Francisco, CA, USA) and Roche (Basel, Switzerland) [152]). Nusinersin and risdiplam act by increasing exon 7 inclusion in SMN2 transcripts while onasemnogene abeparvovec replaces full-length SMN mRNA and protein. Since there is a strong relationship between SMN2 copy number and disease severity, accurate and rapid measurements of SMN2 copy number are often used to identify treatment options and regimens for children with SMA [96,153,154]. Accurate and rapid measurement of SMN2 CN is particularly essential to guide decisions around timing and treatment choice for SMA infants identified by newborn screening. The impact of SMN hybrid genes and partial deletions on the responsiveness of these therapeutics has not yet been determined, but these atypical SMN genes are predicted to effect therapeutic efficacy, especially for nusinersin and risdiplam, as they are dependent on endogenous SMN2.
7. Intragenic Mutations in SMN1 and SMA
As mentioned earlier, approximately 5% of all cases of SMA linked to 5q13 result from intragenic mutations within SMN1 as opposed to the loss of SMN1. Table 1 provides a list of the currently known SMA-associated intragenic mutations in SMN1. The SMA-associated intragenic mutations located within the exons can be either missense, nonsense or frameshift mutations. Additionally, there are intragenic mutations within the intronic regions of SMN1, which can cause aberrant splicing of SMN1 pre-mRNAs.
Table 1.
Intragenic mutations in SMN1 that have been identified in SMA patients. The nucleotide position for the mutation starts relative to the initiation codon for DNA (NM_022874.2) or amino acid for protein (NP_075012.1).
| Type of Mutation | Mutation | Phenotype | Reference(s) |
|---|---|---|---|
| Nonsense mutations | p.E14X | I | [127,155] |
| p.Q15X | I, II, III | [127,144,155,156] | |
| p.Q27X | I | [157] | |
| p.S63X | I | [157] | |
| p.W102X | II, III | [158,159] | |
| p.Q154X | III | [160] | |
| p.Q157X | II | [160,161] | |
| p.W190X | I | [162] | |
| p.L228X | I | [127,155,163,164] | |
| p.Q282X | I | [165] | |
| p.R288X | II | [166] | |
| Frameshift mutations | c.-7-9del | III | [127] |
| c.19delG | I | [164] | |
| c.22dupA | I, II, III | [127,155,163,164,167,168] | |
| c.48_55dupGGATTCCG | I | [87] | |
| c.56delT | II | [127,155,168] | |
| c.81+1dupG | II | [169,170] | |
| c.90_91insT | I, II | [144,162] | |
| c.98delT | I | [170] | |
| c.100delT | N/A | [171] | |
| c.109dupA | N/A | [158] | |
| c.124insT | I | [144] | |
| c.198_214del | N/A | [172] | |
| c.208_209ins4 | III | [144] | |
| c.241-242in4 | III | [144] | |
| c.286delG | I | [166] | |
| c.312dupA | III | [162] | |
| c.314_317dup | III | [172] | |
| c.401_402delAG | I | [164] | |
| c.411delT | I | [162] | |
| c.429_435del | I | [171] | |
| c.430del4 | I, II, III | [173] | |
| c.431delC | I | [171] | |
| c.439_443del | I | [159,174] | |
| c.472del5 | I | [174] | |
| c.509_510delGT | N/A | [158] | |
| c.524delC | N/A | [175] | |
| c.542delGT | II | [176,177] | |
| c.549delC | N/A | [178] | |
| c.551_552insA | I | [164] | |
| c.585dupT | I | [172] | |
| c.591delA | II | [144] | |
| c.627_628ins65 | I | [161] | |
| c.722delC | I | [171] | |
| c.734_735insC | I | [160,175] | |
| c.735_736insA | N/A | [178] | |
| c.740dupC | N/A | [179] | |
| c.744delC | I | [127] | |
| c.770-780dup11 | I | [158,160,162,172,175] | |
| c.773insC | III | [179] | |
| c.811_814dupGGCT | II | [127] | |
| c.813ins/dup11 | I, II | [176,179,180] | |
| c.819_820insT | I | [157,181,182] | |
| Missense mutations | p.A2G | II, III | [127,155,158,160,176] |
| p.A2V | III | [129,157,182] | |
| p.D30N | II | [156] | |
| p.D44V | III | [156] | |
| p.W92S | I | [157,181,182,183] | |
| p.V94F | I | [184] | |
| p.V94G | II | [172] | |
| p.G95R | III | [156,178] | |
| p.Y109C | III | [127] | |
| p.A111G | I | [156] | |
| p.I116F | I | [160,175,185] | |
| p.Y130C | III | [158,186] | |
| p.Y130H | III | [186] | |
| p.E134K | I, II | [127,156,168,187] | |
| p.Q136E | I | [185] | |
| p.S139S | N/A | [158,159] | |
| p.L141P | I | [157] | |
| p.D181G | N/A | [188] | |
| p.A188S | I | [170] | |
| p.P221L | I | [87] | |
| p.S230L | II, III | [127,165,168,171] | |
| p.P244L | III | [171] | |
| p.P245L | III | [189] | |
| p.L260S | II | [172] | |
| p.S262G | III | [156] | |
| p.S262I | III | [65,158,190] | |
| p.M263R | I | [172] | |
| p.M263T | II | [162] | |
| p.S266P | II | [158] | |
| p.M269T | III | [160] | |
| p.Y272C | I, II, III | [144,156,162,164,170,172,189,191] | |
| p.H273R | II | [158] | |
| p.T274I | II, III | [71,144,170,190] | |
| p.G275S | III | [172] | |
| p.Y276H | I | [157,192] | |
| p.Y277C | II, III | [127,129,168,182] | |
| p.G279C | II, III | [178,193] | |
| p.G279V | I | [194] | |
| p.G279D | N/A | [143] | |
| p.F280I | N/A | [178] | |
| p.R288M | I, II | [168,195,196] | |
| Splice-site mutations | c.*3+3A>T | I | [184] |
| c.628-140A>G | N/A | [161] | |
| c.834+2T>G | I | [162] | |
| c.835-1G>A | III | [197] | |
| c.835-2A>G | I | [157,198] | |
| c.835-3C>A | I | [157] | |
| c.835-5T>G | I | [127] | |
| c.867+2T>G | I | [179] | |
| c.868-11del7 | I | [37] | |
| c.888+3delAGAG | I | [37,172] | |
| c.922+3del4 | I | [97] | |
| c.922+6T>G | III | [144] |
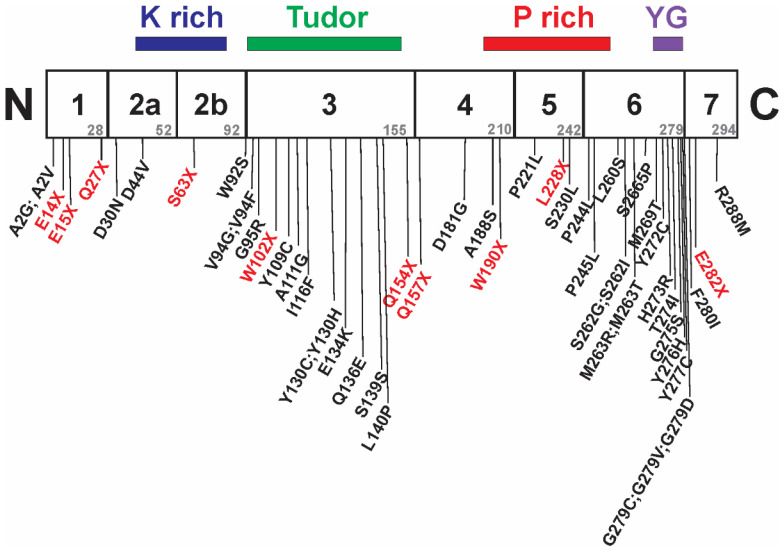
SMN is a highly conserved protein containing 294 amino acids (in humans) with multiple domains (Figure 4). There are three regions within SMN1—located within exons 2A (K rich domain), 3 (tudor domain) and 6 (YG box)—that are highly conserved evolutionarily [58]. Even though SMA mutations have been linked throughout SMN1, a greater proportion of SMA-associated intragenic point mutations are localized within these evolutionarily conserved regions (Figure 4). These conserved regions are required for self-oligomerization (YG box) as well as interactions with Sm proteins (tudor domain) and gemin-2 (K rich domain).
Figure 4.
Location of SMA-associated intragenic missense and nonsense mutations within SMN1 relative to protein domain location. The mutated residues in red are nonsense mutations while those in black are missense mutations. Adapted from [48].
As mentioned previously, the loss of Smn in animal models is embryonically lethal and SMN2 rescues this embryonic lethality but results in an SMA phenotype whose severity depends on SMN2 copy number. Transgenic introduction of SMA intragenic missense mutations—specifically SMN1(A2G) [199], SMN1(A111G) [200], SMN1(D44V) [201], SMN1(T74I) [201] and SMN1(Q282A) [201]—into severe SMA mice (two copies of SMN2 on an mSmn zulligygous background) improves the motor phenotype of severe SMA mice but does not completely ameliorate the SMA phenotype in these mice. These observations suggest that SMN genes harboring these point mutations are partially functional.
On their own, none of these intragenic missense SMN1 mutations can rescue the embryonic lethality of the loss of Smn in mice [199,200,201]. In zebrafish models for SMA where zSmn is knocked down with an antisense morpholino oligonucleotide [202], intragenic SMA missense mutations cannot rescue the motor axon deficits observed in these fish. These observations also support that these patient-derived point mutations are not fully functional. Interestingly, addition of both an N-terminal missense mutation and a C-terminal missense mutation can fully rescue the embryonic lethality caused by the loss of Smn in mice [203]. These intragenic complementation studies demonstrate that SMN must be oligomeric in order to function completely.
8. Silent Carriers and Compound Heterozygotes in SMA
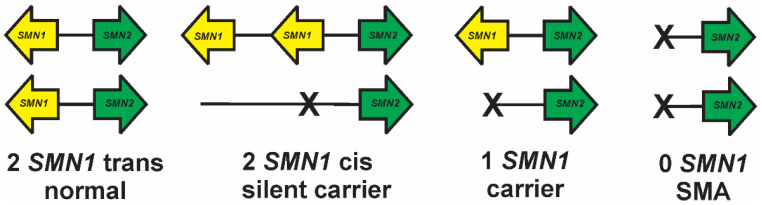
Most parents of children with SMA are both carriers harboring one copy of SMN1. Interestingly, multiple independent studies have identified SMA carriers who have two copies of SMN1 [65,204,205]. In fact, one study identified 4.3% of the SMA carrier parents within their cohort as having two copies of SMN1 [205]. It is hypothesized that these so-called silent carriers have two copies of SMN1 on one allele (i.e. the duplication allele) and zero copies of SMN1 on the other allele (i.e., the deletion allele; Figure 5). In other words, the two copies of SMN1 in a silent carrier have a cis allelic distribution (SMN1:2+0) as opposed to having two alleles each with a single copy of SMN1 (the trans allelic distribution; SMN1:1+1), which would be phenotypically normal.
