Abstract
The Covid-19 European outbreak in February 2020 has challenged the world's health systems, eliciting an urgent need for effective and highly reliable diagnostic instruments to help medical personnel. Deep learning (DL) has been demonstrated to be useful for diagnosis using both computed tomography (CT) scans and chest X-rays (CXR), whereby the former typically yields more accurate results. However, the pivoting function of a CT scan during the pandemic presents several drawbacks, including high cost and cross-contamination problems. Radiation-free lung ultrasound (LUS) imaging, which requires high expertise and is thus being underutilised, has demonstrated a strong correlation with CT scan results and a high reliability in pneumonia detection even in the early stages. In this study, we developed a system based on modern DL methodologies in close collaboration with Fondazione IRCCS Policlinico San Matteo's Emergency Department (ED) of Pavia. Using a reliable dataset comprising ultrasound clips originating from linear and convex probes in 2908 frames from 450 hospitalised patients, we conducted an investigation into detecting Covid-19 patterns and ranking them considering two severity scales. This study differs from other research projects by its novel approach involving four and seven classes. Patients admitted to the ED underwent 12 LUS examinations in different chest parts, each evaluated according to standardised severity scales. We adopted residual convolutional neural networks (CNNs), transfer learning, and data augmentation techniques. Hence, employing methodological hyperparameter tuning, we produced state-of-the-art results meeting F1 score levels, averaged over the number of classes considered, exceeding 98%, and thereby manifesting stable measurements over precision and recall.
Keywords: SARS-CoV-2, Lung ultrasound, Deep learning, LUS Score
Graphical abstract
1. Introduction
SARS-CoV-2, which is the causative agent of the current Covid-19 pandemic, originated in China and abruptly began being transmitted within Europe in February 2020. It rapidly spread throughout the world and is still challenging the world's health systems. It manifests after a long incubation period along with a high contagion rate[1], thus necessitating the development of fast and cheap diagnostic tools to detect infected subjects.
Moreover, Covid-19 can cause bilateral multifocal interstitial pneumonia, which can rapidly evolve into acute respiratory distress syndrome (ARDS), an ominous complication that is responsible for causing hundreds of thousands of deaths worldwide. Subjects infected by SARS-CoV-2 may present an evolving clinical picture ranging from focal to multifocal interstitial pulmonary involvement that may be visualised by LUS in the so-called white lung pattern, as well as by bilateral submantellar-subpleural consolidations[2,3]. The high contagion rate adds a further level of complexity because patient care, according to the highest healthcare standards, must be combined with strict pandemic protocols that need to be followed for the safety of healthcare professionals[4].
Currently, the main diagnostic tools for detecting and isolating infected people include reverse transcription-polymerase chain reactions (RT-PCR) in nasopharyngeal swabs (NPS) and IgM-IgG combined antibody tests[5]. However, both these tools have limitations: the former does not reach a 100% sensitivity, introducing the possibility of false-negative results, one of the causes for the incorrect separation of patient flows in hospitals. Moreover, it is time-consuming and when the number of infected subjects increases, inevitable shortages in reagents and other specific laboratory supplies occur, thereby preventing the completion of tests. IgM-IgG tests not only exhibit the same poor sensitivity as that of the former, with a slight increase only after a certain duration following symptom manifestation, but also may result in false-negative results in the early phases of the infection. Covid-19 begins with mild or no symptoms, and yet can rapidly transform, subjecting patients to extremely critical conditions with possible fatal consequences resulting from multi-organ failure. Therefore, it is critical to promptly and reliably detect infected subjects to apply the appropriate treatments and prevent the virus from spreading. Moreover, no tests can describe the presence or severity of lung engagement. Hence, the need for devices that accommodate increases in resources is an undeniable necessity[[6], [7], [8], [9], [10]].
First-line diagnosis of pneumonia may exploit chest X-rays (CXR) for first-aid treatment of patients exhibiting symptoms of pneumonia[11]. Potential alternatives to CXR include computed tomography (CT) scans and lung ultrasound (LUS)[[12], [13], [14]]. The main conclusions from studies concerning these methodologies state that: first, both LUS and CT scans are significantly better first-line diagnostic tools than CXR, whose main drawback is poor sensitivity; second, although ultrasonography is a cost-effective, radiation-free, and promising tool, it must be performed by a highly skilled radiographer to achieve accurate results. Furthermore, LUS effectively performed at a bedside in approximately 13 min yielded a higher sensitivity than that of CXR. This makes it comparable to other CT imaging tools with its cost being significantly lower than those of the other two solutions. Moreover, LUS is radiation-free, easier to disinfect, and can be repeated even with small time intervals between two observations, while the same is not true for the other methodologies[7,15]. However, it has certain drawbacks, such as operator dependency and high expertise requirements, resulting in underutilisation, and it may not be useful for Covid-19 asymptomatic patients.
