Abstract
The mutation of TP53 gene affects half of all human cancers, resulting in impairment of the regulation of several cellular functions, including cell cycle progression and cell death in response to genotoxic stress. In the recent years additional p53-mediated tumour suppression mechanisms have been described, questioning the contribution of its canonical pathway for tumour suppression. These include regulation of alternative cell death modalities (i.e. ferroptosis), cell metabolism and the emerging role in RNA stability. Here we briefly summarize our knowledge on p53 “canonical DNA damage response” and discuss the most relevant recent findings describing potential mechanistic explanation of p53-mediated tumour suppression.
Keywords: Tumour suppression, DNA damage, Stress response, Cell death
Canonical tumour suppression signalling: apoptotic cell death
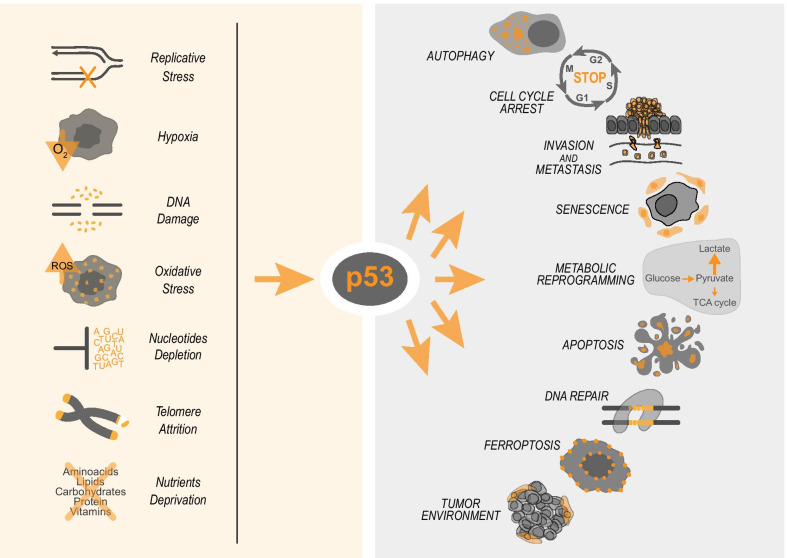
Every second cancer carries an inactivating mutation in the TP53 gene. Nonetheless, despite 40 years of high-quality studies, the p53-regulated tumour suppressive programme is still a puzzle, and cell death/cell cycle arrest are still considered among the most relevant regulatory aspects (Fig. 1). Apoptosis is an ordered and tightly regulated form of cell death and is highly associated to prevention of tumorigenesis. Sequence and homology of p53 are evolutionarily conserved in Drosophila melanogaster (named dmp53) and Caenorhabditis elegans (named cep-1) [1–3], although debate is still open on whether the mammalian p53 family member, p63 better resembles biochemically and functionally the ancestor form of the protein [4–10]. The response to DNA damage, responsible of activating p53-mediated cell cycle arrest and apoptosis, is a dominant mechanism of p53-mediated network and strongly conserved across species, including Drosophila melanogaster and Caenorhabditis elegans. Experimental evidence however emerged over the past decade, questioning the simplistic interpretation that p53 tumour suppression mainly rely on the regulation of cell cycle arrest and apoptosis. Mouse models carrying DNA-damage insensitive p53 forms have shown that p53 can suppress tumour development in absence of cell cycle arrest and/or apoptosis induction [11–15]. In further support of this, p53 can prevent tumour development in the mouse lacking p21, Puma and Noxa, the major p53 downstream targets responsible for cell cycle arrest and apoptosis in DNA damaged cells [16]. Thus, these data have questioned the specific contribution of p53-mediated DNA damage response (here referred as “canonical” p53 tumour suppressive signalling), indicating that p53 network might be more complex and unexplored that previously anticipated [17].
Fig. 1.
