Abstract
Animal manure can be a source of antibiotic-resistant genes (ARGs) and pharmaceutical residues; however, few studies have evaluated the presence of ARG in pasture-raised animal production systems. The objective of this study was to examine changes in microbiome diversity and the presence of antibiotic residues (ABRs) on three farms that contained a diverse range of animal species: pasture-raised poultry (broiler and layer), swine, and beef cattle. Total bacterial communities were determined using 16S rRNA microbiome analysis, while specific ARGs (sulfonamide [Sul; Sul1] and tetracycline [Tet; TetA]) were enumerated by qPCR (real-time PCR). Results indicated that the ARG abundances (Sul1 [P < 0.05] and TetA [P < 0.001]) were higher in layer hen manures (16.5 × 10−4 and 1.4 × 10−4 µg kg−1, respectively) followed by broiler chickens (2.9 × 10−4 and 1.7 × 10−4 µg kg−1, respectively), swine (0.22 × 10−4 and 0.20 × 10−4 µg kg−1, respectively) and beef cattle (0.19 × 10−4 and 0.02 × 10−4 µg kg−1, respectively). Average fecal TetA ABR tended to be greater (P = 0.09) for broiler chickens (11.4 µg kg−1) than for other animal species (1.8 to 0.06 µg kg−1), while chlortetracycline, lincomycin, and oxytetracycline ABRs were similar among animal species. Furthermore, fecal microbial richness and abundances differed significantly (P < 0.01) both among farms and specific species of animal. This study indicated that the microbial diversity, ABR, ARG concentrations, and types in feces varied from farm-to-farm and from animal species-to-animal species. Future studies are necessary to perform detailed investigations of the horizontal transfer mechanism of antibiotic-resistant microorganisms (ARMs) and ARG.
Keywords: animal species, antibiotic resistance, genes, microbiome, pasture-raised animals
Introduction
Antibiotic residues (ABRs) that spread the environment through land application of livestock manure or compost could influence structure and function of microbial communities and stimulate the spread of antibiotic-resistant (AR) microorganisms (ARMs) and AR genes (ARGs). However, no regulations exist for concentration limits of antibiotics in manure, soil, or wastewater. A report from the U.S. Centers for Disease Control and Prevention (CDC, 2013) shows that drug-resistant infections continue to be a major concern (Thiele-Bruhn, 2003; Brandt et al., 2015; Menz et al., 2019). Subtherapeutic (growth promoters) and therapeutic antibiotics (use to treat clinically ill animals) are administrated in commercial animal production systems (Thiele-Bruhn, 2003). A considerable quantity of the antibiotics administered are not adsorbed by the animals (17% to 80%) and are excreted through urine and feces (Halling-Sorensen et al., 1998; Montforts et al., 1999). Bacteria in an animal’s gastrointestinal tract (GI) or in the environment after land application of manure exposed to ABR have been recognized as potential sources of antimicrobials and ARGs (Rothrock et al., 2016), which can affect environmental and animal health (Pruden et al., 2006; Heuer et al., 2011). We need to understand the environmental impact of antibiotic use in animal production; however, a broader knowledge of the microbiota community diversity, ABR, and ARG content in feces from different species of livestock and different production systems is needed. Several studies have shown that ARG increased in environments (e.g., soil and wastewater) where agricultural operations occur (Knapp et al., 2017) and in environments that receive animal manure (Shafiani and Malik, 2003; Luo et al., 2010). Following the lead of the United States, however, an environmental risk assessment of veterinary pharmaceuticals was prescribed in the European Union (EU) in 1998 with the EU directives 81/852/EEC and 92/18/EEC (council regulation) (EMEA, 1997).
Pasture-based domesticated animal production can strengthen small and mid-size farm communities across the United States (Conner et al., 2008; HFAC, 2019) and EU (Stampa et al., 2020). Pasture-raised animal products, such as meat, milk, and eggs, have increased consumer demand (Conner et al., 2008; Stampa et al., 2020). Little is known about the prevalence and characteristics of AR and ARG in the manure and soil. Experimental studies of cattle production systems usually find that cattle from conventional dairies harbor a greater occurrence of ARM compared to organic dairies or beef cattle operations; given that dairies usually use more antimicrobials (Zwald et al., 2004; Sato et al., 2005; Harvey et al., 2009). However, it is interesting to note that no significant difference in resistance to individual antimicrobial agents was observed between organic and conventional dairy farms in various studies (Ray et al., 2006; Noyes et al., 2016). In the face of developing AR and ARGs, it is important to examine reduced susceptibility of microorganisms below resistant breakpoints. Our knowledge of antimicrobial use among the farms in our study is limited to herd-level, farmer-reported antimicrobial agent use, so we were incompetent to observe the direct association between the amount of antimicrobial drug use and the ARM from these herds. This means that there may be a background population of ABR and exchange of ARG between organisms, which may contribute to AR and ARG presence on pasture-raised livestock and poultry farms (Singer et al., 2007). Melendez et al. (2010) collected samples from two pastured poultry farms (n = 164) and retail carcasses (n = 36) in Arkansas and found that Salmonella serotypes isolated from pasture-raised poultry farms (e.g., pens, feed, and water) exhibit AR and class I integrons (presence of an integrase gene [intI] and a proximal primary recombination site [attI]). Consequently, previous studies characterized ARM and ARG in native Nebraskan prairie soils (Durso et al., 2016) and organic livestock farms (Cadena et al., 2018). These works provided information on the most frequently detected ARG, which was for tetracycline (Tet).
The gut microbiome of livestock is complex, dynamic, and variable (Zhu et al., 2002; Ming et al., 2017). The characteristics of the gut microbiota may be explained by different host characteristics, environment, dietary compositions, and use of feed additives (Costa et al., 2017; Kers et al., 2018; Lourenco et al., 2019a). Antibiotics not only act on bacteria that cause infections but also affect the resident microbiota. A better understanding of the richness of ARM, ARGs, and ARGs associated with microbial diversity and pathogenicity in the animal gut will have a major role in reducing the contribution of animal production to this problem. Auffret et al. (2017) reported that 204 genes associated with ARM, colonization, communication, or pathogenicity functions were identified from 4,966 metagenomic genes from beef cattle. Same authors also reported that a high ratio of Proteobacteria to Firmicutes + Bacteroidetes ratio was confirmed as a good indicator for rumen dysbiosis and zoonotic pathogens. Furthermore, addition of plant tannins in the diets increased Firmicutes and Firmicutes/Bacteroidetes ratio in the rumen (Min et al., 2014a, 2014b; Carrasco et al., 2017), which improved average daily gain due to altered rumen fermentation (Min et al., 2019a, 2019b). All of these factors can have negative or positive effects on the overall health and production performance of cattle. Furthermore, Bacteroidetes and Firmicutes are the two prevailing bacterial phyla in the gut of humans, mice, and pigs (Ley et al., 2006; Guo et al., 2008), cattle and camels (Ming et al., 2017), and meat goats (Min et al., 2019a). This phylum is composed of many pathogenic bacteria such as Escherichia coli, and the richness of some of these adaptable pathogens is sensitive to dietary change (Bäumler and Sperandio, 2016). Most published studies have focused on either single animal species or indoor confinement systems. It is important to investigate the effects of pasture-raised animal species and different farms on fecal nutrient profiles, ABR, and fecal microbiome community diversity associated with ARG. The results from the present study represent a preliminary experiment that can begin to inform baseline AR/ARG monitoring studies in the future. The objective of this study was to determine the effects of various animal species and different farm on fecal ABR, microbiome changes, and ARG dynamics in the Southeastern region of the United States.
Materials and Methods
To effectively determine the environmental impact of antibiotic use in animal agriculture, a baseline of AR levels must first be determined. Background levels of AR in pasture-based “no antibiotics ever” poultry and livestock systems were determined from: broiler chickens and layer hens, swine, and beef cattle. Animals were located on three farms within the southeastern United States (Table 1), with farm A being sampled once (n = 5) and farms B and C both being sampled twice (n = 10). This study was exempt from animal ethics approvals since the farmers managed the animals; thus, ethics approval was not required as per applicable institutional and national guidelines and regulations.
Table 1.
Comparison of the three all pastured “no antibiotic ever” farms
| Farm | |||
|---|---|---|---|
| Item | Farm 1 | Farm 2 | Farm 3 |
| Animal types | Layers, broilers, swine, beef cattle | Layers, broilers, swine, beef cattle | Layers, broilers, swine, beef cattle |
| Times sampled | 1 | 2 | 2 |
| Flock/herd size | |||
| Broilers | >500 | 50 | >500 |
| Layers | >500 | 150 | >500 |
| Swine | 35 | 15 | 25 |
| Cattle | 25 | 10 | 25 |
| Breed information | |||
| Broilers | Freedom Ranger | Cornish Cross | Freedom Ranger |
| Layers | Rhode Island Red | Rhode Island Red/Araucana | Rhode Island Red |
| Swine | Tamworth | Ossobaw | Tamworth |
| Cattle | South Poll | Dexter | South Poll |
Farm Sites
This analysis was part of a longitudinal study conducted on 43 flocks of broiler chickens across 11 pastured poultry farms in the southeastern United States from March 2015 to November 2016. Five flocks from three farms were selected for this specific study because the farms also raised pastured layer hens, swine, and/or beef cattle (Table 1). All farms reared their pastured flocks/herds within specific areas of the farms cordoned off by temporary fencing, and the pastured were rotationally grazed. Rotation of flocks/herds depended on livestock species. Broilers and layers were reared in movable pens with temporary fences to allow for extended ranging. Poultry housing was relocated either daily (broiler chickens) or weekly (layer hens). Beef cattle were moved to fresh pasture three to four times a week and swine were moved two to three times weekly. Livestock breed and diet on pasture was at the discretion of the individual farmers and could not be controlled for within the experimental design. A brief description of the size, scale, and animal breed of each farm is contained in Table 1. Additionally, based on information obtained from the participating farmers prior to inclusion in the parent student, no antibiotics were used on farm for any animals during the study, or at any point prior to the study since they owned the farms (at least 5 yr for each participating farm).
Sample collection
Fresh fecal samples were collected from the area of the pasture they were currently grazing at the time of sampling. For each animal, their grazing area was divided into five areas, and within each area at least five fecal samples were combined by weight into a single sampling bag and homogenized by hand (≥25 g of fresh weight). This resulted in five replicate pooled samples for each animal for each sampling time. Due to availability of target animals on farm and ability to sample, farm A was only sampled once, while farms B and C were sampled twice during the experimental period. The participating farms were sampled between 2015 (June 17 and July 20) and 2016 (March 13, May 16, and June 08). Fecal samples were transported back to the laboratory on ice for all downstream analyses.
Chemical analysis
Analytical dry matter (DM) concentrations of fecal samples were determined by oven-drying at 105 °C for 24 h (AOAC, 1998). The fecal pH was determined after mixed with distilled water at 1:2.5 (w/v) solid-to-water ratio for 1 h (Huang et al., 2017). Total carbon (C) and total nitrogen (N), and crude protein (CP) were determined by extracting 10 g of sample with 100 mL distilled water (w/v 1:10) by 18-h end-over-end shaking, followed by membrane filtration of the supernatant using 0.45-µm cellulose acetate filters. The aqueous extracts were calorimetrically analyzed on a SEAL Auto-analyzer (Keeney and Nelson, 1982). Atomic absorption spectroscopy was used to determine concentrations of minerals in samples at the University of Georgia, Athens, GA (Paul et al., 2014).