The Covid-19 pandemic has resulted in renewed attention on these studies and led to medical professionals considering possible solutions for the above-mentioned problems and the procurement of fast, cheap, and efficient diagnostic tools. Covid-19 necessitates certain imperative and strict constraints to avoid cross-contamination, such as through infected staff and infected medical devices, and provide patients with the highest standard of healthcare, such as moving patients around the hospital for treatments, and making diagnostic tools easily available to everyone. These crucial necessities made it impossible to use a stethoscope during hospital operations in infectious disease departments owing to the use of personal protective equipment. Therefore, researchers concluded that both CT scans and LUS are promising diagnostic instruments that are capable of early SARS-CoV-2 pneumonia detection and present highly correlated patterns for different disease stages[3,7,[16], [17], [18], [19]]. Although the former initially served a pivotal function during the pandemic, it exhibits some weaknesses in terms of the previously stated constraints, while the latter does not. Hence, it is beneficial to rely upon an international standardisation of LUS exploitation[3,20], providing not only a medical procedure to be applied by sonographers but also a scoring scale ranging from 0, indicating a healthy patient, to 3, which represents a critical clinical situation (e.g. a patient with a damaged lung, who is almost incapable of breathing without medical treatment).
In this context, researchers have extensively reviewed deep learning (DL)-based biomedical imaging[21,22], highlighting the challenges of using labelled datasets in medical contexts. This technique has been investigated as a promising solution for overcoming the previously stated problems and introduces advantages, including diagnostic speed, reliability, and provision of support to physicians who are managing the emergency. Recent systematic surveys on DL applications for the novel coronavirus revealed that studies mainly focused on CT scans and X-rays[[23], [24], [25], [26], [27]]. Less than half of the investigations employed transfer learning, while none of them evaluated the severity of lung engagement[6,28]. Physicians extensively used LUS to estimate consequences in patients admitted to the emergency department (ED) and to detect Covid-19 pneumonia in subjects who presented a negative swab[7,29,30]. Nevertheless, only a few studies have investigated the application of DL algorithms to LUS data. These studies focused on either detecting B-lines, namely artefacts appearing when patients suffer from pneumonia, or binary classifications of LUS frames into Covid-19 and non-Covid-19[15,31]. Moreover, only a few researchers have exploited data from reliable hospital sources[[32], [33], [34]], indicating the lack of a reliable dataset; several authors have described the inconsistent quality of their data and the need to rely on non-validated sources as limitations of their studies[35]. In addition, some researchers have worked with LUS from only one particular type of probe, thus lacking heterogeneous data to train the neural networks, posing another limitation on the soundness of their conclusions and DL algorithm usage[33]. Only two studies have focused on DL systems for the purpose of detecting Covid-19 pneumonia and assessing the severity of lung engagement[32,34]. The former exploited a spatial transform network[36] developed in 2015, while the latter proposed an original neural network; however, both exhibited poor performance at frame-level scoring for assessing the severity of lung engagement, and neither of them made use of pre-trained or state-of-the-art architectures. Furthermore, the authors of the former study proposed a novel scoring methodology[3] for already validated and researched scales evaluating lung health status[20], which the latter adopted as well. To the best of our knowledge, there has been no investigation on assessing and ranking the lung pleural line health conditions through the application of artificially intelligent systems to LUS data obtained from Covid-19 subjects. Moreover, all investigations regard frame classification, without addressing the entire LUS clip, as the research we propose in this manuscript.
In this study, we propose an innovative artificial intelligence (AI) system based on pre-trained and state-of-the-art residual convolutional neural networks (ResNets, CNNs -[37,38]), to detect SARS-CoV-2 pneumonia patterns in LUS frames and classify the severity of lung engagement. By extensively tuning the architecture's hyperparameters, we improved on previously presented results[32,34]. The quality of the work was also assured through close collaboration with Pavia's San Matteo Hospital ED, whose Ethics Committee granted us access to LUS data from different probes, obtained by several different hospital physicians during the pandemic and evaluated according to two different scales. We extended one of the employed scoring systems, already proven in the literature[20], by adding information regarding the lung's pleural line health condition, which helps in differentiating cardiogenic from non-cardiogenic causes of B-lines[33]. The developed AI-enabled assistant can function both in emergency contexts and in home monitoring of patients. Additionally, it can help detect patients with clear Covid-19 symptoms whose RT-PCR or IgM-IgG blood tests were negative. These AI methods can overcome the limitations, such as inadequate number of available RT-PCR tests, their high costs, and waiting time for test outcomes[25].
We structured the rest of the article as follows: Materials and Methods presents a detailed description of the methodologies, techniques, and data used to conduct the experiments. Results and Discussion present the principal and most representative results essential for comparing our study with state-of-the-art works published by our colleagues, thus highlighting the significance of the results. Finally, the last section presents the main conclusions, and implications that advance the field based on current knowledge and our achievements.