Figure 5.
Allelic organizations of SMN1 copies within normal (SMN1:1+1), silent carrier (SMN1:2+0), carrier (SMN1:1+0) and SMA (SMN1:0+0) patients.
Luo et al. [206] recently identified two small variants within SMN1—SMN1g.27134T>G (also known as SMN1c.*3+80T>G) and SMN1g.27706_27707delAT (also known as SMN1c.*211_*212del)—that are tightly associated with silent carriers. The allelic frequencies of these variants were higher than expected in certain ethnic populations, such as the Ashkenazi Jewish and African American populations [206]. Another group identified an association between either of these variants and silent carriers in about 20% of their cohort, suggesting that there may be other variants in SMN1 associated with silent carriers [128]. Alternatively, some instances of silent SMA carriers may not be linked with any structural variant within SMN1.
It is essential to develop diagnostic tools which can detect silent SMA carriers, as standard assays cannot readily distinguish SMN1:2+0 cases from SMN1:1+1. To facilitate the development of genotyping assays that can identify silent carriers, the Genetic Testing Reference Materials Coordination Program has identified a set of reference samples containing structural variants linked with silent carriers [207]. Recently, targeted next-generation and whole-genome sequencing approaches have identified silent carriers using the SMN1g.27134T>G single nucleotide polymorphism [117,120,208]. A quantitative PCR assay has also been developed to identify silent carriers using this variant [209]. It should be noted that most of these approaches will most likely not identify all silent SMA carriers, as not all of them are associated with these polymorphisms. Other approaches, including long-read PCR and sequencing, may provide additional ways to rapidly identify silent carriers.
Compound heterozygosity, wherein the SMA phenotype results from two different types of genetic event on each allele, can help explain discordant phenotypes within the families of SMA patients with differing phenotypic severities. One of the first cases of compound heterozygosity in SMA was identified by detailed analysis of haplotype markers [210]. In most cases, compound heterozygosity results from the deletion of one SMN1 allele and an intragenic mutation within the other allele [133,155,166,179,192]. There have been cases where two different types of intragenic mutations, i.e., a frameshift mutation and a missense mutation, in SMN1 occur on the same allele (cis) [87]. With new advances in molecular diagnostic tools, the genetics of complex cases of SMA can be resolved with relative ease.
9. Intrafamilial Variation in SMA Clinical Presentation
In some SMA families with more than one affected sibling, intrafamilial variability in clinical presentation has been observed [211,212]. In fact, recent analysis of a patient database curated by Cure SMA found 15.2% of SMA siblings to be discordant with respect to phenotypic severity [213]. These siblings have the same SMN2 copy number but have differing clinical presentations [65,214,215,216,217]. This would suggest that there are additional genetic modifiers of SMA disease severity aside from SMN2. It is important to identify and characterize these novel modifiers for the development of new SMA biomarkers and targets for the development of therapeutic strategies for SMA [218].
Plastin-3 (PLS3) was one of the first SMN2-independent modifier genes identified for SMA. On examining the transcriptomes of SMA families with discordant siblings, PLS3 mRNA levels were found to be higher in females with milder SMA than those siblings with a more severe SMA clinical presentation [219,220,221,222,223,224]. The mechanism by which PLS3 expression is altered in these discordant families remains to be resolved. Reduction in the expression of PLS3-interacting proteins coronin 1C (CORO1C) [225] and calcineurin-like EF-hand protein 1 (CHP1) [226] improves neurite outgrowth in motor neurons from SMA model systems. A link between CORO1C and CHP1 levels and disease severity, however, has yet to be shown in SMA patients. In some families, female siblings with a more severe SMA phenotype had higher PLS3 mRNA levels than their more mildly affected siblings, suggesting that the protective effects of PLS3 on SMA patients may be age- and sex-dependent or incompletely penetrant [222]. In the Smn2B/− mouse model for SMA, SMA mice on a C57bl/6J genetic background lived, on average, over 30% longer than those mice on a FVB/N background [227]. Interestingly, Pls3 levels were elevated in SMA mice on a C57bl/6J genetic background when compared against Smn2B/− mice on a FVB/N genetic background. Ectopic overexpression of PLS3 improved the survival and phenotype of SMA mice in some cases, but not all [228,229,230,231].
By using an approach combining linkage analysis with transcriptomics, Neurocalcin-D (NCALD) was found to be a potential modifier gene within a cohort of discordant SMA cases [232]. Targeted sequencing of the NCALD region identified a 17 bp deletion within the promoter region of this gene in these discordant SMA patients, which led to reduced levels of NCALD mRNA and protein.
Whole-exome sequencing of a discordant SMA family, where one sibling presented a milder SMA phenotype than the other sibling, even through they both had two copies of SMN2, identified point mutations in the Tolloid-like 2 (TLL2) gene in the sibling with the milder phenotype [233]. TLL2 acts as an activator of myostatin (MSTN; growth differentiation factor 8), which inhibits skeletal muscle growth. The TLL2 point mutations identified in the milder sibling are predicted to reduce MSTN activation. MSTN inhibitors (such as SRK-015) have shown therapeutic benefit in mouse models for SMA and are currently in clinical trials with SMA patients [234].
Neuritin 1 (NRN1; cpg15) is an SMN-interacting protein present in neurons which promotes neurite outgrowth. Overexpression of NRN1 in various animal models for SMA showed increased motor neurite outgrowth [235]. Yener et al. [223] recently showed elevated NRN1 mRNA levels in a mildly affected sibling within a discordant family. The molecular basis for increased NRN1 expression in this case remains to be resolved.
10. Conclusions
SMA results from the loss of SMN1, but retention of its paralog SMN2 copy number can modulate disease severity in SMA. SMN2 copy number is becoming an inclusion criterion for many clinical trials for SMA. Additionally, SMN2 copy number can be used to help guide the type of care SMA patients will receive. Because of this relationship, SMN2 is a primary target for the development of therapeutics for SMA [236,237]. Numerous targets of SMN2 gene regulation—including promoter activation, increased inclusion of exon 7 and protein stabilization—are currently being developed to increase SMN2 expression. Given the genomic heterogeneity of SMN1 and SMN2, it will become very important to comprehensively assess the these genes in individual SMA patients, as some of them may harbor SMN1–SMN2 hybrid genes or partial SMN1/2 deletions that may affect the therapeutic efficacy of both current and future therapeutics.
Acknowledgments
I would like to thank the members of the Motor Neuron Diseases Research Laboratory, both present and former, for their work and their insights.
Funding
The APC was funded by the Nemours Foundation.
Conflicts of Interest
The author declares no conflict of interest. The funders had no role in the writing of the manuscript or in the decision to publish the review.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Kolb S.J., Kissel J.T. Spinal muscular atrophy. Neurol. Clin. 2015;33:831–846. doi: 10.1016/j.ncl.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuscó I., Barceló M.J., Soler C., Parra J., Baiget M., Tizzano E. Prenatal diagnosis for risk of spinal muscular atrophy. Br. J. Obstet. Gynaecol. 2002;109:1244–1249. doi: 10.1016/S1470-0328(02)02983-X. [DOI] [PubMed] [Google Scholar]
- 4.Pearn J. Incidence, prevalence and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs A.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: Clinical laboratory analysis of > 72400 specimens. Eur. J. Hum. Genet. 2012;20:27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Shachar S., Orr-Urtreger A., Bardugo E., Shomrat R., Yaron Y. Large-scale population screening for spinal muscular atrophy: Clinical implications. Genet. Med. 2011;13:110–114. doi: 10.1097/GIM.0b013e3182017c05. [DOI] [PubMed] [Google Scholar]
- 7.Su Y.N., Hung C.C., Lin S.Y., Chen F.Y., Chern J.P.S., Tsai C., Chang T.S., Yang C.C., Li H., Ho H.N., et al. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005–2009: A prospective population-based cohort study. PLoS ONE. 2011;6:e17067. doi: 10.1371/journal.pone.0017067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyahyai J., Sbiti A., Barkat A., Ratbi I., Sefiani A. Spinal muscular atrophy carrier frequency and estimated prevalence of the disease in Moroccan newborns. Genet. Test. Mol. Biomark. 2012;16:215–218. doi: 10.1089/gtmb.2011.0149. [DOI] [PubMed] [Google Scholar]
- 9.Sangaré M., Hendrickson B., Sango H.A., Chen K., Nofziger J., Amara A., Dutra A., Schindler A.B., Guindo A., Traoré M., et al. Genetics of low spinal muscular atrophy carrier frequency in sub-Saharan Africa. Ann. Neurol. 2014;75:525–532. doi: 10.1002/ana.24114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaldívar T., Montejo Y., Acevedo A.M., Guerra R., Vargas J., Garofalo N., Alvarez R., Alvarez M.A., Hardiman O. Evidence of reduced frequency of spinal muscular atrophy type I in the Cuban population. Neurology. 2005;65:636–638. doi: 10.1212/01.wnl.0000172860.41953.12. [DOI] [PubMed] [Google Scholar]
