Abstract
Cardiac fibroblasts (CFBs) support heart function by secreting extracellular matrix (ECM) and paracrine factors, respond to stress associated with injury and disease, and therefore are an increasingly important therapeutic target. We describe how developmental lineage of human pluripotent stem cell-derived CFBs, epicardial (EpiC-FB) and second heart field (SHF-FB) impacts transcriptional and functional properties. Both EpiC-FBs and SHF-FBs exhibited CFB transcriptional programs and improved calcium handling in human pluripotent stem cell-derived cardiac tissues. We identified differences including in composition of ECM synthesized, secretion of growth and differentiation factors, and myofibroblast activation potential, with EpiC-FBs exhibiting higher stress induced activation potential akin to myofibroblasts and SHF-FBs demonstrating higher calcification and mineralization potential. These phenotypic differences suggest that EpiC-FBs have utility in modeling fibrotic diseases while SHF-FBs are a promising source of cells for regenerative therapies. This work directly contrasts regional and developmental specificity of CFBs and informs CFB in vitro model selection.
Keywords: Pluripotent stem cell, Cardiac, Fibroblast
Introduction
Cardiovascular disease accounts for one in three deaths in the United States and 48% of all adults over 20 years old suffer from cardiovascular disease1. Fibroblasts are one of the most prevalent cardiac cell types and estimates suggest they comprise approximately 20–60% percent of the total heart cells, in contrast to cardiomyocytes (CMs) which comprise about 30% of the heart2,3. Fibroblasts in many organs serve as support cells by producing extracellular matrix (ECM) and secreting paracrine factors. Under stress associated with injury and disease, fibroblasts produce excess ECM, inflammatory cytokines, and contribute to scar tissue formation. Thus, a better understanding of cardiac fibroblast (CFB) heterogeneity in heart development, maintenance, disease, and response to injury can provide new therapeutic insights to treat heart disease.
During heart development, CFBs arise from four distinct progenitor populations: epicardial cells, endocardial cells, neural crest cells, and second heart field progenitors. Epicardial cells line the surface of the heart at mouse embryonic day E9.5 and undergo epithelial-to-mesenchymal transition to generate epicardial-derived cells that migrate into the myocardium around E12.5. Lineage tracing studies of Tbx18 and Tcf21-expressing cells have shown that epicardial-derived fibroblasts are primarily located in the ventricles but also contribute to the atrioventricular valves in many model organisms including zebrafish, quail, and mice where they comprise approximately 80% of all CFBs4–10. Conversely, endocardial cells, which line the inside of the heart chambers, arise at E8.0 and contribute primarily to fibroblast populations in the ventricles and septum in mice starting around E9.55. Neural crest cells contribute primarily to fibroblasts in the coronary trunk and aorta as demonstrated by Pax3 lineage tracing in mice around E9.55,11. Second heart field progenitors which are present by E8.0 are also thought to differentiate into fibroblasts in the outflow tract as well as the dorsal mesenchymal protrusion, important for atrial septation, thus contributing to atrial fibroblasts12–14. However, despite the results of lineage tracing studies, it is not yet clear whether or how the developmental origin and corresponding developmental timeframe of CFBs influences their subsequent phenotype and function within the developed heart.
In attempts to better characterize cardiac cell diversity, researchers have used single cell transcriptomic profiling to classify and compare populations of cells at different developmental stages. Non-biased clustering of cells of mouse and human hearts have identified 4–6 different fibroblast populations that can be discriminated from other cardiac cell types by high expression of ECM genes and little to no expression of genes encoding sarcomeric proteins15–18. In addition, spatiotranscriptomic approaches, such as fluorescence in situ hybridization (FISH) to target clusters identified from single cell transcriptomics, have been used to trace fibroblast populations to different regions of the developing human heart15. A recent single cell RNA sequencing study of adult human hearts found that CFBs in the atria differentially express genes including CNTN4 and NAV2 while CFBs from the ventricles express genes including BMPER and ADCY117. Additionally, fibroblasts in the left and right sides of the heart differentially expressed genes with links to fibrosis, including CLIP and ITGBL1. Another transcriptomic study identified differences in collagen isoforms and ECM-related transcripts between heart chambers19. Since the majority of CFBs in the ventricles arise from epicardial lineages, this suggests that CFB gene expression and functional phenotype in the four chambers may be influenced by local environmental cues. Thus, although diverse CFB populations populate the heart, we have an incomplete understanding of how developmental trajectory influences CFB heterogeneity and functional consequences.
Human pluripotent stem cells (hPSCs) offer a tool to differentiate cells through developmentally relevant stages and systematically study human CFB function. Over the past ten years, we and others have developed methods to differentiate hPSCs to cardiac cell types through T+ mesoderm to MESP1+ and GATA4+ cardiac mesoderm and further into cardiac progenitors by modulating Wnt signaling20. To differentiate hPSCs into second heart field fibroblasts, cardiac mesoderm progenitors were treated with FGF2 to generate NKX2-5+, ISL1+, HAND2+ and transient TBX1+ and CXCR4+ second heart field progenitors to a TE7+POSTN+MF20− CFBs21. Alternatively, hPSCs can be differentiated to epicardial cells by modulation of Wnt signaling using either recombinant protein WNT3A or small molecule CHIR99021 and further treated with FGF2 to achieve POSTN+ CFBs22–26. These EpiC-FBs have been shown to respond to pro- and anti-fibrotic drugs and have been used to study paracrine signaling implicated in fibrogenesis25. Furthermore, tissue constructs containing hPSC-CMs and hPSC-epicardial cells implanted in a mouse myocardial infarction resulted in epithelial-to-mesenchymal-transition (EMT) of epicardial cells to CFBs, enhanced engraftment, and improved ejection fraction one month later compared to CM monoculture grafts27. These studies highlight the benefits of hPSC-CFBs in cardiac disease modeling and tissue engineering but a clearer understanding of specific functions of different CFB populations is needed to inform CFB in vitro model selection.
Here, we compare and contrast gene and protein expression profiles as well as fibroblast phenotypes of CFBs differentiated through epicardial cell progenitors (EpiC-FB) and second heart field (SHF-FB) to determine how hPSC differentiation through different CFB lineages affects the resulting cell populations. hPSC-CFBs differentiated through epicardial and second heart field progenitors expressed fibroblast markers, expressed transcripts for pan-cardiac marker GATA4, and enhanced calcium handling in cardiac microtissues. Key characteristics of the EpiC-FBs include expression of epicardial-lineage markers TBX18 and TBX20, secretion of fibrosis related factors including Gremlin-1, and higher myofibroblast activation potential in response to TGFβ−1, Angiotensin-II, and serum. Since EpiC-FBs can exhibit fibrotic characteristics induced by stress, this suggests they may be better suited for modeling cardiac fibrosis and anti-fibrotic drug testing. In contrast, SHF-FBs expressed second heart field-lineage markers including TBX1 and SALL1, secretion of WNT5A, lower activation potential, and higher mineralization potential suggesting they may be superior for studying outflow tract development and calcification. Furthermore, we identified striking differences in matrix composition and secreted factors produced by the two CFB populations which may be important in developmental specification. Together, these two hPSC-CFBs provide insight into how distinct developmental lineages of human CFB leads to molecular and functional diversity.
Materials and Methods
Experimental Design
The goal of these experiments is to determine molecular and functional differences between hPSCs differentiated through second heart field progenitors or epicardial cell progenitors to cardiac fibroblasts.
Maintenance of hPSCs
Human pluripotent stem cells (hPSCs) were maintained on Matrigel (Corning, Corning, NY, USA)-coated plates in mTeSR1 (STEMCELL Technologies, Vancouver, Canada) according to previously published methods28. At 80–90% confluency hPSCs were passaged with Versene to maintain colonies. For this study, human embryonic stem cell (hESC) line H9 (WiCell, Madison, WI, USA) and H9-7TGP (Palecek Lab) and hiPSC lines WTC-CAAX-RFP (Allen Institute, Seattle, WA, USA), WTC11-GCaMP (Gladstone, San Francisco, CA, USA)29, and 19-9-11 (WiCell, Madison, WI, USA) were used.
Cardiac progenitor cell differentiation via modulation of canonical Wnt signaling
As previously published in the GiWi protocol to derive cardiac progenitors, hPSCs were singularized with Accutase at 37°C for 5 min, quenched in DMEM/F12, and centrifuged at 200g for 5 min28. hPSCs were seeded at 100,000–600,000 cells/cm2 in mTeSR1 supplemented with 5 μM ROCK inhibitor Y-27632 (Selleckchem, Houston, TX, USA) (day −2) for 24 hrs. The following day (day −1), cells were treated with fresh mTeSR1. At day 0, cells were treated with 6–15 μM CHIR99021 (Selleckchem, Houston, TX, USA) in RPMI1640 supplemented with B27 minus insulin (RPMI/B27−) media. 23–24 hrs later, media was changed to fresh RPMI/B27− (day 1). At day 3, 5 μM IWP2 (Tocris, Bristol, United Kingdom) was added to 1:1 conditioned media to fresh RPMI/B27− media. At day 5, cells were treated with RPMI/B27− media. At day 6, cardiac progenitors were either frozen in cryomedium (60% DMEM/F12, 30% FBS, 10% DMSO) or singularized for further differentiation.
Epicardial cell differentiation via activation of canonical Wnt signaling
Following our previously published protocol for epicardial differentiation, day 6 cardiac progenitors were either singularized in Accutase at 37°C for 10 min or thawed from cryo and seeded onto a gelatin (Sigma-Aldrich, St. Louis, MO, USA) or Matrigel-coated cell culture plate at 20,000–80,000 cells/cm2 (approximately a 1:3 or 1:12 split) in LaSR basal media (500 mL advanced DMEM/F12 (Life Technologies, Carlsbad, CA, USA) with 0.06 g/L L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich, St. Louis, MO, USA) and 6.25 mL GlutaMAX) supplemented with 5 μM ROCK inhibitor Y-2763230. At day 7 and 8, cells were treated with fresh LaSR basal media supplemented with 3 μM CHIR99021. At days 9, 10, and 11, cells were treated with fresh LaSR basal media. At day 12, epicardial cells were singularized with Accutase for 5 min at 37°C and either cryopreserved for later use or replated in LaSR basal media supplemented with A8301 (R&D Systems, Minneapolis, MN, USA) and 5 μM ROCK inhibitor Y-27632. Subsequently, epicardial cells were treated with LaSR supplemented with A8301 daily until 90–100% confluent. Epicardial cells were then passaged using Versene into fresh LaSR basal media supplemented with 0.5 μM A8301 without ROCK inhibitor Y-27632 to maintain colonies, prevent further differentiation, and improve attachment for up to five passages. Alternatively, cells were frozen in cryomedium (60% DMEM/F21, 30% FBS, 10% DMSO). Differentiations were validated to have at least 90% WT1 positive cells by flow cytometry.
Epicardial fibroblast differentiation via bFGF signaling
When epicardial cells reached approximately 100% confluency, they were treated with LaSR supplemented with 5 ng/mL bFGF (Waisman Biomanufacturing, Madison, WI, USA) daily for 10 days. At this point, EpiC-FBs were cryopreserved or passaged at 7,000 cells/cm2 or approximately 1:3–1:6 split with Accutase into FibroGRO (EMD Millipore, Burlington, MA, USA) medium on a cell culture treated plate (FibroGRO basal media with manufacturers supplements, GlutaMAX supplemented for Glutamine, and 2% FBS). Medium was changed every two days until fibroblasts reached approximately 80–90% confluency when they were passaged with Accutase. Differentiations were validated to have be at least 80% double positive for TE-7 and VIM by flow cytometry. All experiments were performed between P1 and 5 unless otherwise noted.
Epicardial smooth muscle cell differentiation via TGFβ signaling
When epicardial cells reached approximately 100% confluency, they were treated with LaSR supplemented with 5 ng/mL TGFβ (Waisman Biomanufacturing, Madison, WI, USA) daily for 6 days30. At this point, smooth muscle cells (SMCs) were used for experiments.