Fecal ABRs
Samples were analyzed for the antibiotic substances shown in Table 4. Multiple ABRs from fecal samples were performed by the Water Sciences Laboratory (University of Nebraska-Lincoln) and quantified according to the modified methods described by Ho et al. (2012). One gram of wet sample was accurately weighed into a 15-mL centrifuge tube and spiked with 25 µL of phosphate-buffered saline (pH 7.4) solutions, 5 mL of extraction buffer (MeOH:acetonitrile:0.1 M ethylenediaminetetraacetic acid:0.2 M Mcilvaine buffer [pH 4.0]; 30:20:25:25 ratios) were added (Ho et al., 2012). The tube was vortexed for 30 s and then placed into an ultrasonic bath for 10 min. The tube was centrifuged at 1,800 × g for 10 min. The supernatant was then decanted into a clean 500-mL plastic bottle and the settled solid was extracted twice more. Decanted supernatant (20 mL) was diluted to 500 mL with ultrapure water. Finally, 250 µL of H2PO4 was added to adjust the pH to approximately 2.3. Prior to solid phase extraction, the extract was filtered through 0.45-μm nylon membrane filter paper (Whatman, United Kingdom) to remove particulate matter. The extract was then freeze-dried, resuspended in 5 mL of deionized water, and stored at −80 °C until analysis. The ABR samples were analyzed two farms only (no statistical different between farms) and then equally mixed from three farms into each animal species prior to analysis, due to the low ABR levels (or not detected). Analyses of selected antibiotics were performed using an Agilent 1260 binary high-pressure liquid chromatography (HPLC) coupled to an Agilent 6410 Triple Quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA). Antibiotics were separated using a HyPURITY C18 HPLC column 250 mm × 2.1 mm ID, 5 µM particle size (Thermo-Scientific, Waltham, MA) at a temperature of 50 °C (D’Alessio et al., 2019).
Table 4.
ABR1 profiles for the four domesticated animal fecal samples
| Item, µg kg−1 | Farm A2 | Farm B2 | SEM | P-value | ||
|---|---|---|---|---|---|---|
| n | 5 | 10 | — | — | ||
| Chlortetracycline | 60.2 | 59.11 | 36.64 | 0.98 | ||
| Lincomycin | 0.69 | 2.35 | 1.26 | 0.39 | ||
| Oxytetracycline | 0.98 | 0.01 | 0.40 | 0.13 | ||
| Ractopamine | < LOD | < LOD | — | — | ||
| Sulfadiazine | < LOD | < LOD | — | — | ||
| Sulfadimethoxine | < LOD | < LOD | — | — | ||
| Sulfamethazine | < LOD | < LOD | — | — | ||
| Sulfamethizole | < LOD | < LOD | — | — | ||
| Sulfamethoxazole | < LOD | < LOD | — | — | ||
| Sulfathiazole | < LOD | < LOD | — | — | ||
| Tetracycline | 1.97 | 2.53 | 1.17 | 0.83 | ||
| Trimethoprim | < LOD | < LOD | — | — | ||
| Item | Animal type | |||||
| Broiler | Layer | Swine | Cattle | SEM | P-value | |
| n | 25 | 25 | 25 | 25 | — | — |
| Chlortetracycline | 98.2 | 8.6 | 27.8 | 21.7 | 37.76 | 0.11 |
| Lincomycin | 0.85 | 0.05 | 0.70 | 2.92 | 1.49 | 0.19 |
| Oxytetracycline | 0.40 | < LOD | 0.42 | < LOD | 0.28 | 0.31 |
| Ractopamine | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfadiazine | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfadimethoxine | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfamethazine | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfamethizole | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfamethoxazole | < LOD | < LOD | < LOD | < LOD | — | — |
| Sulfathiazole | < LOD | < LOD | < LOD | < LOD | — | — |
| Tetracycline | 4.1 | 0.05 | 0.8 | 0.4 | 1.82 | 0.09 |
| Trimethoprim | < LOD | < LOD | < LOD | < LOD | — | — |
1Only ABR samples were equally mixed from three farms into each animal species prior to analysis, due to the low levels (or not detected) of ABRs detected. SEM = standard errors of the means. Number of experimental samples used; n = 5 (one sampling time for farm A), n = 10 (two sampling times for farms B and C).
2The limit of detection (< LOD).
Microbiome analysis
To characterize the fecal microbiomes of pasture-raised animals, the Illumina MiSeq platform was used (Navas-Molina et al., 2013; Rothrock et al., 2014). Fecal samples (0.33 g wet weight) were weighed into a 2-mL lysing Matrix E tube (Fisher Scientific International, Inc., PA) and thoroughly mixed with 825 µL of sodium phosphate buffer and 275 µL of pre-lysis solution using a vortex for 15 s. The material was then centrifuged at 14,000 × g for 5 min. The supernatant was decanted by adding 700 µL of stool lysis buffer (ASL) and vortexed for 5 s. Samples were placed into a MPBio FastPrep 24 instrument (MP Biomedical, Santa Ana, CA), and homogenized at a speed of 6.0 m s−1 for 40 s. The homogenized samples were then centrifuged at 14,000 × g for 5 min and the supernatant transferred sterile 2-mL microcentrifuge tubes. The DNA was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA) using a Synergy 2 Microplate Reader (BioTek Instruments, Winooski, VT). Total 16S rDNA abundances (the V3/4 region of the 16S rRNA gene), an estimate of the total bacterial community contained within a sample, were PCR-amplified with primers containing MiSeq sequencing adapters and Golay barcodes and sequenced on the Illumina MiSeq platform (Caporaso et al., 2011, 2012), followed by sequence analysis using QIIME2 (https://docs.qiime2.org) ver. 2020.2 (Bolyen et al., 2019). All amplicon sequence variants (ASVs) were aligned using mafft via q2-alignment (Katoh et al., 2002) and phylogeny was constructed using q2-phylogeny with fasttree2 (http://www.microbesonline.org/fasttree) (Price et al., 2010). Taxonomy was assigned to ASVs using the q2-feature-classifier (https://github.com/qiime2/q2-feature-classifier) (Bokulich et al., 2018) classify-sklearn naïve Bayes taxonomy classifier against the Greengenes operational taxonomic unit (OUT) reference sequences (McDonald et al., 2012).
Antimicrobial resistance gene analysis
The ARG Sul1 and TetA were selected for qPCR (real-time PCR) analyses of sulfonamide (Sul) and tetracycline (Tet) resistance, respectively. These genes have been closely associated with class I integrons responsible for transfer of ARGs between bacteria (Cadena et al., 2018). DNA extractions were carried out using the Quick-DNA Fecal/Soil Microbe Miniprep Kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol. The qPCR of Sul1 and TetA were carried out using primers presented in Table 2. Primers were designed using AlleleID 7 (Premiere Biosoft, Palo Alto, CA) and were based on Sul1 and TetA nucleotide reference sequences NG_048098.1 and NG_048153.1, respectively, from the Bacterial Antimicrobial Resistance Reference Gene Database (NCBI). Reaction mixtures for qPCR included 10 µL of 2× QuantiTect SYBR Green PCR kit (Qiagen, Carlsbad, CA), 300 nM of each primer, and 1.5 µL of extracted DNA for a 20 µL total reaction volume. Thermocycling conditions were the same for Sul1 and TetA: 95 °C for 15 min; 40 cycles of 94 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s; followed by a final melting from 65 to 95 °C, increased by 0.5 °C every 5 s. A standard curve was generated using serial dilutions (102 to 107 copies) of gBlocks (Integrated DNA Technologies, Coralville, IA) designed from 250 and 500 bp fragments of the Sul1 and TetA genes, respectively (NCBI, 2019).
Table 2.
Sequences, target size, and melting temperature of primers used
| Organisms or group | Target gene | Primer | Primer sequences (5′–3′) | T m 1, °C |
|---|---|---|---|---|
| Sulfonamide (Sul) resistance | Sul1 | Sul1-FW319 | 5′-CGA TCA GAT GCA CCG TGT T-3′ | 60 |
| Sul1-RV430 | 5′-CGC AGG GTC AGG AAA TCC-3′ | |||
| Tetracycline (Tet) resistance | TetA | TetA-FW767 | 5′-CAA CTT GTC GGA CAG GTG -3′ | 60 |
| TetA-RV886 | 5′-GGC GAG TGA ATG CAG AAT-3′ |
1T m melting temperature.
Statistical analysis
Statistical analyses were conducted using the GLM procedure of SAS (SAS Institute, Inc., Cary, NC), with the factors examined being different farms, animal species, and farms × animal species interactions. Data were presented as least-square means, together with the standard error of the mean. Results are reported at both P ≤ 0.05 and P ≤ 0.01 probability levels. Observed bacterial richness (i.e., the number of distinct OTU), diversity indices (Hill et al., 2003; Oksanen et al., 2017), and the relative abundance of each OTU were analyzed by ANOVA using the GLM procedure of the SAS (Figure 1). Finally, UniFrac analysis (Lozupone and Knight, 2005) followed by weighted principal coordinated analysis (PCoA) characterized the diversity of select microbial populations. An unweighted distance-based analysis of molecular variance was used to assess the statistical significance of the spatial separation observed among various farms and animal species of the PCoA plots (Figure 2).
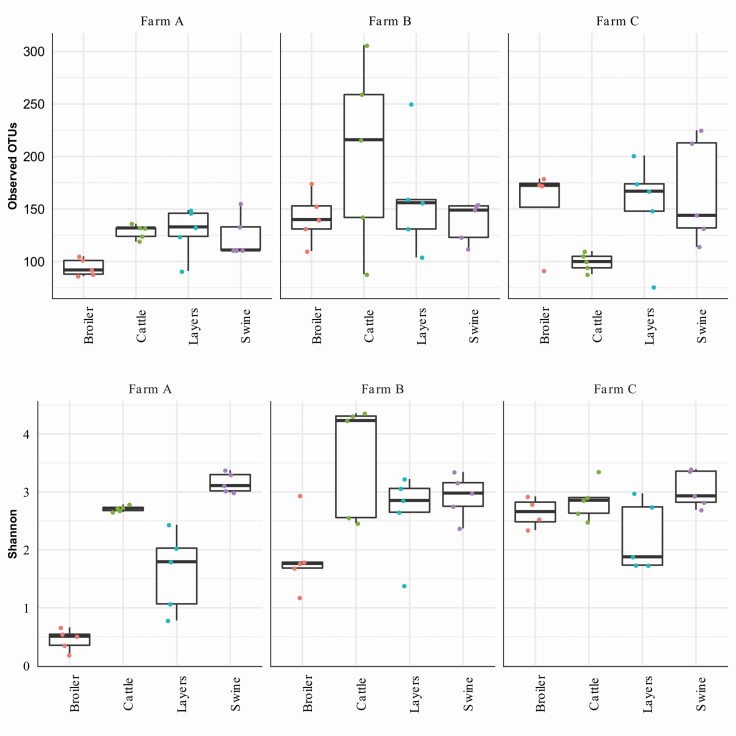
Figure 1.
Alpha diversities (richness and abundance) among farms and livestock species (pairwise comparisons between species) in bacterial community compositions in broilers, cattle, layers, and swine fed mixed pasture-based diets. Richness and diversity were measured by OTUs and Shannon index, respectively. P-value: farm P < 0.01; animal species P < 0.01. Number of experimental samples used; n = 5 (one sampling time for farm A), n = 10 (two sampling times for farms B and C).