P53 drives the cellular adaptation to stress. Different types of cell stressors, such as DNA damage, nutrient deprivation, and replicative stress, can activate p53 which in turn drives specific responses depending on type and magnitude of stress. For example, in mild/moderate reversible DNA damage p53 can stop cell cycle progression and promote the DNA repair. When the DNA damage is severe p53 promotes apoptosis
The “non-canonical” p53 tumour suppressive signalling includes several biological processes that in stressed/damaged cells can influence tumour initiation or progression; these includes metabolic stress, epigenetic reprogramming, regulation of reactive oxygen species (ROS) response, autophagy [18]. Multiple stress signalling can indeed control p53 activation. Conditions such as nutrient deprivation, hypoxia, nucleotide depletion can lead to different degree of p53 activation, impinging on the biology of the cell at different level, with different mechanisms (Fig. 1) [19–23].
Although there is consensus that p53 acts by cooperating with a network of executors of its tumour suppression function, the spectrum and relative importance of individual players and the nature of their overlap and complementarity are unknown. Even less clear is the mechanism of the p53 mutant and its gain-of-function effects [24–28] and how interaction with extrinsic factors, such as microenvironment, microbiota and others, influences its function [29–32]. Thus, the mechanisms underlining the p53-dependent maintenance of genome integrity and tumour suppression remain fundamentally only partially known.
p53 in ferroptotic cell death
Ferroptosis is associated to an iron-dependent peroxidation of membrane lipids [33] and can be triggered by GSH depletion or by direct inactivation of GPX4 [34–37], both involved in reduction of peroxidic species of polyunsaturated fatty acids. p53 has been implicated in both activation/inhibition of this cell death modality. Despite p533KR (K117R/K161R/K162R) mutant is transcriptionally inactive on p53 pro-apoptotic target genes, mice carrying p533KR do not develop tumours [38]. This phenotype was associated to the ability of p533KR to repress the cystine/glutamate antiporter xCT (SLC7A11), responsible for GSH synthesis, promoting ferroptosis. Hence, p53 tumour suppressive function was postulated to rely on the regulation of ferroptosis.
The calcium-independent phospholipase iPLA2β was also recently shown to critically regulate p53-dependent ferroptosis upon reactive oxygen species (ROS)-induced stress, a classic regulator of cellular functions [39–41]. Peroxidized lipids detoxification mediated by iPLA2β is sufficient to suppress p53-driven ferroptosis, while iPLA2β inhibition sensitizes tumour cells to p53-driven ferroptosis, promoting p53-dependent tumour suppression in xenograft mouse models [42].
p53 is also implicated in the metabolism of polyamines. The SAT1 gene is transcriptionally controlled by p53 [43]. SAT1 is an enzyme that regulates the rate of conversion of spermidine and spermine to putrescine. The transcriptional induction of SAT1 correlates with the expression of arachidonate 15-lipoxygenase (ALOX15), that participates in the peroxidation process. A recent study identified a p53-dependent ferroptosis pathway mediated by another member of the lipoxygenase family, ALOX12, an enzyme involved in lipid peroxidation. Following redox stress, p53 downregulates SLC7A11 and indirectly activates the ALOX12, increasing the intracellular levels of lipid peroxides [44]. Also, the p53 family transcriptional target glutaminase-2 (GLS-2) was associated to activation of ferroptosis. GLS2 converts glutamine into glutamate in a process called glutaminolysis [45–48]. GLS2 inhibition affects the ferroptosis process in fibroblasts [49]. How and whether GLS2 regulation of ferroptosis correlates with p53 tumour suppression is not clearly defined.
p53 was also implicated in the opposite role of repressing ferroptosis. A first example is the inhibition of the (dipeptidyl peptidase-4) DPP4 activity. DPP4 is an enzyme expressed on the cell surface where it can activate lipid peroxidation interacting with NADPH oxidase 1 (NOX1). In human colorectal cancer, p53 was shown to form a complex with DPP4, translocating it into the nucleus. Loss of p53 prevents DPP4 accumulation in the nucleus and facilitates plasma-membrane-associated DPP4-dependent lipid peroxidation, which finally results in ferroptosis [50].
p53 metabolic regulation and lipids biosynthesis
The function of p53 in tumour suppression has emerged as a complex integration of multi biological processes [51]. Within these the regulation of cellular metabolism and autophagy in addition to influencing the ferroptosis process strongly impacts the ability of cell to respond and adapt to perturbation, preventing accumulation of damage that can lead to cancer.