Cardiomyocyte differentiation
WTC11-GCaMP6f hiPSCs29 were differentiated into CMs following the GiWi protocol28. Briefly, hPSCs were seeded onto Matrigel-coated (80 μg/mL) plates at a density of 3×104 cells/cm2 in mTeSR medium with 10 μM Rock inhibitor. Once cells reached 100% confluence (~3 days), medium was changed to RPMI/B27− supplemented with 12 μM CHIR99021 (Day 0). Exactly 24 hrs later, cells were fed with fresh RPMI/B27− and on day 3, medium was changed to RPMI/B27− supplemented with 5 μM IWP2. On days 7, 10, and 13, cells were fed with RPMI1640 with B27 supplement containing insulin (RPMI/B27+). On day 15, cells were replated onto Matrigel-coated plates at a density of 2×105 cells/cm2 in RPMI/B27+ containing 15% FBS and 10 μM Rock inhibitor. Medium was changed to fresh RPMI/B27+ on day 16. Enrichment of CMs occurred by feeding cells with lactate purification medium31 (no-glucose Dulbecco’s Modified Eagle Medium with 1X Non Essential Amino Acids, 1X Glutamax, and 4mM Lactate) on days 17 and 20, and cells were returned to RPMI/B27+ on day 23 and used for cardiac microtissues between days 25 and 28.
Second heart field fibroblast differentiation
Second heart field CFBs were differentiated following the GiFGF protocol as previously published32. When hPSCs maintained in mTeSR1 reached approximately 90–100% confluency, they were treated with 6–15 μM CHIR99021 in RPMI/B27− media (day 0). 23–24 hrs later, media was changed to fresh RPMI/B27− (day 1). From day 2 to 20, cells were treated with fresh CFB basal media every two days. At day 20, CFBs were singularized with Accutase for 10 min at room temperature and cryopreserved or replated on tissue cultured plastic in FibroGRO media at approximately 7,000 cells/cm2. Following this, FibroGRO media was changed every two days until the fibroblasts reached approximately 80–90% confluency when they were passaged with Accutase. Differentiations were validated to have be at least 80% double positive for TE-7 and VIM by flow cytometry. All experiments were performed between P1 and 5 unless otherwise noted.
Primary fibroblast cell culture
Primary human adult dFBs (Lonza, Basel, Switzerland), primary human adult ventricular CFBs (Lonza), and primary human fCFBs (Cell Applications, San Diego, CA, USA) were cultured in FibroGRO medium. For these studies, we used dFB, aCFB, and fCFB from a single donor, so we are unable to determine the key attributes are representative of the age group across many genetic backgrounds. Medium was changed every two days until the fibroblasts reached approximately 80–90% confluency when they were passaged with Accutase up to five times.
Immunostaining analysis
As explained previously, cells were fixed with 4% paraformaldehyde for 10 min or ice cold methanol for 5 minutes at room temperature and then blocked in milk buffer (PBS plus 0.4% Triton X-100 and 5% non-fat dry milk or BSA buffer (PBS plus 0.1% Triton X-100 and 0.5% BSA) for one hour at room temperature28. Then, primary antibodies were added, and samples were incubated overnight at 4°C on a shaker. The following day, cells were washed with PBS and stained with secondary antibodies at room temperature for one hour or overnight at 4°C on a shaker. Antibody dilutions and information are provided Data File S3. Hoescht counterstain was added at 5 μg/mL in PBS for five min. For image analysis, an epifluorescence microscope Olympus IX70 or Nikon Ti2 was used.
Flow cytometry analysis
As previously described, cells were singularized with Accutase then fixed with 1% paraformaldehyde for 20 min at room temperature and stained with primary antibodies overnight at 4°C in BSA buffer (PBS plus 0.1% Triton X-100 and 0.5% BSA)28. The following day, cells were washed and stained with secondary antibodies at room temperature for one hr. Antibody dilutions and product information are in Data File S3. At least 10,000 events/sample were collected on a BD Accuri C6 flow cytometer and analyzed using FlowJo. FACS gating was based on a no primary control and negative cell type control.
mRNA extraction, cDNA preparation, and qPCR analysis
Cells were singularized in Accutase, quenched, and centrifuged for 5 min at 200g. Cell pellets were snap-frozen at −80°C until mRNA extraction. Total RNA was isolated using the RNeasy mini kit (Qiagen, Hilden, Germany) and treated with DNase (Qiagen, Hilden, Germany). Extracted mRNA was stored in nuclease-free water at −20°C and 1 μg RNA was reverse transcribed into cDNA using the Omniscript Reverse Transcriptase kit (Qiagen, Hilden, Germany) and Oligo(dT)20 Primers (Life Technologies, Carlsbad, CA, USA). Real-time quantitative PCR with two technical replicates in 25 uL reactions using PowerUP Syber Master Mix (Applied Biosystems, Foster City, CA, USA) on an AriaMx Real-Time PCR System at 60°C (Agilent Technologies, Santa Clara, CA, USA). GAPDH and ZNF384 were used as housekeepers and analysis was performed using the ΔΔCt method. Primer sequences can be found in Data File S3.
Single Cell Sequencing Data Analysis
We obtained count matrices from publicly available single cell sequencing datasets and selected cells from clusters Asp et al. had previously annotated as fibroblasts or fibroblast-like cells15. Violin plots were prepared using clusters identified by the authors using the Seurat package (version 3)33. Differentially expressed genes amongst the fibroblast clusters were identified and the top ten for each cluster were plotted in a heatmap.
RNA Sequencing
Quality and quantity of RNA samples was first analyzed using Nanochip to confirm presence of 18S and 28S ribosomal RNA with appropriate A260/A280 and A260/A230 ratios. Then, RNA was quantified on an Agilent 2100 Bioanalyzer using Qubit prior to library construction and sequencing. Sequencing libraries were constructed using the Illumina TruSeq Stranded mRNA kit (polyA enrichment). Libraries were sequenced on an Illumina NovaSeq6000. Between 62 and 88 million reads were collected per sample.
Raw FASTQ files were mapped to the human genome (hg38) using RNA STAR (version 2.7.5b) implemented on the Galaxy public server at usegalaxy.org34. Gene-level transcript abundances were calculated using featureCounts (version 1.6.4+galaxy2) in Galaxy.
Differential expression analysis was performed in Galaxy using DESeq2 (version 2.11.40.6+galaxy1) using raw counts as an input. Transcript abundances are presented as transcripts per million (TPM). Hierarchical clustering was performed on TPM using the publicly-available online platform Morpheus at https://software.broadinstitute.org/morpheus. For k-means and GO analysis, we used the top 1000 variable genes and performed an average silhouette approach to identify 6 unique clusters among the genes. K-means cluster analysis was performed using 6 clusters and the genes composing the 6 clusters were imported into Metascape [source: https://pubmed.ncbi.nlm.nih.gov/30944313/] for an express pathway analysis. GO analysis was performed on genes with fold change >2 and p<0.05 using http://geneontology.org/35,36. GSEA analysis was performed on preranked list of differentially expressed genes based on log2(FC)*log10(p) using publicly downloadable software from https://www.gsea-msigdb.org/gsea/index.jsp37,38.
Western Blot Analysis
Cells were lysed in RIPA buffer in the presence of Halt Protease Inhibitor Cocktail (ThermoFisher, Waltham, MA, USA). BCA assay was used to determine protein concentration. Equal amounts of lysates were loaded on 4 to 12% tris-glycine gels and transferred to nitrocellulose membranes. Membranes were blocked in tris-buffered saline + 0.1% Tween20 (TBST) + 5% BSA for 1 hour and incubated with primary antibodies (WNT5A and β-actin) overnight at 4°C on a shaker. The following day, membranes were washed with TBST and incubated with secondary antibodies at 1:5000 in 15 mL antibody solution/blot for 1 hr on a shaker. Blots were washed again. Then, blots were incubated with 12 mL of chemiluminescent substrates (Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA) for β-actin blots) and (Supersignal West Femto Maximum Sensitivity Substrate (ThermoFisher, Waltham, MA, USA) for WNT5A blots) for 5 min and analyzed on ChemiDoc XRS+ (Bio-Rad, Hercules, CA, USA). Bands were normalized to β-actin loading control.
Osteogenic Induction
Cells were plated at 7,000k/cm2 in a 12 well plate and cultured in αMEM+10%FBS control medium (Life Technologies, Carlsbad, CA, USA) or control medium supplemented with 50 mg/L L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma, St. Louis, MO, USA), 10 mM β-glycerophosphate disodium salt hydrate (CHEM-IMPEX INT’L INC, Wood Dale, IL, USA), and 10 nM dexamethasone (Sigma, St. Louis, MO, USA) as has commonly been used for osteoinduction39–41. Cells were treated for 3 or 4 weeks with media changes every two days.
ALP Activity Assay
To normalize between samples, the BCA assay was used to quantify protein concentration and equal amounts of protein were used for the alkaline phosphatase diethanolamine activity kit (Sigma-Aldrich, St. Louis, MO, USA). Two technical replicates were performed per sample and absorbance (410 nm) was measured on a Tecan M100 plate reader.
Alizarin Red Staining and Quantification
After four weeks in osteogenic medium or control medium, cells were fixed in 4% paraformaldehyde for 10 min and washed with DI water. Samples were stained with 0.5 mL/well of 40 mM Alizarin red (Sigma, St. Louis, MO, USA) at room temperature on a shaker for 30 min. Then, samples were washed four times with 1 mL/well DI water and imaged on an EVOS XL Core Imaging System. To quantify Alizarin red staining, samples were treated with 300 μL/well 10 w/v% cetylpyridinium chloride (Sigma-Aldrich, St. Louis, MO, USA) at room temperature on a shaker for one hr. 150 μL was transferred into 2 wells of a 96 well plate (for technical replicates) and absorbance (560 nm) was measured on a Tecan M100 plate reader.
High density fibroblast culture deceullarization
High density fibroblast culture and decellularization protocols were modified from previous methods32,42. Fibroblasts were seeded at 7,000 cells/cm2 and cultured in FibroGRO media for 10 days without passaging. At day 10, cells were decellularized using a protocol adapted from literature43,44. Briefly, cells were washed with PBS and then wash buffer 1 (100 mM Na2HPO4, 2 mM MgCl2, 2 mM EDTA). Then, they were lysed in buffer (8 mM Na2HPO4, 1% Triton X-100) and incubated at 37°C for 3 hrs with fresh lysis buffer added after each hr. Finally, matrix was washed with wash buffer 2 (100 mM Na2HPO4, 300 mM KCl) and washed with DI water. Plates were dried overnight in a sterile environment and then stored at −20°C until further sample processing.
High density fibroblast culture Mass Spectrometry Sample Preparation
Decellularized high density fibroblast cultures were prepared for trypsinization by removing plates −20°C for 20 minutes until they reach room temperature (RT). The decellularized protein was dissolved in 75 μL of 6 M urea with 3.75 μL of 200mM dithiothreitol (DTT) and incubated for 1 hr at RT. 15 μL of 200 mM iodoacetamide was added into the existing solution in each well and mixed thoroughly followed by a 1 hr incubation at RT in the dark. An additional 15 μL of DTT was added to each well and mixed thoroughly followed by a 1 hr incubation at RT in the dark. The solution was quenched with 340 μL of 1mM CaCl2 and the pH was adjusted to 7.8–8.7 with NaOH for optimal trypsin activity. Samples were trypsinized for 24 hrs at 37°C with 5 μL of 1 μg/μL Trypsin Gold, Mass Spectrometry Grade (Promega, Madison, WI, USA). The following day, the peptide solution is removed from the well plate and placed in an Eppendorf® LoBind microcentrifuge tube, frozen at −80°C for at least 3 hrs and lyophilized overnight.
Protein purification was done using the ZipTip®C18 (Millipore Sigma, Burlington, MA, USA) protocol as follows. Lyophilized samples were reconstituted in reconstitution solution (5:95 Acetonitrile (ACN):H2O, 0.1% TFA). Sample pH was then adjusted to a pH<3 with 10% TFA. ZipTips®C18 were hydrated by aspirating and expelling hydration solution (50:50 ACN:H2O, 0.1% TFA) from the ZipTip®C18 twice, followed by wash solution (0.1% TFA in H2O) twice. Samples were loaded into the ZipTips®C18 by aspirating and expelling the reconstituted sample from the ZipTip®C18 6-times. Samples were desalted by washing 3-times with wash solution. The purified peptides were then eluted into an Eppendorf® LoBind microcentrifuge tube containing elution solution (60:40 ACN:H2O, 0.1% TFA). The eluted samples were frozen, lyophilized and stored at −80°C until further analysis.
Mass Spectrometry Data Acquisition, Processing and Analysis
Purified peptides from decellularized high density fibroblast cultures were reconstituted and analyzed using 1D capillary mass spectrometry on the Thermo Orbitrap Velos. Using Proteome Discover™ Software the raw mass spectrometry data was run through the human UniPprot database for both cellular and extra cellular proteins. Proteins detected from cellular debris were excluded. Proteins with a sum of PEP score below 2 were also excluded to avoid false positives for protein detection. Raw excel files generated via Proteome Discover™ are in Data File S2. Molar percent of ECM and secreted proteins present were calculated using the exponentially modified protein abundance index (emPAI) as follows: where the emPAIA is the emPAI of the protein of interest and emPAItot is the sum of the emPAI of all ECM and secreted proteins45.