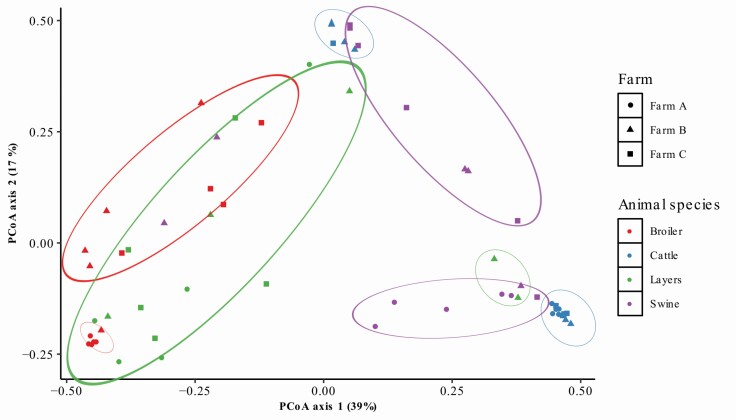
Figure 2.
Principal co-ordinated analysis (PCoA) of 16S bacterial profiles (□-diversity indices) with pairwise comparisons between taxa from fecal collected from broilers, layers, swine, and cattle across the farms. P-value: farm P < 0.01; animal species P < 0.01. Number of experimental samples used; n = 5 (one sampling time for farm A), n = 10 (two sampling times for farms B and C).
Results and Discussion
Chemical composition of pasture-raised animal manure
The average fecal properties across animal types and different farms are presented in Table 3. An average fecal sample in cattle contained low levels of DM, while a significant higher pH was detected in cattle fecal samples (7.6) compared to other animal species (6.2 to 7.4) which is similar to other studies (Huang et al., 2017; Muhsen and Al-Autaish, 2017). There was a significant interaction (P < 0.01) between farms and animal species for fecal pH. Fecal pH has a substantial impact on the sorption of ABR by changing the charge state of the antibiotics and sorbents (Figueroa et al., 2004; Chang et al., 2015). It should also be noted that under acidic conditions, like most other high-use antibiotics, tetracycline and oxytetracycline become negatively charged and can sorb to soil or clay minerals by anionic exchange or electrostatic adsorption (Figueroa and Mackay, 2005). However, under alkaline conditions, the majority of tetracycline is present in anionic form which causes electrostatic repulsion with the +/− of ABR with manure and soil C (organic matter–ABR complexes), reducing sorption (Figueroa et al., 2004; Figueroa and Mackay, 2005; Gu and Karthikeyan, 2005). Consequently, ABR sorption gradually decreases with pH increase due to a decreased cation exchange capacity (CEC) (Chang et al., 2015). In the present study, pH of cattle manure was higher (P < 0.01) than other animals studied (Table 3), with significant interactions between farms and livestock variables. Sorption of tetracycline, chlortetracycline, and oxytetracycline differ due to specific on soil properties, such as pH, clay content, soil type, CEC, anion exchange capacity, and organic C contents (Sassman and Lee, 2005). However, at present, Kd (solid–water sorption coefficients) values for antibiotics and other veterinary pharmaceuticals must be obtained experimentally (Sassman and Lee, 2005).
Table 3.
Nutrients and elements composition in fecal samples from various livestock and poultry
| Item1 | Farm A | Farm B | Farm C | P-value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | L | S | C | B | L | S | C | B | L | S | C | SEM | Farm | Anim. | INT | |
| n | 5 | 5 | 5 | 5 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | — | — | — | |
| DM | 25.8b | 29.8ab | 31.3a | 11.1c | 26.4b | 43.9a | 21.8b | 15.9c | 22.6c | 28.0b | 36.0a | 19.4c | 3.29 | 0.05 | 0.01 | 0.10 |
| pH | 6.3c | 7.1b | 6.6c | 7.6a | 6.3c | 7.1bc | 8.2a | 7.5b | 6.1c | 7.6a | 7.4b | 7.7a | 0.22 | 0.01 | 0.001 | 0.01 |
| Nutrient composition, % DM | ||||||||||||||||
| Total C | 11.2a | 9.5b | 12.4a | 6.4c | 9.9a | 9.8a | 9.9a | 8.8b | 11.3a | 8.0b | 11.7a | 7.2b | 0.79 | 0.85 | 0.001 | 0.01 |
| Total N | 1.1a | 1.0a | 0.8ab | 0.3b | 1.6a | 1.0b | 0.4c | 0.3c | 1.1a | 1.0a | 0.8ab | 0.2b | 0.22 | 0.97 | 0.001 | 0.56 |
| C/N ratio | 10.8 | 10.7 | 14.7 | 22.0 | 9.3b | 12.1b | 25.9a | 27.0a | 11.3b | 10.2b | 14.4b | 50.4a | 6.21 | 0.43 | 0.001 | 0.14 |
| Minerals, % | ||||||||||||||||
| Ca | 0.68b | 1.33a | 0.50b | 0.12b | 0.38b | 2.21a | 0.39b | 0.17b | 0.66b | 1.97a | 0.43b | 0.22b | 0.174 | 0.46 | 0.001 | 0.13 |
| Fe | 0.01 | 0.13 | 0.22 | 0.04 | 0.04b | 0.11b | 0.42a | 0.03b | 0.02b | 0.04b | 0.18a | 0.03b | 0.059 | 0.16 | 0.001 | 0.47 |
| K | 0.42ab | 0.53a | 0.21b | 0.14b | 0.63a | 0.58a | 0.53a | 0.25b | 0.51a | 0.49a | 0.27ab | 0.18b | 0.050 | 0.01 | 0.001 | 0.30 |
| Mg | 0.11 | 0.12 | 0.15 | 0.08 | 0.09b | 0.18a | 0.14ab | 0.12b | 0.13b | 0.12b | 0.19a | 0.10b | 0.022 | 0.33 | 0.001 | 0.01 |
| P | 0.34a | 0.35a | 0.29a | 0.07b | 0.22b | 0.41a | 0.23a | 0.15b | 0.32a | 0.35a | 0.31a | 0.12b | 0.054 | 0.73 | 0.001 | 0.18 |
| S | 0.10a | 0.07ab | 0.08ab | 0.04b | 0.07b | 0.12a | 0.08b | 0.05b | 0.12a | 0.10ab | 0.09b | 0.06b | 0.001 | 0.01 | 0.001 | 0.01 |
| Al | 0.01c | 0.2b | 0.4a | 0.05c | 0.04 | 0.1b | 0.3a | 0.03c | 0.01b | 0.02b | 0.3a | 0.04b | 0.07 | 0.16 | 0.001 | 0.75 |
| Zn | 84.7b | 72.7b | 172.5a | 17.9c | 127.2a | 115.8ab | 67.9c | 31.2c | 252.5a | 122.2b | 137.4b | 16.9c | 29.01 | 0.01 | 0.001 | 0.001 |
| Cr, ppm | 1.0b | 3.2ab | 5.2a | 1.2b | 1.1b | 2.3ab | 4.4a | 1.0b | b1.2 | 3.2a | 3.1a | 1.0b | 0.97 | 0.67 | 0.001 | 0.73 |
| Cu, ppm | 8.8b | 6.5b | 19.4a | 18.7a | 21.1a | 12.8b | 10.1b | 5.0c | 16.9a | 8.3b | 17.6a | 5.0b | 1.85 | 0.67 | 0.001 | 0.01 |
| Pb, ppm | 5.6b | 7.6a | 5.6b | 5.6b | 1.9b | 10.3a | 6.0b | 4.1b | 0.4 | 0.5 | 1.2 | 0.5 | 1.07 | 0.001 | 0.001 | 0.01 |
1Anim. = animal. B, L, S, C = broilers, layers, swine, and cattle, respectively. INT = interaction between farm and animal species. Total C = total carbon, total N = total nitrogen, C/N ratio = carbon/nitrogen ratio. Number of experimental samples used; n = 5 (one sampling time for farm A), n = 10 (two sampling times for farms B and C).
a–cMeans within row treatment within a farm or between farms (average only) with a different superscript differ at P < 0.05. Values without asterisks are not significantly different (P < 0.05).
The primary factors that affect nutrient composition of livestock fecal matter are dependent on livestock type (ruminant vs. monogastric) and stage of growth and feeding management (ration, feed sources, quality of feeds, supplement use; Sheppard and Sanipelli, 2012; Huang et al., 2017; Ali et al., 2021). In the present study, feces from broiler chickens and layer hens contained high levels of N (1.0% to 1.3%) and potassium (K; 0.52% to 0.53%), compared to other species (Table 3) which is similar to other studies (Sheppard and Sanipelli, 2012; Huang et al. 2017). According to Sheppard and Sanipelli (2012), poultry manure (broilers and layers; 2.5% to 2.9%) exhibited greater K content than swine manure (3.42% to 8.32%), with the layer manures had the highest concentrations of most of the mineral elements (e.g., Ca, Cu, Mg, Mn, Mo, Na, P, Pb, etc.) compared to swine manure, demonstrating that a larger mineral nutrient input is required for layers. This is probably related to the high egg and especially egg-shell production that are both physiological sinks for mineral elements.
Cattle feces had significantly lower (P < 0.01) levels of total C, total N, calcium (Ca), K, phosphate (P), aluminum (Al), and chromium (Cr); however, cattle feces had a higher C:N ratio than other animals tested. These results are consistent with other data (Sheppard and Sanipelli, 2012; Huang et al., 2017). Their research reported a higher C/N ratio in cattle manure (10.7) than swine (8.0) and chicken (5.1) manures. The average C/N ratios for the broiler chickens (10.5:1) and layer hens (11.0:1) feces were lower (P < 0.01) than for cattle (33.2:1), but with a higher N content compared to others, likely due to diets containing high levels of C-rich cellulose and lignin (Kerr et al., 2006; MAFRD, 2015). The C/N ratio can be used as an index of fecal stability, as C/N ratios decrease during composting to values approaching 10:1 (Larney and Hao, 2007). Kerr et al. (2006) reported that increasing dietary cellulose increased manure C content as a percentage of nutrient intake. According to Sheppard and Sanipelli (2012), comparing manure concentrations of beef (n = 20) and dairy (n = 30), for every mineral element the concentrations for beef were lower than those of dairy. This is may be because beef cattle are fed more roughage and even lower quality feed than are dairy, with the result that more undigested organic matter is passed to the manure thus diluting the elemental concentrations. Beef cattle may also receive fewer mineral supplements.
Average amounts of fecal total C, iron (Fe), sulfate (S), Al, and Cr were greater (P < 0.01) for swine than for other animal species. There were significant interactions (P < 0.01) between farms and animal species for fecal total C, magnesium (Mg), S, copper (Cu), Zinc (Zn), and lead (Pb) concentrations, indicating that individual farm and animal type influenced fecal nutrient and elemental composition across animal types. Similar to other studies (Kerr et al., 2006; Larney and Hao, 2007; MAFRD, 2015), the present study showed that total C content varied significantly (P < 0.01) from 7.5% DM in cattle to 11.4% DM in swine, with a mean of 9.7% DM across all animal species. High total C in swine (11.4% DM) and broiler (10.8% DM) feces suggests high levels of undigested dietary carbohydrate than in feces from cattle and layer hens. Total fecal N and CP values were greater (P < 0.01) for broilers than for other animals, which has been related to the feeding of high CP diets and excess N excretion (Kerr et al., 2006; Spiehs et al., 2021).