- 11.Labrum R., Rodda J., Krause A. The molecular basis of spinal muscular atrophy (SMA) in South African black patients. Neuromuscul. Disord. 2007;17:684–692. doi: 10.1016/j.nmd.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson B.C., Donohoe C., Akmaev V.R., Sugarman E.A., Labrousse P., Boguslavskiy L., Flynn K., Rohlfs E.M., Walker A., Allitto B., et al. Differences in SMN1 allele frequencies among ethnic groups within North America. J. Med. Genet. 2009;46:641–644. doi: 10.1136/jmg.2009.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swoboda K.J., Prior T.W., Scott C.B., McNaught T.P., Wride M.C., Reyna S.P., Bromberg M.B. Natural history of denervation in SMA: Relation to age, SMN2 copy number and function. Ann. Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munsat T.L., Davies K.E. International SMA Consortium meeting. Neuromuscul. Disord. 1992;2:423–428. doi: 10.1016/S0960-8966(06)80015-5. [DOI] [PubMed] [Google Scholar]
- 15.Russman B.S. Spinal muscular atrophy: Clinical classification and disease heterogeneity. J. Child Neurol. 2007;22:946–951. doi: 10.1177/0883073807305673. [DOI] [PubMed] [Google Scholar]
- 16.Shababi M., Lorson C.L., Rudnik-Schöneborn S. Spinal muscular atrophy: A motor neuron disorder or a multi-organ disease? J. Anat. 2014;224:15–28. doi: 10.1111/joa.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton G., Gillingwater T.H. Spinal muscular atrophy: Going beyond the motor neuron. Trends Mol. Med. 2012;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Arnold A.S., Gueye M., Guettier-Sigrist S., Courdier-Fruh I., Coupin G., Poindron P., Gies J.P. Reduced expression of nicotinic AChRs in myotubes from spinal muscular atrophy I patients. Lab. Investig. 2004;84:1271–1278. doi: 10.1038/labinvest.3700163. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Hernández R., Soler-Botija C., Also E., Alias L., Caselles L., Gich I., Bernal S., Tizzano E.F. The developmental pattern of myotubes in spinal muscular atrophy indicates prenatal delay of muscle maturation. J. Neuropathol. Exp. Neurol. 2009;68:474–481. doi: 10.1097/NEN.0b013e3181a10ea1. [DOI] [PubMed] [Google Scholar]
- 20.Rudnik-Schöneborn S., Goebel H.H., Schlote W., Molaian S., Omran H., Ketelsen U., Korinthenberg R., Wenzel D., Lauffer H., Kreiβ-Nachtsheim M., et al. Classical infantile spinal muscular atrophy with SMN deficiency causes sensory neuronopathy. Neurology. 2003;60:983–987. doi: 10.1212/01.WNL.0000052788.39340.45. [DOI] [PubMed] [Google Scholar]
- 21.Yonekawa T., Komaki H., Saito Y., Sugai K., Sasaki M. Peripheral nerve abnormalities in pediatric patients with spinal muscular atrophy. Brain Dev. 2013;35:161–171. doi: 10.1016/j.braindev.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Shishikura K., Hara M., Sasaki Y., Misugi K. A neuropathological study of Werdnig-Hoffmann disease with special reference to the thalamus and posterior roots. Acta Neuropathol. 1983;60:99–106. doi: 10.1007/BF00685353. [DOI] [PubMed] [Google Scholar]
- 23.Ito Y., Kumada S., Uchiyama A., Saito K., Osawa M., Yagishita A., Kurata K., Hayashi M. Thalamic lesions in a long-surviving child with spinal muscular atrophy type I: MRI and EEG findings. Brain Dev. 2003;26:53–56. doi: 10.1016/S0387-7604(03)00075-5. [DOI] [PubMed] [Google Scholar]
- 24.Rudnik-Schöneborn S., Vogelgesang S., Armbrust S., Graul-Neumann L., Fusch C., Zerres K. Digital necroses and vascular thrombosis in severe spinal muscular atrophy. Muscle Nerve. 2010;42:144–147. doi: 10.1002/mus.21654. [DOI] [PubMed] [Google Scholar]
- 25.Palladino A., Passamano L., Taglia A., D’Ambrosio P., Scutifero M., Cecio M.R., Picillo E., Viggiano E., Torre V., De Luca F., et al. Cardiac involvement in patients with spinal muscular atrophies. Acta Myol. 2011;30:175–178. doi: 10.1016/j.nmd.2011.06.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araujo A.Q.C., Araujo M., Swoboda K.J. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J. Pediatr. 2009;155:292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowerman M., Swoboda K.J., Michalski J.P., Wang G.S., Reeks C., Beauvais A., Murphy K., Woulfe J., Screaton R.A., Scott F.W., et al. Glucose metabolism and pancreatic defects in spinal muscular atrophy. Ann. Neurol. 2012;72:256–268. doi: 10.1002/ana.23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley R.I., Sladky J.T. Dicarboxylic aciduria in an infant with spinal muscular atrophy. Ann. Neurol. 1986;20:734–736. doi: 10.1002/ana.410200615. [DOI] [PubMed] [Google Scholar]
- 29.Tein I., Sloane A.E., Donner E.J., Lehotay D.C., Millington D.S., Kelley R.I. Fatty acid oxidation abnormalities in childhood-onset spinal muscular atrophy: Primary or secondary defect(s)? Pediatr. Neurol. 1995;12:21–30. doi: 10.1016/0887-8994(94)00100-G. [DOI] [PubMed] [Google Scholar]
- 30.Crawford T.O., Sladky J.T., Hurko O., Besner-Johnston A., Kelley R.I. Abnormal fatty acid metabolism in childhood spinal muscular atrophy. Ann. Neurol. 1999;45:337–343. doi: 10.1002/1531-8249(199903)45:3<337::AID-ANA9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Kölbel H., Hauffa B.P., Wudy S.A., Bouikidis A., Della Marina A., Schara U. Hyperleptinemia in children with autosomal recessive spinal muscular atrophy type I-III. PLoS ONE. 2017;12:e0173144. doi: 10.1371/journal.pone.0173144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandt S. Hereditary factors in infantile progressive muscular atrophy; study of 112 cases in 70 families. Am. J. Dis. Child. 1949;78:226–236. doi: 10.1001/archpedi.1949.02030050237007. [DOI] [PubMed] [Google Scholar]
- 33.Morrison K.E. Advances in SMA research: Review of gene deletions. Neuromuscul. Disord. 1996;6:397–408. doi: 10.1016/S0960-8966(96)00368-9. [DOI] [PubMed] [Google Scholar]
- 34.Schmutz J., Martin J., Terry A., Couronne O., Grimwood J., Lowry S., Gordon L.A., Scott D., Xie G., Huang W., et al. The DNA sequence and comparative analysis of human chromosome 5. Nature. 2004;431:268–274. doi: 10.1038/nature02919. [DOI] [PubMed] [Google Scholar]
- 35.Courseaux A., Richard F., Grosgeorge J., Ortola C., Viale A., Turc-Carel C., Dutrillaux B., Gaudray P., Nahon J.L. Segmental duplications in euchromatic regions of human chromosome 5: A source of evolutionary instability and transcriptional innovation. Genome Res. 2003;13:369–381. doi: 10.1101/gr.490303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochette C.F., Gilbert N., Simard L.R. SMN gene duplication and emergence of the SMN2 gene occured in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- 37.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 38.Roy N., Mahadevan M.S., McLean M., Shutler G., Yaraghi Z., Farahani R., Baird S., Besner-Johnston A., Lefebvre C., Kang X., et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 39.Carter T.A., Bönnemann C.G., Wang C.H., Obici S., Parano E., De Fatima Bonaldo M., Ross B.A., Penchaszadeh G.K., MacKenzie A., Soares M.B., et al. A multicopy transcription-repair gene, BTF2p44, maps to the SMA region and demonstrates SMA associated deletions. Hum. Mol. Genet. 1997;6:229–236. doi: 10.1093/hmg/6.2.229. [DOI] [PubMed] [Google Scholar]
- 40.Bürglen L., Seroz T., Miniou P., Lefebvre S., Burlet P., Munnich A., Pequignot E.V., Egly J.M., Melki J. The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig-Hoffmann disease. Am. J. Hum. Genet. 1997;60:72–79. [PMC free article] [PubMed] [Google Scholar]
- 41.Scharf J.M., Endrizzi M.G., Wetter A., Huang S., Thompson T.G., Zerres K., Dietrich W.F., Wirth B., Kunkel L.M. Identification of a candidate modifying gene for spinal muscular atrophy by comparative genomics. Nat. Genet. 1998;20:83–86. doi: 10.1038/1753. [DOI] [PubMed] [Google Scholar]
- 42.Butchbach M.E.R., Burghes A.H.M. Perspectives on models of spinal muscular atrophy for drug discovery. Drug Discov. Today Dis. Models. 2004;1:151–156. doi: 10.1016/j.ddmod.2004.07.001. [DOI] [Google Scholar]
- 43.Butchbach M.E.R. Copy number variations in the Survival Motor Neuron genes: Implications for spinal muscular atrophy and other neurodegenerative diseases. Front. Mol. Biosci. 2016;3:7. doi: 10.3389/fmolb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrigues N.R., Owen N., Talbot K., Patel S., Muntoni F., Ignatius J., Dubowitz V., Davies K.E. Gene deletions in spinal muscular atrophy. J. Med. Genet. 1996;33:93–96. doi: 10.1136/jmg.33.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burlet P., Bürglen L., Clermont O., Lefebvre S., Viollet L., Munnich A., Melki J. Large scale deletions of the 5q13 region are specific to Werdnig-Hoffmann disease. J. Med. Genet. 1996;33:281–283. doi: 10.1136/jmg.33.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wirth B., Hahnen E., Morgan K., DiDonato C.J., Dadze A., Rudnik-Schöneborn S., Simard L.R., Zerres K., Burghes A.H.M. Allelic association and deletions in autosomal recessive proximal spinal muscular atrophy: Association of marker genotype with disease severity and candidate cDNAs. Hum. Mol. Genet. 1995;4:1273–1284. doi: 10.1093/hmg/4.8.1273. [DOI] [PubMed] [Google Scholar]
- 47.Velasco E., Valero C., Valero A., Moreno F., Hernández-Chico C. Molecular analysis of the SMN and NAIP genes in Spanish spinal muscular atrophy (SMA) families and correlation between number of copies of c BCD541 and SMA phenotype. Hum. Mol. Genet. 1996;5:257–263. doi: 10.1093/hmg/5.2.257. [DOI] [PubMed] [Google Scholar]
- 48.Burghes A.H.M., Beattie C.E. Spinal muscular atrophy: Why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H.M. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 50.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 51.Kolb S.J., Gubitz A.K., Olszewski R.F., Jr., Ottinger E., Sumner C.J., Fischbeck K.H., Dreyfuss G. A novel cell immunoassay to measure survival of motor neurons protein in blood cells. BMC Neurol. 2006;6:6. doi: 10.1186/1471-2377-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simard L.R., Bélanger M.C., Morissette S., Wride M., Prior T.W., Swoboda K.J. Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA. Neurology. 2007;68:451–456. doi: 10.1212/01.wnl.0000252934.70676.ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sumner C.J., Kolb S.J., Harmison G.G., Jeffries N.O., Schadt K., Finkel R.S., Dreyfuss G., Fischbeck K.H. SMN mRNA and protein levels in peripheral blood. Biomarkers for SMA clinical trials. Neurology. 2006;66:1067–1073. doi: 10.1212/01.wnl.0000201929.56928.13. [DOI] [PubMed] [Google Scholar]