Cardiac microtissue formation
Lactate-purified CMs, SHF-FBs, EpiC-FBs, and primary human fCFBs (fCFBs; Cell Applications) were dissociated with 0.25% Trypsin for 5–10 min and then mixed together at a ratio of 3:1 CMs:FBs in RPMI/B27+ medium with 10 μM Rock inhibitor. The heterotypic cell mixtures were seeded into 400 μm inverted pyramidal agarose microwells at a density of 2000 cells per microwell and incubated overnight to allow cells to self-assemble into 3D microtissues46. 18–24 hrs later, the microtissues were removed from the microwells and transferred to low-attachment plates in RPMI/B27+ medium. Microtissues were maintained in rotary suspension culture at a density of 8000 tissues per 10 cm plate for 10 days and fed every 2–3 days with RPMI/B27+ medium.
Calcium imaging
Cardiac microtissues cultured for 10 days were incubated in Tyrode’s solution (137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.2 mM Na2HPO4, 12 mM NaHCO3, 5.5 mM D-glucose, 1.8 mM CaCl2) for 30 minutes at 37°C immediately prior to imaging. A Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-Flash 4.0 camera was used for image acquisition. Electrical field stimulation of 1 Hz was applied to the samples by placing electrodes in the Tyrode’s bath containing the microtissues (MyoPacer, IonOptix, Westwood, MA, USA). Calcium transient videos were acquired using Zen Professional software (v.2.0.0.0) at 10 ms exposure and 100 frames per sec. Circular regions of interest (65-pixel diameter) were selected at the center of each microtissue and mean fluorescence intensity values were plotted against time. Metrics of calcium transient kinetics, such as amplitude, time-to-peak, upstroke and downstroke velocities, and beat rate, were analyzed using a custom python script47 (source code available at https://github.com/david-a-joy/multilineage-organoid).
Sectioning and staining of cardiac microtissues.
Microtissues were fixed in 10% Neutral Buffered Formalin (VWR, Radnor, PA, USA) for 1 hr at room temperature and embedded in HistoGel Specimen Processing Gel (ThermoFisher, Waltham, MA, USA) prior to paraffin processing. Five micron sections were cut and adhered to positively charged glass slides. Slides were deparaffinized with xylene and re-hydrated through a series of decreasing ethanol concentrations (100%, 100%, 95%, 80%, 70%). Epitope retrieval was performed by submersing slides in Citrate Buffer pH 6.0 (Vector Laboratories, Burlingame, CA, USA) in a 95°C water bath for 35 min. Slides were cooled at room temperature for 20 min and washed with PBS. Samples were permeabilized in 0.2% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min, blocked in 1.5% normal donkey serum (Jackson Immunoresearch, Westgrove, PA, USA) for 1 hr, and probed with primary and secondary antibodies (1:400) against cTnT and VIM and counterstained with Hoechst (1:10000) (antibody information in Data File S3). Coverslips were mounted with anti-fade mounting medium (ProlongGold, Life Technologies, Carlsbad, CA, USA) and samples were imaged on a Zeiss Axio Observer Z1 inverted microscope equipped with a Hamamatsu ORCA-Flash 4.0 camera.
Statistics
All experiments were conducted using at least three technical replicates (e.g., three 12-wells) from the same differentiation. All experiments were replicated (independent differentiations) at least three times with one replicate in the 19-9-11 hiPSC line and one replicate in the H9 hESC line except where otherwise indicated. Statistical significance was evaluated using Student’s t-test, one-way analysis of variance (ANOVA), two-way ANOVA, or three-way ANOVA followed by post hoc tests used for multiple comparisons, Dunnett’s test for comparing experimental groups to a control group or Tukey’s test for comparing between all experimental groups. P<0.05 was considered statistically significant. Statistical analysis was performed using GraphPad Prism 8.0 or JMP PRO 15 software.
Data and Code Availability
Sequencing data is available at Gene Expression Omnibus GSE168380. All processed data are available in the paper or supplementary information.
Results
1. Molecular Characterization of hPSC-CFBs Reveals Distinct CFB Signatures
hPSCs were differentiated to CFBs through WT1+ epicardial cell progenitors treated with FGF2 (EpiC-FB) or through TBX1+HAND2+ second heart field progenitors (SHF-FBs) via the GiFGF protocol, as shown in Fig. 1a. Resulting CFBs from both protocols were maintained in FibroGRO media (containing 2% FBS) and passaged at ~80% confluency. EpiC-CFBs have a similar morphology, growth rate, and time to senescence compared to SHF-FBs (Fig. S1)21.
Fig. 1. Comparison of fibroblast markers and cardiac transcription factors.

(A) Schematic of hPSC differentiation to epicardial-derived and second heart field progenitor-derived cardiac fibroblasts. (B) qPCR analysis of cardiac transcription factor expression in hPSCs (D0), EpiC-FB at P1, and SHF-FBs at P3 relative to GAPDH. The y-axis corresponds to relative fold change, 2^(–ddCt) and is a log scale. Each color represents an independent differentiation and each dot represents a the average of two technical replicates. Samples having expression below the limit of detection are reported as not detected (N.D.). Statistics are *P<0.05 and **P<0.01 using Student’s t-test comparing the averages of three well replicates from three or four independent SHF-FB and EpiC-FB differentiations. See also Figure S1–2.
First, we compared expression of fibroblast markers in EpiC-FBs and SHF-FBs immediately after differentiation (P1). By flow cytometry, we observed that the majority of EpiC-FBs and SHF-FBs expressed TE7, CD90, and VIM, and 20–40% of EpiC-FBs and SHF-FBs expressed FSP1, similar to primary adult dermal fibroblasts (dFBs) (Fig. S1)48. Surprisingly, WT1+ EpiCs also expressed canonical fibroblast markers FSP1, TE7, CD90, and VIM at similar levels to EpiC-FBs and SHF-FBs (N.S., p>0.05) (Fig. S1). There has been significant debate as to whether these markers are specific to fibroblasts48. However, the hPSC-derived CFBs did not express the epicardial marker WT1 (p<0.01 in comparison to EpiCs) or high levels of the smooth muscle cell marker calponin, consistent with a fibroblast molecular signature. Immunocytochemistry for these markers demonstrated expected nuclear localization of WT1 in EpiCs, striated patterns of calponin in EpiC-smooth muscle cells (SMCs)49, localization of VIM to filaments in hPSC-CFBs, cell-surface localization of CD90 in hPSC-CFBs, and cytoplasmic localization of TE7 and FSP1 in hPSC-CFBs (Fig. S1).
Next, we compared expression of a panel of cardiac transcription factors in differentiated SHF-FB and EpiC-FB (Fig. 1b). Dual cardiac heart field markers ISL1 and GATA4 were expressed in EpiC-FB and SHF-FB to a greater extent than in hPSCs which aligns with previous data demonstrating expression of these key cardiac proteins in SHF-FB21. TBX genes have previously been used to spatiotemporally mark different lineages of the mouse heart and are thought to regulate cardiac regionalization through crosstalk with other cardiac transcription factors. For example, TBX1 marks second heart field progenitors and is later expressed in the outflow tract and non-chamber regions of the heart, while TBX18 marks epicardial progenitors and is later expressed in the sinus horns and left ventricle50. Spatial expression patterns of TBX gene expression have not yet been explored in developing human hearts, so we analyzed the expression of cardiac transcription factors and fibroblast markers in single cell sequencing data from fetal human hearts which also contained spatial data (Fig. S2)15. Asp et al. identified five fibroblast clusters based upon their high expression of ECM proteins and lack of sarcomeric genes. We verified that these fibroblast clusters contained cells which expressed high levels of the cardiac transcription factor GATA4 and fibroblast marker THY1 and VIM and then proceeded using these clusters for our analysis. We found that TBX2 was expressed in cells the authors identified as related to larger vessel development by gene ontology analysis (Cluster 8), TBX3 was expressed in cells within the outflow tract (Cluster 5) but not those associated with the base of the outflow tract (Cluster 2), and TBX18 was expressed in the atrioventricular sub-epicardial mesenchyme (Cluster 3). Interestingly, HAND2, a canonical SHF marker which is also expressed in the developing epicardium, was expressed in Clusters 2, 3, 4 and 5.
Our qPCR analysis showed that EpiC-FBs expressed significantly higher levels of TBX18 (p<0.01) and TBX20 (p<0.01), which are expressed in the epicardium and epicardial-derived cells, compared to the SHF-FBs (Fig. 1b)51,52. EpiC-FBs also expressed higher levels of TBX3 (p<0.01), which is important in conduction system development53,54, and TBX2 (p<0.01), a marker associated with the outflow tract and atrioventricular canal development55. TBX1 is a transient transcription factor expressed during second heart field development, and has previously been shown to be upregulated in SHF progenitors during differentiation to SHF-FBs21. Compared to EpiC-FBs, differentiated SHF-FBs also expressed higher levels TBX1 (p<0.05)56. HAND2 has often been used to distinguish FHF and SHF lineages. Additionally, we observed higher levels of HAND2 in the EpiC-FB compared to the SHF-FB (p<0.01), which was more than 10–100 fold higher than expression in hPSCs. This suggests the hPSC differentiation protocols can generate fibroblast populations expressing distinct sets of markers which are representative of in vivo cardiac fibroblast populations.
To further probe molecular similarities and differences between CFB populations, we performed bulk RNA sequencing on SHF-FBs, EpiC-FBs, primary human fetal cardiac fibroblasts (fCFBs), primary human adult cardiac fibroblasts (aCFBs), and primary adult dFBs (Data File S1). We used three independent differentiation replicates in H9 hESCs for the hPSC-derived cell types and three technical replicates of the primary cell types. Although we observed significant batch-to-batch variation between the SHF-FB differentiations, all samples expressed fibroblast markers VIM, THY1, and DDR2, and the cardiac transcription factor GATA4 to similar levels, which aligns with our previously described differentiation quality control metrics. Between the different fibroblast populations, we observed differential expression of cardiac transcription factors and ECM related proteins (Fig. S3), which emphasizes fibroblast transcriptomic heterogeneity amongst and within tissues57. The data suggest that fCFB and aCFB samples were primarily from the epicardial lineage based on high expression of TBX3 and TBX18, and therefore, these samples likely predominantly represent one CFB subtype. However, both SHF-FBs and EpiC-FBs expressed pan-cardiac markers ISL1 and GATA4 at a similar level to primary CFBs, suggesting that the hPSC-CFB populations recapitulated some degree of the primary CFB molecular signature.
We next performed k-means clustering on the top 1,000 differentially expressed genes and identified clusters of genes enriched in each cell type (Fig. S4). Cluster 2 (CM-specific) included sarcomeric genes such as MYH6, MYH7, TNNT2, and TTN which demonstrate that CMs are distinct from the fibroblast samples as expected. Additionally, COL6A1 and COL6A3 were enriched in the dFBs (Cluster 4) which suggested a different composition of ECM compared to CFB. Cluster 1 (genes enriched in SHF-FB) included genes such as GJA1, known to be important in fibroblast-CM interconnectivity, and MCM7, important in cell cycle regulation58,59. Cluster 3 (genes enriched in EpiC-FBs) included ECM proteins FN1, COL3A1, COL1A1, and COL1A2, suggesting a distinct composition of ECM between the EpiC-FBs and SHF-FBs. Clusters 4, 5, and 6 were genes enriched in primary CFBs and dFBs and included FB markers such as VIM, DCN, and POSTN. Gene ontology analysis of differentially expressed genes between primary CFBs and hPSC-CFBs yielded the term “active transmembrane transporter activity” enriched in primary CFBs and “adenylate cyclase inhibiting G protein-coupled glutamate receptor activity” and “GABA-gated chloride ion channel activity” enriched in hPSC-CFBs. This suggests that hPSC-CFBs may be representative of an earlier stage in development than primary CFB samples.