Concentration of mineral elements in feces differed among animal species (Table 3). Generally, Ca and P were higher (P < 0.01) for layer hens than for other animals, but the Zn and Cu contents were greater (P < 0.01) for broiler chickens and swine than other animals, indicating that those animals are often fed Zn and Cu to support intestinal function (MAFRD, 2015). In the present study, the Pb content was greater (P < 0.001) for layer hens than other animals. Some trace elements such as cadmium (Cd), Pb, and mercury (Hg) concentrations have no biological function in plants and animals and application of these metals to soil has no positive effects on crops and can have problematic impacts when added in excess (Sheppard et al., 2009; MAFRD, 2015). Sheppard and Sanipelli (2012) studied about 60 elements in 124 manure or fecal samples from broilers, layers, turkey, swine, dairy, and beef operations in Manitoba, Canada. In general, same authors reported that the manure from young and growing animals often had greater trace element concentrations than the manure from grown animals. The fecal samples from beef cattle had lesser concentrations of trace elements than were conducted in the swine or poultry manures. Many trace elements, and especially the heavy metals, are sturdily retained by soils and can lead to problematic concentrations for animal or human consumption (Sheppard et al. 2009). Fitzgerald and Racz (2001) and Sheppard et al. (2009) reported that concentrations of some unwanted metals, such as Cd, nickel (Ni), Hg, and Pb, were associated with elements added as dietary supplements or for disease suppression, suggesting the Cd, Ni, and Pb were most possible pollutants in the mineral supplements. The occurrence of many of these undesirable elements in manure can be improved by altering the source of mineral supplements.
Antibiotic residues
Intensive animal agriculture has relied on antibiotics as feed additives to promote growth, disease treatment, and prevent diseases in healthy animals considered to be at risk and to enhance growth (Wegener et al., 1999; Boyd, 2001; Teillant et al., 2015). However, intensive feedlot cattle systems are restricted to the United States, the EU, Brazil, and Argentina (Millen et al. 2011). Yet, most antibiotics are still used for growth promotion and prophylaxis in intensive pig and poultry operations in much of the world (Acar et al., 2000; Teillant et al. 2015). ABRs used by the animal industry can enter the environment either directly by excretion from grazing animals or by the spreading of manure as crop fertilizer (Hamscher et al., 2002; Bergmann et al., 2011). There are also occurrences in surface water and ground water that can be recognized to veterinary uses (Hannappel et al., 2014; Bailey et al., 2015), but concentrations frequently drop into the very low levels of range (ng kg−1) and thus are considerably lower than those found in soils (Burke et al., 2016). The most serious consequence of ABR entering the environment is the potential transfer of ARM to humans (Teillant et al., 2015). In addition, ABR may cause other various side effects, including allergies, reproductive disorders, immunopathological effects, mutagenicity, nephropathy (gentamicin), hepatotoxicity, and even carcinogenicity (Bacanli and Basaran, 2019).
The World Health Organization (WHO) list of antimicrobials of importance to human medicine contains 32 drug classes (260 individual drugs) listed as important, highly important, or critical for human medicine (WHO, 2011). Of the 260 drugs on the WHO list of antimicrobial agents important for human medicine, only 39 are suggested or recorded for use in cattle, swine, and poultry in the United States (Durso and Cook, 2014). Veterinary ABRs have been noticed frequently in livestock manure, surface water, and manure-amended soil (Martinez-Carballo et al., 2007; Harms and Bauer, 2011) and reached concentrations in soil varied from 2.56 µg kg−1 of sulfadimidine to 1,590.16 µg kg−1 of chlortetracycline (An et al., 2015). Numerous veterinary antibiotics are poorly adsorbed in animal gastrointestinal organs, subsequent in as much as 30% to 90% of the antibiotic compounds being defecated through feces or urine (Halling-Sorense et al., 1998; Alcock et al. 1999; Aust et al. 2008). In the present study, there was no significant different between farms, but chlortetracycline was the major ABR identified (Table 4). It was the highest in broiler manures (98.2 µg kg−1), followed by swine (27.8 µg kg−1), then cattle (21.7 µg kg−1), and layer hens (8.6 µg kg−1). There were no ABR detected for ractopamine, sulfadiazine, sulfadimethoxine, sulfamethazine, sulfamethizole, sulfamethoxazole, and trimethoprim in the feces of the four species of pasture-raised animals. Both chlortetracycline and tetracycline ABR tended to be greater (P = 0.10 to 0.09) in feces from broiler chickens than from layer hens, swine, and cattle. Lincomycin (0.05 to 0.85 µg kg−1) and oxytetracycline (0.0 to 0.42 µg kg−1) ABR were similar among animal species.
The greater chlortetracycline and lesser lincomycin, and oxytetracycline ABR detected in the manures of free-range poultry and grazing animals in this study was probably due to the several factors, such as antibiotic-containing manure used as a fertilizer or occur naturally in soils and water (D’Costa et al., 2006, 2007; Durso et al., 2012, 2016). A major reason for this finding may be that organic manure is commonly used as fertilizer in crop fields. The concentration of antibiotics in vegetable soil ranged from 0.29 to 1,590.16 µg kg−1. Earlier studies indicated the manure applied to vegetable fields is often measured to be the most significant sources of antibiotics to soil (Hamscher et al. 2002; Brambilla et al. 2007; Karci and Balcioglu 2009). Likewise, studies investigated during the last two decades show that more than 40 different drugs can be found in river water, in groundwater, and even in drinking water sources from the nanogram per liter to the microgram per liter range (Jorgensen and Halling-Sorensen, 2000; Kummerer, 2001). Furthermore, An et al. (2015) reported that the concentrations of tetracyclines and sulfonamides including sulfadiazine, sulfamerazine, sulfadimidine, and sulfamethoxazole were in the range of the concentrations observed in different research areas, according to the previous literatures in manure, soil, and sludge. Additionally, Zho et al. (2010) reported that fecal samples collected from chickens (n = 54) and cows in confinement (n = 28) had oxytetracycline concentrations of 59,060 and 59,590 μg kg−1, and chlortetracycline concentrations of 21,060 and 27,590 μg kg−1 for chickens and cattle, respectively. In a study conducted in China, pig manure (n = 30), chicken manure (n = 20), and soil samples (n = 30) had chlortetracycline, oxytetracycline, and tetracycline concentrations of 46,000, 29,000, and 23,000 μg kg−1, respectively (Martinez-Carballo et al., 2007). Another study reported that the most frequently detected ABRs in pig manure were doxycycline, sulfadiazine, and lincomycin (Rasschaert et al., 2020). In the present study, feces from pasture-raised animals had a much lower average range of tetracycline-like ABR (0.96 to 98.2 µg kg−1) when compared to average ranges of ABR in confinement animal farms in Austria (23,000 to 46,000 µg kg−1) and China (21,06 to 59,590 µg kg−1; Montforts et al., 1999; Martinez-Carballo et al., 2007; Zho et al., 2010). This phenomenon is mainly caused by the extensive production and overuse of antibiotics in China due to no restrictions on its use in animal feed (An et al., 2015).
ARG prevalence in fecal microbiota
AR and ARG challenge the effectiveness of antibiotic treatments for both humans and animals (Spellberg and Gilbert, 2014). Resistant microbes and ARG can circulate among humans, animals, food, water, and the environment. Because many antibiotics commonly used in subtherapeutic concentrations are identical or similar to antibiotics used in human medicine, the development of AMR and their transmission from animals to humans could reduce antibiotic effectiveness in humans (Marshall and Levy, 2011). The commonly identified ARG classes in domesticated animal manure include tetracycline (tet), sulfonamides (sul), β-lactams (bla), macrolide–lincosamide–streptogramin (erm), and fluoroquinolone (fca) (Qian et al., 2018; Hurst et al., 2019), which correspond to the major classes of antibiotics used by the animal industry (Durso and Cook, 2014; Checcucci et al., 2020). Of these five ARG classes, tet (102 to 1012 copies per gram) and sul (108 to 1011 copies per gram) are the most abundant ARG appearing in manure from confinement swine and poultry farms (He et al., 2020) along with organic farming operations in Nebraska, United States (Cadena et al., 2018). Substantial changes in the abundances of ARGs in livestock waste among livestock species have been observed (He et al., 2020), which may be due to varying antibiotic usage and dosing patterns. In this study, the average occurrence of Sul1 and TetA ARG were the highest in the feces of layers (16.5 × 104 and 1.4 × 104 copies per gram), followed by broilers (2.9 × 104 and 1.7 × 104 copies per gram), swine (0.22 × 104 and 0.20 × 104 copies per gram), and beef cattle (0.19 × 104 and 0.02 × 104 copies per gram) (Table 5) which is similar to other studies (Yu et al., 2005; Selvam et al., 2012; Cheng et al., 2013).
Table 5.
Bacterial phylum profiles and antimicrobial resistance genes (ARGs) for the four domesticated animal metagenome samples
| Item1 | Farm A | Farm B | Farm C | P-value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | L | S | C | B | L | S | C | B | L | S | C | SEM | Farm | Anim | INT | |
| n | 5 | 5 | 5 | 5 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | ||||
| DNA concentration2, ng µL−1 | 20.2 | 34.1 | 36.0 | 23.7 | 21.9 | 24.2 | 34.9 | 22.1 | 21.7 | 14.2 | 16.9 | 19.9 | 5.64 | — | — | — |
| Bacterial phylum | ||||||||||||||||
| Firmicutes (F) | 98.9a | 83.8ab | 71.0b | 75.8b | 72.3a | 73.5a | 47.4b | 51.1b | 84.8a | 82.5a | 74.7b | 59.3c | 4.77 | 0.001 | 0.001 | 0.19 |
| Bacteroidetes (B) | 0.2d | 3.5c | 17.5b | 19.3a | 4.3c | 9.0c | 16.0b | 26.8a | 2.8b | 4.6b | 16.1ab | 23.1a | 3.52 | 0.37 | 0.001 | 0.95 |
| Actinobacteria | 0.3 | 1.3 | 1.4 | 0.1 | 6.9b | 3.3b | 17.6a | 8.3b | 3.4 | 2.6 | 2.4 | 0.5 | 2.56 | 0.001 | 0.21 | 0.12 |
| Proteobacteria | 0.5 | 10.6 | 0.7 | 0.4 | 16.1 | 7.8 | 14.9 | 7.9 | 8.4 | 6.8 | 2.9 | 13.2 | 3.64 | 0.03 | 0.90 | 0.12 |
| F/B ratio3 | 1.0 | 1.0 | 0.7 | 0.7 | 0.9a | 0.8ab | 0.6b | 0.5b | 0.9a | 0.9a | 0.8ab | 0.6b | 0.07 | 0.01 | 0.01 | 0.91 |
| ARGs, ×104 | ||||||||||||||||
| Sul1 | 0.7 | 0.4 | 0.08 | 0.2 | 7.7b | 48.5a | 0.5c | 0.2c | 0.4 | 0.7 | 0.1 | 0.1 | 0.871 | 0.04 | 0.12 | 0.04 |
| TetA | 2.6a | 1.4ab | 0.1b | 0.04b | 2.0a | 2.3a | 0.2b | 0.02b | 0.7 | 0.4 | 0.4 | 0.07 | 0.456 | 0.08 | 0.001 | 0.21 |
| Average | Farm | Animal type | ||||||||||||||
| A | B | C | SEM | P-value | B | L | S | C | SEM | P-value | ||||||
| DNA concentration2, ng µL−1 | 28.5 | 25.8 | 18.2 | 2.82 | — | 21.3 | 24.2 | 29.3 | 21.9 | 3.76 | — | |||||
| Bacterial phylum | ||||||||||||||||
| Firmicutes (F) | 82.4a | 61.1c | 75.3b | 2.29 | 0.01 | 85.3a | 80.0a | 64.4b | 62.1b | 3.06 | 0.01 | |||||
| Bacteroidetes (B) | 10.1 | 14.0 | 11.6 | 1.69 | 0.18 | 2.4c | 5.7c | 16.5b | 23.0a | 2.28 | 0.001 | |||||
| Actinobacteria | 0.8b | 9.0b | 2.2a | 1.30 | 0.01 | 3.6ab | 2.4b | 7.1a | 3.0ab | 1.73 | 0.05 | |||||
| Proteobacteria | 3.0b | 11.7a | 7.8ab | 1.84 | 0.01 | 8.3 | 8.4 | 6.2 | 7.2 | 2.46 | 0.53 | |||||
| F/B ratio3 | 0.9a | 0.7b | 0.8ab | 0.05 | 0.01 | 1.0a | 0.9a | 0.7b | 0.6b | 0.05 | 0.01 | |||||
| ARGs, ×104 | ||||||||||||||||
| Sul1 | 0.4b | 1.4a | 0.3b | 0.41 | 0.05 | 2.9b | 16.5a | 0.22b | 0.19b | 0.552 | 0.04 | |||||
| TetA | 1.1a | 1.1a | 0.4b | 0.24 | 0.02 | 1.7a | 1.4a | 0.2b | 0.02b | 0.32 | 0.001 |
1Anim. = animal. B, L, S, C = broilers, layers, swine, and cattle, respectively. INT = interaction between farm and animal species. n = number of experimental samples used. Sulfonamide resistance (Sul1; 104), tetracycline resistance (TetA; 104). Number of experimental samples used; n = 5 (one sampling time for farm A), n = 10 (two sampling times for farms B and C).