- 54.Crawford T.O., Paushkin S., Kobayashi D.T., Forrest S.J., Joyce C.L., Finkel R.S., Kaufmann P., Swoboda K.J., Tiziano F., Lomastro R., et al. Evaluation of SMN protein, transcript and copy number in the Biomarkers for Spinal Muscular Atrophy (BforSMA) clinical study. PLoS ONE. 2012;7:e33572. doi: 10.1371/journal.pone.0033572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vezain M., Saugier-Veber P., Melki J., Toutain A., Bieth E., Husson M., Pedespan J.M., Viollet L., Pénisson-Besnier I., Fehrenbach S., et al. A sensitive assay for measuring SMN mRNA levels in peripheral blood and in muscle samples of patients affected with spinal muscular atrophy. Eur. J. Hum. Genet. 2007;15:1054–1062. doi: 10.1038/sj.ejhg.5201885. [DOI] [PubMed] [Google Scholar]
- 56.Tiziano F.D., Pinto A.M., Fiori S., Lomastro R., Messina S., Bruno C., Pini A., Pane M., D’Amico A., Ghezzo A., et al. SMN transcript levels in leukocytes of SMA patients determined by absolute real-time PCR. Eur. J. Hum. Genet. 2010;18:52–58. doi: 10.1038/ejhg.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh R.N., Howell M.D., Ottesen E.W., Singh N.N. Diverse role of survival motor neuron protein. Biochim. Biophys. Acta. 2017;1860:299–315. doi: 10.1016/j.bbagrm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H.M., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 61.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 62.Cho S., Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burnett B.G., Muñoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le T.T., Pham L.T., Butchbach M.E.R., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H.M. SMNΔ7, the major product of the centromeric survival motor neuron gene (SMN2), extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 65.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H.M. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prior T.W., Swoboda K.J., Scott H.D., Hejmanowski A.Q. Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am. J. Med. Genet. 2005;130A:307–310. doi: 10.1002/ajmg.a.30251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wirth B., Brichta L., Schrank B., Lochmüller H., Blick S., Baasner A., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 68.Elsheikh B., Prior T., Zhang X., Miller R., Kolb S.J., Moore D., Bradley W., Barohn R., Bryan W., Gelinas D., et al. An analysis of disease severity based on SMN2 copy number in adults with spinal muscular atrophy. Muscle Nerve. 2009;40:652–656. doi: 10.1002/mus.21350. [DOI] [PubMed] [Google Scholar]
- 69.Tiziano F.D., Bertini E., Messina S., Angelozzi C., Pane M., D’Amico A., Alfieri P., Fiori S., Battini R., Berardinelli A., et al. The Hammersmith functional score correlates with the SMN2 copy number: A multicentric study. Neuromuscul. Disord. 2007;17:400–403. doi: 10.1016/j.nmd.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 70.Stabley D.L., Harris A.W., Holbrook J., Chubbs N.J., Lozo K.W., Crawford T.O., Swoboda K.J., Funanage V.L., Wang W., Mackenzie W., et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol. Genet. Genomic Med. 2015;3:248–257. doi: 10.1002/mgg3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mailman M.D., Heinz J.W., Papp A.C., Snyder P.J., Sedra M.S., Wirth B., Burghes A.H.M., Prior T.W. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 72.Qu Y., Ge X., Bai J., Wang L., Cao Y., Lu Y., Jin Y., Wang H., Song F. Association of copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein gene with the natural history in a Chinese spinal muscular atrophy cohort. J. Child Neurol. 2014;30:429–436. doi: 10.1177/0883073814553271. [DOI] [PubMed] [Google Scholar]
- 73.Amara A., Adala L., Ben Charfeddine I., Mamaï O., Mili A., Ben Lazreg T., H-Mida D., Amri F., Salem N., Boughammura L., et al. Correlation of SMN2, NAIP, p44, H4F5 and Occludin genes copy number with spinal muscular atrophy phenotype in Tunisian patients. Eur. J. Paediatr. Neurol. 2012;16:167–174. doi: 10.1016/j.ejpn.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Brkusanin M., Kosac A., Jovanovic V., Pesovic J., Brajuskovic G., Dimitrijevic N., Todorovic S., Romac S., Milic Rasic V., Savic-Pavicevic D. Joint effect of the SMN2 and SERF1A genes on childhood-onset types of spinal muscular atrophy in Serbian patients. J. Hum. Genet. 2015;60:723–728. doi: 10.1038/jhg.2015.104. [DOI] [PubMed] [Google Scholar]
- 75.Van der Steege G., Grootscholten P.M., van der Vlies P., Draaijers T.G., Osinga J., Cobben J.M., Scheffer H., Buys C.H.C.M. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. Lancet. 1995;345:985–986. doi: 10.1016/S0140-6736(95)90732-7. [DOI] [PubMed] [Google Scholar]
- 76.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anhuf D., Eggermann T., Rudnik-Schöneborn S., Zerres K. Determination of SMN1 and SMN2 copy number using TaqMan technology. Hum. Mutat. 2003;22:74–78. doi: 10.1002/humu.10221. [DOI] [PubMed] [Google Scholar]
- 78.Gómez-Curet I., Robinson K.G., Funanage V.L., Crawford T.O., Scavina M., Wang W. Robust quantification of the SMN gene copy number by real-time TaqMan PCR. Neurogenetics. 2007;8:271–278. doi: 10.1007/s10048-007-0093-1. [DOI] [PubMed] [Google Scholar]
- 79.Taylor J.E., Thomas N.H., Lewis C.M., Abbs S.J., Rodrigues N.R., Davies K.E., Mathew C.G. Correlation of SMNt and SMNc gene copy number with age of onset and survival in spinal muscular atrophy. Eur. J. Hum. Genet. 1998;6:467–474. doi: 10.1038/sj.ejhg.5200210. [DOI] [PubMed] [Google Scholar]
- 80.Gérard B., Ginet N., Matthijs G., Evrard P., Baumann C., Da Silva F., Gérard-Blanleut M., Mayer M., Grandchamp B., Elion J. Genotype determination at the survival motor neuron locus in a normal population and SMA carriers using competitive PCR and primer extension. Hum. Mutat. 2004;16:253–263. doi: 10.1002/1098-1004(200009)16:3<253::AID-HUMU8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 81.Su Y.N., Hung C.C., Li H., Lee C.N., Cheng W.F., Tsao P.N., Chang M.C., Yu C.L., Hsieh W.S., Lin W.L., et al. Quantitative analysis of SMN1 and SMN2 genes based on DHPLC: A highly efficient and reliable carrier-screening test. Hum. Mutat. 2005;25:460–467. doi: 10.1002/humu.20160. [DOI] [PubMed] [Google Scholar]
- 82.Scarciolla O., Stuppia L., De Angelis M.V., Murru S., Palka C., Giuliani R., Pace M., Di Muzio A., Torrente I., Morella A., et al. Spinal muscular atrophy genotyping by gene dosage using multiple ligation-dependent probe amplification. Neurogenetics. 2006;7:269–276. doi: 10.1007/s10048-006-0051-3. [DOI] [PubMed] [Google Scholar]
- 83.Arkblad E.L., Darin N., Berg K., Kimber E., Brandberg G., Lindberg C., Holmberg E., Tulinius M., Nordling M. Multiplex ligation-dependent probe amplification improves diagnostics in spinal muscular atrophy. Neuromuscul. Disord. 2006;16:830–838. doi: 10.1016/j.nmd.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 84.Huang C.H., Chang Y.Y., Chen C.H., Kuo Y.S., Hwu W.L., Gerdes T., Ko T.M. Copy number analysis of survival motor neuron genes by multiplex ligation-dependent probe amplification. Genet. Med. 2007;9:241–248. doi: 10.1097/GIM.0b013e31803d35bc. [DOI] [PubMed] [Google Scholar]
- 85.Alías L., Bernal S., Barceló M.J., Also-Rallo E., Martínez-Hernández R., Rodríguez-Alvarez F.J., Hernández-Chico C., Baiget M., Tizzano E.F. Accuracy of marker analysis, quantitative real-time polymerase chain reaction and multiple ligation-dependent probe amplification to determine SMN2 copy number in patients with spinal muscular atrophy. Genet. Test. Mol. Biomark. 2011;15:587–594. doi: 10.1089/gtmb.2010.0253. [DOI] [PubMed] [Google Scholar]
- 86.Fang P., Li L., Zhou W.J., Wu W.Q., Zhong Z.Y., Yan T.Z., Xie J.S., Huang J., Lin L., Zhao Y., et al. Molecular characterization and copy number of SMN1, SMN2 and NAIP in Chinese patients with spinal muscular atrophy and unrelated healthy controls. BMC Musculoskelet. Disord. 2015;16:11. doi: 10.1186/s12891-015-0457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirwin S.M., Vinette K.M.B., Gonzalez I.L., Al Abdulwahed H., Al-Sannaa N., Funanage V.L. A homozygous double mutation in SMN1: A complicated genetic diagnosis of SMA. Mol. Genet. Genomic Med. 2013;1:113–117. doi: 10.1002/mgg3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobrowolski S.F., Pham H.T., Pouch-Downes F., Prior T.W., Naylor E.W., Swoboda K.J. Newborn screening for spinal muscular atrophy by calibrated short-amplicon melt profiling. Clin. Chem. 2012;58:1033–1039. doi: 10.1373/clinchem.2012.183038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang C.C., Jong Y.J., Chang J.G., Chen Y.L., Wu S.M. Universal fluorescent multiplex PCR and capillary electrophoresis for evaluation of gene conversion between SMN1 and SMN2 in spinal muscular atrophy. Anal. Bioanal. Chem. 2010;397:2375–2383. doi: 10.1007/s00216-010-3761-1. [DOI] [PubMed] [Google Scholar]
- 90.Wang C.C., Chang J.G., Chen Y.L., Jong Y.J., Wu S.M. Multi-exon genotyping of SMN gene in spinal muscular atrophy by universal fluorescent PCR and capillary electrophoresis. Electrophoresis. 2010;31:2396–2404. doi: 10.1002/elps.201000124. [DOI] [PubMed] [Google Scholar]