2. Materials and Methods
This section provides an in-depth description of the data, exploratory analysis, collection, and annotation processes, together with the selection, design, and training of the CNN architectures, which we selected for our diagnostic purposes. In particular, we focused on data augmentation, transfer learning, and training options, as well as the hyperparameters used to train and fine-tune deep networks.
2.1. Lung ultrasound score
To better highlight the reliability of the results and the ability of the deep architecture to detect Covid-19 pneumonia patterns, we first describe the employed ranking scales and compare them with the one used by other authors[3,32,34], revealing their differences. Doing so explains the implications of the deep residual networks, and also emphasises that exploiting and extending a different scoring measurement system[20], which has already been presented and validated in the literature, contributes to outperforming the state-of-the-art results attained by our colleagues. The differences between the LUS score used to rank our data in this study and that used by our colleagues[3,32,34] are examined in Table 1 .
Table 1.
Scoring comparison Soldati et al. (2020) and S. Mongodi et al. Modified Score.
| Severity Score | Soldati et al. | Modified Score |
|---|---|---|
| Score 0 | A-lines | A-lines with at most two B-lines |
| Score 0* | Not defined | A-lines, and at most two B-lines, with a slightly irregular pleural line |
| Score 1 | An irregular or damaged pleural line along with visible vertical artefacts | Artefacts occupy at most 50% of the pleura |
| Score 1* | Not defined | Artefacts occupy at most 50% of the pleura and present a damaged pleural line |
| Score 2 | Broken pleural line with either small or broad consolidated areas with wide vertical artefacts below (white lung) | Artefacts occupy more than 50% of the pleura, while consolidated areas may be visible |
| Score 2* | Not defined | Artefacts occupy more than 50% of the pleura, while consolidated areas may be visible. The pleura is either damaged or irregular |
| Score 3 | Dense and broadly visible white lung with or without larger consolidations | Tissue-like pattern |
We began evaluating ultrasound data with Score 0, in which the pleural line was continuous and regular, and A-lines were present as horizontal artefacts owing to the high reflectance of the aerated lung surface. Hence, multiple reflections appeared between the probe and lung surface. We evaluated this level of data in a similar manner to that in Reference 3.
Next, we defined Score 0* as any image evaluated as Score 0 but with an irregular or slightly damaged pleural line.
Furthermore, we increased the level of severity when either vertical areas of white or consolidations were visible (Score 1). These white regions were due to local alterations in the acoustic properties of the lung when the previously aerated lung volume transformed into tissue or water-like aggregates. This process clearly explains the appearance of vertical artefacts. While Reference 3 assigned an ultrasound recording with Score 1 when the pleural line was flawed with any visible vertical area of white, we ranked a recording with this level of severity when artefacts were present and occupied less than 50% of the pleura. The two constraints imposed allowed for a more structural hierarchy in our classification spectrum, thus improving our classification performance.
In addition to the introduction of Score 0*, herein, we further introduced another classification, defining Score 1* as a recording that would have typically been assigned Score 1 but had an irregular or damaged pleural line. Clearly, the higher the score, the greater the damage detected upon examination of the pleura.
In general, specialists evaluate a patient's lung as Score 2 if larger consolidated regions (dark areas) appear along associated areas of white below solidifications. This pattern typically leads to what is commonly referred to as the white lung. Dark and dense sections in the lung suggest a transition in the lung tissue and its acoustic properties toward a condition observed when examining soft tissue. However, the appearance of white and large areas implies that the lung is not fully ventilated; air inclusions are still present but embedded in tissue-like compounds. These high scattering conditions can explain this specific pattern. Therefore, Reference 3 assigned the level of severity 2 when a pleural line was broken, along with either small or large dense fields with broad and white vertical artefacts below. In contrast, we marked a recording with this severity level when the vertical artefacts occupied more than 50% of the pleura and both small and bounded consolidations indicating a more critical stage of illness were visible[20].
Similarly, we assigned Score 2* when the pleura was either irregular or damaged in a lung that would have typically received Score 2.
Finally, a lung characterised by dense and broadly extended white lung areas with abundant consolidations was usually assigned Score 3. Although this description fits well with the scoring methodology presented in Reference 3, we assigned this severity level when the lung presented tissue-like patterns, i.e. widely dense and dark consolidations.