A direct comparison of SHF-FB and EpiC-FB transcriptomes (Fig. 2a) highlights key molecular differences which have been linked to specific developmental lineages. For example, MYH10 was more highly enriched in the EpiC-FBs than in SHF-FBs, and has been shown to inhibit epithelial-to-mesenchymal transition of epicardial cells during mammalian development60. Additionally, Sall1 is expressed in precardiac mesoderm and expression is maintained in the murine second heart field, and Wnt7b is expressed in the remodeling atrioventricular and mitral valve at E16.561,62. We validated differential expression of SALL1 in three independent differentiations and demonstrated that indeed this gene is a marker of the SHF-FB (Fig. S4c). Additionally, comparing key markers of CFB subtypes from a human spatiotranscriptomic dataset (Fig. S2), we identified that DCN, LUM, PENK, and ASPN were enriched in the EpiC-FBs and NDUFA4L2 was enriched in the SHF-FBs. GSEA analysis of differentially expressed genes using KEGG pathways identified terms such as “cell cycle” upregulated in SHF-FBs and “gap junctions” in EpiC-FBs (Fig. 2c). Taken together, these data suggest that hPSC-derived SHF-FBs and EpiC-FBs are transcriptionally similar to primary CFBs and express distinct molecular signatures corresponding to their developmental lineages.
Fig. 2. Transcriptomic analysis of hPSC-CFBs.

(A) Volcano plot comparing EpiC-FB and SHF-FB at P1 using three samples collected from three independent differentiations in the H9 line. (B) GSEA KEGG Pathway Enrichment plot on preranked list of genes based on −log10(P)*sign*log2(FC). See also Figure S3–4 and Data File S1.
2. Mass Spectrometry of Decellularized Matrix reveals hPSC-CFB Lineage Leads to a Distinct Matrix Composition
Although fibroblasts are not the only cell type in the heart to produce ECM, they are thought to contribute a significant proportion of the matrix and that changes in matrix composition occur as a function of changes in fibroblast matrix secretion. Our transcriptomic analysis identified differentially-expressed ECM and ECM-related genes, however transcription is upstream of matrix secretion and accumulation. Therefore, to compare ECM composition secreted by EpiC-FBs and SHF-FBs on a protein level, we performed mass spectrometry on decellularized matrix after ten days of high-density culture. We validated that the matrix was decellularized by absence of Hoechst staining (Fig. S5a). To compare the composition of deposited ECM, proteins were divided into ten distinct categories (Fig. 3a). Notably, fCFBs and the hPSC-CFBs showed similar relative percentages of all ten ECM categories, suggesting that the hPSC-CFBs have a more fetal-like phenotype. The most prevalent ECM category for fCFBs and hPSC-CFBs was linking ECM, composed primarily of fibronectin which correlates with expectations of developing heart matrix63. By contrast, the most prevalent ECM category in aCFBs was matricellular proteins, ECM components not typically involved in structural support but that instead interact directly with bioeffector molecules. We observed dFB matrices were most distinct from CFB due to higher accumulation of fibrillar proteins, including collagen III and VI, consistent with transcriptomic analysis (Fig. S5b). Collagen VI is abundant in the native dFB matrix and is important in dFB matrix assembly and regulating cell motility, whereas in the heart, collagen VI is only present under high stress conditions64,65. dFB matrices also exhibited differences in remodeling proteins including high proportions of serine protease HTRA1.
Figure 3. ECM composition and secreted factors.
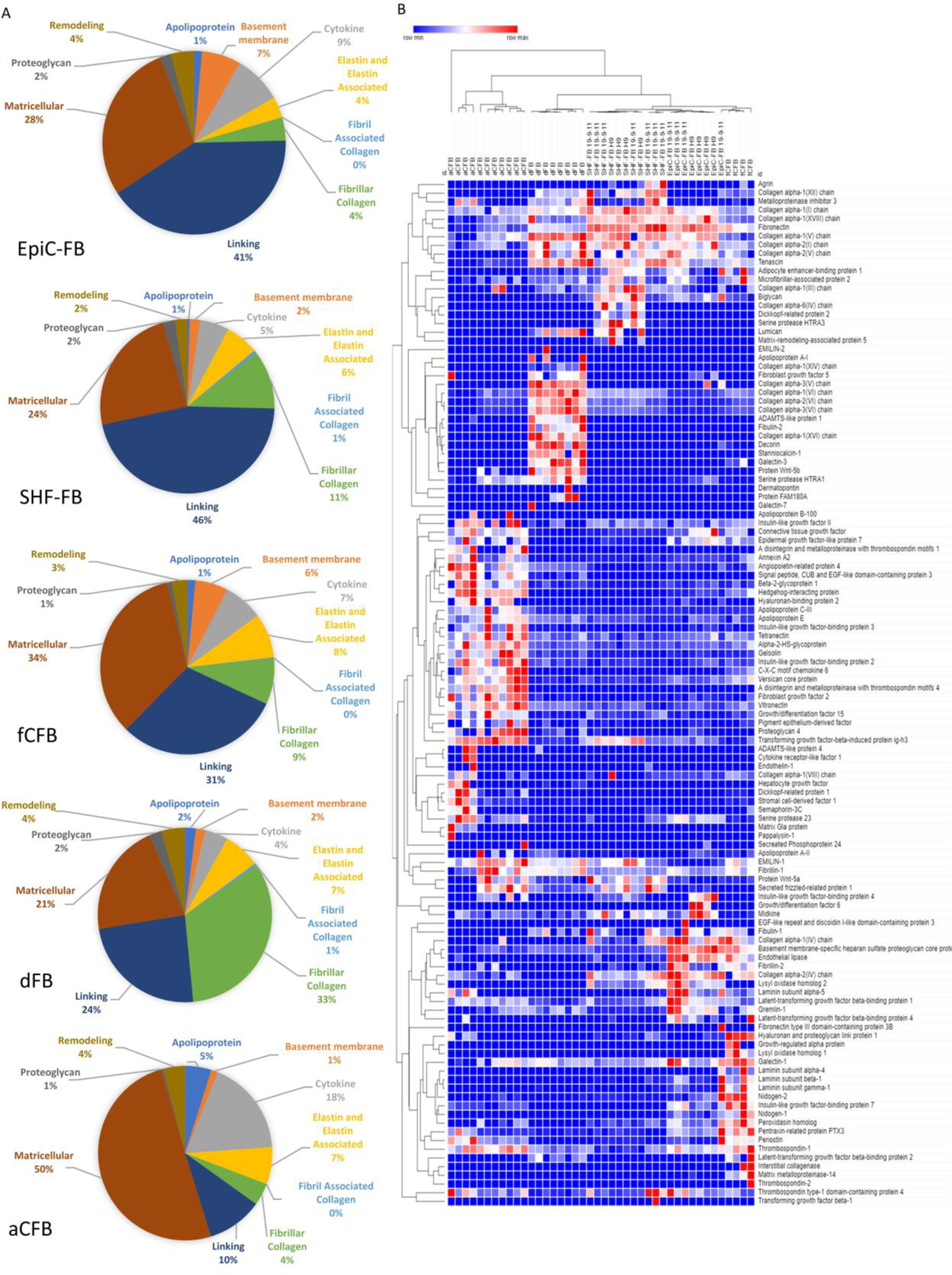
Mass spectrometry proteomics comparing decellularized matrix deposited by EpiC-FBs, SHF-FBs, dermal FBs, primary adult CFBs, and primary fetal CFBs at P3–5 with 4–11 samples. EpiC-FB and SHF-FB samples were collected from three independent differentiations in either the H9 or 19-9-11 line. (A) Pie charts displaying fractional compositions of extracellular matrix components. (B) Heat map comparing matrix and secreted factors from fibroblasts. See also Figure S5–6 and Data File S2.
Hierarchical clustering of all 118 proteins detected showed low sample technical and biological variation and no dependence on hPSC line used to generate the CFBs (H9 hESC or 19-9-11 hiPSC). hPSC-CFB matrices clustered with fCFB matrices (Fig. 3B), consistent with the ECM category analysis of Fig. 3A. fCFBs and hPSC-CFB matrices clustered separately from both aCFB and dFB matrices based largely upon deposition of greater amounts of fibronectin (Fig. S5b, Data File S2). In the developing mouse heart, fibronectin has been shown to regulate cardiovascular morphogenesis through integrin signaling, and in vitro coculture of CFBs and CMs isolated from E12.5-E13.5 hearts promoted CM proliferation through fibronectin synthesis66,67. Surprisingly, the dFB matrix clustered more closely to fCFB and hPSC-CFB matrices than the aCFB matrix (Fig. 3B). Among various matricellular proteins that were detected at high levels in the aCFB matrix were insulin-like growth factor binding proteins and angiopoietin related protein 4 (Fig S5b). Notably, insulin-like growth factor binding proteins have been studied as biomarkers for cardiovascular disease risk in adults68–70.
Comparing the compositions of matrices deposited by the fCFBs and hPSC-CFBs with aCFBs from our dataset we observed increased levels of fibronectin (p<0.01), collagen I (p<0.01 comparing hPSC-CFB to aCFB) and tenascin (p<0.01) in the hPSC-CFBs (Fig S5b). Additionally, we observed decreased lysyl oxidase, a key remodeling protein that is integral for elastin and collagen crosslinking and fibrillogenesis, in aCFB matrices relative to fCFB (p<0.01), SHF-FB (p<0.05) and EpiC-FB (p<0.01) matrices71,72. This is consistent with the substantial amount of collagen and elastin synthesis and remodeling that occur early in development and in the first few weeks postnatally73,74. We observed higher levels of collagen I in the hPSC-CFB matrices compared to aCFB matrices (p<0.01) while overall low levels of collagen III were detected across all CFB matrices. Taken together, these results demonstrate that CFB matrix composition changes as CFBs mature. hPSC-CFB and fCFB matrices contained higher levels of fibronectin (p<0.01), lysyl oxidase (p<0.01), and higher levels of collagen I (p<0.01) compared to the aCFB matrices suggesting they are more fetal than adult-like.
Additionally, there are a few key differences between the EpiC-FB and fCFB matrices relative to the SHF-FB matrices indicating a lineage-dependent matrix composition for EpiC-FB and SHF-FB. One major difference, as seen in Fig. 3a, is the large amount of basement membrane proteins present in EpiC-FB and fCFB matrices. This difference can be partially attributed to a slightly larger of abundance of laminins and collagen IV in EpiC-FB and dFB matrices, but is mainly due to large percentage of basement membrane-specific heparan sulfate proteoglycan core protein, also known as perlecan, present in EpiC-FB and fCFB matrices (p<0.01 in comparison to all other matrices). Perlecan has been shown play an integral role in development of the coronary vasculature, heart stability as well as cardiomyocyte organization and sarcomere structure75–77. These data suggest that EpiC-FB may play a role in perlecan synthesis during development. Other matrix components detected in larger proportions in EpiC-FB and fCFB matrices include pentraxin related protein PTX3 (N.S. EpiC-FB matrix compared to aCFB matrix, all other comparisons p<0.01) and Nidogen 1 and 2 (p<0.05) (Fig. S5b). Interestingly, periostin is present in larger proportions in aCFB matrices (p<0.01) as well as fCFB (p<0.01) matrices in comparison to SHF-FB matrices suggesting a lineage specific role of periostin in development and adult cardiac matrix homeostasis78.
3. hPSC-CFBs Secrete Lineage Specific Factors
CFBs secrete signaling factors that have been shown to alter CM contraction through ion channel remodeling and cardiac hypertrophy in vitro79. We identified several differentially upregulated secreted factors in the aCFB matrices compared to the fCFB matrices, including C-X-C motif Chemokine 6 (p<0.01) and Growth/differentiation factor 15 (p<0.01) (Fig. S5b). These factors were higher in the aCFB matrices compared to hPSC-CFB matrices (C-X-C motif Chemokine 6 p<0.01, stromal cell-derived factor 1 p<0.05, Dickkopf-related protein 1 p<0.01, and Growth/differentiation factor 15 p<0.01) and contributed to distinct clustering of aCFB matrices relative to other CFB matrices again suggesting that hPSC-CFBs are more fetal-like than adult-like. There were high proportions of secreted hedgehog-interacting protein, an inhibitor of the hedgehog signaling pathway, associated with aCFB and fCFB matrices (p<0.01 in comparison to all other matrices). SHF-FB matrices contained low levels of Gremlin-1 (p<0.01 in comparison to EpiC-FB matrices), epidermal growth factor-like protein 7 (p<0.01 in comparison to EpiC-FB and aCFB matrices), and connective tissue growth factor (p<0.01 in comparison to EpiC-FB and aCFB matrices) which have been shown to be upregulated during cardiac fibrosis, compared to EpiC-FB and aCFB matrices80,81.
