Abstract
Background
LncRNAs play an important role in tumor initiation and development. However, the underlying involvement of lncRNA expression in colorectal carcinoma remains to be clarified.
Methods
All analyses were performed in R software v4.0, SPSS v13.0, and GraphPad Prism 8. The “limma” package was used to identify differentially expressed lncRNAs between two groups with the threshold of |logFC| >1 and P <0.05. The “Survival” package was used to conduct survival analysis. HCT8 and SE480 cell lines were used to conduct further phenotype experiments, including transwell, wound-healing, CCK8 and colony formation assay. Gene set enrichment analysis was used to explore the biological pathway difference in high and low IGFL2-AS1 patients.
Results
The lncRNA IGFL2-AS1 was highly expressed in colon adenocarcinoma (COAD) tissue and cell lines (HCT116, HCT8, HCT129, and SW480). The COAD patients with high IGFL2-AS1 were associated with a worse prognosis. Meanwhile, the knockdown of IGFL2-AS1 could significantly suppress the proliferation and invasion of COAD cells. Gene set enrichment analysis showed that the top five biological pathways involving IGFL2-AS1 were angiogenesis, epithelial–mesenchymal transition, KRAS signaling, myogenesis, and coagulation. Western blot results showed that the inhibition of IGFL2-AS1 could significantly reduce the N-cadherin, HIF1A and KRAS protein expression, yet increase the E-cadherin protein level. IGFL2-AS1 was also positively correlated with M0 macrophages, M2 macrophages, and neutrophils but negatively correlated with CD4+ memory T cells and CD8+ T cells.
Conclusion
IGFL1-AS1 could seriously worsen patient outcomes and facilitate COAD progression, thus serving as an independent tumor marker.
Keywords: IGFL2-AS1, lncRNA, colon cancer, immune
Introduction
Colorectal cancer (CRC) is the third most prevalent cancer and the fourth leading cause of cancer-related deaths worldwide, where colon adenocarcinoma (COAD) is the primary subtype.1 Recently, CRC incidence and mortality have been steadily declining because of early detection and lifestyle change.2 From the 1970s to 2012, the five-year survival rate of CRC in all stages has increased from 51% to 66%.3 Nowadays, surgical resection combined with adjuvant therapy, such as radiotherapy, chemotherapy, and immunotherapy, remains the only curative option for CRC.4 However, with different therapy combinations, the long-term survival rate is still suboptimal for advanced-stage CRC.5 Therefore, new targeted therapies and markers of CRC need to be identified and developed urgently.
Long-chain non-coding RNA (lncRNAs) are transcripts of more than 200 nucleotides that are not translated into proteins, and most of them were formerly regarded as useless in the human genome.6 However, accumulated evidence over the last decade has indicated their crucial regulatory function in various diseases, such as cis-/trans-transcriptional regulation, organization of nuclear domains, protein interaction, and RNA molecule regulation.7 Researchers have also extensively investigated the role of lncRNAs in cancers and found a tight relation between lncRNAs and tumor development.8 For example, He et al revealed that the lncRNA PCGEM1 is highly expressed in prostate cancer and could facilitate the proliferation of tumor cells through binding with miR-145.9 Gutschner et al demonstrated that the lncRNA MALAT1 is a prognosis marker and could promote tumor metastasis through a MALAT1 knockout model.10 In CRC, KCNQ1OT1 is associated with the response to fluoropyrimidine-based chemotherapy and could predict disease-free survival in the early stages.11
Currently, the rapid development of bioinformatics and next-generation sequencing brings great convenience for relevant biological research. In parallel, the massive data generated provide comprehensive information on disease occurrence and development. In this study, we first identified IGFL2-AS1 as a candidate gene through a series of bioinformatics analysis. A high level of IGFL2-AS1 was found associated with worse clinical features and patient survival. In vitro experiments showed that the knockdown of IGFL2-AS1 could suppress the proliferation and invasion of COAD cells. Bioinformatics analysis showed that IGFL2-AS1 might affect the COAD tumor microenvironment.
Methods
Acquisition of Public Data
The bulk sequence data of colon cancer patients were obtained from TCGA portal sites (https://portal.gdc.cancer.gov/; TCGA-COAD), including 41 healthy samples and 473 tumor samples.12 The original ENSMBLE IDs were annotated using the latest reference file, GRCh38.gtf. The gene expression profile was normalized, and missing values were removed. Clinical information, including survival data, was downloaded in the form of “bcr xml” file and collated by Perl code. The differentially expressed lncRNAs (DELs) were identified using “limma package” with the threshold of |logFC| >1 and P <0.05.