In conclusion, we further refined the classification task, extending an already validated standardised LUS score from Reference 20 consisting of four classes, to a novel and more complex version with seven classes. We added three more classes to indicate whether the lung's pleural line was affected by pneumonia. We inserted these classes between the already existing classes. While the original pneumonia severity classification scale comprised scores ranging from 0 to 3, we improved it by inserting the scores 0*, 1*, and 2*, respectively, between scores 0–1, 1–2, and 2–3. As the severity level Score 3 describes a lung almost incapable of breathing, there is no need to define Score 3*. A lung rated as Score 3 indicates that the pleural line is affected by the illness. The pleural line is defined as the interface between the fluid-rich soft tissues of the wall and the gas-rich lung tissue[39]. Therefore, by adding three classes to indicate its condition, we can provide useful information regarding the severity of the disease, thereby assisting in discriminating cardiogenic from non-cardiogenic causes of B-lines[33]. Therefore, a patient who would typically have received a score of 0, indicating a healthy lung, could be assigned a score of 0*, informing the specialist that the patient may have a lung injury. Understanding that a subject might be suffering from Covid-19 before the lung approaches a more severe condition not only results in providing immediate and appropriate treatments but also improves survival rates in critical situations and the possibility of quick healing.
We adopted the two aforementioned scales to understand whether DL architectures would benefit from a hierarchical labelling extension in classifying input data or cause a clustering performance degradation; hospital physicians could regularly monitor the pleura without any particular additional effort while still retrieving valuable information regarding the patient's health condition. This necessitates the replication of the same procedure using a computer-aided system.
2.2. Data collection and annotation
Since March 2020, the San Matteo Hospital's ED has been collecting LUS data to assess the health conditions of patients who contracted Covid-19. The medical personnel used the ultrasound machine Aloka Arietta V70 (Hitachi Medical Systems), equipped with both convex and linear probes, at 5 MHz and 12 MHz, respectively. They standardised the acquisition procedure through abdominal settings, focusing on the pleural line, reaching a depth of 10 cm with the convex probe. Moreover, they adjusted the gain so as to attain the best possible imaging of the pleura, vertical artefacts, and peripheral consolidations with or without air bronchograms. Physicians conducted both longitudinal and transversal scans to explore the broader pleural length, disabling all harmonics and artefact-erasing software.
Physicians performed LUS on people with a clear clinical picture[7], owing to the RT-PCR test introducing many false negatives. Namely, artefacts reported in the earlier section were formed by either pulmonary oedema or non-cardiac causes of interstitial syndromes[33]. Despite presenting a negative RT-PCR test, subjects manifesting lung involvement have a high probability of being Covid-19 positive. Physicians are accustomed to differentiating suspicious subjects from healthy subjects following a triaging procedure involving LUS investigation.
Hereafter, we define a clip as the result of an LUS examination. It consists of a set of frames, namely, the images used in our study. The proposed definition is intended to produce continuity regarding observations in other similar works[33].
The hospital's medical personnel collected 12 clips for each patient, all assigned with a standardised LUS score[20,40]. The ED collected data from 450 patients whose clinical information is presented in Table 2 , treated in Pavia, consequently gathering a total of 5400 clips. Table 2 lists the subjects, who were classified as Covid-19 positive and negative, and the clinical data through median and 25th–75th percentile values. The LUS Score entry indicates the sum of the values collected for each patient who received 12 examinations, as reported in Section 2.1.
Table 2.
Fondazione IRCCS San Matteo Hospital patients’ clinical information.
| Negative (172 patients) | Positive (278 patients) | Total (450 patients) | ||||
|---|---|---|---|---|---|---|
| Median | 25 - 75 P | Median | 25 - 75 P | Median | 25 - 75 P | |
| Age (years) | 54 | 37.0–67.5 | 63 | 51.0–75.0 | 60 | 47.0–73.0 |
| Systolic blood pressure (mmHg) | 135 | 125.0–150.0 | 130 | 115.5–144.0 | 130 | 120.0–145.0 |
| Diastolic blood pressure (mmHg) | 80 | 70.0–90.0 | 80 | 70.0–85.8 | 80 | 70.0–90.0 |
| Respiratory rate | 20 | 16.0–22.0 | 20 | 16.0–26.0 | 20 | 16.0–24.0 |
| Oxygen saturation (%) | 97 | 94.0–98.0 | 94 | 90.0–97.0 | 95 | 91.0–98.0 |
| Body temperature (°C) | 36.7 | 36.2–37.6 | 37.1 | 36.5–38.0 | 37 | 36.3–37.9 |
| Hemoglobin (g/dL) | 13.5 | 12.2–14.9 | 13.9 | 12.8–14.9 | 13.7 | 12.6–14.9 |
| White blood cell (10∧9/L) | 8.2 | 6.3–11.5 | 6.3 | 4.8–8.1 | 6.92 | 5.1–9.2 |
| Lymphocytes (10∧9/L) | 1.555 | 0.9–2.2 | 0.8 | 0.6–1.1 | 1 | 0.7–1.6 |
| Platelets (10∧9/L) | 224.5 | 179.5–272.5 | 184 | 146.0–239.0 | 204 | 157.0–256.7 |
| C-reactive protein (mg/dL) | 1.325 | 0.1–10.5 | 7.97 | 2.6–15.2 | 5.29 | 0.9–14.4 |
| Lactate dehydrogenase (U/L) | 222 | 182.0–290.0 | 326 | 243.5–428.0 | 286 | 211.2–399.7 |
| Creatine phosphokinase (U/L) | 86 | 51.0–143.0 | 113 | 68.0–293.5 | 99 | 62.0–217.7 |
| PH | 7.4 | 7.4–7.4 | 7.4 | 7.4–7.4 | 7.44 | 7.4–7.4 |
| PaO2/FiO2 | 392.1 | 317.5–462.9 | 299.5 | 226.4–352.7 | 323.8 | 256.0–405.8 |
| Alveolar-arterial gradient of O2 (mmHg) | 22.4 | 9.5–42.5 | 47.3 | 33.6–93.1 | 40.4 | 20.8–60.8 |
| LUS Score | 2 | 0.0–7.5 | 11 | 6.0–16.0 | 7 | 2.0–13.0 |
However, not all clips received an LUS score from the same medical practitioner. Therefore, we further reviewed the collection to validate the classifications and avoid incorrect severity-scoring problems. This process was mandatory to ensure that each clip had a standardised LUS score and there were no discrepancies in the scores assigned to different clips, which are problems stressed in other studies[32].