- 91.Wang C.C., Shih C.J., Jong Y.J., Wu S.M. Universal fluorescent tri-probe ligation equipped with capillary electrophoresis for targeting SMN1 and SMN2 genes in diagnosis of spinal muscular atrophy. Anal. Chim. Acta. 2014;833:40–47. doi: 10.1016/j.aca.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y., He J., Zhang Y., Li L., Tang X., Wang L., Guo J., Jin C., Tighe S., Zhang Y., et al. The analysis of the association between the copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein genes and the clinical phenotypes in 40 patients with spinal muscular atrophy: Observational study. Medicine. 2020;99:e18809. doi: 10.1097/MD.0000000000018809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stabley D.L., Holbrook J., Scavina M., Crawford T.O., Swoboda K.J., Robbins K.M., Butchbach M.E.R. Detection of SMN1 and SMN2 gene conversion events and partial SMN1 deletions using array digital PCR. Neurogenetics. 2021;22:53–64. doi: 10.1007/s10048-020-00630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calucho M., Bernal S., Alías L., March F., Venceslá A., Rodríguez-Álvarez F.J., Aller E., Fernández R.M., Borrego S., Millán J.M., et al. Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord. 2018;28:208–215. doi: 10.1016/j.nmd.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 95.Müller-Felber W., Vill K., Schwartz O., Gläser D., Nennstiel U., Wirth B., Burggraf S., Röschinger W., Becker M., Durner J., et al. Infants diagnosed with spinal muscular atrophy and 4 SMN2 copies through newborn screening—Opportunity or burden? J. Neuromuscul. Dis. 2020;7:109–117. doi: 10.3233/JND-200475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cuscó I., Bernal S., Blasco-Pérez L., Calucho M., Alias L., Fuentes-Prior P., Tizzano E.F. Practical guidelines to manage discordant situations of SMN2 copy number in patients with spinal muscular atrophy. Neurol. Genet. 2020;6:e530. doi: 10.1212/NXG.0000000000000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DiDonato C.J., Chen X.N., Noya D., Korenberg J.R., Nadeau J.H., Simard L.R. Cloning, characterization and copy number of the murine survival motor neuron gene: Homolog of the spinal muscular atrophy-determining gene. Genome Res. 1997;7:339–352. doi: 10.1101/gr.7.4.339. [DOI] [PubMed] [Google Scholar]
- 98.Viollet L., Bertrandy S., Beuno Brunialti A.L., Lefebvre S., Burlet P., Clermont O., Cruaud C., Guénet J.L., Munnich A., Melki J. cDNA isolation, expression and chromosomal localization of the mouse survival motor neuron gene (Smn) Genomics. 1997;40:185–188. doi: 10.1006/geno.1996.4551. [DOI] [PubMed] [Google Scholar]
- 99.Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cifuentes-Diaz C., Nicole S., Velasco M.E., Borra-Cebrian C., Panozzo C., Frugier T., Millet G., Roblot N., Joshi V., Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum. Mol. Genet. 2002;11:1439–1447. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 101.Nicole S., Desforges B., Millet G., Lesbordes J., Cifuentes-Diaz C., Vertes D., Cao M.L., De Backer F., Languille L., Roblot N., et al. Intact satellite cells lead to remarkable protection against Smn gene defect in differentiation skeletal muscle. J. Cell Biol. 2003;161:571–582. doi: 10.1083/jcb.200210117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F., et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn-/- mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 104.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 105.Michaud M., Arnoux T., Bielli S., Durand E., Rotrou Y., Jablonka S., Robert F., Giraudon-Paoli M., Riessland M., Mattei M.G., et al. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol. Dis. 2010;38:125–135. doi: 10.1016/j.nbd.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 106.Boon K.L., Xiao S., McWhorter M.L., Donn T., Wolf-Saxon E., Bohnsack M.T., Moens C.B., Beattie C.E. Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects. Hum. Mol. Genet. 2009;18:3615–3625. doi: 10.1093/hmg/ddp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hao Le t.h.i., Burghes A.H.M., Beattie C.E. Generation and characterization of a genetic zebrafish model of SMA carrying the human SMN2 gene. Mol. Neurodegener. 2011;6:24. doi: 10.1186/1750-1326-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wadman R.I., Jansen M.D., Stam M., Wijngaarde C.A., Curial C.A.D., Medic J., Sodaar P., Schouten J., Vijzelaar R., Lemmink H.H., et al. Intragenic and structural variation in the SMN locus and clinical variability in spinal muscular atrophy. Brain Commun. 2020;2:fcaa075. doi: 10.1093/braincomms/fcaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prior T.W., Nagan N., Sugarman E.A., Batish S.D., Braastad C. Technical standards and guidelines for spinal muscular atrophy testing. Genet. Med. 2011;13:686–694. doi: 10.1097/GIM.0b013e318220d523. [DOI] [PubMed] [Google Scholar]
- 110.Vijzelaar R., Senetsalaar R., Clausen M., Mason A.G., Rinsma M., Zegers M., Molleman N., Boschloo R., Yilmaz R., Kuilboer R., et al. The frequency of SMN gene variants lacking exon 7 and 8 is highly population dependent. PLoS ONE. 2019;14:e0220211. doi: 10.1371/journal.pone.0220211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhong Q., Bhattacharya S., Kotsopoulos S., Olson J., Taly V., Griffiths A.D., Link D.R., Larson J.W. Multiplex digital PCR: Breaking the one target per color barrier of quantitative PCR. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- 112.Stabley D.L., Holbrook J., Harris A.W., Swoboda K.J., Crawford T.O., Sol-Church K., Butchbach M.E.R. Establishing a reference dataset for the authentication of spinal muscular atrophy cell lines using STR profiling and digital PCR. Neuromuscul. Disord. 2017;27:439–446. doi: 10.1016/j.nmd.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vidal-Folch N., Gavrilov D., Raymond K., Rinaldo P., Tortorelli S., Matern D., Oglesbee D. Multiplex droplet digital PCR method applicable to newborn screening, carrier status and assessment of spinal muscular atrophy. Clin. Chem. 2018;64:1753–1761. doi: 10.1373/clinchem.2018.293712. [DOI] [PubMed] [Google Scholar]
- 114.Park S., Lee H., Shin S., Lee S.T., Lee K.A., Choi J.R. Analytical validation of the droplet digital PCR assay for the diagnosis of spinal muscular atrophy. Clin. Chim. Acta. 2020;510:787–789. doi: 10.1016/j.cca.2020.09.024. [DOI] [PubMed] [Google Scholar]
- 115.Jiang L., Lin R., Gallagher S., Zayac A., Butchbach M.E.R., Hung P. Development and validation of a 4-color multiplexing spinal muscular atrophy (SMA) genotyping assay on a novel integrated digital PCR instrument. Sci. Rep. 2020;10:19892. doi: 10.1038/s41598-020-76893-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Larson J.L., Silver A.J., Chan D., Borroto C., Spurrier B., Silver L.M. Validation of a high resolution NGS method for detecting spinal muscular atrophy carriers among phase 3 participants in the 1000 Genomes Project. BMC Med. Genet. 2015;16:100. doi: 10.1186/s12881-015-0246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Feng Y., Ge X., Meng L., Scull J., Li J., Tian X., Zhang T., Jin W., Cheng H., Wang X., et al. The next generation of population-based spinal muscular atrophy carrier screening: Comprehensive pan-ethnic SMN1 copy-number and sequence variant analysis by massively parallel sequencing. Genet. Med. 2017;19:936–994. doi: 10.1038/gim.2016.215. [DOI] [PubMed] [Google Scholar]
- 118.Lopez-Lopez D., Loucera C., Carmona R., Aquino V., Salgado J., Pasalodos S., Miranda M., Alonso Á., Dopazo J. SMN1 copy-number and sequence variant analysis from next-generation sequencing data. Hum. Mutat. 2020;41:2073–2077. doi: 10.1002/humu.24120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tan C.A., Westbrook M.J., Truty R., Kvitek D.J., Kennemer M., Winder T.L., Shieh P.B. Incorporating spinal muscular atrophy analysis by next-generation sequencing into a comprehensive multigene panel for neuromuscular diseases. Genet. Test. Mol. Biomark. 2020;24:616–624. doi: 10.1089/gtmb.2019.0282. [DOI] [PubMed] [Google Scholar]
- 120.Chen X., Sanchis-Juan A., French C.E., Connell A.J., Delon I., Kingsbury Z., Chawla A., Halpern A.L., Taft R.J., BioResource N.I.H.R., et al. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet. Med. 2020;22:945–953. doi: 10.1038/s41436-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Burghes A.H.M. When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Campbell L., Potter A., Ignatius J., Dubowitz V., Davies K. Genomic variation and gene conversion in spinal muscular atrophy: Implications for disease process and clinical phenotype. Am. J. Hum. Genet. 1997;61:40–50. doi: 10.1086/513886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.DiDonato C.J., Ingraham S.E., Mendell J.R., Prior T.W., Lenard S., Moxley R.T., III, Florence J., Burghes A.H.M. Deletion and conversion in spinal muscular atrophy: Is there a relationship to severity? Ann. Neurol. 1997;41:230–237. doi: 10.1002/ana.410410214. [DOI] [PubMed] [Google Scholar]
- 124.Hahnen E., Schönling J., Rudnik-Schöneborn S., Zerres K., Wirth B. Hybrid survival motor neuron genes in patients with autosomal recessive spinal muscular atrophy: New insights into molecular mechanisms responsible for the disease. Am. J. Hum. Genet. 1996;59:1057–1065. [PMC free article] [PubMed] [Google Scholar]
- 125.Van der Steege G., Grootscholten P.M., Cobben J.M., Zappata S., Scheffer H., den Dunnen J.T., van Ommen G.J.B., Brahe C., Buys C.H.C.M. Apparent gene conversions involving the SMN gene in the region of the spinal muscular atrophy locus at chromosome 5. Am. J. Hum. Genet. 1996;59:834–838. [PMC free article] [PubMed] [Google Scholar]
- 126.Devriendt K., Lammens M., Schollen E., Van Hole C., Dom R., Devlieger H., Cassiman J.J., Fryns J.P., Matthijs G. Clinical and molecular genetic features of congenital spinal muscular atrophy. Ann. Neurol. 1996;40:731–738. doi: 10.1002/ana.410400509. [DOI] [PubMed] [Google Scholar]
- 127.Qu Y.J., Bai J.L., Cao Y.Y., Wang H., Jin Y.W., Du J., Ge X.S., Zhang W.H., Li Y., He S.X., et al. Mutation spectrum of the survival of motor neuron 1 and functional analysis of variants in Chinese spinal muscular atrophy. J. Mol. Diagn. 2016;18:741–752. doi: 10.1016/j.jmoldx.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 128.Alías L., Bernal S., Calucho M., Martínez E., March F., Gallano P., Fuentes-Prior P., Abuli A., Serra-Juhe C., Tizzano E.F. Utility of two SMN1 variants to improve spinal muscular atrophy carrier diagnosis and genetic counselling. Eur. J. Hum. Genet. 2018;26:1554–1557. doi: 10.1038/s41431-018-0193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kubo Y., Nishio H., Saito K. A new method for SMN1 and hybrid SMN gene analysis in spinal muscular atrophy using long-range PCR followed by sequencing. J. Hum. Genet. 2015;60:233–239. doi: 10.1038/jhg.2015.16. [DOI] [PubMed] [Google Scholar]