Tissue Specimens and Cell Lines
Eight paired COAD tissue were collected from the Affiliated Hospital of YouJiang Medical University For Nationalities in accordance with the Declaration of Helsinki. All patients signed the informed consent before the operation, and the ethics committees approved the consent procedure. Colon cancer cell lines HCT116, HCT8, HCT129, and SW480 and normal colonic epithelial cell line NCM460 were purchased from iCell (Shanghai, China). All these cell lines were cultured under standard conditions (37°C, 5% CO2).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA of tissues and cell lines was extracted using the RNA simple Total RNA kit (TIANGEN, Beijing, China). cDNA was reversely transcribed using the Fast Quant RT kit (TIANGEN, Beijing, China). The mRNA level of target genes was detected in 96-well plates by using Super Real PreMix Plus SYBR Green (TIANGEN, Beijing, China). The primers used were as follows: IGFL2-AS1, forward primer, 5ʹ-AGCCTATTTCCAGACAACT-3ʹ, reverse primer, 5ʹ-AGAATCAACGACCTCTACAT-3ʹ; GAS1RR, forward primer, 5ʹ-TGGCATTTGGCTAGGTTC-3ʹ, reverse primer, 5ʹ-AACAGGGTTGGGAGTGGGTATTTGC-3ʹ; AC011352.3, forward primer, 5ʹ-AGGCTATTGAGGCAGATT-3ʹ, reverse primer, 5ʹ-TTCCTGTCCAGCAAAGAT-3ʹ; HOTAIR, forward primer, 5ʹ-GGCAAATGTCAGAGGGTT-3ʹ, reverse primer, 5ʹ-CTTAAATTGGGCTGGGTC-3ʹ; AC093895.1, forward primer, 5ʹ-CCCCAGTCTTGTGCCTAT-3ʹ, reverse primer, 5ʹ-CACCACGAAGCGAGATAA-3ʹ; AC007785.1, forward primer, 5ʹ-AAACTGACCCTCCTTCTT-3ʹ, reverse primer, 5ʹ-CTCCTTGGCTCCTATACTC-3ʹ; AC104809.2, forward primer, 5ʹ-TCTGCTGAAAGACCCGTTAT-3ʹ, reverse primer, 5ʹ-TCGGTGGAGTGATTGGTTAG-3ʹ.
Western Blot
Total protein of control and IGFL2-AS1 knockdown cells were collected with RIPA lysis buffer at 4°C for 1.5 h. Western blot was performed using 10% SDS-polyacrylamide gel electrophoresis (PAGE) to semi-quantitatively show protein level. The primary antibody used were all purchased from proteintech (Proteintech Group, Inc), including anti-E-cadherin, anti-N-cadherin, anti-HIF1A, anti-KRAS. The GAPDH antibody were obtained from laboratory stocks.
Immunofluorescence
The tissue of high and low IGFL2-AS1 expression patients was frozen and sectioned at a thickness of 5 μm. These tissue section slides were fixed, washed, and blocked with PBS containing BSA and FBS. CD206 primary antibody was purchased from proteintech (Proteintech Group, Inc). The second antibody was goat anti-mouse IgG heavy and light chain. After the immunofluorescence, cells were photoed using a immunofluorescence microscopy.
Cell Transfection
IGFL2-AS1 small interfering RNAs (siRNA; 5′-GCCCAGATCAACAGAATCA-3′) and negative control (NC) siRNA were purchased from Thermo Fisher Scientific. Lipofectamine 2000 (Invitrogen) was used to perform the transfection of siRNA-LINC00662 and siRNA-NC. The transfection efficiency was measured by qRT-PCR assay.
Transwell Invasion and Migration Assays
Transwell assay was performed using 24 transwell chambers (Corning) to assess the invasion and migration of cancer cells. Cells at a density of 4×103 were seeded into each upper chamber with a serum-free conditioned medium. Bottom chambers were filled with a culture medium containing 10% fetal bovine serum. After 24 h, the cells migrated to the lower chamber were stained with 0.1% crystal violet.
Wound‐Healing Assay
Cells were plated into six-well plates to 90% confluence with serum-free conditioned medium. Cell gaps were scratched in each well using a 100 µL tip set to indicate cell boundaries at 0 and 24 h.
Cell Proliferation Assays
CCK8 and colony formation assays were used to test cell proliferation. CCK8 assay was conducted using a CCK8 Kit in accordance with the manufacturer’s protocol (Dojindo, Shanghai, China). Colony formation assay was performed with cells seeded into six-well plates with 300 cell per well. The medium was replaced every 4 days. Cell counts were conducted after 14 days.
Flow Cytometry
Cell apoptosis was evaluated by flow cytometry. In brief, the cells were resuspended with phosphate-buffered saline and stained with Annexin V/propidium iodide at room temperature for 20 min (Invitrogen, Inc., Carlsbad, CA, USA). The resulting fluorescence was detected using flow cytometry.
Pathway Enrichment Analysis
Gene set enrichment analysis (GSEA) was performed between patients with high and low IGFL2-AS1 to explore the underlying biological pathway of IGFL2-AS1.13 The gene set database was “Hallmark gene set v7.2,” the number of permutations was 1000, the metric for ranking genes was “Signal2Noise,” the normalization mode was “meandiv,” and the randomization mode was “no_balance.” GO analysis was conducted using the “Clusterprofiler” package in R software, which included three modules, namely, biological process (BP), cellular component (CC), and molecular function (MF).