Fondazione IRCCS Policlinico San Matteo Emergency Room's physicians observed the methodological procedure and ensured that the labelling was correct. During the first part of the collection and annotation process, they manually selected all clips from each patient, assessed the quality of each clip, and either proceeded to evaluate it according to the two scoring methodologies or discarded it. They reviewed each clip to assign a score and verify that SARS-CoV-2 pneumonia patterns, described in Section 2.1, were present. The scoring was based on the first ranking scale[20] with four classes or its extended version with seven classes. Second, from among the many frames belonging to a clip, they selected the ones containing such patterns manually, as other frames might be related either to a healthy lung's portion or noisy and blurred due to incorrect probe movements or respiration-induced dynamic motions, thereby altering the imaging quality across the clip[34]. In a blinded and random process, to avoid biasing the final results of our experiments, physicians examined an extracted clip and selected frames in which lung patterns were visible. The higher the score assigned, the fewer the time instants required for classifying a clip. For instance, a patient assigned a score of 1 might have only a few frames containing B-lines. Because DL architectures must be trained to detect and classify pneumonia patterns, we need to identify and extract such patterns. The number of frames selected is not equal for each clip. The blind selection process avoids retrieving all clips from a patient with the same pattern in most lung portions, while discarding clips exhibiting other manifestations. Therefore, both the number of patients from whom we clipped the frames and the number of images used for each subject are unknown. Although the acquisition process had been standardised[40], it was conducted during contingency periods; thus, not all patients underwent 12 examinations. Some subjects might have received fewer examinations than others because of the detection of severe lung engagement in the early stages of the procedure.
The entire annotation and collection procedure lasted for longer than one month, resulting in a set of 676 gathered clips based on 5400 starting clips. As physicians performed LUS investigations employing different probes with slightly different settings, clips were of different sizes in terms of pixels. Therefore, we resized all the clips such that each frame had dimensions of 224 × 224, which is compliant with the input size for a DL architecture.
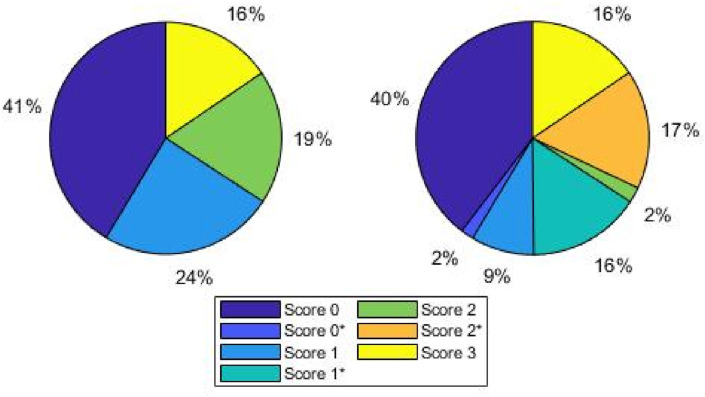
As the medical personnel had to continuously meet the demanding and urgent pandemic requirements, and the aforementioned process is demanding and time-consuming, we considered the collection and labelling process to be completed when the DL architectures began yielding promising results for the validation and test sets, as described below, and the dataset was said to be well-balanced. Hence, we started with a smaller set of collected frames and, finally, 2908 frames were carefully selected to train the CNNs from among more than 60000 frames. Fig. 1 shows the percentage of images assigned to each score for both diagnostic tasks; pleural line involvement is highly likely and more severe when a frame is assigned a high score. For instance, this explains why most frames that belonged to group with Score 2 were assigned to the group with Score 2*, while the same is not true for lower score values.