We also detected WNT signaling factors, which play key roles in cardiac development62, associated with fibroblast matrices. Interestingly, we only detected WNT5B in dFB matrix, and more intriguingly, we discovered higher levels of WNT5A and secreted frizzled-related protein 1 in the SHF-FB matrices compared to the EpiC-FB (p<0.01) and fCFB matrices (p<0.05). In mouse development, Wnt5a expression in second heart field progenitors causes polarized second heart field morphogenesis and migration via non-canonical Wnt signaling and is required for proper outflow tract development82–85. Thus, to further investigate if WNT5A is upregulated by SHF-FBs, we compared WNT5A transcripts by qPCR in the SHF-FBs and EpiC-FBs (Fig. S5c). We did not observe any statistically-significant changes across three differentiations. Since WNT5A undergoes post-translational modifications, we also compared WNT5A protein expression by Western blot and observed approximately two-fold higher increase in WNT5A protein expression in SHF-FBs compared to EpiC-FBs (p<0.05) (Fig. S5d). Thus, hPSC SHF-FBs exhibit secretion of a distinct set of matricellular proteins that may affect heart development and function.
4. Fibroblast Activation Reveals Greater Activation Potential in EpiC-FBs than SHF-FBs
Extracellular matrix production is a key function of CFBs, however they also play key roles in tissue stress, disease, and repair. Thus, we performed two functional assays to ascertain the ability of hPSC-CFBs to become activated in response to stress.
Fibroblasts, when stressed in vivo, transition to a myofibroblast state characterized by an increased cell size and increased expression of smooth muscle actin (SMA). To test if hPSC-derived SHF-FBs and EpiC-FBs exhibit differential activation in vitro, we treated them with TGFβ1-, Angiotensin-II-, and FBS- supplemented media for 2 days and then compared by flow cytometry relative cell size assessed by forward scatter area (FSC-A) and SMA expression. We used undifferentiated hPSCs and no primary antibody conditions as negative flow gating controls and EpiC-SMCs49 as a positive control. We then individually tested the effects of TGFβ1, Angiotensin-II, and serum. Across multiple differentiations, we observed fibroblast activation as demonstrated by SMA induction and increased FSC-A in EpiC-FBs (F+10 ng/mL TGFβ1 N.S, all others p<0.01 for change in FSC-A and SMA expression), SHF-FBs (F+10 ng/mL TGFβ1 N.S change in FSC-A and p<0.05 change in SMA expression, D p<0.05 change in FSC-A and N.S. change in SMA expression, D+10 ng/mL TGFβ1 and D+1000 ng/mL Angiotensin-II p<0.01 change in FSC-A and N.S. change in SMA expression, D+100 ng/mL TGFβ1 p<0.01 change in FSC-A and p<0.01 change in SMA expression), and aCFBs (F+10 ng/mL TGFβ1 N.S. change in FSC-A, all other conditions p<0.01 change in FSC-A) and not dFBs (N.S. change in FSC-A or SMA expression) under serum or serum with the addition of TGFβ1 and Angiotensin-II (Fig. 4, Fig. S7). We observed a high variability in basal SMA activation between hPSC-CFBs differentiated from different stem cell lines which may result from genetic variability as previously demonstrated86, however we also observed differentiation-to-differentiation variability. Regardless, we consistently observed higher activation levels in EpiC-FBs compared to SHF-FBs (fold change SMA percentage p<0.01, fold change FSC-A p<0.01) which suggests that EpiC-FBs may be preferred for studying transition to myofibroblast-like cells in a stress response or screening for compounds that induce fibroblast activation (Fig. 4d).
Fig. 4. Fibroblast stress activation by addition of TGFβ−1, Angiotensin-II, or serum.
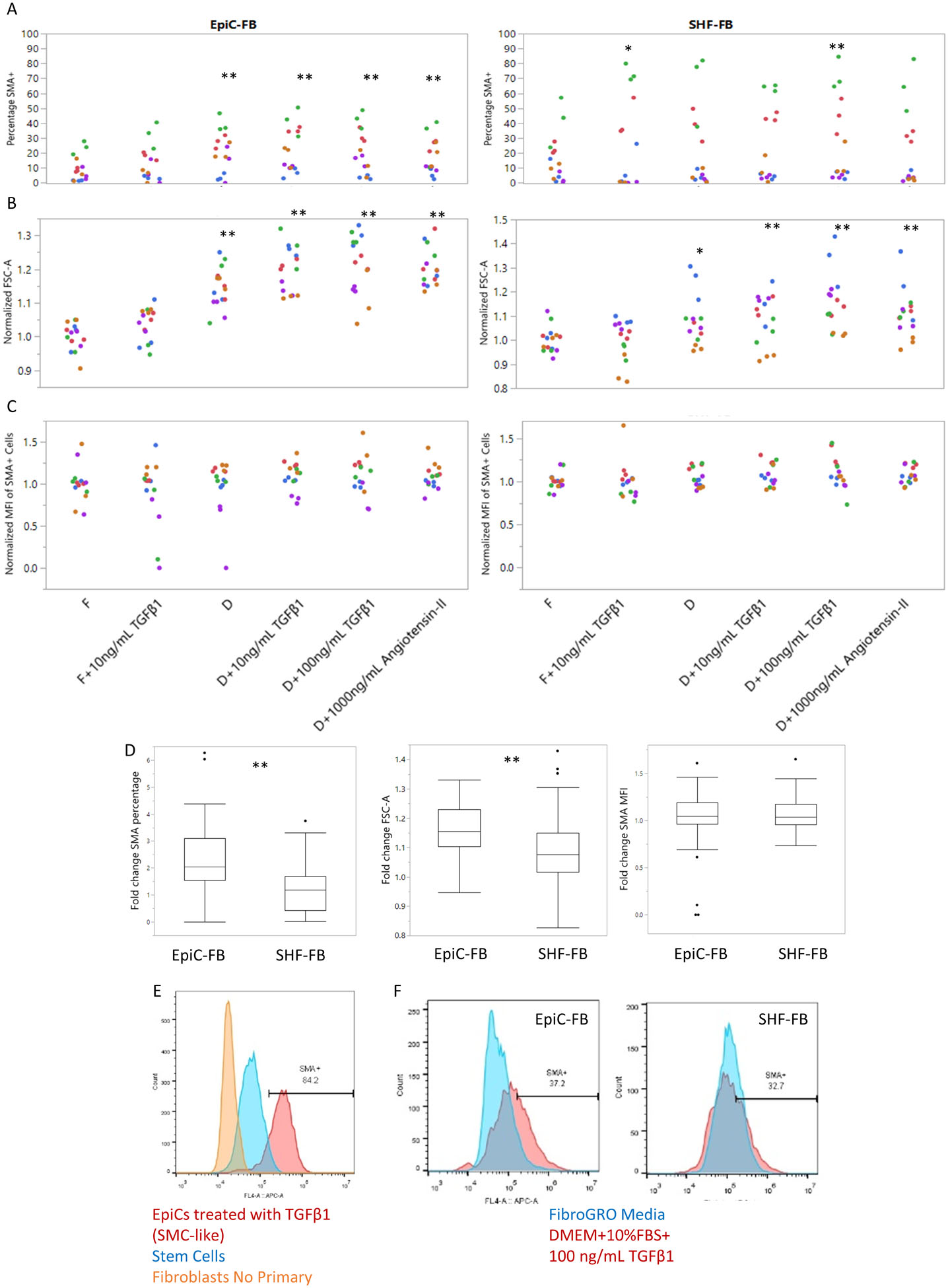
Flow cytometry analysis of EpiC-FBs and SHF-FBs treated with FibroGRO or DMEM+10%FBS media (F=FibroGRO, D=DMEM+10%FBS) and small molecule additions of 10 ng/mL TGFβ1, 100 ng/mL TGFβ1, or 1000 ng/mL Angiotensin-II to induce fibroblast activation. EpiC-FBs and SHF-FBs were from five independent differentiations at P1–5 in either the H9 line or the 19-9-11 line. (A) Percentage SMA+ (B) Median FSC comparison as a relative analysis of cell size. (C) SMA normalized mean fluorescence intensity (MFI) as a sign of fibroblast activation. Each color represents an independent differentiation and each dot represents a well replicate. Statistics are *P<0.05 and **P<0.01 using ANOVA comparing cell treatments. (D) Box and whisker plots of fold change SMA+ percentage, FSC-A, and SMA MFI compared to FibroGRO media condition for each experiment and cell type. Statistics are *P<0.05 and **P<0.01 using 3-way ANOVA controlling for cell treatment and experiment. (E,F) Example flow gating plots for SMA expression. Controls include EpiCs treated with TGFβ1 (SMC-like), undifferentiated hPSCs, and fibroblasts with no primary antibody. See also Figure S7.
5. Fibroblast Mineralization Reveals Greater Potential in SHF-FB than EpiC-FB
Heart calcification results in mineral deposits in the heart valves, coronary arteries, myocardium, and pericardium and is a common pathology associated with diseases including heart blocks87 and human CFBs in vitro were shown to mineralize when exposed to osteogenic medium. Thus, we hypothesized mineralization potential may differ between CFB subtypes.
To test whether EpiC-FBs and SHF-FBs have different mineralization potential, we cultured primary dFBs, fCFBs, aCFBs, EpiC-FBs, and SHF-FBs in osteogenic medium containing L-ascorbic acid, β-glycerophosphate, and dexamethasone for four weeks and compared changes in ALP activity and Alizarin red staining. We observed increased ALP activity upon addition of osteogenic factors in all cell types except the EpiC-FBs (Fig. 5a). Additionally, the hPSC-CFBs had lower mineralization potential, as measured by percentage change in ALP activity with addition of the osteogenic factors, compared to primary fibroblasts (p<0.05), and across four differentiations, EpiC-FBs had lower osteogenic potential than SHF-FBs (p<0.01) (Fig. 5b). To assay mineralization, we stained with Alizarin red and quantified fluorescence. Staining was highest in the aCFBs when exposed to osteogenic factors, and no difference was observed between SHF-FBs and EpiC-FBs (Fig. 5d–f). Overall, this suggests that SHF-FBs may have a higher calcification potential compared to EpiC-FBs but lower than aCFBs.
Fig. 5. Mineralization of CFBs.

Fibroblasts were treated with osteogenic medium for 4 weeks to induce calcification and mineralization. SHF-FBs and EpiC-FBs were from three or four independent differentiations at P1–5 in either the H9 line or the 19-9-11 line. (A) Relative ALP activity in dFBs, fCFBs, aCFBs, EpiC-FBs, and SHF-FBs. Red bars represent control medium and blue bars represent osteogenic medium. Plotted are the mean ±SEM of three well replicates, *P<0.05 and **P<0.01 comparing control medium and osteogenic medium for each cell type using Student’s t-test. Graph is representative of four independent differentiations. (B) Percentage change in ALP activity of fibroblasts in osteogenic medium compared to control medium. Plotted are the mean percentage change of three well replicates with error bars calculated by propagation of error, *P<0.05 and **P<0.01 comparing between cell types using ANOVA. Graph is representative of four independent differentiations. (C) Relative ALP activity of CFBs treated with αMEM+10%FBS for four weeks, *P<0.05 and **P<0.01 comparing between cell types using ANOVA. (D) Alizarin red staining to depict fibroblast mineralization in osteogenic medium. Scale bar is 100 μm. (E) Quantification of Alizarin red staining. Plotted are the mean percentage change of three well replicates with error bars calculated by propagation of error, *P<0.05 and **P<0.01 comparing between cell types using ANOVA. Graph is representative of three independent differentiations. (F) Percentage change in Alizarin red staining of fibroblasts in osteogenic medium compared to control medium. Plotted are the mean percentage change of three well replicates with error bars calculated by propagation of error, *P<0.05 comparing between cell types using ANOVA. Graph is representative of three independent differentiations. (G) Alizarin red staining of CFBs treated with αMEM+10%FBS for four weeks, *P<0.05 and **P<0.01 comparing between cell types using ANOVA.
Calcification, mineralization, and nodule formation have been used to analyze valve interstitial cell activation level88. To compare if hPSC-CFBs have differential response to activation, we treated the cells in serum-supplemented media, which caused fibroblast activation, for four weeks. We observed higher ALP activity (p<0.05) and Alizarin red staining (p<0.01) in the SHF-FBs compared to EpiC-FBs (Fig. 5c, g). Taken together with the previous results, this suggests that the SHF-FBs have higher mineralization and calcification potential in response to serum with or without additional osteogenic factors which may be indicative of regional specific differences in vivo.