Immune Infiltration Analysis
CIBERSORT algorithms were used to calculate the proportion of 22 immune cells in tumor microenvironment, a deconvolution algorithm designed to quantify the abundance of specific cell types based on a standardized gene expression profile. The expression profile was processed with the following steps before CIBERSORT analysis: deletion of normal samples, removal of low abundance genes, and data correlation. In brief, for the parameters of CIBERSORT, the number of permutations was set as “100,” the kappa was set as “999,” and the q-value was set as “0.3”. The reference file and data source were uploaded in https://figshare.com/articles/dataset/CIBERSORT/14741916.
Statistical Analysis
All analyses were performed in R software v4.0, SPSS v13.0 and GraphPad Prism 8. The P-value used in the analysis was two-sided, and <0.05 was regarded as statistically significant. Student paired t-test was used to compare the level of variables between two groups. Wilcox test was used to analyze statistical significance between multiple groups. All experiments were repeated three times. Data are presented as mean ± standard deviation (SD).
Results
Identification of Differentially Expressed lncRNAs
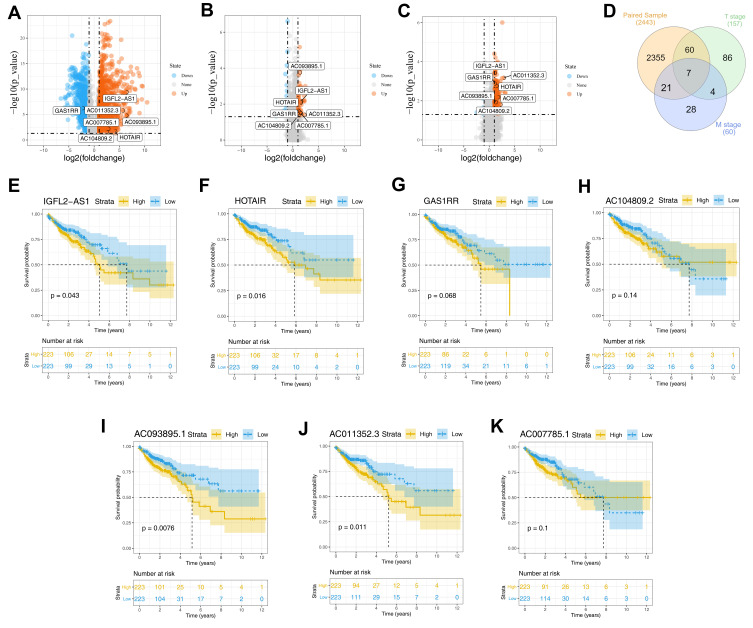
Differential analysis showed that 2443 DELs were identified between COAD and normal tissue, of which 1526 were upregulated and 921 were downregulated (Figure 1A). Similarly, 46 upregulated and 14 downregulated lncRNAs were differentially expressed between the metastatic and non-metastatic groups (Figure 1B); 149 upregulated and eight downregulated DELs were identified between the high (T3 and T4) and low T (T1 and T2) classification groups (Figure 1C). We next intersected these DELs and found seven shared lncRNAs, namely, IGFL2-AS1, GAS1RR, AC011352.3, HOTAIR, AC093895.1, AC007785.1, and AC104809.2 (Figure 1D and Table 1). Then, we conduct survival analysis to explore the underlying effect of these seven lncRNAs on patients survival. Kaplan–Meier survival curve indicated that the patients with high levels of the seven lncRNAs were associated with worse overall survival (OS) (Figure 1E–K).
Figure 1.
Identification of DELs in different groups.
Notes: (A) The volcano plot of DELs between tumor and normal tissue with the threshold of |logFC| >1 and P.value <0.05 (1526 up-regulated and 921 down-regulated); (B) The volcano plot of DELs between metastatic and non-metastatic groups with the threshold of |logFC| >1 and P.value <0.05 (46 up-regulated and 14 down-regulated); (C) The volcano plot of DELs between high and low T classification groups with the threshold of |logFC| >1 and P.value <0.05 (149 up-regulated and 8 down-regulated); (D) The venn plot of intersected lncRNAs between three groups mentioned above; (E–K) The Kaplan-Meier survival curves of seven intersected lncRNAs, IGFL2-AS1, GAS1RR, AC011352.3, HOTAIR, AC093895.1, AC007785.1 and AC104809.2.
Abbreviation: DELs, differentially expressed lncRNAs.
Table 1.