p53 supports mitochondrial respiration by limiting glycolysis. p53 represses expression of the glucose transporters GLUT1, GLU3 and GLUT4 [52–54]. Moreover, p53 reduces expression of TIGAR, a regulator of glucose breakdown [55], and of PDK2 (pyruvate dehydrogenase kinase 2), which inactivates the pyruvate dehydrogenase complex [56], regulating access of pyruvate into the Krebs cycle.
p53 plays also in the metabolism of fatty acids and cholesterol. p53 promotes the oxidation of fatty acids, making them no longer available to cancer cells. In fact, it induces the expression of Lipin-1 which activates other genes involved in the oxidation of fatty acids [57]. Moreover, p53 blocks maturation of SREBP2 (sterol regulatory element binding protein 2) the master transcriptional regulator of biosynthesis of cholesterol and nonsterol isoprenoids. By using a mouse model of liver cancer the groups of Carol Prives and Scott Lowe demonstrated that downregulating mevalonate pathway p53 suppresses tumorigenesis of premalignant hepatocytes [58].
p53 control of mRNAs processing via Zmat3
More recent genomic techniques are helping in dissecting the functional p53 tumour suppressing network. Through a shRNA in vivo screen, 166 known p53 target genes were silenced [59]. This experiment demonstrated that extensive functional overlap of several p53-regulated processes delineates a defence against cancer development and in particular DNA repair plays an essential role. Depletion of the DNA repair gene Mlh1 could recapitulate lymphoma in wt mice and enforced expression of Mlh1 was able to delay the tumour phenotype of p53−\− mice. From this approached also Zmat3 was implicated in development of lymphoma/leukaemia, but only when also Puma and Cdkn1a were depleted. Knockdown of Zmat3 in p53−/− hematopoietic stem/progenitor cells did not accelerate lymphoma in mice, reinforcing the hypothesis that this gene acts downstream of p53 [59]. Also, lack of Zmat3 per se is not sufficient to cause lymphoma/leukaemia, confirming that its loss can only promote tumour development when p53-dependent apoptosis or cell cycle arrest (via Puma and p21 respectively) are impaired [59].
A second in vivo genetic screening using RNAi and CRISPR technologies against 87 p53 targets, confirmed the importance of Zmat3 in p53-dependent cancer biology [60]. Induction of tumour in KrasG12D-driven mouse lung adenocarcinoma and hepatocellular carcinoma depleted for Zmat3 leads to larger tumour and greater tumour burden than in control mice, only when p53 was expressed. No alterations of the tumour size were observed in the same cancer models with p53 depletion [60]. By analysing a cohort of breast cancer patients, it was observed that low level of ZMAT3 correlates with a reduction in the survival, only in wild-type TP53 tumours. Furthermore, mutations and deletions in ZMAT3 locus are mutually exclusive with mutations and deletions in TP53 gene in uterine corpus endometrial carcinoma [60].