Fig. 1.
Percentage distribution of frames for each classification task. Left: four class scenario; right: seven class scenario. The percentage of images assigned to each score for both diagnostic tasks is depicted; pleural line involvement is highly likely and more severe when a frame is assigned a high score.
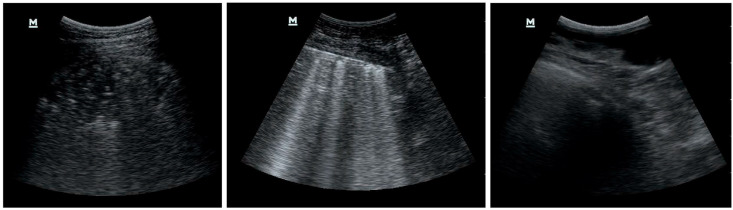
Fig. 2 depicts examples of the selected and discarded frames. The first two images represent a score of 3 and 2, respectively, while we rejected the third image as being too noisy due to probe movements during the bedside ultrasound exam. This process is mandatory and time-consuming, as the first and third images may appear to be significantly similar to an untrained physician's eye. Nonetheless, the same consideration does not hold for a neural network trying to classify the third and noisy frame; because we did not assign any label to the discarded frames, it would try to classify it as belonging to one of the considered classes. However, this would result in an almost random scoring, and is beyond the scope of the manuscript's goal of recognising and classifying Covid-19 patterns in LUS frames.
Fig. 2.
Examples of selected and rejected frames.
Finally, we randomly split the data into training (75%), validation (15%), and test (10%) sets, adopting these percentages in accordance with common DL methodologies[28] and maintaining the training set size as low as possible to avoid overfitting problems. Furthermore, we employed data augmentation techniques, as explained in Section 2.3. Finally, we collected 17448, 436, and 291 frames for the training, validation, and test sets, respectively.
2.3. Deep learning architectures
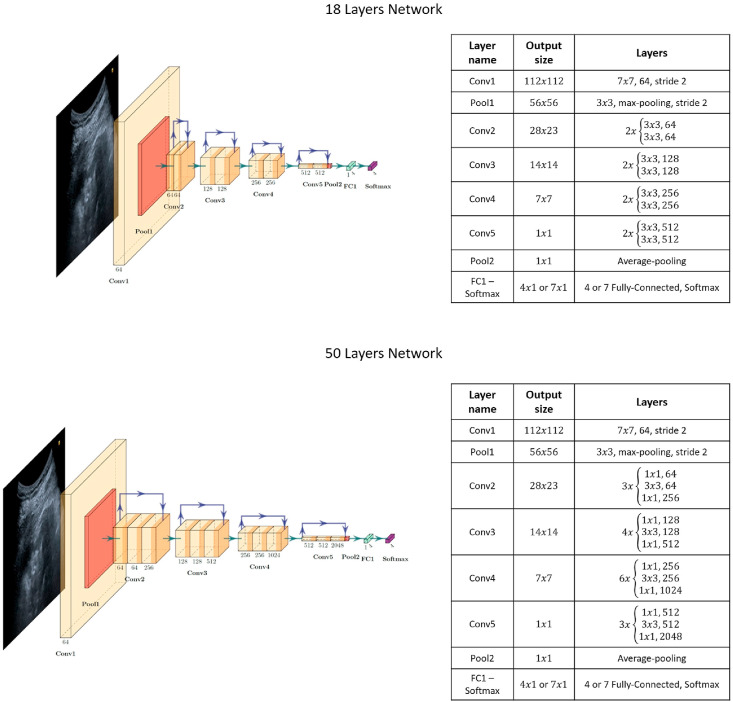
In this study, we adopted deep residual networks to achieve the best and reliable classification performance, avoiding vanishing gradient problems and allowing for deeper architectures than the commonly used ones, which do not exploit residual connections. Researchers have described the process of using already proven models as a more rational approach for initiating DL model development from scratch[25]. In particular, we selected two residual networks with 18 and 50 layers each and structured them as reported in the original paper[38]. In addition, we extensively exploited a commonly known methodology, transfer learning[37], to significantly improve the classification results by exploiting features belonging to pre-trained networks. This methodology has been confirmed to improve Covid-19 detection[35]. The best common practice is to use DL architectures that have been pre-trained on similar domains to overcome small-sized dataset problems and poor classification performances. Therefore, we selected ResNet18 and ResNet50 architectures, which had already undergone optimisation based on the ImageNet dataset[41]. However, we made a few modifications to these networks before using them; we changed the last fully connected layers because they had as many neurons as the number of classes to be detected. The classification problem to be solved involves the detection of the lung patterns described in Section 2.1. Consequently, we designed four different architectures, which are the two ResNets for solving the two clustering queries; the first comprises four categories[20], whereas the second is represented by the seven classes obtained by broadening the first scale, providing information regarding the pleural line integrity.