The Expression Level of Identified Seven lncRNAs in Tumor and Normal Tissue
| LncRNA | conMean | treatMean | logFC | pValue | fdr |
|---|---|---|---|---|---|
| IGFL2-AS1 | 0.03 | 2.17 | 6.38 | <0.01 | <0.01 |
| GAS1RR | 0.42 | 0.09 | −2.25 | <0.01 | <0.01 |
| AC011352.3 | <0.01 | 0.25 | 5.65 | <0.01 | <0.01 |
| HOTAIR | 0.07 | 1.22 | 4.16 | 0.02 | 0.03 |
| AC093895.1 | 0.01 | 0.42 | 6.36 | <0.01 | <0.01 |
| AC007785.1 | 0.09 | 0.62 | 2.79 | 0.01 | 0.01 |
| AC104809.2 | 0.06 | 0.33 | 2.51 | 0.03 | 0.04 |
Clinical Correlation of IGFL2-AS1
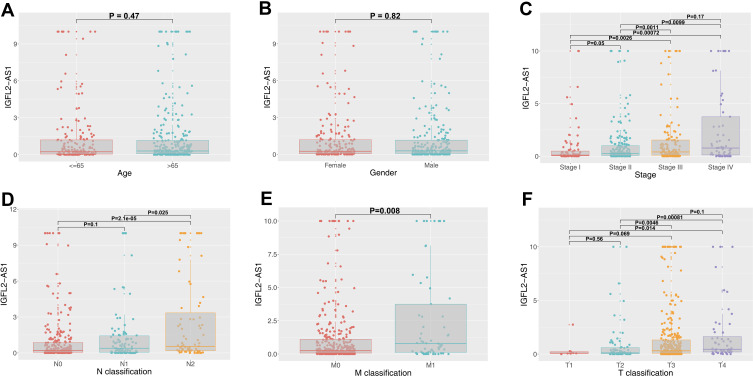
The lncRNA IGFL2-AS1 had the highest expression level in COAD tissue and therefore was used in further study (treatMean = 2.17; logFC = 6.38; Table 1). Also, the relative expression level of the other six lncRNAs was assessed with qRT-PCR and shown in Figure S1. Clinical correlation analysis of IGFL2-AS1 showed no significant difference in different age and gender groups (Figure 2A and B). Notably, a higher level of IGFL2-AS1 was associated with worse clinical features, including stage, T classification, N classification, and M classification (Figure 2C–F).
Figure 2.
Clinical correlation of IGFL2-AS1.
Notes: (A) The expression value of IGFL2-AS1 in different age groups (≤65 and >65); (B) The expression value of IGFL2-AS1 in male and female groups; (C) The expression value of IGFL2-AS1 in different clinical-stage group (Stage I, Stage II, Stage III and Stage IV); (D) The expression value of IGFL2-AS1 in different N classification groups (N0, N1 and N2); (E) The expression value of IGFL2-AS1 in different M classification groups (M0 and M1); (F) The expression value of IGFL2-AS1 in different T classification groups (T1, T2, T3 and T4).
IGFL2-AS1 is Highly Expressed in COAD Tissue and Cell Lines
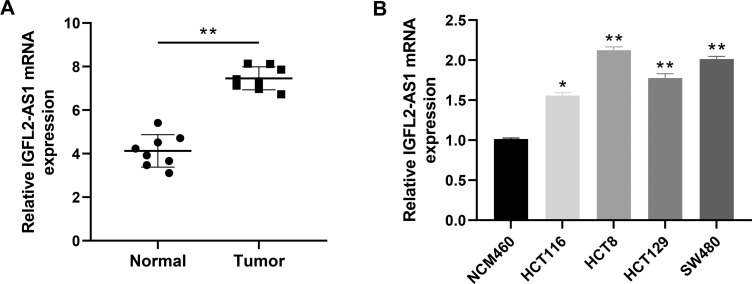
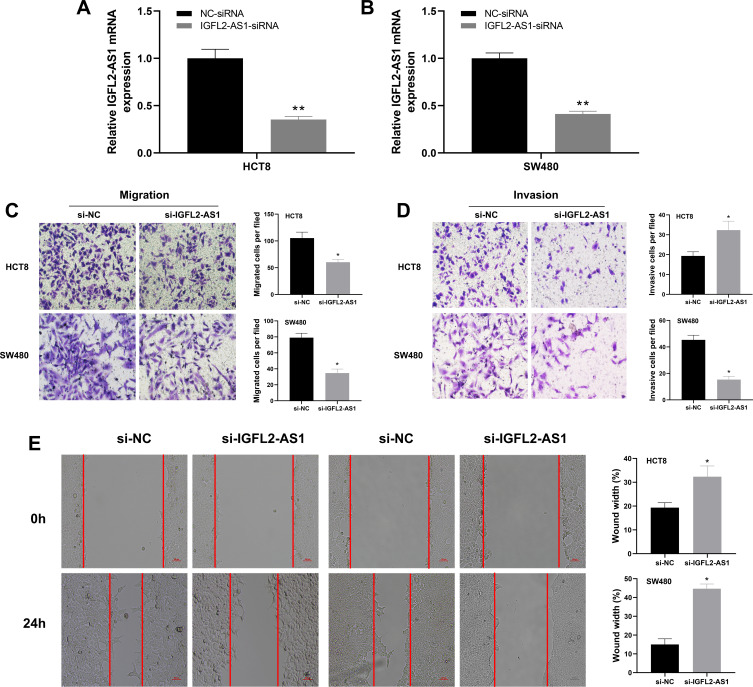
Next, we performed a qRT-PCR analysis based on the total RNA extracted from eight paired tumor and normal tissue. The result revealed a higher IGFL2-AS1 mRNA level in COAD tissue than in healthy colon epithelial tissues (Figure 3A). In parallel, a high IGFL2-AS1 level was also observed in COAD cell lines, including HCT116, HCT8, HCT129, and SW480 (Figure 3B). To explore the biological role of IGFL2-AS1 in COAD, we performed cell transfection to interfere with endogenous IGFL2-AS1 expression. In addition, the transfection efficiency of IGFL2-AS1 was validated by qRT-PCR (Figure 4A and B).