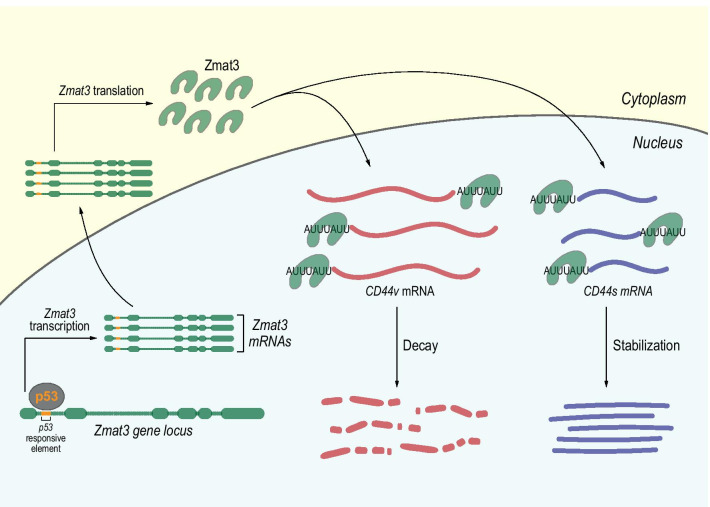
Mechanistically, Zmat3 locus contains a perfectly matched p53 responsive element (RE) in the first intron of both human and mouse gene, and it is directly bound by p53 in ChIP-seq data [61, 62] (Fig. 2). The disruption of the p53 RE by using specific sgRNAs in E1A;HrasG12V;Cas9 MEFs, significantly reduces either p53 binding to Zmat3 locus and the levels of its RNA and protein [60]. ZMAT3 is a 32 KDa zinc-finger protein containing an RNA binding domain highly conserved across the evolution [63–65]. It localizes in the nucleus, where interacting with AU-rich 3′ untranslated regions, it can stabilize the mRNA targets [66] or promotes their decay [67]. One of the best characterized ZMAT3-target is CD44, potent oncogene and stem cell marker [68]. ZMAT3 silencing led to the upregulation of the oncogenic and longer CD44v variant and downregulation of the standard, non-oncogenic, CD44s isoform (Fig. 2). Furthermore, TP53 silencing recapitulates this phenotype, suggesting an important role for TP53 in regulating the splicing of CD44 in cancer biology. These studies underline the importance of Zmat3 in the p53-dependent tumour biology, but above all, underline the complexity behind p53 function(s), which is still only partially understood.
Fig. 2.
p53 in the control of mRNA stability. p53 can transcriptionally regulate Zmat3 by binding a p53-responsive element in the first intron of Zmat3 gene. Zmat3 is localised in the nucleus and can bind AU-rich region of CD44 mRNAs. The binding on the long and oncogenic CD44 isoform (CD44v) promotes its degradation by mRNA decay process. Conversely, by binding the shorter and non-oncogenic isoform (CD44s) Zmat3 promotes the mRNA stabilization. In this context, modulating the expression of Zmat3, p53 regulates the balance of CD44 isoforms
Conclusion
Despite over 40 years of studies dedicated to understanding the role of p53 in cancer, there are still key unanswered questions. These include for example the exact mechanisms by which p53 protects integrity of the somatic cell genome. Based on the dispensable role for tumour suppression of p53 mediated DNA damage response, the mechanism by which genomic instability is prevented remains elusive. The last decade has seen a massive expansion of cancer genomics studies, including development of predictive model of gene network [69–75] and improvement of experimental model of studies, including effective genetic editing techniques and 3D cell culture models [76–79]. With the support of the expansion of cancer genomics studies [70, 80–86] and methodological improvements [77, 87, 88], the determination of the p53 mediated gene network can lead not only to a greater understanding of tumour biology but also to design of more accurate anti-cancer therapies.
Acknowledgements
This work has been supported by the Associazione Italiana per la Ricerca contro il Cancro (AIRC) to IA (AIRC Start-Up ID 23219; 2020-2024) and GM (IG#20473; 2018-2022).
Authors' contributions
EP and CZ wrote the first draft, IA and GM revised the draft, IA conceived the article. All authors read and approved the final version.
Availability of data and materials
Available upon requests.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Emanuele Panatta, Email: panatta.emanuele@gmail.com.
Carlotta Zampieri, Email: carlottazampieri20@gmail.com.
Gerry Melino, Email: melino@uniroma2.it.
Ivano Amelio, Email: ivano.amelio@uniroma2.it.