The two architectures employed in this study are depicted in Fig. 4. ResNet18 and ResNet50 take input images with dimensions of 224 × 3. All of them undergo a first step consisting of a 7 × 7 convolution with a feature size of 64 and a stride of 2, followed by a 3 × 3 max-pooling step with the same stride. Next, each of the following layers performs either 3 × 3 or 1 × 1 convolutions with a fixed feature map dimension for the first residual network, namely , and with an increasingly repeated pattern for the second residual network, that is, withfollowing the fixed feature map order mentioned above. The input is bypassed every two convolutions for ResNet18 and every three convolutions for the other residual architecture. Both width and height remain constant throughout the section because the padding and stride are set to 1 during the operations, thereby allowing the connection to be skipped. The residual models exploit batch normalisation to improve regularisation together with the pooling layers. ReLu is selected as the activation function. Finally, the 18-layer residual network has 11.174 M parameters, while the 50-layer network consists of 23.521 M parameters.
Fig. 4.
– Residual Network Structure Diagrams: plot of each ResNet employed together with their structure and exploited layers.
Table 3 presents the training options and hyperparameters for each network, to address the two different detection problems solved in our study. The training process relies on the pseudo-random selection of both the mini-batches and the initial weights of the models. Hence, we set the random seed to 19 for all experiments. This setting makes the experiments reproducible and provide a clear view of the improvements derived from tuning the training options and hyperparameters.
Table 3.
Training Options and Hyperparameters.
| Options and Hyper-parameters | Four Classes |
Seven Classes |
||
|---|---|---|---|---|
| ResNet18 | ResNet50 | ResNet18 | ResNet50 | |
| Initial Learning Rate | 0.0005 | 0.0001 | 0.0001 | 0.0001 |
| Learning Rate's Drop Factor | 0.05 | 0.05 | 0.05 | 0.05 |
| Learning Rate's Drop Period (Epochs) | 2 | 3 | 3 | 3 |
| Batch Size | 128 | 64 | 128 | 64 |
| L2 – Regularisation | 0.4 | 0.75 | 0.3 | 0.3 |
| Epochs | 15 | 12 | 15 | 12 |
| Environment | Multi-GPU | Multi-GPU | Multi-GPU | Multi-GPU |
| Optimiser | Adam | Adam | Adam | Adam |
| Loss Function | Cross-Entropy | Cross-Entropy | Cross-Entropy | Cross-Entropy |
Before describing the hyperparameter tuning procedure, it is worth explaining some of the rows listed in Table 3 whose names may be misleading considering the commonly encountered nomenclature in articles focusing on DL, such as L2-regularisation, number of epochs, and mini-batch size. The learning rate drop factor implies that we steadily decreased each predetermined number of epochs, in a piecewise manner, at the same pace at which we were updating the network weights. We reduced the learning step by multiplying it by the dropping factor. Second, we selected Adam Gradient Descent[42] as the optimisation algorithm for the two residual architecture weights. During training, we adopted a validation set for tuning the hyperparameters and predicting the behaviour of the models with a new test set, revealing the robustness of our results.
First, we heuristically determined the initial learning rate, which allows for a desirable classification performance level, evaluated over both the training and validation sets. Then, we selected the learning rate's drop factor in a similar manner, enabling the optimal achievement of the cost function's minimum with elapsing epochs from the start of training. Additionally, we selected the number of epochs after which the learning rate was expected to decrease. Reducing it too early may lead to almost no update to the networks' weights after a few iterations; however, waiting too long may cause the weights to continuously leap near the cost function's minimum while never reaching it. On completion of these steps, we focused on L2-regularisation, batch size, and the number of training epochs. L2-regularisation reduces overfitting and the architectures' batch normalisation and pooling layers. It acts by adding a cost function term equal to the sum of all network weights squared and multiplied by the L2 constant. The larger the constant, the smaller the weights, thereby reducing overfitting while introducing the risk of underfitting. Therefore, we determined the best L2-regularisation constant by observing the network behaviour for the validation and test sets. We employed this latter procedure in selecting the mini-batch size and number of epochs. Finally, we set the squared gradient decay factor and gradient decay factor to 0.999 and 0.98, respectively. Researchers commonly adopt this default decision for Adam optimisation.
Once the tuning process ended, satisfying the classification performances for the test and validation sets, we turned the random seed off and repeated all experiments seven times to display all performance metrics as a mean and standard deviation, and to reject the possibility of the results being biased.