Figure 3.
The lncRNA IGFL2-AS1 is highly expressed in COAD tumor tissue and cells.
Notes: (A) IGFL2-AS1 is highly expressed in COAD tissue (eight paired tumor and normal tissue), *P<0.05, **P<0.01; (B) IGFL2-AS1 is highly expressed in COAD cell lines, HCT116, HCT8, HCT129 and SW480, *P<0.05, **P<0.01.
Figure 4.
IGFL2-AS1 promotes migration and invasion of COAD cells.
Notes: (A and B) RT-qPCR of IGFL2-AS1 in si-NC and si-IGFL2-AS1 groups, *P<0.05, **P<0.01; (C and D) Downregulation of IGFL2-AS1 reduced the number of migration and invasion cells in the transwell assay (HCT8 and SW480 cell lines), *P<0.05, **P<0.01; (E) Wound-healing assay revealed that downregulation of IGFL2-AS1 significantly reduced the migration rate (HCT8 and SW480 cell lines), *P<0.05, **P<0.01.
Knockdown of IGFL2-AS1 Inhibits the Invasion and Proliferation of COAD Cells
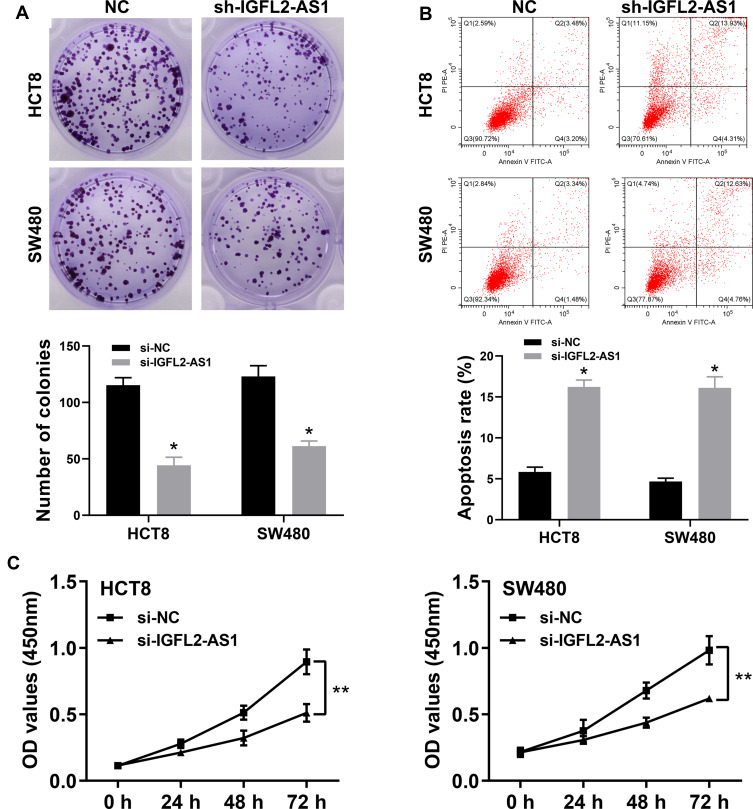
The cell phenotype experiments were then performed to assess the impact of IGFL2-AS1 on cell malignant biological behaviors, including transwell, wound-healing, colony formation and CCK8 assay. The transwell assay showed that IGFL2-AS1 knockdown could significantly decrease the number of migration and invasion cells, indicating that IGFL2-AS1 promotes tumor metastasis (Figure 4C and D). The wound-healing assay also validated this phenotype, in which a prominent wound healing area was observed in the si-IGFL2-AS1 group (Figure 4E). Colony formation assay suggested that IGFL2-AS1 knockdown reduced the colony number in HCT8 and SE480 cell lines (Figure 5A). Meanwhile, we found a higher apoptosis rate in the si-IGFL2-AS1 group than in the si-NC group (Figure 5B). CCK8 assay also demonstrated that IGFL2-AS1 knockdown can significantly inhibit the proliferation of COAD cells (Figure 5C).
Figure 5.
IGFL2-AS1 promotes proliferation of COAD cells.
Notes: (A) Downregulation of endogenous IGFL2-AS1 reduced the mean colony number in the colony formation assay (HCT8 and SW480 cell lines), * = P<0.05, **P<0.01; (B) Flow cytometry showed that the downregulation of endogenous IGFL2-AS1 significantly increased cell apoptosis (HCT8 and SW480 cell lines), *P<0.05, **P<0.01. (C) CCK8 assays revealed that downregulation of endogenous IGFL2-AS1 significantly reduced the cell viability (HCT8 and SW480 cell lines).