References
- 1.Jin S, et al. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 3.Panchin AY, Aleoshin VV, Panchin YV. From tumors to species: a SCANDAL hypothesis. Biol Direct. 2019;14:3. doi: 10.1186/s13062-019-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gebel J, et al. p63 uses a switch-like mechanism to set the threshold for induction of apoptosis. Nat Chem Biol. 2020;16:1078–1086. doi: 10.1038/s41589-020-0600-3. [DOI] [PubMed] [Google Scholar]
- 5.Tuppi M, et al. Oocyte DNA damage quality control requires consecutive interplay of CHK2 and CK1 to activate p63. Nat Struct Mol Biol. 2018;25:261–269. doi: 10.1038/s41594-018-0035-7. [DOI] [PubMed] [Google Scholar]
- 6.Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition mechanism of p63 by the E3 ligase Itch: novel strategy in the study and inhibition of this interaction. Cell Cycle. 2012;11:3638–3648. doi: 10.4161/cc.21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lena AM, et al. Skn-1a/Oct-11 and DeltaNp63alpha exert antagonizing effects on human keratin expression. Biochem Biophys Res Commun. 2010;401:568–573. doi: 10.1016/j.bbrc.2010.09.102. [DOI] [PubMed] [Google Scholar]
- 8.Lena AM, et al. The p63 C-terminus is essential for murine oocyte integrity. Nat Commun. 2021;12:383. doi: 10.1038/s41467-020-20669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vikhreva P, Melino G, Amelio I. p73 Alternative Splicing: exploring a biological role for the C-terminal isoforms. J Mol Biol. 2018;430:1829–1838. doi: 10.1016/j.jmb.2018.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellomaria A, Barbato G, Melino G, Paci M, Melino S. Recognition of p63 by the E3 ligase ITCH: effect of an ectodermal dysplasia mutant. Cell Cycle. 2010;9:3730–3739. doi: 10.4161/cc.9.18.12933. [DOI] [PubMed] [Google Scholar]
- 11.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, et al. Olig2 SUMOylation protects against genotoxic damage response by antagonizing p53 gene targeting. Cell Death Differ. 2020;27:3146–3161. doi: 10.1038/s41418-020-0569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radine C, et al. The RNA-binding protein RBM47 is a novel regulator of cell fate decisions by transcriptionally controlling the p53–p21-axis. Cell Death Differ. 2020;27:1274–1285. doi: 10.1038/s41418-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, et al. Transient inhibition of p53 homologs protects ovarian function from two distinct apoptotic pathways triggered by anticancer therapies. Cell Death Differ. 2019;26:502–515. doi: 10.1038/s41418-018-0151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank T, et al. Cell cycle arrest in mitosis promotes interferon-induced necroptosis. Cell Death Differ. 2019;26:2046–2060. doi: 10.1038/s41418-019-0298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valente LJ, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–1345. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutelle AM, Attardi LD. p53 and tumor suppression: it takes a network. Trends Cell Biol. 2021;31:298–310. doi: 10.1016/j.tcb.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valente LJ, et al. p53 deficiency triggers dysregulation of diverse cellular processes in physiological oxygen. J Cell Biol. 2020 doi: 10.1083/jcb.201908212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitolli C, et al. p53-mediated tumor suppression: DNA-damage response and alternative mechanisms. Cancers (Basel) 2019;11:198. doi: 10.3390/cancers11121983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amelio I, et al. p53 mutants cooperate with HIF-1 in transcriptional regulation of extracellular matrix components to promote tumor progression. Proc Natl Acad Sci USA. 2018;115:E10869–E10878. doi: 10.1073/pnas.1808314115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40:425–434. doi: 10.1016/j.tibs.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Shao J, et al. Derepression of LOXL4 inhibits liver cancer growth by reactivating compromised p53. Cell Death Differ. 2019;26:2237–2252. doi: 10.1038/s41418-019-0293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, et al. Gain-of-function mutant p53 in cancer progression and therapy. J Mol Cell Biol. 2020 doi: 10.1093/jmcb/mjaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitolli C, et al. Do Mutations Turn p53 into an Oncogene? Int J Mol Sci. 2019;20:6241. doi: 10.3390/ijms20246241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ham SW, et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019;26:409–425. doi: 10.1038/s41418-018-0126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amelio I, Melino G. Context is everything: extrinsic signalling and gain-of-function p53 mutants. Cell Death Discov. 2020;6:16. doi: 10.1038/s41420-020-0251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celardo I, Melino G, Amelio I. Commensal microbes and p53 in cancer progression. Biol Direct. 