- 130.Chen T.H., Tzeng C.C., Wang C.C., Wu S.M., Chang J.G., Yang S.N., Hung C.H., Jong Y.J. Identification of bidirectional gene conversion between SMN1 and SMN2 by simultaneous analysis of SMN dosage and hybrid genes in a Chinese population. J. Neurol. Sci. 2011;308:83–87. doi: 10.1016/j.jns.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 131.Ogino S., Gao S., Leonard D.G.B., Paessler M., Wilson R.B. Inverse correlation between SMN1 and SMN2 copy numbers: Evidence for gene conversion from SMN2 to SMN1. Eur. J. Hum. Genet. 2003;11:275–277. doi: 10.1038/sj.ejhg.5200957. [DOI] [PubMed] [Google Scholar]
- 132.Mazzei R., Gambardella A., Conforti F.L., Magariello A., Patitucci A., Gabiele A.L., Sprovieri T., Labate A., Valentino P., Bono F., et al. Gene conversion events in adult-onset spinal muscular atrophy. Acta Neurol. Scand. 2004;109:151–154. doi: 10.1034/j.1600-0404.2003.00181.x. [DOI] [PubMed] [Google Scholar]
- 133.Cuscó I., Barceló M.J., del Rio E., Martín Y., Hernández-Chico C., Bussaglia E., Baiget M., Tizzano E.F. Characterization of SMN hybrid genes in Spanish SMA patients: De novo, homozygous and compound heterozygous cases. Hum. Genet. 2001;108:222–229. doi: 10.1007/s004390000452. [DOI] [PubMed] [Google Scholar]
- 134.Wirth B. Spinal muscular atrophy: In the challenge lies a solution. Trends Neurosci. 2021;44:306–322. doi: 10.1016/j.tins.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 135.Ruhno C., McGovern V.L., Avenarius M.R., Snyder P.J., Prior T.W., Nery F.C., Muhtaseb A., Roggenbuck J.S., Kissel J.T., Sansone V.A., et al. Complete sequencing of the SMN2 gene in SMA patients detect SMN gene deletion junctions and variants in SMN that modify the SMA phenotype. Hum. Genet. 2019;138:241–256. doi: 10.1007/s00439-019-01983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blasco-Pérez L., Paramonov I., Leno J., Bernal S., Alías L., Fuentes-Prior P., Cuscó I., Tizzano E.F. Beyond copy number: A new, rapid and versatile methods for sequencing the entire SMN2 gene in SMA patients. Hum. Mutat. 2021;42:787–795. doi: 10.1002/humu.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu X., Wang S.H., Sun J., Krainer A.R., Hua Y., Prior T.W. A-44G transition in SMN2 intron 6 protects patients with spinal muscular atrophy. Hum. Mol. Genet. 2017;26:2768–2780. doi: 10.1093/hmg/ddx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kashima T., Rao N., Manley J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Prior T.W., Krainer A.R., Hua Y., Swoboda K.J., Snyder P.C., Bridgeman S.J., Burghes A.H.M., Kissel J.T. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am. J. Hum. Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vezain M., Saukkonen A.M., Goina E., Touraine R., Manel V., Toutain A., Fehrenbach S., Frébourg T., Pagani F., Tosi M., et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum. Mutat. 2010;31:E1110–E1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- 141.Bernal S., Alías L., Barceló M.J., Also-Rallo E., Martínez-Hernández R., Gámez J., Guillén-Navarro E., Rosell J., Hernando I., Rodríguez-Alvarez F.J., et al. The c.859G>C variant in the SMN2 gene is associated with types II and III SMA and originates from a common ancestor. J. Med. Genet. 2010;47:640–642. doi: 10.1136/jmg.2010.079004. [DOI] [PubMed] [Google Scholar]
- 142.Gambardella A., Mazzei R., Toscano A., Annesi G., Pasqua A., Annesi F., Quattrone F., Oliveri R.L., Valentino P., Bono F., et al. Spinal muscular atrophy due to an isolated deletion of exon 8 of the telomeric survival motor neuron gene. Ann. Neurol. 1998;44:836–839. doi: 10.1002/ana.410440522. [DOI] [PubMed] [Google Scholar]
- 143.Thauvin-Robinet C., Drunat S., Saugier Veber P., Cantereau D., Cossée M., Cassini C., Soichot P., Masurel-Paulet A., De Monléon J.V., Sagot P., et al. Homozygous SMN1 exons 1-6 deletion: Pitfalls in genetic counseling and general recommendations for spinal muscular atrophy molecular diagnosis. Am. J. Med. Genet. 2012;158:1735–1741. doi: 10.1002/ajmg.a.35402. [DOI] [PubMed] [Google Scholar]
- 144.Wirth B., Herz M., Wetter A., Moskau S., Hahnen E., Rudnik-Schöneborn S., Wienker T., Zerres K. Quantitative analysis of survival motor neuron copies: Identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation and implications for genetic counseling. Am. J. Hum. Genet. 1999;64:1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jedličková I., Přistoupilová A., Nosková L., Majer F., Stránecký V., Hartmannová H., Hodaňová K., Trešlová H., Hýblová M., Solár P., et al. Spinal muscular atrophy caused by a novel Alu-mediated deletion of exons 2a-5 in SMN1 undetectable with routine genetic testing. Mol. Genet. Genomic Med. 2020;8:e1238. doi: 10.1002/mgg3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Deininger P. Alu elements: Know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ottesen E.W., Seo J., Singh N.N., Singh R.N. A multilayered control of the human Survival Motor Neuron gene expression by Alu elements. Front. Microbiol. 2017;8:2252. doi: 10.3389/fmicb.2017.02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sen S.K., Han K.S., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 150.Mercuri E., Darras B.T., Chiriboga C.A., Day J.W., Campbell C., Connolly A.M., Iannaccone S.T., Kirschner J., Kuntz N.L., Saito K., et al. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 2018;378:625–635. doi: 10.1056/NEJMoa1710504. [DOI] [PubMed] [Google Scholar]
- 151.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 152.Baranello G., Darras B.T., Day J.W., Deconinck N., Klein A., Masson R., Mercuri E., Rose K., El-Khairi M., Gerber M., et al. Risdiplam in type 1 spinal muscular atrophy. N. Engl. J. Med. 2021;384:915–923. doi: 10.1056/NEJMoa2009965. [DOI] [PubMed] [Google Scholar]
- 153.Glascock J., Sampson J., Haidet-Phillips A., Connolly A., Darras B., Day J., Finkel R., Howell R.R., Klinger K.W., Kuntz N., et al. Treatment algorithm for infants diagnosed with spinal muscular atrophy through newborn screening. J. Neuromuscul. Dis. 2018;5:145–158. doi: 10.3233/JND-180304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Glascock J., Sampson J., Connolly A.M., Darras B.T., Day J.W., Finkel R., Howell R.R., Klinger K.W., Kuntz N., Prior T., et al. Revised recommendations for the treatment of infants diagnosed with spinal muscular atrophy via newborn screening who have 4 copies of SMN2. J. Neuromuscul. Dis. 2020;7:97–100. doi: 10.3233/JND-190468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bai J.L., Qu Y.J., Cao Y.Y., Li E.Z., Wang L.W., Li Y., Zhu Y.L., Zhang W.H., Jin Y.W., Wang H., et al. Subtle mutation detection of SMN1 gene in Chinese spinal muscular atrophy patients: Implications of molecular diagnostic procedure for SMN1 gene mutations. Genet. Test. Mol. Biomark. 2014;18:546–551. doi: 10.1089/gtmb.2014.0002. [DOI] [PubMed] [Google Scholar]
- 156.Sun Y., Grimmler M., Schwarzer V., Schoenen F., Fischer U., Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum. Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- 157.Wijaya Y.O.S., Ar Rochmah M., Niba E.T.E., Morisada N., Noguchi Y., Hidaka Y., Ozasa S., Inoue T., Shimazu T., Takahashi Y., et al. Phenotypes of SMA patients retaining SMN1 with intragenic mutation. Brain Dev. 2021;43:745–758. doi: 10.1016/j.braindev.2021.03.006. [DOI] [PubMed] [Google Scholar]
- 158.Prior T.W. Spinal muscular atrophy diagnostics. J. Child Neurol. 2007;22:952–956. doi: 10.1177/0883073807305668. [DOI] [PubMed] [Google Scholar]
- 159.Sossi V., Giuli A., Vitali T., Tiziano F., Mirabella M., Antonelli A., Neri G., Brahe C. Premature termination mutations in exon 3 of the SMN1 gene are associated with exon skipping and a relatively mild SMA phenotype. Eur. J. Hum. Genet. 2001;9:113–120. doi: 10.1038/sj.ejhg.5200599. [DOI] [PubMed] [Google Scholar]
- 160.Mendonça R.H., Matsui Jr C., Polido G.J., Silva A.M.S., Kulikowski L., Torchio Dias A., Zanardo E.A., Solla D.J.F., Gurgel-Giannetti J., de Moura A.C.M.L., et al. Intragenic variants in the SMN1 gene determine the clinical phenotype in 5q spinal muscular atrophy. Neurol. Genet. 2020;6:e505. doi: 10.1212/NXG.0000000000000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brichta L., Garbes L., Jedrzejowska M., Grellscheid S.N., Holker I., Zimmermann K., Wirth B. Nonsense-mediated messenger RNA decay of survival motor neuron 1 causes spinal muscular atrophy. Hum. Genet. 2008;123:141–153. doi: 10.1007/s00439-007-0455-7. [DOI] [PubMed] [Google Scholar]
- 162.Alías L., Bernal S., Fuentes-Prior P., Barceló M.J., Also E., Martínez-Hernández R., Rodríguez-Alvarez F.J., Martín Y., Aller E., Grau E., et al. Mutation update of spinal muscular atrophy in Spain: Molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 163.Tsai C.H., Jong Y.J., Hu C.J., Chen C.M., Shih M.C., Chang C.P., Chang J.G. Molecular analysis of SMN, NAIP and P44 genes of SMA patients and their families. J. Neurol. Sci. 2001;190:35–40. doi: 10.1016/S0022-510X(01)00574-3. [DOI] [PubMed] [Google Scholar]
- 164.Xu Y., Xiao B., Liu Y., Qu X.X., Dai M.Y., Ying X.M., Jiang W.T., Zhang J.M., Liu X.Q., Chen Y.W., et al. Identification of novel SMN1 subtle mutations using an allele-specific RT-PCR. Neuromuscul. Disord. 2020;30:219–226. doi: 10.1016/j.nmd.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 165.He J., Zhang Q.J., Lin Q.F., Chen Y.F., Lin X.Z., Lin M.T., Murong S.X., Wang N., Chen W.J. Molecular analysis of SMN1, SMN2, NAIP, GTF2H2 and H4F5 genes in 157 Chinese patients with spinal muscular atrophy. Gene. 2013;518:325–329. doi: 10.1016/j.gene.2012.12.109. [DOI] [PubMed] [Google Scholar]
- 166.Ganji H., Nouri N., Salehi M., Aryani O., Houshmand M., Basiri K., Fazel-Najafabadi E., Sedghi M. Deletion of intragenic SMN1 mutations in spinal muscular atrophy patients with a single copy of SMN1. J. Child Neurol. 2015;30:558–562. doi: 10.1177/0883073814521297. [DOI] [PubMed] [Google Scholar]