6. Cardiac Fibroblasts Promote Robust Microtissue Formation and Enhance Calcium Cycling
Although cell level analysis provides insight into fibroblast function, it does not address how CFBs impacts overall cardiac tissues. Therefore, we formed engineered cardiac microtissues from a heterotypic mixture of hPSC-CMs and CFBs in order to study how EpiC-FBs and SHF-FBs supported CM function compared to primary human fCFBs. The same pool of lactate-enriched CMs was used for all microtissue groups, suggesting that any differences in tissue formation and function were due to the different sources of fibroblasts and their ability to influence CM physiology. The population of highly-enriched CMs (>90% cTnT+) did not form into uniform microtissues when aggregated alone (without CFBs), but instead clumped into small clusters (Fig. 6a). In contrast, all microtissues that contained CFBs self-assembled into 3D spheroids within 24 hrs of seeding into microwell molds (Fig. 6ai), indicating that a stromal population is necessary to facilitate robust microtissue assembly. The heterotypic cardiac microtissues compacted after removal from the microwell molds, as demonstrated by the more distinct tissue boundaries and increasingly spheroidal shape 3 days after tissue formation compared to 1 day after, and stably persisted throughout the 10 days of microtissue culture. Sectioning of cardiac microtissues shows CFBs dispersed amongst CMs by staining for VIM and cTnT, respectively (Fig. 6b).
Fig. 6. Cardiac microtissue formation and calcium handling.
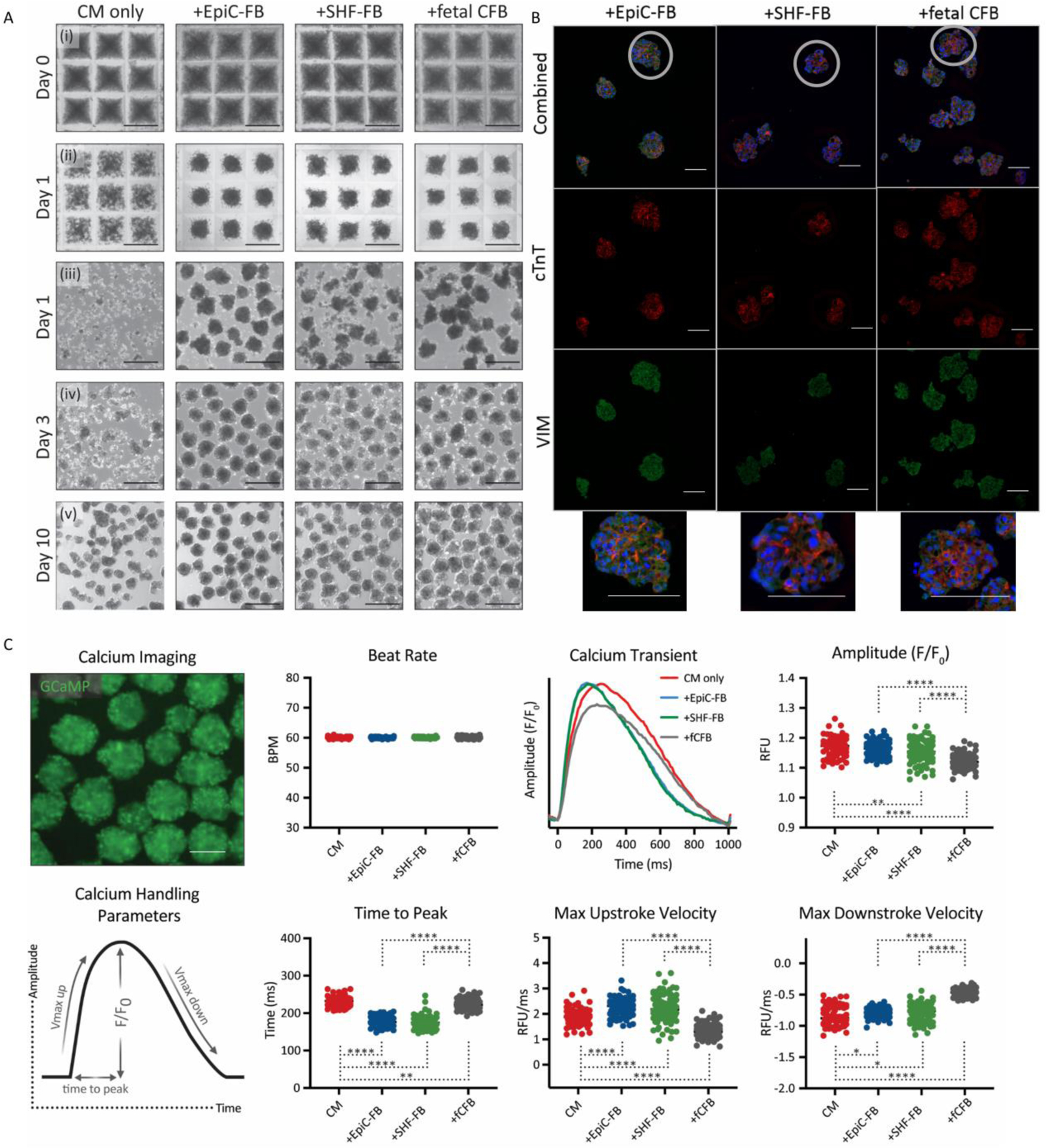
Enriched hPSC-CMs were seeded alone or in combination with EpiC- or SHF-derived CFBs or with primary human fetal CFBs (3:1 ratio) at 2000 cells per microwell. (A) Bright field images of microtissues (i). 1 day later, the heterotypic cell mixtures had robustly self-assembled into 3D spheroids while the CM only cells had not (ii- in microwells; iii- immediately after removing from microwells). Heterotypic cardiac microtissues compacted over the next few days (iv) and remained stable throughout 10 days of culture (v). Scale bar is 200 μm. (B) Immunocytochemistry of sectioned aggregates for DAPI, cTnT, and VIM. Scale bar is 100 μm. (C) GCaMP fluorescence of calcium flux in cardiac microtissues (top) and definitions of calcium handling parameters (bottom). Scale bar is 100 μm. Cardiac microtissues were subjected to 1Hz electrical field stimulation and calcium transient amplitude, time-to-peak, and stroke velocities were quantified for each microtissue condition. *p < 0.05, **p < 0.01, ****p < 0.0001. hPSC-CMs were differentiated in the WTC11-GCaMP6f line. EpiC-FBs and SHF-FBs were differentiated in the 19-9-11 line.
Calcium handling properties of the engineered cardiac microtissues containing SHF-FBs or EpiC-FBs were assessed in order to determine whether different CFB subtypes alter CM function. Day 10 microtissues were subjected to 1 Hz electrical field stimulation to eliminate intrinsic differences in beat rate between the individual tissues (Fig. 6c). Video-based imaging of the fluorescence intensity of the genetically-encoded calcium sensor GCaMP6f in the CMs enabled quantification of the kinetic parameters of the microtissue calcium transients. There was no statistical significance between any of the calcium handling properties from microtissues made with EpiC-FBs compared to those made with SHF-FBs, indicating that the hPSC-CFB subtypes similarly support hPSC-CM function. The calcium transient amplitudes of the +EpiC-FB and +SHF-FB microtissues were similar to those of the hPSC-CM only (N.S. +EpiC-FB to CM, p<0.01 +SHF-FB to CM) microtissues and slightly higher than the values of the microtissues containing primary fCFs (p<0.0001) (Fig. 6c). The microtissues comprised of the hPSC-CFBs displayed the fastest upstroke kinetic properties, taking the shortest time to reach the peak of the calcium transient (p<0.001 in comparison to CM or +fCFB) and exhibiting the fastest maximum upstroke velocities (p<0.001 in comparison to CM or +fCFB). The maximum downstroke velocities followed the same trend as the amplitude values, where the CM only, CM+EpiC-FB, and CM+SHF-FB microtissues exhibited similar velocity values and were all faster than the CM+fCFB microtissues (p<0.0001).
Since heterotypic cardiac microtissues containing hPSC-CFBs demonstrated faster kinetic properties compared to microtissues containing fCFBs, this reflects that the ‘age-matching’ or ‘source-matching’ between heterotypic cell types facilitated improved performance. Previous studies from our lab demonstrated that non-cardiac specific hPSC-FBs did not support heterotypic cardiac microtissue and function suggesting that the cardiac-specific derivation process of the hPSC-FBs is necessary for robust culture of engineered cardiac tissues89. Thus, these phenotypic assays suggest that SHF-FBs and EpiC-FBs contribute to the generation of microtissues and support similarly CM function.
Overall, this study demonstrated that SHF-FBs and EpiC-FBs mimic in vivo CFB equivalents at the transcriptional level, in addition to its differential development nature, EpiC-FBs and SHF-FBs have distinct extracellular matrix compositions and fibrotic potential indicating that the EpiC-FB may be preferred for fibrotic disease modeling studies and the SHF-FB for tissue engineering and cell therapies.
Discussion
Fibroblasts are present in many organs and play a key support role by producing ECM, secreting paracrine factors, and responding to injury and other stresses. Presumably during development, lineage specification and niche environments manifest in functional differences amongst fibroblasts at the tissue level, although our understanding of tissue specificity of fibroblasts remains limited. CFBs arise from multiple developmental lineages, which have been shown to contribute to different regions of the heart. A human heart spatiotranscriptomics study has identified five fibroblast populations localized to spatially distinct regions which can characterized by differential expression of markers including ASPN, LUM, DCN, PENK, and NDUFA4L215. Additionally, CFBs from the left and right side of the heart have been shown to differentially express genes related to fibrosis suggesting that fibroblast specification may result in functional differences17.
In this study, we compared hPSC-CFBs that differentiate through epicardial and second heart field progenitors (EpiC-FBs and SHF-FBs, respectively) to model CFB subtypes observed in vivo. Both populations expressed canonical fibroblast markers VIM, TE7, and CD90, and the pan-cardiac transcription factor GATA4, and enhanced formation and calcium handling properties in cardiac microtissues containing hPSC-derived CFBs and CMs, indicating that both EpiC-FB and SHF-FB are beneficial in cardiac tissue engineering and regenerative medicine applications.
Epicardial-derived CFBs represent the majority of all CFBs in the adult human heart and are identifiable by expression of epicardial-lineage markers TBX3, TBX18, and TBX205. As expected, hPSC-derived EpiC-FBs similarly expressed these markers. Phenotypically, EpiC-FBs have higher stress-induced activation potential to transition to a myofibroblast-like state than SHF-FBs and they secrete Gremlin-1, epidermal growth factor-like protein 7, and connective tissue growth factor, which have been implicated in fibrosis80,81. Thus, one should consider using EpiC-FBs in modeling cardiac fibrosis and anti-fibrotic drug testing where CFB activation and fibrosis are important.
SHF-FBs represent a portion of CFBs which arise from the second heart field and can be identified by expression of lineage markers TBX1 and SALL1. hPSC-derived SHF-FBs also expressed second heart field markers as well as noncanonical WNT signaling pathway factors including WNT5A, which developmentally is secreted from second heart field progenitors and is required for proper outflow tract development82–85. SHF-FBs have a higher mineralization potential than EpiC-FBs, suggesting they may be better for modeling of cardiac tissue mineralization. Additionally, they lack expression of factors identified in the EpiC-FBs as associated with fibrosis, and therefore may be ideal for cell therapy into an injured or diseased heart, or for other applications where activation is undesired.
The developmental stage of CFB maturation has been shown to influence CM proliferation and contraction and endothelial cell tubule formation in vitro66,90–93. fCFBs compared to aCFBs have distinct transcriptional and epigenetic signatures92,94,95. Furthermore, cardiac matrix changes through development, and we demonstrated that human CFB ECM matrix composition changes from fetal to adult matrices63,93,96. EpiC-FBs and SHF-FBs are more fetal-like than adult-like in ECM synthesis and deposition. These fetal properties may have advantages in development modeling and in regenerative therapies, but development of strategies to functionally mature the hPSC-derived CFBs to a more adult-like state with increased mineralization potential and altered ECM composition may be important for advancing later stage development and disease modeling applications.
Furthermore, CFBs from different anatomical locations, such as the valves and chambers, are thought to have differential potential roles in extracellular matrix production, response to injury, and crosstalk interactions with CMs. Recent single cell sequencing studies have demonstrated CFB diversity but were unable to link these results to developmental trajectories15,97. We identified striking differences between EpiC-FB and SHF-FB matrix compositions including higher expression of pentraxin related protein PTX3, perlecan, and nidogen 1 and 2 in the EpiC-FB matrices compared to the SHF-FB matrices. Additionally, we observed incorporation of signaling factors involved in cardiac heart patterning and development such as WNT5A in the SHF-FB matrices. The differences in matrix protein synthesis by EpiC-FBs and SHF-FBs suggests that CFB specification early in development may lead to regional specific differences in matrix composition and paracrine signaling necessary for proper heart development.
hPSC-CFBs offer an alternative to human primary CFBs, and provide a consistent donor source. Although many components of primary CFB matrix are not synthesized by hPSC-CFBs, hPSC-CFBs express key cardiac ECM proteins such as fibronectin and remodeling protein lysyl oxidase and lack dFB fibrillar collagens. hPSC-CFBs also enhance cardiac microtissue formation and calcium handling in a manner similar to fCFBs. Additionally, incorporating hPSC-EpiC-FB with hPSC-endothelial cells and hPSC-CMs in tissue constructs resulted in improved sarcomere structures and enhanced contractility demonstrating that non-myocytes can improve tissue level function98.