2DNA concentration was measured before normalization for amplicon sequencing.
3Firmicutes/Bacteroides ratio = F/B ratio. Overall, 26 phyla were detected, but only four phyla were found in all animals as dominant phyla (cutoff: >1.0%).
a–cMeans within row treatment within a farm or between farms (average only) with a different superscript differ at P < 0.05. Values without asterisks are not significantly different (P < 0.05).
All the three farms were positive for ARGs, demonstrating that ARGs are common in agricultural manure and soils, even in the absence of routine antibiotic drug or pesticide use. These data support other work done in organic farming operations examining ARGs in organic poultry, swine, and cattle production, where ARGs were also identified even when antibiotic drugs were not directed to animals (Stanton et al., 2011; Rothrock et al., 2016). It was not unexpected to identify Sul1 and TetA ARGs at every farm sampled, as they occur naturally in soils, and have been detected in soils and water from around the globe (D’Costa et al., 2006, 2007; Allen et al., 2010; Durso et al., 2012, 2016). The abundance of total ARGs in untreated livestock waste varies from 106 to 1011 copies per gram dry weight or 106 to 1012 copies per mL (absolute abundance), and 10−3 to 10−1 copies per 16S ribosomal RNA (rRNA; relative abundance) (He et al., 2020). Similarly, Yu et al. (2005) and Cheng et al. (2013) reported that manure from poultry and swine samples contained greater abundance of Sul and Tet genes than cattle or sheep manure samples, respectively. Generally, chicken and swine waste show higher ARG abundances than cow waste (He et al., 2020). These variances may be as a result of the more intensive use of antibiotics on chicken (148 mg kg−1) and swine (172 mg kg−1) farms in comparison to cattle (45 mg kg−1) farm (Van Boeckel et al., 2015).
There was a significant interaction (P < 0.04) between farm and animal species for Sul1, indicating that the Sul1 ARG was more prevalent in layer hen feces from farm 2 than from other animal species and farms 1 and 3. However, all farms were positive for at least three ARGs, demonstrating the prevalence of these genes in the feces of animals that did not routinely receive antibiotics. Similarly, Stanton et al. (2011) and Rothrock et al. (2016) reported that ARGs were positively detected in the feces of organically reared cattle, swine, and poultry, where ARGs were detected even when antibiotics were not administrated to animals (Cadena et al., 2018). These results are consistent with other studies (Allen et al., 2010; Durso et al., 2016). However, more research is needed to fully assess the prevalence of ARG in pasture-raised animal manure across farms, geophysical conditions (e.g., soil types), and seasons (e.g., weather).
Microbial community diversity
The alpha- (richness and abundance) and beta-diversities (variation of microbial communities between samples) of bacterial communities were measured by OTUs, Shannon index, and PCoA analyses, respectively, among farms and animal species (Figures 1 and 2). Results indicated that observed OTU richness and Shannon index (the variety and abundance of species) differed significantly (P < 0.01) in samples collected from different farms and across animal species (Figure 1). Besides, the variety and abundance of bacterial species measured by the Shannon index, and the bacterial composition of the broiler chickens and layer hen feces were significantly different (P < 0.01) among farms. The number of OTU (bacterial richness) in the layer hen and swine feces was not different (P > 0.10) among farms, but OTUs in the broiler and cattle feces were significantly different (P < 0.01) among farms. This agrees with data of Wongsaroj et al. (2021), who reported that the greater dissimilarity in bacterial communities was found generally between the manures from different animal species (chicken, cattle, deer, swine, rabbits, and goats), while the slight variation in bacterial communities was found between the animal breeds (beef vs. dairy cattle, black-borne chickens vs. yellow-feather chickens), or the feeding diets (Pangola grass vs. Napier grass).
Furthermore, PCoA (multidimensional scaling; a similarity matrix) was conducted to compare the bacterial profiles among farms and animal types (Figure 2). As shown in Figure 2, PCoA axes 1 and 2 accounted for 39.0% and 17.0%, respectively. Fecal bacterial communities have a high degree of variability (P < 0.01; two distinct clusters across the farms) among farms and animal species (Figure 2) through PCoA axes 1 and 2, which indicated farm locations and animal species impacted the fecal microbial community in pasture-raised animals. Although this occurrence may have been noticed by researchers in previous scientific research studies, prior to our experiment, no formal report on the individual variations among animal species has been published. Results indicate that broiler, layer, swine, and cattle fecal samples have a wide degree of variability in microbial community composition across the pasture-raised, “no antibiotics ever” farms. Similar to our findings, a previous study followed the successional changes in the fecal microbiome of poultry and livestock and identified the effect of animal species, age, and dietary composition on PCoA clustering in manure from poultry (Lourenco et al., 2019a, b), beef cattle (Durso et al., 2010), dairy cattle (Mao et al., 2015), and swine (Guevarra et al., 2018).
When classified at the phylum level, there were 26 phyla observed but only four phyla had an abundance greater than 0.5% to 1.0% in one or more of the animal species (Table 5). Across animal species, Firmicutes (62.1% to 85.3%), Bacteroidetes (2.4% to 23.0%), Actinobacteria (2.4% to 7.1%), and Proteobacteria (6.2% to 8.3%) represented the major phyla in the fecal samples, which was consistent with previous reports (Mosites et al., 2017; Wongsaroj et al., 2021). Mosites et al. (2017) reported that the most abundant phyla among all samples (poultry, cattle, and human) were Firmicutes and Bacteroidetes, followed by Proteobacteria (56.4%, 27.7%, and 5.1% average abundance, respectively). Interestingly, Firmicutes were the most prevalent phyla in all animal species (62% to 85%), which is similar trend to other studies (Videnska et al., 2014; Ming et al., 2017). Regardless of diet, Firmicutes were the predominant bacterial phyla in the pasture-raised broiler chickens (63%; Lourenco et al., 2019a, b), Mongolian cattle (51.2%; Ming et al., 2017), camels (63%; Ming et al., 2017), feedlot cattle (65% to 70%; Bessegatto et al., 2017), and dairy cattle (60%; Hagey et al., 2019). This agrees with data of Mote et al. (2019), who reported a considerably larger number (up to 90%) of Firmicutes and Bacteroidetes phyla in steers grazing tall fescue (Festuca arundinacea). Results from the present experiment indicate that the microbial composition of feces from broiler and layer chickens were dominated by Firmicutes (85% to 80%) followed by Proteobacteria (8.3%). The ratio of Firmicutes to Bacteroidetes was a greater (P < 0.01) for broiler chickens than for other animal species. Bacteroidetes formed only 2.4% to 5.7% of the total fecal microbiota of broilers and layers, respectively, which is similar to what Videnska et al. (2014) reported. However, the Bacteroidetes (23.2%) population was greater (P < 0.001) in the feces from cattle than broilers (2.4%), layers (5.7%), and swine (16.5%) (Table 5). Bacteroidetes are increasingly regarded as specialists for the breakdown of organic matter (i.e., proteins and carbohydrates) (Thomas et al., 2011). Fecal Bacteroidetes were nearly 9-fold greater from cattle (23.0%) than broilers (2.4%), and this phylum had the highest abundance in the cattle manure (26.8%; Table 5), compared to other animals. In the current study, Proteobacteria were less variable (6.2% to 8.4%) and were the third most abundant phylum across the animal types. Actinobacteria were the fourth most prevalent phylum and varied across the animal types. Actinobacteria were more abundant in the layer hen feces (7.1%) than in feces from the other animal species (2.4% to 3.6%). Mosites et al. (2017) also reported that fecal samples from cattle were dominated by Firmicutes and Bacteroidetes, although a minor division of cow samples (13 out of 123) were dominated instead by Proteobacteria and Actinobacteria OTUs.
At the genus level, the most predominant were Lactobacillus (56.7%, 40.1%, 7.5%, and 0.2%), Clostridium (0.1%, 0.5%, 2.2%, and 1.5%), Ruminococcus (0.1%, 1.7%, 5.3%, and 16.9%), Bacteroides (0.5%, 2.2%, 0.5%, and 0.3%), Faecalibacterium (0.1%, 0.3%, 0.2%, and 0.01%), and unclassified bacteria derived from Clostridiales (1.3%, 1.1%, 3.6%, and 1.8%) in broiler, layer, swine, and cattle fecal samples, respectively. In mature chickens, Lactobacillus (35% to 60%) has been found to be the dominant bacterial genus in the GI (Gong et al., 2007; Xiao et al., 2017), which helps explain the results from the present study. Other studies have also found that the chicken GI was dominated by Lactobacilli and Lactobacillales (Bjer-rum et al., 2006; Dumonceaux et al., 2006; Mohd Shaufi et al., 2015). However, in the current study on grazing-based diets, the most abundance genera were Ruminococcaceae (16.9%). This result is supported by a previous study (Mote et al., 2019), indicating that Ruminococcaceae were core genera for pasture-raised cattle. Mote et al. (2019) reported that the relative abundances of both Ruminococcaceae and Lachnospiraceae families significantly increased in steers grazing tall fescue pasture over a 28-d period. Both bacterial families include cellulose- and hemicellulose-degrading bacteria and contribute to butyrate production (La Reau et al., 2016; La Reau and Suen, 2018). Butyrate is a main microbial fermentation product in the GI of humans and all animal species and contributes to the daily metabolizable energy requirement of ruminants and humans (Bergman, 1990). Production of volatile fatty acids, especially butyrate, in the gut microbiome is required for optimal health but is frequently limited by the lack of fermentable fiber in the diet (Baxter et al., 2019). These data provide insight into the composition of the core fecal microbiota and ARG of commercial farms and different animal types.