2020;15:25. doi: 10.1186/s13062-020-00281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadosh E, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586:133–138. doi: 10.1038/s41586-020-2541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lennon JT, Locey KJ. More support for Earth's massive microbiome. Biol Direct. 2020;15:5. doi: 10.1186/s13062-020-00261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang WS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc(.) Cell Death Differ. 2020;27:662–675. doi: 10.1038/s41418-019-0380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Li S, Li F, Lv C, Yang QK. High-fat diet impairs ferroptosis and promotes cancer invasiveness via downregulating tumor suppressor ACSL4 in lung adenocarcinoma. Biol Direct. 2021;16:10. doi: 10.1186/s13062-021-00294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angelucci S, et al. Purification and characterization of glutathione transferases from the sea bass (Dicentrarchus labrax) liver. Arch Biochem Biophys. 2000;373:435–441. doi: 10.1006/abbi.1999.1569. [DOI] [PubMed] [Google Scholar]
- 40.Mauretti A, et al. Design of a novel composite H2 S-releasing hydrogel for cardiac tissue repair. Macromol Biosci. 2016;16:847–858. doi: 10.1002/mabi.201500430. [DOI] [PubMed] [Google Scholar]
- 41.Pallucca R, et al. Specificity of epsilon and non-epsilon isoforms of arabidopsis 14-3-3 proteins towards the H+-ATPase and other targets. PLOS ONE. 2014;9:e90764. doi: 10.1371/journal.pone.0090764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen D, et al. iPLA2beta-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4. Nat Commun. 2021;12:3644. doi: 10.1038/s41467-021-23902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci USA. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu B, et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat Cell Biol. 2019;21:579–591. doi: 10.1038/s41556-019-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velletri T, et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle. 2013;12:3564–3573. doi: 10.4161/cc.26771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amelio I, et al. p73 regulates serine biosynthesis in cancer. Oncogene. 2014;33:5039–5046. doi: 10.1038/onc.2013.456. [DOI] [PubMed] [Google Scholar]
- 47.Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aceto A, et al. Identification of an N-capping box that affects the alpha 6-helix propensity in glutathione S-transferase superfamily proteins: a role for an invariant aspartic residue. Biochem J. 1997;322(Pt 1):229–234. doi: 10.1042/bj3220229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 51.Li X, et al. Competitive ubiquitination activates the tumor suppressor p53. Cell Death Differ. 2020;27:1807–1818. doi: 10.1038/s41418-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 53.Lonetto G, et al. Mutant p53-dependent mitochondrial metabolic alterations in a mesenchymal stem cell-based model of progressive malignancy. Cell Death Differ. 2019;26:1566–1581. doi: 10.1038/s41418-018-0227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amelio I, et al. TAp73 promotes anabolism. Oncotarget. 2014;5:12820–12934. doi: 10.18632/oncotarget.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 56.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012;72:560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 57.Assaily W, et al. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol Cell. 2011;44:491–501. doi: 10.1016/j.molcel.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 58.Moon SH, et al. p53 Represses the Mevalonate Pathway to Mediate Tumor Suppression. Cell. 2019;176:564–580e.519. doi: 10.1016/j.cell.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janic A, et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med. 2018;24:947–953. doi: 10.1038/s41591-018-0043-5. [DOI] [PubMed] [Google Scholar]
- 60.Bieging-Rolett KT, et al. Zmat3 Is a Key Splicing Regulator in the p53 Tumor Suppression Program. Mol Cell. 2020;80:452–469.e459. doi: 10.1016/j.molcel.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Younger ST, Kenzelmann-Broz D, Jung H, Attardi LD, Rinn JL. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic Acids Res. 2015;43:4447–4462. doi: 10.1093/nar/gkv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Israeli D, et al. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J. 1997;16:4384–4392. doi: 10.1093/emboj/16.14.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hellborg F, Wiman KG. The p53-induced Wig-1 zinc finger protein is highly conserved from fish to man. Int J Oncol. 2004;24:1559–1564. [PubMed] [Google Scholar]