Although our study investigated how developmental lineage influences gene expression and functional phenotypes in hPSC-derived CFBs, we only compared EpiC-FBs (thought to account for more than 80% of all CFBs in the mouse heart4–10) and SHF-FBs, but we did not compare CFBs from other lineages such as endocardial or neural crest. Furthermore, it is possible that the developmental microenvironment, which is not recapitulated in hPSC differentiation in vitro, has significant effects on CFB phenotypes. Lastly, one should consider that it is possible that the hPSC-CFB populations used in this study represent a heterogenous mixture of fibroblasts and further exploration of heterogeneity within a differentiation or between differentiations may identify subpopulations with unique gene expression profiles and phenotypes.
CFB subtype specification demonstrates the diversity and complexity of cardiac cell subtypes. By developing well characterized protocols to differentiate hPSCs to subtype specific CFBs, we have identified that CFB differentiation lineage leads to distinct molecular and functional phenotypes.
Supplementary Material
Acknowledgments
The authors would like to thank Benjamin Gastfriend and Koji Foreman for their assistance in designing figures and statistical analysis. The authors thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing RNA sequencing facilities and services. The authors also thank the Gladstone Institutes Histology and Light Microscopy Core for paraffin processing of microtissue samples and the Gladstone Stem Cell Core (Roddenberry Stem Cell Foundation) and Ronald Manlapaz for stem cell culture and cardiomyocyte differentiation support.
M.E.F, O.B.M., T.D.M., A.B.S., J.Z., T.C.M., T.J.K., and S.P.P would like to acknowledge support from the NSF Engineering Research Center for Cell Manufacturing Technologies (CMaT; NSF EEC-1648035), NSF grant CBET-1066311, and NIH grant R01 EB007534. O.B.M. was a National Science Foundation Graduate Research Fellow (1650113) and O.B.M., A.C.S., and T.C.M. acknowledge funding support from the California Institute for Regenerative Medicine (LA1-08015). C.M.K. was funded by NIH T32 HL007260 and NIH F31 HL154665 and is an NIH predoctoral fellow (1F31HL154665). Y.M. would like to acknowledge the support from NIH (R01 HL133308). B.M.O. would like to acknowledge support from NIH (R01 HL137204). S.E.G. would like to acknowledge support from the National Science Foundation Graduate Research Fellowship Program (2019272038).
Abbreviations
- aCFB
adult cardiac fibroblast
- CFB
cardiac fibroblast
- CM
cardiomyocyte
- dFB
dermal fibroblast
- ECM
extracellular matrix
- EpiC
epicardial cell
- EpiC-FB
epicardial-derived fibroblast
- EMT
epithelial-to-mesenchymal-transition
- fCFB
fetal cardiac fibroblast
- FISH
fluorescence in situ hybridization
- FSC-A
forward scatter area
- hESC
human embryonic stem cell
- hPSC
human pluripotent stem cell
- SHF-FB
second heart field-derived fibroblast
- SMC
smooth muscle cell
Footnotes
Declaration of Interests
J.Z. and T.J.K. hold US20180094245 patent on the method of differentiating SHF-FB.
References
- 1.Benjamin Emelia J., Muntner Paul, Alonso Alvaro, et al. Heart Disease and Stroke Statistics—2019 Update: A Report From the American Heart Association. Circulation. 2019;139(10):e56–e528. doi: 10.1161/CIR.0000000000000659 [DOI] [PubMed] [Google Scholar]
- 2.Zhou P, Pu WT. Recounting cardiac cellular composition. Circ Res. 2016;118(3):368–370. doi: 10.1161/CIRCRESAHA.116.308139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto AR, Ilinykh A, Ivey MJ, et al. Revisiting Cardiac Cellular Composition. Circ Res. 2016;118(3):400–409. doi: 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acharya A, Baek ST, Huang G, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Dev Camb Engl. 2012;139(12):2139–2149. doi: 10.1242/dev.079970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali SR, Ranjbarvaziri S, Talkhabi M, et al. Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circ Res. 2014;115(7):625–635. doi: 10.1161/CIRCRESAHA.115.303794 [DOI] [PubMed] [Google Scholar]
- 6.Cai C-L, Martin JC, Sun Y, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454(7200):104–108. doi: 10.1038/nature06969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, Poss KD. The epicardium as a hub for heart regeneration. Nat Rev Cardiol. 2018;15(10):631–647. doi: 10.1038/s41569-018-0046-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82(10):1043–1052. doi: 10.1161/01.res.82.10.1043 [DOI] [PubMed] [Google Scholar]
- 9.Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ Res. 2012;111(2):165–169. doi: 10.1161/CIRCRESAHA.112.273946 [DOI] [PubMed] [Google Scholar]
- 10.Wessels A, van den Hoff MJB, Adamo RF, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol. 2012;366(2):111–124. doi: 10.1016/j.ydbio.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore-Morris T, Guimarães-Camboa N, Banerjee I, et al. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest. 2014;124(7):2921–2934. doi: 10.1172/JCI74783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacGrogan D, Luxán G, Driessen-Mol A, Bouten C, Baaijens F, de la Pompa JL. How to Make a Heart Valve: From Embryonic Development to Bioengineering of Living Valve Substitutes. Cold Spring Harb Perspect Med. 2014;4(11). doi: 10.1101/cshperspect.a013912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018;15(11):705–724. doi: 10.1038/s41569-018-0086-9 [DOI] [PubMed] [Google Scholar]
- 14.Snarr BS, Wirrig EE, Phelps AL, Trusk TC, Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev Dyn Off Publ Am Assoc Anat. 2007;236(5):1287–1294. doi: 10.1002/dvdy.21074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asp M, Giacomello S, Larsson L, et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell. 2019;179(7):1647–1660.e19. doi: 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- 16.Cui Y, Zheng Y, Liu X, et al. Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart. Cell Rep. 2019;26(7):1934–1950.e5. doi: 10.1016/j.celrep.2019.01.079 [DOI] [PubMed] [Google Scholar]
- 17.Tucker NR, Chaffin M, Fleming SJ, et al. Transcriptional and Cellular Diversity of the Human Heart. Circulation. Published online May 14, 2020. doi: 10.1161/CIRCULATIONAHA.119.045401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skelly DA, Squiers GT, McLellan MA, et al. Single-Cell Transcriptional Profiling Reveals Cellular Diversity and Intercommunication in the Mouse Heart. Cell Rep. 2018;22(3):600–610. doi: 10.1016/j.celrep.2017.12.072 [DOI] [PubMed] [Google Scholar]
- 19.Litviňuková M, Talavera-López C, Maatz H, et al. Cells of the adult human heart. Nature. Published online September 24, 2020:1–10. doi: 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin G, Palecek SP. Chapter 6 - Inductive factors for generation of pluripotent stem cell-derived cardiomyocytes. In: Fernandes TG, Diogo MM, Cabral JMS, eds. Engineering Strategies for Regenerative Medicine. Academic Press; 2020:177–242. doi: 10.1016/B978-0-12-816221-7.00006-9 [DOI] [Google Scholar]
- 21.Zhang J, Tao R, Campbell KF, et al. Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat Commun. 2019;10(1):2238. doi: 10.1038/s41467-019-09831-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao X, Lian X, Hacker TA, et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng. 2016;1. doi: 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer D, Gambardella L, Bernard WG, et al. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Dev Camb Engl. 2015;142(8):1528–1541. doi: 10.1242/dev.119271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witty AD, Mihic A, Tam RY, et al. Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol. 2014;32(10):1026–1035. doi: 10.1038/nbt.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Tian L, Shen M, et al. Generation of Quiescent Cardiac Fibroblasts from Human Induced Pluripotent Stem Cells for In Vitro Modeling of Cardiac Fibrosis. Circ Res. Published online July 10, 2019. doi: 10.1161/CIRCRESAHA.119.315491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Cao H, Tian L, et al. Efficient Differentiation of TBX18+/WT1+ Epicardial-Like Cells from Human Pluripotent Stem Cells Using Small Molecular Compounds. Stem Cells Dev. 2017;26(7):528–540. doi: 10.1089/scd.2016.0208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bargehr J, Ong LP, Colzani M, et al. Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nat Biotechnol. 2019;37(8):895–906. doi: 10.1038/s41587-019-0197-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian X, Zhang J, Azarin SM, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandegar MA, Huebsch N, Frolov EB, et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18(4):541–553. doi: 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao X, Lian X, Qian T, Bhute VJ, Han T, Palecek SP. Directed differentiation and long-term maintenance of epicardial cells derived from human pluripotent stem cells under fully defined conditions. Nat Protoc. 2017;12(9):1890–1900. doi: 10.1038/nprot.2017.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tohyama S, Hattori F, Sano M, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137. doi: 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Tao R, Campbell KF, et al. Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat Commun. 2019;10(1):2238. doi: 10.1038/s41467-019-09831-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afgan E, Baker D, Batut B, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 38.Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles | PNAS. Accessed September 30, 2020. https://www.pnas.org/content/102/43/15545.abstract [DOI] [PMC free article] [PubMed]
- 39.Masjedi S, Amarnath A, Baily KM, Ferdous Z. Comparison of calcification potential of valvular interstitial cells isolated from individual aortic valve cusps. Cardiovasc Pathol. 2016;25(3):185–194. doi: 10.1016/j.carpath.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 40.Kostina A, Shishkova A, Ignatieva E, et al. Different Notch signaling in cells from calcified bicuspid and tricuspid aortic valves. J Mol Cell Cardiol. 2018;114:211–219. doi: 10.1016/j.yjmcc.2017.11.009 [DOI] [PubMed] [Google Scholar]
- 41.Pillai ICL, Li S, Romay M, et al. Cardiac fibroblasts adopt osteogenic fates and can be targeted to attenuate pathological heart calcification. Cell Stem Cell. 2017;20(2):218–232.e5. doi: 10.1016/j.stem.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmuck EG, Mulligan JD, Ertel RL, et al. Cardiac fibroblast-derived 3D extracellular matrix seeded with mesenchymal stem cells as a novel device to transfer cells to the ischemic myocardium. Cardiovasc Eng Technol. 2014;5(1):119–131. doi: 10.1007/s13239-013-0167-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Murray A, Segal R, Bushnell A, Walsh M. Studies on intercellular LETS glycoprotein matrices. Cell. 1978;14(2):377–391. [DOI] [PubMed] [Google Scholar]
- 44.Harris GM, Raitman I, Schwarzbauer JE. Cell-derived decellularized extracellular matrices. Methods Cell Biol. 2018;143:97–114. doi: 10.1016/bs.mcb.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishihama Y, Oda Y, Tabata T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics MCP. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 46.Hookway TA, Butts JC, Lee E, Tang H, McDevitt TC. Aggregate formation and suspension culture of human pluripotent stem cells and differentiated progeny. Methods San Diego Calif. 2016;101:11–20. doi: 10.1016/j.ymeth.2015.11.027 [DOI] [PubMed] [Google Scholar]
- 47.Silva A, Matthys O, Joy D, et al. Developmental co-emergence of cardiac and gut tissues modeled by human iPSC-derived organoids. bioRxiv. Published online 2020. [Google Scholar]
- 48.Floy ME, Mateyka TD, Foreman KL, Palecek SP. Human pluripotent stem cell-derived cardiac stromal cells and their applications in regenerative medicine. Stem Cell Res. 2020;45:101831. doi: 10.1016/j.scr.2020.101831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao X, Lian X, Hacker TA, et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng. 2016;1. doi: 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91(2):212–222. doi: 10.1093/cvr/cvr112 [DOI] [PubMed] [Google Scholar]