Conclusions
Given the diversity of alternative animal management styles, the results from this study cannot represent the AR landscape in all “no antibiotic ever” farming systems. Such a feat would take multiple comprehensive surveys covering different geographical regions, which was beyond the scope of this preliminary study. Our study concluded that fecal samples from pasture-raised animals can serve as a reference for determining accurate target levels of ARG in animal production. This suggests that Sul1 and TetA occurrence could serve as a valuable indicator of recent manure-borne resistance in the environment, and that there is possible advantage in monitoring this gene over time when manures are land-applied. They also deliver valuable evidence for studies examining the ecology of AR on farms and in fields. This study indicated that the microbial diversity, ABR, ARG concentrations, and types in feces varied from farm-to-farm and from animal species-to-animal species. Further controlled studies are needed to more fully understand the influence of both animal types and farm management practices (e.g., dietary components) on antibiotic resistomes (resistance genes and their precursors) and its interactions if these relationships are broadly applicable across different dimensional and sequential scales.
Acknowledgments
The authors would like to thank Lee-Rutherford Laura, Cheryl Gresham-Pearson, Tori McIntosh, and Aude Locatelli for assistance in sample acquisition and processing. The mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Glossary
Abbreviations
- ABR
antibiotic residue
- AR
antibiotic resistance
- ARG
antibiotic-resistant gene
- ARM
antibiotic-resistant microorganism
- ASV
amplicon sequence variant
- CEC
cation exchange capacity
- CP
crude protein
- DM
dry matter
- EU
European Union
- GI
gastrointestinal tract
- HPLC
high-pressure liquid chromatography
- OTU
operational taxonomic unit
- PCoA
principal coordinated analysis
- Sul
Sul1, sulfonamide
- Tet
TetA, tetracycline
- WHO
World Health Organization
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Literature Cited
- Acar, J., Casewell M., Freeman J., Friis C., and Goossens H.. . 2000. Avoparcin and virginiamycin as animal growth promotors: a plea for science in decision making. Clin. Microbiol. Inf. 6:477–482. doi: 10.1046/j.1469-0691.2000.00128.x [DOI] [PubMed] [Google Scholar]
- Alcock, R. E., Sweetman A., and Jones K. C.. . 1999. Assessment of organic contaminant fate in waste water treatment plants. I: selected compounds and physicochemical properties. Chemosphere 38:2247–2262. doi: 10.1016/s0045-6535(98)00444-5 [DOI] [PubMed] [Google Scholar]
- Ali, A. I. M., Wassie S. E., Joergensen R. G., Kori D., Goopy J. P., Bahl K. B., Merbold L., Dickhoefer U., and Schecht E.. . 2021. Feed quality and feeding level effects on faecal composition in East African cattle farming systems. Animals 11(564):1–12. doi: 10.3390/ani11020564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., and Handelsman J.. . 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259. doi: 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- An, J., Chen H., Wei S., and Gu J.. . 2015. Antibiotic contamination in animal manure, soil, and sewage sludge in Shenyang, northeast China. Environ. Earth Sci. 74:5077–5086. doi: 10.1007/s12665-015-4528-y [DOI] [Google Scholar]
- AOAC. 1998. Association of official methods of analysis. 16th ed. Gaithersburg (MD): AOAC International. [Google Scholar]
- Auffret, M. D., Dewhurst R. J., Duthie C. A., Rooke J. A., John Wallace R., Freeman T. C., Stewart R., Watson M., and Roehe R.. . 2017. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 5:159. doi: 10.1186/s40168-017-0378-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust, M. O., Godlinski F., Travis G. R., Hao X., McAllister T. A., Leinweber P., and Thiele-Bruhn S.. . 2008. Distribution of sulfamethazine, chlortetracycline and tylosin in manure and soil of Canadian feedlots after subtherapeutic use in cattle. Environ. Pollut. 156:1243–1251. doi: 10.1016/j.envpol.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Bacanli, M., and Basaran N.. . 2019. Importance of anitibiotic residues in animal food. Food Chem. Toxicol. 125:462–466. [DOI] [PubMed] [Google Scholar]
- Bailey, C., Spielmeyer A., Frings R. M., Hamscher G., and Schüttrumpf H.. . 2015. From agricultural fields to surface water systems: the overland transport of veterinary antibiotics. J. Soil. Sediment. 15:1630–1634. [Google Scholar]
- Bäumler, A. J., and Sperandio V.. . 2016. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535:85–93. doi: 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, N. T., Schmidt A. W., Venkataraman A., Kim K. S., Waldron C., and Schmidt T. M.. . 2019. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. Am. Soc. Microbiol. 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, E. N. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590. doi: 10.1152/physrev.1990.70.2.567 [DOI] [PubMed] [Google Scholar]
- Bergmann, A., Fohrmann R., and Weber F.. . 2011. Compilation of monitoring data on environmental concentrations of medicinal products [in German]. In: Federal Environment Agency. Dessau-Roßlau (Germany): Umwelt Bundes Amt; p. 1–99. http://www.uba.de/uba-info-medien/4188.html. [Google Scholar]
- Bessegatto, J. A., Paulino L. R., Lisboa J. A. N., Alfieri A. A., Monteor C. H., Mederios L. P., Kobayashi R. K. T., Weese J. S., and Costa M. C.. . 2017. Changes in the fecal microbiota of beef cattle caused by changes in management and the use of virginiamycin as a growth promotor. Res. Vet. Sci. 114:355–362. doi: 10.1016/j.rvsc.2017.06.011 [DOI] [PubMed] [Google Scholar]
- Bokulich, N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., Huttley G. A., and Gregory Caporaso J.. . 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., Alexander H., Alm E. J., Arumugam M., Asnicar F., . et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:852–857. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, W. 2001. Making meat: science, technology, and American poultry production. Technol. Cult. 42:631–664. doi: 10.1353/tech.2001.0150 [DOI] [Google Scholar]
- Brambilla, G., Patrizii M., De Filippis S. P., Bonazzi G., Mantovi P., Barchi D., and Migliore L.. . 2007. Oxytetracycline as environmental contaminant in arable lands. Anal. Chim. Acta 586:326–329. doi: 10.1016/j.aca.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Brandt, K. K., Amézquita A., Backhaus T., Boxall A., Coors A., Heberer T., Lawrence J. R., Lazorchak J., Schönfeld J., Snape J. R., . et al. 2015. Ecotoxicological assessment of antibiotics: a call for improved consideration of microorganisms. Environ. Int. 85:189–205. doi: 10.1016/j.envint.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Burke, V., Richter D., Greskowiak J., Mehrtens A., Schulz L., and Massmann G., . 2016. Occurrence of antibiotics in surface and ground water of a drinking water catchment area in Germany. Water Environ. Res. 88:652–659. doi: 10.2175/106143016X14609975746604 [DOI] [PubMed] [Google Scholar]
- Cadena, M., Durso L. M., Miller D. N., Waldrip H. M., Castleberry B. L., Drijber R. A., and Wortmann C.. . 2018. Tetracycline and sulfonamide antibiotic resistance genes in soils from Nebraska organic farming operations. Front. Microbiol. 9:1283. doi: 10.3389/fmicb.2018.01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., . et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., and Knight R.. . 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, J. M., Cabral C., Redondo L. M., Pin Viso N. D., Colombatto D., Farber M. D., and Fernández Miyakawa M. E.. . 2017. Impact of chestnut and quebracho tannins on rumen microbiota of bovines. Biomed. Res. Int. 2017:9610810. doi: 10.1155/2017/9610810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. 2013. Antibiotic resistance threats in the United States, 2013. CS239559. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. http://www.cdc.gov/drugresistance/pdf/ar-threats-2013–508.pdf. [Google Scholar]
- Chang, P. H., Jiang W. T., Li Z., Jean J. S., and Kuo C. Y.. . 2015. Antibiotic tetracycline in the environments—a review. Res. Rev. J. Pharma. Anal. 4:15–40. [Google Scholar]
- Checcucci, A., Trevisi P., Luise D., Modesto M., Blasioli S., Braschi I., and Mattarelli P.. . 2020. Exploring the animal waste resistome: the spread of antimicrobial resistance genes through the use of livestock manure. Front. Microbiol. 11:1416. doi: 10.3389/fmicb.2020.01416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W., Chen H., Su C., and Yan S.. . 2013. Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ. Int. 61:1–7. doi: 10.1016/j.envint.2013.08.023 [DOI] [PubMed] [Google Scholar]
- Conner, D. S., Campbell-Arval V., and Hamm M. W.. . 2008. Value in the values: pasture-raised livestock products offer opportunities for connecting producers and consumers. Renew. Agric. Food Sys. 23:61–69. [Google Scholar]
- Costa, M. C., Bessegatto J. A., Alfieri A. A., Weese J. S., Filho A. B., and Oba A.. . 2017. Different antibiotic growth promotors induce specific changes in the cecal microbiota membership of broiler chicken. PLoS One 12:e0171642. doi: 10.1371/journal.Pone.017642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio, M., Durso L. M., Miller D. N., Woodbury B., and Ray C.. . 2019. Snow environmental fate and microbial effects of monensin, lincomycin, and sulfamethazine residues in soil. Environ. Pollut. 246:60–68. [DOI] [PubMed] [Google Scholar]
- D’Costa, V. M., Griffiths E., and Wright G. D.. . 2007. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10:481–489. doi: 10.1016/j.mib.2007.08.009 [DOI] [PubMed] [Google Scholar]
- D’Costa, V. M., McGrann K. M., Hughes D. W., and Wright G. D.. . 2006. Sampling the antibiotic resistome. Science 311:374–377. doi: 10.1126/science.1120800 [DOI] [PubMed] [Google Scholar]
- Dumonceaux, T. J., Hill J. E., Hemmingsen S. M., and Van Kessel A. G.. . 2006. Characterization of intestinal microbiota and response to dietary virginiamycin supplementation in the broiler chicken. Appl. Environ. Microbiol. 72:2815–2823. doi: 10.1128/AEM.72.4.2815-2823.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso, L. M., and Cook K. L.. . 2014. Impacts of antibiotic use in agriculture: what are the benefits and risks? Curr. Opin. Microbiol. 19:37–44. doi: 10.1016/j.mib.2014.05.019 [DOI] [PubMed] [Google Scholar]
- Durso, L. M., Harhay G. P., Smith T. P., Bono J. L., Desantis T. Z., Harhay D. M., Andersen G. L., Keen J. E., Laegreid W. W., and Clawson M. L.. . 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl. Environ. Microbiol. 76:4858–4862. doi: 10.1128/AEM.00207-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso, L. M., Miller D. N., and Wienhold B. J.. . 2012. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes. PLoS One 7:e48325. doi: 10.1371/journal.pone.0048325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durso, L. M., Wedin D. A., Gilley J. E., Miller D. N., and Marx D. B.. . 2016. Assessment of selected antibiotic resistances in ungrazed native Nebraska prairie soils. J. Environ. Qual. 45:454–462. doi: 10.2134/jeq2015.06.0280 [DOI] [PubMed] [Google Scholar]
- EMEA (The European Agency for the Evaluation of Medicinal Products). 1997. Veterinary medicinal products other than GMO-containing and immunological products. Canary Wharf, London (UK): Committee for Veterinary and Medicinal Products; p. 1–10. [Google Scholar]
- Figueroa, R. A., Leonard A., and MacKay A. A.. . 2004. Modeling tetracycline antibiotic sorption to clays. Environ. Sci. Technol. 38:476–483. doi: 10.1021/es0342087 [DOI] [PubMed] [Google Scholar]
- Figueroa, R. A., and MacKay A. A.. . 2005. Sorption of oxytetracycline to iron oxides and iron oxide-rich soils. Environ. Sci. Technol. 39:6664–6671. doi: 10.1021/es048044l [DOI] [PubMed] [Google Scholar]
- Fitzgerald, M. M., and Racz G. J.. . 2001. Long term effects of hog manure on soil quality and productivity. Report to the Agri-Food Research and Development Initiative (ARDI). The Department of Soil Science, Faculty of Agricultural and Food Sciences, The University of Manitoba. http://manure.mb.ca/projects/pdfs/Racz%20and%20Fitzgerald%20Volume%201%20-%20Properties.pdf. [Google Scholar]
- Gong, J., Weiduo S., Forster J. F. R., Huang R. R., Yu H., Yin Y., Yang C., and Han Y.. . 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 59:147–157. [DOI] [PubMed] [Google Scholar]
- Gu, C., and Karthikeyan K. G.. . 2005. Sorption of the antimicrobial ciprofloxacin to aluminum and iron hydrous oxides. Environ. Sci. Technol. 39:9166–9173. doi: 10.1021/es051109f [DOI] [PubMed] [Google Scholar]