- 167.Zeng J., Lin Y., Yan A., Ke L., Zhu Z., Lan F. Establishment of a molecular diagnostic system for spinal muscular atrophy. Experience from a clinical laboratory in China. J. Mol. Diagn. 2011;13:41–47. doi: 10.1016/j.jmoldx.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Qu Y., Du J., Li E., Bai J., Jin Y., Hong W., Wang H., Song F. Subtle mutations in the SMN1 gene in Chinese patients with SMA: p.Arg288Met mutation causing SMN1 transcript exclusion of exon7. BMC Med. Genet. 2012;13:86. doi: 10.1186/1471-2350-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Skordis L.A., Dunckley M.G., Burglen L., Campbell L., Talbot K., Patel S., Melki J., Davies J.E., Dubowitz V., Muntoni F. Characterisation of novel point mutations in the survival motor neuron gene SMN, in three patients with SMA. Hum. Genet. 2001;108:356–357. doi: 10.1007/s004390100497. [DOI] [PubMed] [Google Scholar]
- 170.Zapletalová E., Hedvicáková P., Kozák L., Vondrácek P., Gaillyová R., Maríková T., Kalina Z., Jüttnerová V., Fajkus J., Fajkusová L. Analysis of point mutations in the SMN1 gene in SMA patients bearing a single SMN1 copy. Neuromuscul. Disord. 2007;17:476–481. doi: 10.1016/j.nmd.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 171.Jedrzejowska M., Gos M., Zimowski J.G., Kostera-Pruszczyk A., Ryniewicz B., Hausmanowa-Petrusewicz I. Novel point mutations in survival motor neuron 1 gene expand the spectrum of phenotypes observed in spinal muscular atrophy patients. Neuromuscul. Disord. 2014;24:617–623. doi: 10.1016/j.nmd.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 172.Clermont O., Burlet P., Benit P., Chanterau S., Saugier-Veber P., Munnich A., Cusin V. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum. Mutat. 2004;24:417–427. doi: 10.1002/humu.20092. [DOI] [PubMed] [Google Scholar]
- 173.Bussaglia E., Clermont O., Tizzano E., Lefebvre S., Bürglen L., Cruaud C., Urtizberea J.A., Colomer J., Munnich A., Baiget M., et al. A frame-shift deletion in the survival motor neuron gene in Spanish spinal muscular atrophy patients. Nat. Genet. 2004;11:335–337. doi: 10.1038/ng1195-335. [DOI] [PubMed] [Google Scholar]
- 174.Brahe C., Clermont O., Zappata S., Tiziano F., Melki J., Neri G. Frameshift mutation in the survival motor neuron gene in a severe case of SMA type I. Hum. Mol. Genet. 1996;5:1971–1976. doi: 10.1093/hmg/5.12.1971. [DOI] [PubMed] [Google Scholar]
- 175.Gonçalves-Rocha M., Oliveira J.R.M., Rodrigues L., Santos R. New approaches in molecular diagnosis and population carrier screening for spinal muscular atrophy. Genet. Test. Mol. Biomark. 2011;15:319–325. doi: 10.1089/gtmb.2010.0164. [DOI] [PubMed] [Google Scholar]
- 176.Parsons D.W., McAndrew P.E., Iannaccone S.T., Mendell J.R., Burghes A.H.M., Prior T.W. Intragenic telSMN mutations: Frequency, distribution, evidence of a founder effect and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am. J. Hum. Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Parsons D.W., McAndrew P.E., Allinson P.S., Parker W.D., Jr., Burghes A.H.M., Prior T.W. Diagnosis of spinal muscular atrophy in an SMN non-deletion patient with quantitative PCR screen and mutation analysis. J. Med. Genet. 1998;35:674–676. doi: 10.1136/jmg.35.8.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sharifi Z., Taheri M., Fallah M.S., Abiri M., Golnabi F., Begherian H., Zeinali R., Farahzadi H., Alborji M., Tehrani P.G., et al. Comprehensive mutation analysis and report of 12 novel mutations in a cohort of patients with spinal muscular atrophy in Iran. J. Mol. Neurosci. 2021 doi: 10.1007/s12031-020-01789-0. in press. [DOI] [PubMed] [Google Scholar]
- 179.Martín Y., Valero A., del Castillo E., Pascual S.I., Hernández-Chico C. Genetic study of SMA patients without homozygous SMN1 deletions: Identification of compound heterozygotes and characterisation of novel intragenic SMN1 mutations. Hum. Genet. 2002;110:257–263. doi: 10.1007/s00439-002-0681-y. [DOI] [PubMed] [Google Scholar]
- 180.Parsons D.W., McAndrew P.E., Monani U.R., Mendell J.R., Burghes A.H.M., Prior T.W. An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: Further evidence for SMN as the primary SMA-determining gene. Hum. Mol. Genet. 1996;5:1727–1732. doi: 10.1093/hmg/5.11.1727. [DOI] [PubMed] [Google Scholar]
- 181.Takarada T., Ar Rochmah M., Harahap N.I.F., Shinohara M., Saito T., Saito K., Lai P.S., Bouike Y., Takeshima Y., Awano H., et al. SMA mutations in SMN Tudor and C-terminal domains destabilize the protein. Brain Dev. 2017;39:606–612. doi: 10.1016/j.braindev.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 182.Yamamoto T., Sato H., Lai P.S., Nurputra D.K., Harahap N.I.F., Morikawa S., Nishimura N., Kurashige T., Ohshita T., Nakajima H., et al. Intragenic mutations in SMN1 may contribute more significantly to clinical severity than SMN2 copy numbers in some spinal muscular atrophy (SMA) patients. Brain Dev. 2014;36:914–920. doi: 10.1016/j.braindev.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 183.Kotani T., Sutomo R., Sasongko T.H., Sadewa Gunadi A.H., Minato T., Fujii E., Endo S., Lee M.J., Ayaki M., Harada Y., et al. A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J. Neurol. 2007;254:624–630. doi: 10.1007/s00415-006-0410-x. [DOI] [PubMed] [Google Scholar]
- 184.Zhao X., Wang Y., Mei S., Chen C.H., Liu L., Wang C., Zhao G., Kong X. Identification of two novel SMN1 point mutations associated with a very severe SMA-I phenotype. Eur. J. Med. Genet. 2020;63:104006. doi: 10.1016/j.ejmg.2020.104006. [DOI] [PubMed] [Google Scholar]
- 185.Cuscó I., Barceló M.J., del Río E., Baiget M., Tizzano E.F. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63:146–149. doi: 10.1212/01.WNL.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- 186.Fraidakis M.J., Drunat S., Maisonobe T., Gerard B., Pradat P.F., Meininger V., Salachas F. Genotype-phenotype relationship in 2 SMA III patients with novel mutations in the Tudor domain. Neurology. 2012;78:551–556. doi: 10.1212/WNL.0b013e318247ca69. [DOI] [PubMed] [Google Scholar]
- 187.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 188.Wadman R.I., Stam M., Gijzen M., Lemmink H.H., Snoeck I.N., Wijngaarde C.A., Braun K.P.J., Schoenmakers M.A.G.C., van den Berg L.H., Dooijes D., et al. Association of motor milestones, SMN2 copy and outcome in spinal muscular atrophy types 0-4. J. Neurol. Neurosurg. Psychiatry. 2017;88:365–367. doi: 10.1136/jnnp-2016-314292. [DOI] [PubMed] [Google Scholar]
- 189.Rochette C.F., Surh L.C., Ray R.N., McAndrew P.E., Prior T.W., Burghes A.H.M., Vanasse M., Simard L.R. Molecular diagnosis of non-deletion SMA patients using quantitative PCR of SMN exon 7. Neurogenetics. 1997;1:141–147. doi: 10.1007/s100480050021. [DOI] [PubMed] [Google Scholar]
- 190.Hahnen E., Schönling J., Rudnik-Schöneborn S., Raschke H., Zerres K., Wirth B. Missense mutations in exon 6 of the survival motor neuron gene in patients with spinal muscular atrophy (SMA) Hum. Mol. Genet. 1997;6:821–825. doi: 10.1093/hmg/6.5.821. [DOI] [PubMed] [Google Scholar]
- 191.Sumner C.J., Huynh T.N., Markowitz J.A., Perhac J.S., Hill B., Coovert D.D., Schussler K., Chen X., Jarecki J., Burghes A.H.M., et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 192.Ar Rochmah M., Awano H., Awaya T., Harahap N.I.F., Morisada N., Bouike Y., Saito A., Kubo Y., Saito K., Lai P.S., et al. Spinal muscular atrophy carriers with two SMN1 copies. Brain Dev. 2017;39:851–860. doi: 10.1016/j.braindev.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 193.Wang C.H., Papendick B.D., Bruinsma P., Day J.K. Identification of a novel missense mutation of the SMNT gene in two siblings with spinal muscular atrophy. Neurogenetics. 1998;1:273–276. doi: 10.1007/s100480050040. [DOI] [PubMed] [Google Scholar]
- 194.Talbot K., Ponting C.P., Theodosiou A.M., Rodrigues N.R., Surtees R., Mountford R., Davies K.E. Missense mutation clustering in the survival motor neuron gene: A role for conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum. Mol. Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 195.Kang S.H., Cho S.I., Chae J.H., Chung K.N., Ra E.K., Kim S.Y., Seong M.W., Kim J.Y., Park S.S. False homozygous deletions of SMN1 exon 7 using Dra I PCR-RFLP caused by a novel mutation in spinal muscular atrophy. Genet. Test. Mol. Biomark. 2009;13:511–513. doi: 10.1089/gtmb.2008.0158. [DOI] [PubMed] [Google Scholar]
- 196.Qu Y.J., Song F., Yang Y.L., Jin Y.W., Bai J.L. Compound heterozygous mutation in two unrelated cases of Chinese spinal muscular atrophy patients. Chin. Med. J. 2011;124:385–389. [PubMed] [Google Scholar]
- 197.Zhu S.Y., Xiong F., Chen Y.J., Yan T.Z., Zeng J., Li L., Zhang Y.N., Chen W.Q., Bao X.H., Zhang C., et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur. J. Hum. Genet. 2010;18:978–984. doi: 10.1038/ejhg.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Eggermann T., Eggermann K., Elbracht M., Zerres K., Rudnik-Schöneborn S. A new splice site mutation in the SMN1 gene causes discrepant results in SMN1 deletion screening approaches. Neuromuscul. Disord. 2008;18:146–149. doi: 10.1016/j.nmd.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 199.Monani U.R., Pastore M.T., Gavrilina T.O., Jablonka S., Le T.T., Andreassi C., DiCocco J.M., Lorson C., Androphy E.J., Sendtner M., et al. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J. Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Workman E., Saieva L., Carrel T.L., Crawford T.O., Liu D., Lutz C., Beattie C.E., Pellizzoni L., Burghes A.H.M. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2115–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Iyer C.C., Corlett K.M., Massoni-Laporte A., Duque S.I., Madabusi N., Tisdale S., McGovern V.L., Le T.T., Zaworksi P.G., Arnold W.D., et al. Mild SMN missense alleles are only functional in the presence of SMN2 in mammals. Hum. Mol. Genet. 2018;27:3404–3416. doi: 10.1093/hmg/ddy251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H.M., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McGovern V.L., Kray K.M., Arnold W.D., Duque S.I., Iyer C.C., Massoni-Laporte A., Workman E., Patel A., Battle D.J., Burghes A.H.M. Intragenic complementation of amino and carboxy terminal SMN missense mutations can rescue Smn null mice. Hum. Mol. Genet. 2020;29:3493–3503. doi: 10.1093/hmg/ddaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mailman M.D., Hemingway T., Darsey R.L., Glasure C.E., Huang Y., Chadwick R.B., Heinz J.W., Papp A.C., Snyder P.J., Sedra M.S., et al. Hybrids monosomal for human chromosome 5 reveal the presence of a spinal muscular atrophy (SMA) carrier with two SMN1 copies on one chromosome. Hum. Genet. 2001;108:109–115. doi: 10.1007/s004390000446. [DOI] [PubMed] [Google Scholar]