- 65.Gallo M, et al. Identification of a conserved N-capping box important for the structural autonomy of the prion alpha 3-helix: the disease associated D202N mutation destabilizes the helical conformation. Int J Immunopathol Pharmacol. 2005;18:95–112. doi: 10.1177/039463200501800111. [DOI] [PubMed] [Google Scholar]
- 66.Vilborg A, et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci USA. 2009;106:15756–15761. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bersani C, Xu LD, Vilborg A, Lui WO, Wiman KG. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14–3-3sigma. Oncogene. 2014;33:4407–4417. doi: 10.1038/onc.2013.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 69.Salehi S, et al. Clonal fitness inferred from time-series modelling of single-cell cancer genomes. Nature. 2021 doi: 10.1038/s41586-021-03648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Consortium, I. T. P.-C. A. o. W. G. Pan-cancer analysis of whole genomes. Nature 2020; 578, 82–93, doi:10.1038/s41586-020-1969-6 [DOI] [PMC free article] [PubMed]
- 71.Khairi S, et al. Outcome of clinical genetic testing in patients with features suggestive for hereditary predisposition to PTH-mediated hypercalcemia. Horm Cancer. 2020;11:250–255. doi: 10.1007/s12672-020-00394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu L, et al. Computational identification and characterization of glioma candidate biomarkers through multi-omics integrative profiling. Biol Direct. 2020;15:10. doi: 10.1186/s13062-020-00264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han Y, et al. Integration of molecular features with clinical information for predicting outcomes for neuroblastoma patients. Biol Direct. 2019;14:16. doi: 10.1186/s13062-019-0244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han Y, et al. Integrative analysis based on survival associated co-expression gene modules for predicting Neuroblastoma patients' survival time. Biol Direct. 2019;14:4. doi: 10.1186/s13062-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee PMY, et al. Heterogeneous associations between obesity and reproductive-related factors and specific breast cancer subtypes among Hong Kong Chinese women. Horm Cancer. 2020;11:191–199. doi: 10.1007/s12672-020-00386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sprangers J, Zaalberg IC, Maurice MM. Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ. 2021;28:95–107. doi: 10.1038/s41418-020-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Funata M, Nio Y, Erion DM, Thompson WL, Takebe T. The promise of human organoids in the digestive system. Cell Death Differ. 2021;28:84–94. doi: 10.1038/s41418-020-00661-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamastra FR, et al. Polymer composite random lasers based on diatom frustules as scatterers. Rsc Adv. 2014;4:61809–61816. doi: 10.1039/c4ra12519c. [DOI] [Google Scholar]
- 79.Sidhaye J, Knoblich JA. Brain organoids: an ensemble of bioassays to investigate human neurodevelopment and disease. Cell Death Differ. 2021;28:52–67. doi: 10.1038/s41418-020-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amelio I, et al. Cancer predictive studies. Biol Direct. 2020;15:18. doi: 10.1186/s13062-020-00274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oktay K, et al. A computational statistics approach to evaluate blood biomarkers for breast cancer risk stratification. Horm Cancer. 2020;11:17–33. doi: 10.1007/s12672-019-00372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mihaylov I, Kandula M, Krachunov M, Vassilev D. A novel framework for horizontal and vertical data integration in cancer studies with application to survival time prediction models. Biol Direct. 2019;14:22. doi: 10.1186/s13062-019-0249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Insabato L, et al. Elevated expression of the tyrosine phosphatase SHP-1 defines a subset of high-grade breast tumors. Oncology. 2009;77:378–384. doi: 10.1159/000276765. [DOI] [PubMed] [Google Scholar]
- 84.Pinto MP, et al. Chilean registry for neuroendocrine tumors: a Latin American perspective. Horm Cancer. 2019;10:3–10. doi: 10.1007/s12672-018-0354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amelio I, et al. Liquid biopsies and cancer omics. Cell Death Discov. 2020;6:131. doi: 10.1038/s41420-020-00373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pekic S, et al. Familial cancer clustering in patients with prolactinoma. Horm Cancer. 2019;10:45–50. doi: 10.1007/s12672-018-0348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seidlitz T, Koo BK, Stange DE. Gastric organoids-an in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ. 2021;28:68–83. doi: 10.1038/s41418-020-00662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bova L, Billi F, Cimetta E. Mini-review: advances in 3D bioprinting of vascularized constructs. Biol Direct. 2020;15:22. doi: 10.1186/s13062-020-00273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Available upon requests.