- Guevarra, R. B., Hong S. H., Cho J. H., Kim B. R., Shin J., Lee J. H., Kang B. N., Kim Y. H., Wattanaphansak S., Isaacson R. E., . et al. 2018. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J. Anim. Sci. Biotechnol. 9:54. doi: 10.1186/s40104-018-0269-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Xia X., Tang R., Zhou J., Zhou H., and Wang K.. . 2008. Development of a real time PCR method for Firmicutes and Bacteroidetes in faces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 47:367–373. doi: 10.1111/j.1472-765X.2008.02408.x [DOI] [PubMed] [Google Scholar]
- Hagey, J. V., Bhatnagar S., Heguy J. M., Karle B. M., Price P. L., Meyer D., and Maga E. A.. . 2019. Fecal microbial communities in a large representative cohort of California dairy cows. Front. Microbiol. 10:1093. doi: 10.3389/fmicb.2019.01093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling-Sørensen, B., Nors Nielsen S., Lanzky P. F., Ingerslev F., Holten Lützhøft H. C., and Jørgensen S. E.. . 1998. Occurrence, fate and effects of pharmaceutical substances in the environment–a review. Chemosphere 36:357–393. doi: 10.1016/s0045-6535(97)00354-8 [DOI] [PubMed] [Google Scholar]
- Hamscher, G., Sczesny S., Höper H., and Nau H.. . 2002. Determination of persistent tetracycline residues in soil fertilized with liquid manure by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. Anal. Chem. 74:1509–1518. doi: 10.1021/ac015588m [DOI] [PubMed] [Google Scholar]
- Hannappel, S., Balzer F., Groeneweg J., Zuehlke S., and Schulz D.. . 2014. Incidence of veterinary drugs in near-surface groundwater below sites with high livestock density in Germany [in German]. Hydrol. Wasserbewirts. 58:208–220. doi: 10.5675/HyWa_2014,4_1 [DOI] [Google Scholar]
- Harms, K., and Bauer J.. . 2011. Detection and occurrence of antibiotics and their metabolites in pig manure in Bavaria (Germany). In: Keen P. L. and Montforts M. H. M. M., editors, Antimicrobial resistance in the environment. Hoboken (NJ): John Wiley & Sons, Inc.; p. 291–307. [Google Scholar]
- Harvey, R., Funk J., Wittum T. E., and Hoet A. E.. . 2009. A metagenomic approach for determining prevalence of tetracycline resistance genes in the fecal flora of conventionally raised feedlot steers and feedlot steers raised without antimicrobials. Am. J. Vet. Res. 70:198–202. doi: 10.2460/ajvr.70.2.198 [DOI] [PubMed] [Google Scholar]
- He, Y., Yuan Q., Mathieu J., Stadler L., Senehi N., Sun S., and Alvarez P. J. J.. . 2020. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water. 3:1–11. [Google Scholar]
- Heuer, H., Solehati Q., Zimmerling U., Kleineidam K., Schloter M., Müller T., Focks A., Thiele-Bruhn S., and Smalla K.. . 2011. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 77:2527–2530. doi: 10.1128/AEM.02577-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HFAC. 2019. Humane Farm Animal Care (HFAC). The Australian Competition and Consumer Commission (ACCC) [accessed November 13, 2019]. www.certifiedhumane.org. [Google Scholar]
- Hill, T. C., Walsh K. A., Harris J. A., and Moffett B. F.. . 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x [DOI] [PubMed] [Google Scholar]
- Ho, Y. B., Zakaria M. P., Latif P. A., and Saari N.. . 2012. Simultaneous determination of veterinary antibiotics and hormone in broiler manure, soil and manure compost by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 1262:160–168. doi: 10.1016/j.chroma.2012.09.024 [DOI] [PubMed] [Google Scholar]
- Huang, J., Yu Z., Gao H., Yan X., Chang J., Wang C., Hu J., and Zhang L.. . 2017. Chemical structures and characteristics of animal manures and composts during composting and assessment of maturity indices. PLoS One. 12(6):1–16. doi: 10.1371/journal.pone.0178110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, J. J., Oliver J. P., Schueler J., Gooch C., Lansing S., Crossette E., Wigginton K., Raskin L., Aga D. S., and Sassoubre L. M.. . 2019. Trends in antimicrobial resistance genes in manure blend pits and long-term storage across dairy farms with comparisons to antimicrobial usage and residual concentrations. Environ. Sci. Technol. 53:2405–2415. doi: 10.1021/acs.est.8b05702 [DOI] [PubMed] [Google Scholar]
- Jørgensen, S. E., and Halling-Sørensen B.. . 2000. Drugs in the environment. Chemosphere 40:691–699. doi: 10.1016/s0045-6535(99)00438-5 [DOI] [PubMed] [Google Scholar]
- Karci, A., and Balcioglu I. A.. . 2009. Investigation of the tetracycline, sulfonamide, and fluoroquinolone antimicrobial compounds in animal manure and agricultural soils in Turkey. Sci. Total Environ. 407:4652–4664. doi: 10.1016/j.scitotenv.2009.04.047 [DOI] [PubMed] [Google Scholar]
- Katoh, K., Misawa K., Kuma K., and Miyata T.. . 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, D. R., and Nelson D. W.. . 1982. Inorganic forms. In: Methods of soil analysis. Part 2 - chemical and microbiological properties. 2nd ed. Madison (WI): ASA and SSSA; p. 672–679. [Google Scholar]
- Kerr, B. J., Ziemer C. J., Trabue S. L., Crouse J. D., and Parkin T. B.. . 2006. Manure composition of swine as affected by dietary protein and cellulose concentrations. J. Anim. Sci. 84:1584–1592. doi: 10.2527/2006.8461584x [DOI] [PubMed] [Google Scholar]
- Kers, J. G., Velkers F. C., Fischer E. A. J., Hermes G. D. A., Stegeman J. A., and Smidt H.. . 2018. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 9:235. doi: 10.3389/fmicb.2018.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp, C., Callan A. C., Aitken B., Shearn R., Koenders A., and Hinwood A.. . 2017. Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ. Sci. Pollut. Res. 24:2484–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer, K. 2001. Pharmaceutical residues in Northern European environments: consequences and perspectives. In: Kummerer K., editor. Pharmaceuticals in the environment: sources, fate, effects and risks. Berlin: Springer-Verlag; p. 61–74. [Google Scholar]
- La Reau, A. J., Meier-Kolthoff J. P., and Suen G.. . 2016. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb. Genom. 2:e000099. doi: 10.1099/mgen.0.000099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Reau, A. J., and Suen G.. . 2018. The ruminococci: key symbionts of the gut ecosystem. J. Microbiol. 56:199–208. doi: 10.1007/s12275-018-8024-4 [DOI] [PubMed] [Google Scholar]
- Larney, F. J., and Hao X.. . 2007. A review of composting as a management alternative for beef cattle feedlot manure in southern Alberta, Canada. Bioresour. Technol. 98:3221–3227. doi: 10.1016/j.biortech.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Ley, R. E., Peterson D. A., and Gordon J. I.. . 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Lourenco, J. M., Rothrock M. J., Fluharty F. L., and Callaway T. R.. . 2019b. The successional changes in the gut microbiome of pasture-raised chickens fed soy-containing and soy-free diets. Front. Sustain. Food Syst. 3:1–8. doi: 10.3389/fsufs.2019.00035 [DOI] [Google Scholar]
- Lourenco, J. M., Rothrock M. J., Sanad Y. M., and Callaway T. R.. . 2019a. The effects of feeding a soybean-based or a soy-free diet on the gut microbiome of pasture-raised chickens throughout their lifecycle. Front. Sustain. Food Syst. 3:1–12. doi: 10.3389/fsufs.2019.00036 [DOI] [Google Scholar]
- Lozupone, C., and Knight R.. . 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y., Mao D., Rysz M., Zhou Q., Zhang H., Xu L., and Alvarez P. J. J.. . 2010. Trends in antibiotic resistance genes occurrences in the Haihe River, China. Environ. Sci. Technol. 44:7220–7225. doi: 10.1021/es100233w [DOI] [PubMed] [Google Scholar]
- MAFRD. 2015. Properties of manure. Manitoba: Manitoba Agriculture, Food and Rural Development (MAFRD); p. 1–37. https://www.gov.mb.ca/agriculture/environment/nutrient-management/pubs/properties-of-manure.pdf. [Google Scholar]
- Mao, S., Zhang M., Liu J., and Zhu W.. . 2015. Characterizing the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function. Sci. Rep. 5:1–14. doi: 10.1038/srep16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, B. M., and Levy S. B.. . 2011. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24:718–733. doi: 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Carballo, E., González-Barreiro C., Scharf S., and Gans O.. . 2007. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 148:570–579. doi: 10.1016/j.envpol.2006.11.035 [DOI] [PubMed] [Google Scholar]
- McDonald, D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., and Hugenholtz P.. . 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez, S. N., Hanning I., Han J., Nayak R., Clement A. R., Wooming A., Hererra P., Jones F. T., Foley S. L., and Ricke S. C.. . 2010. Salmonella enterica isolates from pasture-raised poultry exhibit antimicrobial resistance and class I integrons. J. Appl. Microbiol. 109:1957–1966. doi: 10.1111/j.1365-2672.2010.04825.x [DOI] [PubMed] [Google Scholar]
- Menz, J., Olsson O., and Kümmerer K.. . 2019. Antibiotic residues in livestock manure: does the EU risk assessment sufficiently protect against microbial toxicity and selection of resistant bacteria in the environment? J. Hazard. Mater. 379:120807. doi: 10.1016/j.jhazmat.2019.120807 [DOI] [PubMed] [Google Scholar]
- Millen, D. D., Pacheco R. D. L., Meyer P. M., Rodrigues P. H. M., and Arrigoni M. D. B.. . 2011. Current outlook and future perspectives of beef production in Brazil. Anim. Front. 1(2):46–52. [Google Scholar]
- Min, B. R., Castleberry L., Allen H., Parker D., Waldrop H., Brauer D., and Willis W.. . 2019a. Associative effect of wet distillers’ grains plus solubles and tannin-rich peanut skin supplementation on in vitro rumen fermentation, greenhouse gas emissions, and microbiome changes. J. Anim. Sci. 97:4668–4681. doi: 10.1093/jas/skz317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, B. R., Gurung N., Shange R., and Solaiman S.. . 2019b. Potential role of rumen microbiota in altering average daily gain and feed efficiency in meat goats fed simple and mixed pastures using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 97:3523–3534. doi: 10.1093/jas/skz193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, B. R., Solaiman S., Shange R., and Eun J. S.. . 2014b. Gastrointestinal bacterial and methanogenic archaea diversity dynamics associated with condensed tannins-containing pine bark diet in goats using 16S rDNA amplicon pyrosequencing. Int. J. Micro. 2014:1–11. doi: 10.1155/2014/141909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, B. R., Wright C., Ho P., Eun J. S., Gurung N., and Shange S.. . 2014a. The effect of phytochemical tannins-containing diet on rumen fermentation characteristics and microbial diversity dynamics in goats using 16S rDNA amplicon pyrosequencing. Agri. Food Anal. Bacteriol. 4:195–211. [Google Scholar]
- Ming, L., Sirguleng L. Y., Hasi S., He J., Hai L., Wang Z., Guo F., Qiao X., and Jirimutu. 2017. Comparative analysis of fecal microbial communities in cattle and Bactrian camels. PLoS One 2017:1–11. doi: 10.137/journal.pone.0173062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Shaufi, M. A., Sieo C. C., Chong C. W., Gan H. M., and Ho Y. W.. . 2015. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 7:4. doi: 10.1186/s13099-015-0051-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montforts, M., Kalf D., Vlaardings P., and Linders J.. . 1999. The exposure assessment for veterinary medical products. Sci. Total Environ. 225:119–133. [DOI] [PubMed] [Google Scholar]
- Mosites, E., Sammons M., Otiang E., Eng A., Noecker C., Manor O., Hilton S., Thumbi S. M., Onyango C., Garland-Lewis G., . et al. 2017. Microbiome sharing between children, livestock and household surfaces in western Kenya. PLoS One 12:e0171017. doi: 10.1371/journal.pone.0171017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mote, R. S., Hill N. S., Joseph H., Skarlupka D., Turner B. Z., Sanders Z. P., Jones D. P., Suen G., and Filipov M. N.. . 2019. Response of beef cattle fecal microbiota to grazing on toxic tall fescue. Appl. Environ. Microbiol. 85:1–17. doi: 10.1128/AEM.00032-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhsen, R., and Al-Autaish H. H. N.. . 2017. Fecal pH and fecal score in local cattle. J. Kerbala Agri. Sci. 2014:44–47. Proc. Third Sci. Conf. Faculty of Vet. Medicine. University of Kerbala on 10th April 2017. p. 44–47. [Google Scholar]
- Navas-Molina, J. A., Peralta-Sánchez J. M., González A., McMurdie P. J., Vázquez-Baeza Y., Xu Z., Ursell L. K., Lauber C., Zhou H., Song S. J., . et al. 2013. Advancing our understanding of the human microbiome using QIIME. In: DeLong E. F., editor. Methods in enzymology. Academic Press. p. 371–444. doi: 10.1016/B978-0-12-407863-5.00019-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI National Center for Biotechnology Information (NCBI). 2019. Bacterial antimicrobial resistance reference gene database [database].https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/.