- 205.Alías L., Barceló M.J., Bernal S., Martínez-Hernández R., Also-Rallo E., Vázquez C., Santana A., Millán J.M., Baiget M., Tizzano E.F. Improving detection and genetic counseling in carriers of spinal muscular atrophy with two copies of the SMN1 gene. Clin. Genet. 2014;85:470–475. doi: 10.1111/cge.12222. [DOI] [PubMed] [Google Scholar]
- 206.Luo M., Liu L., Peter I., Zhu J., Scott S.A., Zhao G., Eversley C., Kornreich R., Desnick R.J., Edelmann L. An Ashkenazi Jewish SMN1 haplotype specific to duplication alleles improves pan-ethnic carrier screening for spinal muscular atrophy. Genet. Med. 2014;16:149–156. doi: 10.1038/gim.2013.84. [DOI] [PubMed] [Google Scholar]
- 207.Prior T.W., Bayrak-Toydemir P., Lynnes T.C., Mao R., Metcalf J.D., Muralidharan K., Iwata-Otsubo A., Pham H.T., Pratt V.M., Qureshi S.A., et al. Characterization of reference materials for spinal muscular atrophy genetic testing: A Genetic Testing Reference Materials Coordination Program collaborative project. J. Mol. Diagn. 2021;23:103–110. doi: 10.1016/j.jmoldx.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ceylan A.C., Erdem H.B., Şahin İ., Agarwal M. SMN1 gene copy number analysis for spinal muscular atrophy (SMA) in a Turkish cohort by CODE-SEQ technology, an integrated solution for detection of SMN1 and SMN2 copy numbers and the “2+0” genotype. Neurol. Sci. 2020;41:2575–2584. doi: 10.1007/s10072-020-04365-x. [DOI] [PubMed] [Google Scholar]
- 209.Azad A.K., Huang C.K., Jin H., Zou H., Yanakakis L., Du J., Fiddler M., Naeem R., Goldstein Y. Enhanced carrier screening for spinal muscular atrophy: Detection of silent (SMN1:2+0) carriers utilizing a novel TaqMan genotyping method. Lab. Med. 2020;51:408–415. doi: 10.1093/labmed/lmz088. [DOI] [PubMed] [Google Scholar]
- 210.Talbot J., Rodrigues N., Bernert G., Bittner R., Davies K. Evidence for compound heterozygositiy causing mild and severe forms of autosomal recessive spinal muscular atrophy. J. Med. Genet. 1996;33:1019–1021. doi: 10.1136/jmg.33.12.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Muqit M.M.K., Moss J., Sewry C., Lane R.J.M. Phenotypic variability in siblings with type III spinal muscular atrophy. J. Neurol. Neurosurg. Psychiatry. 2004;75:1762–1764. doi: 10.1136/jnnp.2003.018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Arkblad E., Tulinius M., Kroksmark A.K., Henricsson M., Darin N. A population-based study of genotypic and phenotypic variability in children with spinal muscular atrophy. Acta Paediatr. 2009;98:865–872. doi: 10.1111/j.1651-2227.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 213.Jones C.C., Cook S.F., Jarecki J., Belter L., Reyna S.P., Staropoli J., Farwell W., Hobby K. Spinal muscular atrophy (SMA) subtype concordance in siblings: Findings from the Cure SMA cohort. J. Neuromuscul. Dis. 2020;7:33–40. doi: 10.3233/JND-190399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Burghes A.H.M., Ingraham S.E., Kóte-Jarai Z., Rosenfeld S., Herta N., Nadkarni N., DiDonato C.J., Carpten J., Hurko O., Florence J., et al. Linkage mapping of the spinal muscular atrophy gene. Hum. Genet. 1994;93:305–312. doi: 10.1007/BF00212028. [DOI] [PubMed] [Google Scholar]
- 215.Hahnen E., Forkert R., Marke C., Rudnik-Schöneborn S., Schönling J., Zerres K., Wirth B. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: Evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum. Mol. Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 216.Cobben J.M., van der Steege G., Grootscholten P., de Visser M., Scheffer H., Buys C.H.C.M. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am. J. Hum. Genet. 1995;57:805–808. [PMC free article] [PubMed] [Google Scholar]
- 217.Cuscó I., Barceló M.J., Rojas-García R., Illa I., Gámez J., Cervera C., Pou A., Izquierdo G., Baiget M., Tizzano E.F. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J. Neurol. 2006;253:21–25. doi: 10.1007/s00415-005-0912-y. [DOI] [PubMed] [Google Scholar]
- 218.Wirth B., Garbes L., Riessland M. How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic applications. Curr. Opin. Genet. Dev. 2013;23:330–338. doi: 10.1016/j.gde.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 219.Oprea G.E., Kröber S., McWhorter M.L., Rossoll W., Müller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Stratigopoulos G., Lanzano P., Deng L., Guo J., Kaufmann P., Darras B., Finkel R., Tawil R., McDermott M.P., Martens W., et al. Association of plastin 3 expression with disease severity in spinal muscular atrophy only in postpubertal females. Arch. Neurol. 2010;67:1252–1256. doi: 10.1001/archneurol.2010.239. [DOI] [PubMed] [Google Scholar]
- 221.Yanyan C., Yujin Q., Jinli B., Yuwei J., Hong W., Fang S. Correlation of PLS3 expression with disease severity in children with spinal muscular atrophy. J. Hum. Genet. 2014;59:24–27. doi: 10.1038/jhg.2013.111. [DOI] [PubMed] [Google Scholar]
- 222.Bernal S., Also-Rallo E., Martínez-Hernández R., Alías L., Rodríguez-Alvarez F.J., Millán J.M., Hernández-Chico C., Baiget M., Tizzano E.F. Plastin 3 expression in discordant spinal muscular atrophy (SMA) siblings. Neuromuscul. Disord. 2011;21:413–419. doi: 10.1016/j.nmd.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 223.Yener I.H., Topaloglu H., Erdem-Özdamar S., Dayangac-Erdem D. Transcript levels of plastin 3 and neuritin 1 modifer genes in spinal muscular atrophy siblings. Pediatr. Int. 2017;59:53–56. doi: 10.1111/ped.13052. [DOI] [PubMed] [Google Scholar]
- 224.Wadman R.I., Jansen M.D., Curial C.A.D., groen E.J.N., Stam M., Wijngaarde C.A., Medic J., Sodaar P., van Eijk K.R., Huibers M.M.H., et al. Analysis of FUS, PFN2, TDP-43 and PLS3 as potential disease severity modifiers in spinal muscular atrophy. Neurol. Genet. 2020;6:e386. doi: 10.1212/NXG.0000000000000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hosseinibarkooie S., Peters M., Torres-Benito L., Rastetter R.H., Hupperich K., Hoffmann A., Mendoza-Ferreira N., Kaczmarek A., Janzen E., Milbradt J., et al. The power of human protective modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am. J. Hum. Genet. 2016;99:647–665. doi: 10.1016/j.ajhg.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Janzen E., Mendoza-Ferreira N., Hosseinibarkooie S., Schneider S., Hupperich K., Tschanz T., Grysko V., Riessland M., Hammerschmidt M., Rigo F., et al. CHP1 reduction ameliorates spinal muscular atrophy pathology by restoring calcineurin activity and endocytosis. Brain. 2018;141:2343–2361. doi: 10.1093/brain/awy167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Eshraghi M., McFall E., Gibeault S., Kothary R. Effect of genetic background on the phenotype of the Smn2B/- mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2016;25:4494–4506. doi: 10.1093/hmg/ddw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ackermann B., Kröber S., Torres-Benito L., Borgmann A., Peters M., Barkooie S.M.H., Tejero R., Jakubik M., Schreml J., Milbradt J., et al. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum. Mol. Genet. 2013;22:1328–1347. doi: 10.1093/hmg/dds540. [DOI] [PubMed] [Google Scholar]
- 229.McGovern V.L., Massoni-Laporte A., Wang X., Le T.T., Le H.T., Beattie C.E., Rich M.M., Burghes A.H.M. Plastin 3 expression does not modify spinal muscular atrophy severity in the Δ7 SMA mouse. PLoS ONE. 2015;10:e0132364. doi: 10.1371/journal.pone.0132364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kaifer K.A., Villalón E., Osman E.Y., Glascock J.J., Arnold L.L., Cornelison D.D.W., Lorson C.L. Plastin-3 extends survival and reduces severity in mouse models of spinal muscular atrophy. JCI Insight. 2017;2:e89970. doi: 10.1172/jci.insight.89970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Alrafiah A., Karyka E., Coldicott I., Iremonger K., Lewis K.E., Ning K., Azzouz M. Plastin 3 promotes motor axonal growth and extends survival in a mouse model of spinal muscular atrophy. Mol. Ther. Methods Clin. Dev. 2018;9:81–89. doi: 10.1016/j.omtm.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Riessland M., Kaczmarek A., Schneider S., Swoboda K.J., Löhr H., Bradler C., Grysko V., Dimitriadi M., Hosseinibarkooie S., Torres-Benito L., et al. Neurocalcin Delta suppression protects against spinal muscular atrophy in humans and acress species by restoring impaired endocytosis. Am. J. Hum. Genet. 2017;100:297–315. doi: 10.1016/j.ajhg.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jiang J., Huang J., Gu J., Cai X., Zhao H., Lu H. Genomic analysis of a spinal muscular atrophy (SMA) discordant family identifies a novel mutation in TLL2, an activator of growth differentiation factor 8 (myostatin): A case report. BMC Med. Genet. 2019;20:204. doi: 10.1186/s12881-019-0935-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Long K.K., O’Shea K.M., Khairallah R.J., Howell K., Pauschkin S., Chen K.S., Cote S.M., Webster M.T., Stains J.P., Treece E., et al. Specific inhibition of myostatin activation is beneficial in mouse models of SMA therapy. Hum. Mol. Genet. 2019;28:1076–1089. doi: 10.1093/hmg/ddy382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Akten B., Kye M.J., Hao Le t.h.i., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E., et al. Interaction of survival of motor neuron (SMN) with HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cherry J.J., Kobayashi D.T., Lynes M.M., Naryshkin N.N., Tiziano F.D., Zaworski P.G., Rubin L.L., Jarecki J. Assays for the identification and prioritization of drug candidates for spinal muscular atrophy. Assay Drug Dev. Technol. 2014;12:315–341. doi: 10.1089/adt.2014.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Arnold W.D., Burghes A.H.M. Spinal muscular atrophy: Development and implementation of potential therapeutics. Ann. Neurol. 2013;74:348–362. doi: 10.1002/ana.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]