Pathway Enrichment Analysis of IGFL2-AS1
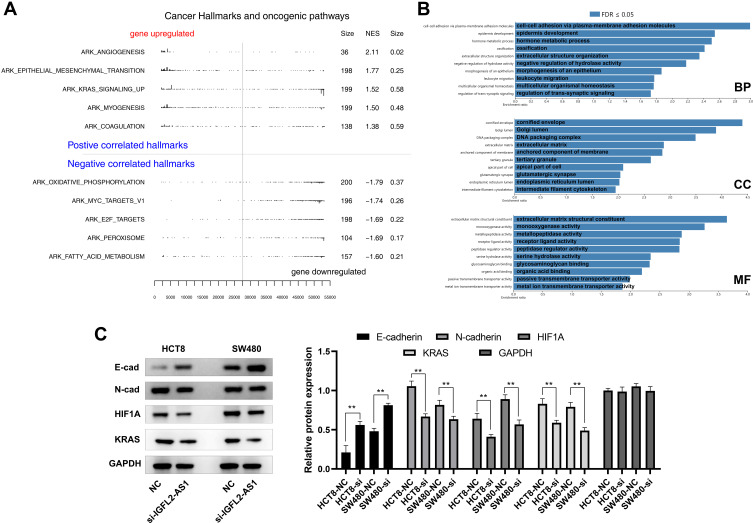
We conducted GSEA analysis to understand the underlying biological function of IGFL2-AS1 in COAD. Based on the Hallmark gene set, in the high IGFL2-AS1 phenotype, the top five upregulated oncogenic pathways were “angiogenesis,” “epithelial–mesenchymal transition,” “KRAS signaling,” “myogenesis,” and “coagulation” (Figure 6A). GO analysis showed that the main enriched terms for BP were “cell-cell adhesion,” “epidermis development,” and “Hormone metabolic process.” For CC, the IGFL2-AS1 was markedly enriched in “cornified envelope,” “Golgi lumen,” and “DNA packaging complex.” Changes in CC were prominently enriched in “extracellular matrix structural,” “monooxygenase activity,” and “metallopeptidase activity” (Figure 6B). To validate the underlying effect of IGFL2-AS1 on the angiogenesis, EMT and KRAS signaling, we performed Western blot assay in control and IGFL2-AS1 knockdown cells. The result showed that the inhibition of IGFL2-AS1 could significantly reduce the N-cadherin, HIF1A and KRAS protein expression, yet increase the E-cadherin protein level (Figure 6C).
Figure 6.
GSEA pathway enrichment analysis of IGFL2-AS1.
Notes: (A) GSEA analysis to explore the biological pathway IGFL2-AS1 involved in (the upper part were the pathways up-regulated in the IGFL2-AS1 high expression patients, the lower part were the pathways down-regulated in the IGFL2-AS1 high expression patients); (B) GO analysis of IGFL2-AS1 with FDR ≤0.05 (three part, BP, biological process; CC, cellular component and MF, molecular function); (C) Western blot of E-cad, N-Cad, HIF1A, KRAS and GAPDH in control and IGFL2-AS1 knockdown cells, *P<0.05, **P<0.01.
Abbreviations: GSEA, gene set enrichment analysis; GO, gene ontology.
Immune Infiltration Analysis
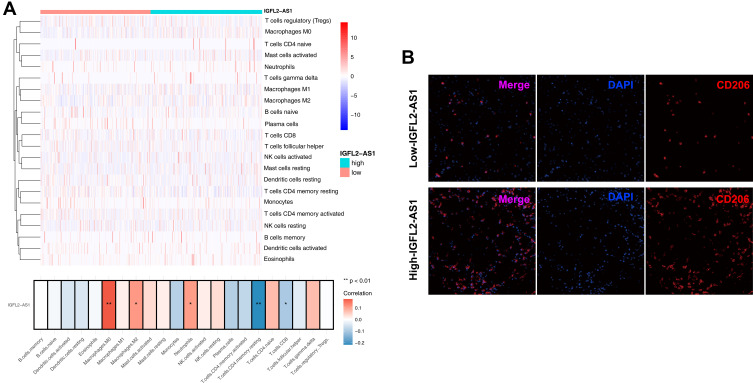
Considering the pivotal role of immune cells in tumor microenvironment, we explored the association of IGFL2-AS1 with 22 immune cells by using the CIBERSORT algorithm. Results revealed that IGFL2-AS1 was positively correlated with M0 macrophages, M2 macrophages, and neutrophils but negatively correlated with CD4+ memory T cells and CD8+ T cells (Figure 7A).
Figure 7.
Immune infiltration analysis performed by CIBERSORT algorithms in high and low IGFL2-AS1 group.
Notes: (A) The upper part is the heatmap overview of immune cell in the high and low IGFL2-AS1 patients; the lower part showed that the IGFL2-AS1 was positively correlated with M0 macrophages, M2 macrophages and neutrophils, while negatively correlated with CD4+ memory T cells and CD8+ T cells, *P<0.05, **P<0.01; (B) Representative immunofluorescence image indicated different CD206 level in high and low IGFL2-AS1 tissue.