- 51.Greulich F, Rudat C, Farin HF, Christoffels VM, Kispert A. Lack of Genetic Interaction between Tbx18 and Tbx2/Tbx20 in Mouse Epicardial Development. PLOS ONE. 2016;11(6):e0156787. doi: 10.1371/journal.pone.0156787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George RM, Firulli AB. Hand Factors in Cardiac Development. Anat Rec Hoboken NJ 2007. 2019;302(1):101–107. doi: 10.1002/ar.23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally EM, Svensson EC. Setting the pace: Tbx3 and Tbx18 in cardiac conduction system development. Circ Res. 2009;104(3):285–287. doi: 10.1161/CIRCRESAHA.109.193680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakker ML, Boukens BJ, Mommersteeg MTM, et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102(11):1340–1349. doi: 10.1161/CIRCRESAHA.107.169565 [DOI] [PubMed] [Google Scholar]
- 55.Harrelson Z, Kelly RG, Goldin SN, et al. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Dev Camb Engl. 2004;131(20):5041–5052. doi: 10.1242/dev.01378 [DOI] [PubMed] [Google Scholar]
- 56.Rana MS, Théveniau-Ruissy M, De Bono C, et al. Tbx1 coordinates addition of posterior second heart field progenitor cells to the arterial and venous poles of the heart. Circ Res. 2014;115(9):790–799. doi: 10.1161/CIRCRESAHA.115.305020 [DOI] [PubMed] [Google Scholar]
- 57.Muhl L, Genové G, Leptidis S, et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun. 2020;11(1):3953. doi: 10.1038/s41467-020-17740-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McArthur L, Chilton L, Smith GL, Nicklin SA. Electrical consequences of cardiac myocyte: fibroblast coupling. Biochem Soc Trans. 2015;43(3):513–518. doi: 10.1042/BST20150035 [DOI] [PubMed] [Google Scholar]
- 59.Zheng D, Ye S, Wang X, et al. Pre-RC Protein MCM7 depletion promotes mitotic exit by Inhibiting CDK1 activity. Sci Rep. 2017;7(1):2854. doi: 10.1038/s41598-017-03148-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridge LA, Mitchell K, Al-Anbaki A, et al. Non-muscle myosin IIB (Myh10) is required for epicardial function and coronary vessel formation during mammalian development. PLoS Genet. 2017;13(10):e1007068. doi: 10.1371/journal.pgen.1007068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita Y, Andersen P, Hotta A, et al. Sall1 transiently marks undifferentiated heart precursors and regulates their fate. J Mol Cell Cardiol. 2016;92:158–162. doi: 10.1016/j.yjmcc.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alfieri CM, Cheek J, Chakraborty S, Yutzey KE. Wnt signaling in heart valve development and osteogenic gene induction. Dev Biol. 2010;338(2):127. doi: 10.1016/j.ydbio.2009.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwach Passier. Native cardiac environment and its impact on engineering cardiac tissue. Biomater Sci. 2019;7(9):3566–3580. doi: 10.1039/c8bm01348a [DOI] [PubMed] [Google Scholar]
- 64.Naugle JE, Olson ER, Zhang X, et al. Type VI collagen induces cardiac myofibroblast differentiation: implications for postinfarction remodeling. Am J Physiol-Heart Circ Physiol. 2006;290(1):H323–H330. doi: 10.1152/ajpheart.00321.2005 [DOI] [PubMed] [Google Scholar]
- 65.Theocharidis G, Drymoussi Z, Kao AP, et al. Type VI Collagen Regulates Dermal Matrix Assembly and Fibroblast Motility. J Invest Dermatol. 2016;136(1):74–83. doi: 10.1038/JID.2015.352 [DOI] [PubMed] [Google Scholar]
- 66.Ieda M, Tsuchihashi T, Ivey KN, et al. Cardiac Fibroblasts Regulate Myocardial Proliferation through β1 Integrin Signaling. Dev Cell. 2009;16(2):233–244. doi: 10.1016/j.devcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mittal A, Pulina M, Hou S-Y, Astrof S. Fibronectin and integrin alpha 5 play requisite roles in cardiac morphogenesis. Dev Biol. 2013;381(1):73–82. doi: 10.1016/j.ydbio.2013.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrela M, Qiao Q, Koistinen R, et al. High serum insulin-like growth factor binding protein-1 is associated with increased cardiovascular mortality in elderly men. Horm Metab Res Horm Stoffwechselforschung Horm Metab. 2002;34(3):144–149. doi: 10.1055/s-2002-23198 [DOI] [PubMed] [Google Scholar]
- 69.Watanabe S, Tamura T, Ono K, et al. Insulin-like growth factor axis (insulin-like growth factor-I/insulin-like growth factor-binding protein-3) as a prognostic predictor of heart failure: association with adiponectin. Eur J Heart Fail. 2010;12(11):1214–1222. doi: 10.1093/eurjhf/hfq166 [DOI] [PubMed] [Google Scholar]
- 70.Undén A-L, Elofsson S, Brismar K. Gender differences in the relation of insulin-like growth factor binding protein-1 to cardiovascular risk factors: a population-based study. Clin Endocrinol (Oxf). 2005;63(1):94–102. doi: 10.1111/j.1365-2265.2005.02306.x [DOI] [PubMed] [Google Scholar]
- 71.Mäki JM, Sormunen R, Lippo S, Kaarteenaho-Wiik R, Soininen R, Myllyharju J. Lysyl Oxidase Is Essential for Normal Development and Function of the Respiratory System and for the Integrity of Elastic and Collagen Fibers in Various Tissues. Am J Pathol. 2005;167(4):927–936. doi: 10.1016/S0002-9440(10)61183-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mienaltowski MJ, Birk DE. Structure, physiology, and biochemistry of collagens. Adv Exp Med Biol. 2014;802:5–29. doi: 10.1007/978-94-007-7893-1_2 [DOI] [PubMed] [Google Scholar]
- 73.Hanson KP, Jung JP, Tran QA, et al. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: a blueprint for regeneration. Tissue Eng Part A. 2013;19(9–10):1132–1143. doi: 10.1089/ten.TEA.2012.0316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marijianowski MM, van der Loos CM, Mohrschladt MF, Becker AE. The neonatal heart has a relatively high content of total collagen and type I collagen, a condition that may explain the less compliant state. J Am Coll Cardiol. 1994;23(5):1204–1208. doi: 10.1016/0735-1097(94)90612-2 [DOI] [PubMed] [Google Scholar]
- 75.Sasse P, Malan D, Fleischmann M, et al. Perlecan is critical for heart stability. Cardiovasc Res. 2008;80(3):435–444. doi: 10.1093/cvr/cvn225 [DOI] [PubMed] [Google Scholar]
- 76.Costell M, Carmona R, Gustafsson E, González-Iriarte M, Fässler R, Muñoz-Chápuli R. Hyperplastic conotruncal endocardial cushions and transposition of great arteries in perlecan-null mice. Circ Res. 2002;91(2):158–164. doi: 10.1161/01.res.0000026056.81424.da [DOI] [PubMed] [Google Scholar]
- 77.Zoeller JJ, McQuillan A, Whitelock J, Ho S-Y, Iozzo RV. A central function for perlecan in skeletal muscle and cardiovascular development. J Cell Biol. 2008;181(2):381–394. doi: 10.1083/jcb.200708022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Norris RA, Moreno-Rodriguez R, Hoffman S, Markwald RR. The many facets of the matricelluar protein periostin during cardiac development, remodeling, and pathophysiology. J Cell Commun Signal. 2009;3(3–4):275–286. doi: 10.1007/s12079-009-0063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mayourian J, Ceholski DK, Gonzalez DM, et al. Physiologic, Pathologic, and Therapeutic Paracrine Modulation of Cardiac Excitation-Contraction Coupling. Circ Res. 2018;122(1):167–183. doi: 10.1161/CIRCRESAHA.117.311589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iwabu A, Murakami T, Kusachi S, et al. Concomitant expression of heparin-binding epidermal growth factor-like growth factor mRNA and basic fibroblast growth factor mRNA in myocardial infarction in rats. Basic Res Cardiol. 2002;97(3):214–222. doi: 10.1007/s003950200014 [DOI] [PubMed] [Google Scholar]
- 81.Mueller KAL, Tavlaki E, Schneider M, et al. Gremlin-1 identifies fibrosis and predicts adverse outcome in patients with heart failure undergoing endomyocardial biopsy. J Card Fail. 2013;19(10):678–684. doi: 10.1016/j.cardfail.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 82.Li D, Sinha T, Ajima R, Seo H-S, Yamaguchi TP, Wang J. Spatial regulation of cell cohesion by Wnt5a during second heart field progenitor deployment. Dev Biol. 2016;412(1):18–31. doi: 10.1016/j.ydbio.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sinha T, Li D, Théveniau-Ruissy M, Hutson MR, Kelly RG, Wang J. Loss of Wnt5a disrupts second heart field cell deployment and may contribute to OFT malformations in DiGeorge syndrome. Hum Mol Genet. 2015;24(6):1704–1716. doi: 10.1093/hmg/ddu584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schleiffarth JR, Person AD, Martinsen BJ, et al. Wnt5a is required for cardiac outflow tract septation in mice. Pediatr Res. 2007;61(4):386–391. doi: 10.1203/pdr.0b013e3180323810 [DOI] [PubMed] [Google Scholar]
- 85.Chen L, Fulcoli FG, Ferrentino R, Martucciello S, Illingworth EA, Baldini A. Transcriptional control in cardiac progenitors: Tbx1 interacts with the BAF chromatin remodeling complex and regulates Wnt5a. PLoS Genet. 2012;8(3):e1002571. doi: 10.1371/journal.pgen.1002571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park S, Ranjbarvaziri S, Lay FD, et al. Genetic Regulation of Fibroblast Activation and Proliferation in Cardiac Fibrosis. Circulation. 2018;138(12):1224–1235. doi: 10.1161/CIRCULATIONAHA.118.035420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ahmed T, Ahmad M, Mungee S. Cardiac Calcifications. In: StatPearls. StatPearls Publishing; 2020. Accessed November 17, 2020. http://www.ncbi.nlm.nih.gov/books/NBK557689/ [PubMed] [Google Scholar]
- 88.Rodriguez KJ, Piechura LM, Masters KS. Regulation of Valvular Interstitial Cell Phenotype and Function by Hyaluronic Acid in 2-D and 3-D Culture Environments. Matrix Biol J Int Soc Matrix Biol. 2011;30(1):70–82. doi: 10.1016/j.matbio.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Phenotypic Variation Between Stromal Cells Differentially Impacts Engineered Cardiac Tissue Function. - PubMed - NCBI. Published July 11, 2019. Accessed July 11, 2019. https://www.ncbi.nlm.nih.gov/pubmed/30968748 [DOI] [PMC free article] [PubMed]
- 90.Li Y, Asfour H, Bursac N. Age-dependent Functional Crosstalk Between Cardiac Fibroblasts and Cardiomyocytes in a 3D Engineered Cardiac Tissue. Acta Biomater. 2017;55:120–130. doi: 10.1016/j.actbio.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liau B, Jackman CP, Li Y, Bursac N. Developmental stage-dependent effects of cardiac fibroblasts on function of stem cell-derived engineered cardiac tissues. Sci Rep. 2017;7:42290. doi: 10.1038/srep42290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vidal R, Wagner JUG, Braeuning C, et al. Transcriptional heterogeneity of fibroblasts is a hallmark of the aging heart. JCI Insight. 2019;4(22). doi: 10.1172/jci.insight.131092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu C-C, Jeratsch S, Graumann J, Stainier D. Modulation of Mammalian Cardiomyocyte Cytokinesis by the Extracellular Matrix. Circ Res. Published online June 19, 2020. doi: 10.1161/CIRCRESAHA.119.316303 [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Yao F, Wang L, et al. Single-cell analysis of murine fibroblasts identifies neonatal to adult switching that regulates cardiomyocyte maturation. Nat Commun. 2020;11(1):2585. doi: 10.1038/s41467-020-16204-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jonsson MKB, Hartman RJG, Ackers-Johnson M, et al. A Transcriptomic and Epigenomic Comparison of Fetal and Adult Human Cardiac Fibroblasts Reveals Novel Key Transcription Factors in Adult Cardiac Fibroblasts. JACC Basic Transl Sci. 2016;1(7):590–602. doi: 10.1016/j.jacbts.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams C, Quinn KP, Georgakoudi I, Black LD. Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta Biomater. 2014;10(1):194–204. doi: 10.1016/j.actbio.2013.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tucker NR, Chaffin M, Fleming SJ, et al. Transcriptional and Cellular Diversity of the Human Heart. Circulation. Published online May 14, 2020. doi: 10.1161/CIRCULATIONAHA.119.045401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giacomelli E, Meraviglia V, Campostrini G, et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell. 2020;0(0). doi: 10.1016/j.stem.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data is available at Gene Expression Omnibus GSE168380. All processed data are available in the paper or supplementary information.