- Noyes, N. R., Yang X., Linke L. M., Magnuson R. J., Cook S. R., Zaheer R., . et al. 2016. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 24645:1–12. doi: 10.1038/srep24645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’Hara R. B., Simpson G. L., Solymos P., . et al. 2017. Vegan: community ecology package. R package. GPL-2. p. 22–146. Vegan: Community Ecology Package. https://github.com/vegandevs/vegan. [Google Scholar]
- Paul, B. N., Chanda S., Das S., Singh P., Pandey B. K., and Giri S. S.. . 2014. Mineral assay in atomic absorption spectroscopy. Beats Nat. Sci. 1:1–16. [Google Scholar]
- Price, M. N., Dehal P. S., and Arkin A. P.. . 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruden, A., Pei R., Storteboom H., and Carlson K. H.. . 2006. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40:7445–7450. doi: 10.1021/es060413l [DOI] [PubMed] [Google Scholar]
- Qian, X., Gu J., Sun W., Wang X. J., Su J. Q., and Stedfeld R.. . 2018. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344:716–722. doi: 10.1016/j.jhazmat.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Rasschaert, G., Elst D. V., Colson L., Herman L., Ferreira H. C., Dewulf J., Decrop J., Meirlaen J., Heydrickx M., and Daeseleire E.. . 2020. Antibiotic residues and antibiotic-resistant bacteria in pig slurry used to fertilize agricultural fields. Antibiotics 9(34):1–15. doi: 10.3390/antibiotics9010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K. A., Warnick L. D., Mitchell R. M., Kaneene J. B., Ruegg P. L., Wells S. J., Fossler C. P., Halbert L. W., and May K.. . 2006. Antimicrobial susceptibility of Salmonella from organic and conventional dairy farms. J. Dairy Sci. 89:2038–2050. doi: 10.3168/jds.S0022-0302(06)72271-8 [DOI] [PubMed] [Google Scholar]
- Rothrock, M. J., K. L., Hiett, Gamble J., Caudill A. C., Cicconi-Hogan K. M., and Caporaso J. G.. . 2014. A hybrid DNA extraction method for the qualitative and quantitative assessment of bacterial communities from poultry production samples. J. Vis. Exp. 94:1–7. doi: 10.3791/52161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock, M. J., Hiett K. L., Guard J. Y., and Jackson C. R.. . 2016. Antibiotic resistance patterns of major zoonotic pathogens from all-natural, antibiotic-free, pasture-raised broiler flocks in the southeastern United States. J. Environ. Qual. 45:593–603. doi: 10.2134/jeq2015.07.0366 [DOI] [PubMed] [Google Scholar]
- Sassman, S. A., and Lee L. S.. . 2005. Sorption of three tetracyclines by several soils: assessing the role of pH and cation exchange. Environ. Sci. Technol. 39:7452–7459. doi: 10.1021/es0480217 [DOI] [PubMed] [Google Scholar]
- Sato, K., Bartlett P. C., and Saeed M. A.. . 2005. Antimicrobial susceptibility of Escherichia coli isolates from dairy farms using organic versus conventional production methods. J. Am. Vet. Med. Assoc. 226:589–594. doi: 10.2460/javma.2005.226.589 [DOI] [PubMed] [Google Scholar]
- Selvam, A., Xu D., Zhao Z., and Wong J. W.. . 2012. Fate of tetracycline, sulfonamide and fluoroquinolone resistance genes and the changes in bacterial diversity during composting of swine manure. Bioresour. Technol. 126:383–390. doi: 10.1016/j.biortech.2012.03.045 [DOI] [PubMed] [Google Scholar]
- Shafiani, S., and Malik A.. . 2003. Tolerance of pesticides and antibiotic resistance in bacteria isolated from wastewater irrigated soil. World J. Microbiol. Biotechnol. 19:897–901. doi: 10.1023/B:WIBI.0000007290.94694.4F [DOI] [Google Scholar]
- Sheppard, S. C., Grant C. A., Sheppard M. I., de Jong R., and Long J.. . 2009. Risk indicator for agricultural inputs of trace elements to Canadian soils. J. Environ. Qual. 38:919–932. doi: 10.2134/jeq2008.0195 [DOI] [PubMed] [Google Scholar]
- Sheppard, S. C., and Sanipelli B.. . 2012. Trace elements in feed, manure, and manure soils. J. Environ. Qual. 41:1846–1856. doi: 10.2134/jeq2012.0133 [DOI] [PubMed] [Google Scholar]
- Singer, R. S., Ward M. P., and Maldonado G.. . 2007. Can landscape ecology untangle the complexity of antibiotic resistance? Nat. Rev. Microbiol. 4:943–952. [DOI] [PubMed] [Google Scholar]
- Spellberg, B., and Gilbert D. N.. . 2014. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin. Infect. Dis. 59(Suppl 2):S71–S75. doi: 10.1093/cid/ciu392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiehs, M., Waldrip H., and Woodbury B.. . 2021. The co-product conundrum: environmental challenges of feeding ethanol co-products. DGGS in cattle diets for 3rd triennial report to congress on biofuels. In press. [Google Scholar]
- Stampa, E., Shipmann-Schwarze C., and Hamm U.. . 2020. Consumer perceptions, preferences, and behavior regarding pasture-raised livestock products: a review. Food Qual. Prefer. 82:1–15. doi: 10.1016/j.foodqual.2020.103872 [DOI] [Google Scholar]
- Stanton, T. B., Humphrey S. B., and Stoffregen W. C.. . 2011. Chlortetracycline-resistant intestinal bacteria in organically raised and feral Swine. Appl. Environ. Microbiol. 77:7167–7170. doi: 10.1128/AEM.00688-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillant, A., Brower C. H., and Laxminarayan R.. . 2015. Economics of antibiotic growth promotors in livestock. Annu. Rev. Resour. Econ. 7:17.1–17.26. doi: 10.1146/annurev-resource-100814-125015 [DOI] [Google Scholar]
- Thiele-Bruhn, S. 2003. Pharmaceutical antibiotic compounds in soils – a review. J. Plant Nutr. Soil Sci. 166:145–167. doi: 10.1002/jpln.200390023 [DOI] [Google Scholar]
- Thomas, F., Hehemann J. H., Rebuffet E., Czjzek M., and Michel G.. . 2011. Environmental and gut Bacteriodetes: the food connection. Front. Microbiol. 2:1–16. doi: 10.3389/fmicb.2011.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel, T. P., Brower C., Gilbert M., Grenfell B. T., Levin S. A., Robinson T. P., Teillant A., and Laxminarayan R.. . 2015. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 112:5649–5654. doi: 10.1073/pnas.1503141112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videnska, P., Rahman M. M., Faldynova M., Babak V., matulova M. E., Radiovcic E. P., Krizek I., Smple-Mozinas S., Kovac J., Szmolka A., Nagy B., Sedlar K., Cejkova D., and Rychlik I.. . 2014. Characterization of egg laying hen and broiler fecal microbiota in poultry farms in Croatia, Czech Republic, Hungary and Slovenia. PLoS One 9:1–7. doi: 10.1376/journal.pone.0110076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener, H. C., Aarestrup F. M., Jensen L. B., Hammerum A. M., and Bager F.. . 1999. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg. Infect. Dis. 5:329–335. doi: 10.3201/eid0503.990303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2011. Critically important antimicrobials for human medicine. 3rd rev. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). http://apps.who.int/iris/bitstream/handle/10665/77376/9789241504485_eng.pdf;sequence=1. [Google Scholar]
- Wongsaroj, L., Chanabun R., Tunsakul N., Prombutara P., Panha S., and Somboonna N.. . 2021. First reported quantitative microbiota in different livestock manures used as organic fertilizers in the Northeast of Thailand. Sci. Rep. 11:102. doi: 10.1038/s41598-020-80543-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., Xiang Y., Zhou W., Chen J., Li K., and Yang H.. . 2017. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 96:1387–1393. doi: 10.3382/ps/pew372 [DOI] [PubMed] [Google Scholar]
- Yu, Z., F. C.Michel, Jr, Hansen G., Wittum T., and Morrison M.. . 2005. Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 71:6926–6933. doi: 10.1128/AEM.71.11.6926-6933.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zho, L., Dong Y. H., and Wang H.. . 2010. Residues of veterinary antibiotics in manure from feedlot livestock in eight provinces of China. Sci. Total Environ. 408:1069–1075. doi: 10.1016/j.scitotenv.2009.11.014 [DOI] [PubMed] [Google Scholar]
- Zhu, X. Y., Zhong T., Pandya Y., and Joerger R. D.. . 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124–137. doi: 10.1128/aem.68.1.124-137.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwald, A. G., Ruegg P. L., Kaneene J. B., Warnick L. D., Wells S. J., Fossler C., and Halbert L. W.. . 2004. Management practices and reported antimicrobial usage on conventional and organic dairy farms. J. Dairy Sci. 87:191–201. doi: 10.3168/jds.S0022-0302(04)73158-6 [DOI] [PubMed] [Google Scholar]