The impact of IGFL2-AS1 on M2 macrophages was selected to further validation. Furthermore, in immunofluorescence slides, the tumor tissue with high IGFL2-AS1 expression showed a prominent high M2 macrophage infiltration compared with the tissue with low IGFL2-AS1 (Figure 7B).
Discussion
As the three most common malignant tumors globally, COAD is still the major cause of cancer death worldwide. Despite remarkable advancement in COAD therapy, the five-year survival rate of later stages is only approximately 10% because of the recurrent disease and chemotherapeutic drug resistance. Therefore, searching for novel cancer and therapeutic markers for COAD is crucial.
To the best of our knowledge, this study is the first to analyze the role of IGFL2-AS1 in COAD to the best of our knowledge. We first identified lncRNA IGFL2-AS1 for its underlying cancer-promoting role in patients with COAD. We also first validated the high expression level of IGFL2-AS1 in COAD tissue and cell lines through a qRT-PCR assay. In vitro experiment demonstrated that IGFL2-AS1 can significantly facilitate the proliferation and invasion but inhibit the apoptosis of COAD cells. We further explored the biological pathway and immune infiltration difference between patients with high and low IGFL2-AS1 expression levels.
Nowadays, lncRNAs are widely involved in cancer initiation and progression, especially in epigenetic regulation, protein interaction, and RNA metabolism. For example, Yue et al found that lncRNA CYTOR could form a positive feed-forward loop with Wnt/β-Catenin signaling and thus promote the metastasis of COAD.14 Cheng et al demonstrated that lncRNA LINC00662 could regulate CLDN8/IL22 co-expression through binding with miR-140-5p, which might activate the ERK signaling pathway and promote tumor development.15 In gastric cancer, Feng et al suggested that lncRNA HOXA-AS3 could activate NF-κB signaling with a competitive endogenous RNA (ceRNA) mechanism (miR-29a-3p/LTβR).16 In another aspect, the lncRNA MALAT1 is up-regulated in multiple types of malignant tumors and could interact with the SR protein family of splicing regulators.17 Meanwhile, Ma et al found that IGFL2-AS1 could promote gastric cancer development through binding to miR-802 with a ceRNA mechanism.18 The present study showed that IGFL2-AS1 is highly expressed in COAD and could promote tumor development. Moreover, patients with high IGFL2-AS1 expression appear to have a poor prognosis, indicating that IGFL2-AS1 might be a novel tumor marker in patients with COAD.
We conducted GSEA analysis to explore the biological pathway of IGFL2-AS1. Results showed that the top five activated pathways were angiogenesis, epithelial-mesenchymal transition (EMT), KRAS signaling, myogenesis, and coagulation. The chaotic vascular system in the local microenvironment is a character of malignant tumors.19 The high permeability of upstream vascular could lead to a low blood perfusion level because of blood and oxygen loss. This hypoxic microenvironment favors the colonies that are more adapted to harsh environments and thus enhances the proliferation and metastatic potential.20 During EMT, epithelial cells are transdifferentiated into motile mesenchymal cells, which plays a vital role in cancer development.21 For instance, Feng et al indicated that CDC42EP3 could facilitate the malignant phenotype of colorectal cancer cells by regulating EMT.22 In addition, as a common mutant oncogene in carcinoma, KRAS plays an essential role in tumor progression. Buscail et al comprehensively reviewed the role of KRAS mutation in pancreatic cancer and found that the existence of KRAS mutant in serum and plasma is associated with a worse prognosis of pancreatic cancer.23 Lal et al revealed that KRAS mutation in COAD decreases the infiltration of neutrophils and cytotoxic cells, resulting in advanced disease progression.24
The heterogeneity of tumor microenvironment affects the progression of tumor cells significantly.12 We subsequently performed immune infiltration analysis to explore the difference in tumor immune microenvironment between patients with high and low IGFL2-AS1 expression levels. Results showed that IGFL2-AS1 increased the contents of M0 macrophages, M2 macrophages, and neutrophils but decreased the contents of CD4 memory T cells and CD8 T cells. In general, M0 macrophages could differentiate into M1 and M2 macrophages under different induced conditions. Zhang et al found that apoptotic SKOV3 cells could stimulate M0 macrophages to differentiate into M2 macrophages by activating the ERK signaling pathway, thus promoting the proliferation and migration of ovarian cancer cells.25 Neutrophils are involved in different tumor development stages, including tumor initiation, proliferation, and metastasis.26 Commonly, the reactive oxygen species, reactive nitrogen species, or proteases released by neutrophils could induce angiogenesis and promote tumor metastasis.27 Furthermore, the suppression of neutrophils on the immune system could facilitate tumor proliferation. For example, the anti-tumor effect mediated by CD8+ T lymphocytes can be suppressed by small molecules released by neutrophils.28,29 CD8+ T cells are the preferred choice targeting cancer therapy. The systemic review performed by Farhood et al concluded that effector CD8+ cytotoxic T lymphocytes can infiltrate the core site of the tumor tissue and exert a killing effect.30
Despite our findings, this study has several limitations. First, the public data included in our analysis were predominantly western people. Thus, whether or not the research results hold true for other populations needs to be further verified. Second, as clinical features records of some patients are incomplete, our conclusions affected by this issue may exhibit potential bias. Third, the in-depth mechanism by which IGFL2-AS1 promotes tumor development still needs to be validated through in vitro and in vivo experiments. Therefore, based on the direction provided in this analysis, we will focus on the underlying mechanism of IGFL2-AS1 in COAD through related biological experiments and cohort validation.
Conclusion
Considering its close relation to clinical features and patient survival, we first identified IGFL2-AS1 as a target gene through a series of bioinformatics analyses. In vitro experiments showed that IGFL2-AS1 was highly expressed in COAD tissue and could promote the proliferation and invasion of cancer cells (HCT8 and SW480). GSEA analysis indicated that the top three biological pathways involving IGFL2-AS1 were angiogenesis, EMT, and KRAS signaling. CIBERSORT analysis showed that IGFL2-AS1 was positively correlated with M0 macrophages, M2 macrophages, and neutrophils but negatively correlated with CD4+ memory T cells and CD8+ T cells.
Funding Statement
This work was supported by Special Funding for Guangxi Special Experts (GRCT2019-13); Guangxi Medical High-level Leading Talents Training “139” Project (GWKJ2018-22).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–573. doi: 10.1002/cncr.24760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. [DOI] [PubMed] [Google Scholar]
- 4.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11(1):164. doi: 10.3390/nu11010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miles A, van Duijnhoven F, McQueen A, Oliphant R. Colorectal cancer: advances in prevention and early detection. Biomed Res Int. 2015;2015:518068. doi: 10.1155/2015/518068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu D, Shi Y, Liu JB, et al. Targeting long non-coding RNA to therapeutically regulate gene expression in cancer. Mol Ther Nucleic Acids. 2020;21:712–724. doi: 10.1016/j.omtn.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9(9):1932–1956. doi: 10.1002/cmdc.201300534 [DOI] [PubMed] [Google Scholar]
- 9.He JH, Zhang JZ, Han ZP, Wang L, Lv YB, Li YG. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer Res. 2014;33(1):72. doi: 10.1186/s13046-014-0072-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapucci A, Perrone G, Di Paolo A, et al. PNN and KCNQ1OT1 can predict the efficacy of adjuvant fluoropyrimidine-based chemotherapy in colorectal cancer patients. Oncol Res. 2021;28(6):631–644. doi: 10.3727/096504020X16056983169118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA). Methods Mol Biol. 2016;1418:111–141. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–1298. doi: 10.1016/j.ymthe.2018.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng B, Rong A, Zhou Q, LncRNA LW. LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J Exp Clin Cancer Res. 2020;39(1):5. doi: 10.1186/s13046-019-1510-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Q, Zhu B, Hu YL, Mao QS, Feng Y. LncRNA HOXA-AS3 promotes gastric cancer progression by regulating miR-29a-3p/LTβR and activating NF-κB signaling. Cancer Cell Int. 2021;21(1):118. doi: 10.1186/s12935-021-01827-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ankö ML, Neugebauer KM. RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci. 2012;37(7):255–262. doi: 10.1016/j.tibs.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 18.Ma Y, Liu Y, Pu YS, et al. LncRNA IGFL2-AS1 functions as a ceRNA in regulating ARPP19 through competitive binding to miR-802 in gastric cancer. Mol Carcinog. 2020;59(3):311–322. doi: 10.1002/mc.23155 [DOI] [PubMed] [Google Scholar]
- 19.Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–426. doi: 10.1007/s10456-017-9562-9 [DOI] [PubMed] [Google Scholar]
- 20.Reymond N, D’água BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13(12):858–870. doi: 10.1038/nrc3628 [DOI] [PubMed] [Google Scholar]
- 21.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Xu D, Zhou M, et al. CDC42EP3 promotes colorectal cancer through regulating cell proliferation, cell apoptosis and cell migration. Cancer Cell Int. 2021;21(1):169. doi: 10.1186/s12935-021-01845-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17(3):153–168. doi: 10.1038/s41575-019-0245-4 [DOI] [PubMed] [Google Scholar]
- 24.Lal N, White BS, Goussous G, et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer. 2018;24(1):224–233. doi: 10.1158/1078-0432.CCR-17-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Li H, Mao Y, et al. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int J Mol Med. 2020;45(1):10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 27.Antonio N, Bønnelykke-Behrndtz ML, Ward LC, et al. The wound inflammatory response exacerbates growth of pre-neoplastic cells and progression to cancer. EMBO J. 2015;34(17):2219–2236. doi: 10.15252/embj.201490147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fridlender ZG, Sun J, Kim S, et al. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell. 2009;16(3):183–194. doi: 10.1016/j.ccr.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodogai M, Moritoh K, Lee-Chang C, et al. Immunosuppressive and prometastatic functions of myeloid-derived suppressive cells rely upon education from tumor-associated B cells. Cancer Res. 2015;75(17):3456–3465. doi: 10.1158/0008-5472.CAN-14-3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234(6):8509–8521. doi: 10.1002/jcp.27782 [DOI] [PubMed] [Google Scholar]