We increased the statistical assortment in the training set by adopting data augmentation techniques, which helped the networks focus on meaningful information. We applied geometric, filtering, random centre cropping, and colour transformations to the training frames. This method, proven to work when applied to Covid-19[43], produces effective results in DL classification tasks, significantly reducing overfitting[44]. Furthermore, we added salt-and-pepper white noise to enlarge the training set. Pre-trained architectures accept images of the size 224 × 3. Therefore, we treated the grey-scale ultrasound frames as RGB images, both to avoid modifying the input layers and to allow for colour augmentation. Data augmentation numerically modifies the training images, introducing statistically diverse samples, and allowing the architectures to robustly classify new frames: moving the point of interest in the frame and slightly modifying its shape or colour together with noise, which prepares the models not to expect relevant features in the same spot. Moreover, the models learn to reject disturbances such as probe sensor measurement errors. Therefore, we applied all augmentations to all training images, independent of the probe employed for the LUS investigation.
First, we applied one of the augmentations listed in Table 4 to the training set. Hence, we created a new set by unifying the original images and the transformed images. Second, we applied a second transformation to the new set. Finally, we recursively applied this procedure to broaden the training set exponentially.
Table 4.
Data augmentation operations used during the investigations. We list both the augmentations names and descriptions.
| Augmentation Name | Augmentation Description |
|---|---|
| Image noise | Adds salt-and-pepper noise to image. Namely, random pixels get randomly coloured towards white. Spreading power of modified pixels can be set by a parameter; hence, different augmentations can be considered as being more or less noisy. |
| Colour jittering | Adjusts the colour of RGB image I with a randomly selected value of hue, saturation, brightness, and contrast from the HSV colour space. Specify the range of each type of adjustment using name-value pair arguments. Four augmentations can be retrieved. |
| Flip | Images are flipped either from left to right or upside down. |
| Centre cropping | Images are centre cropped using a 150 × 150 window to ensure that Covid-19 patterns are selected during operation. |
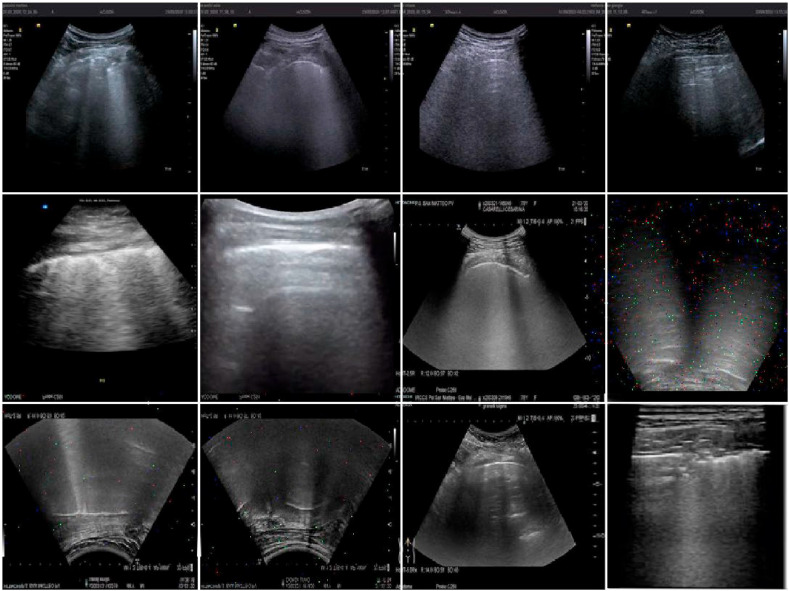
Fig. 3 depicts a set of 12 augmented examples: introducing such small alterations into the training set allowed the CNN architectures to develop invariance to translations, viewpoints, sizes, illumination, and noise, resulting in a more regularised training process[44]. The validation and test sets did not receive such augmentation processes to prevent the final results from being biased.
Fig. 3.
Augmented training set images: augmentations described in this section have been applied to the training images and are shown in this figure.
To further assess the classification reliability, we used both class activation mapping (CAM) and Grad-CAM techniques[45,46]. These processes allow for the interpretation of the decision-making task model. When applied to image classification problems, they highlight the parts that are decisive for the assignment of a rank by the network through a heat map. DL models can focus on points that the human eye may not see. Therefore, emphasising what networks recognise may assist physicians’ perceptions. In particular, we assessed whether the networks correctly highlighted either B-lines or pleural line discontinuities, when present, and all other patterns described in Section 2.1. This pioneering idea allows for a comparison of different prototypes to determine the best one. Moreover, it is a cost-effective way to avoid increasing dataset preparation times by manually creating segmentation maps for detecting Covid-19 pneumonia boundaries. Although we intend to highlight the presence of patterns, we do not focus on exposing their exact profiles. Some researchers have attempted and validated this method by applying it to Covid-19, achieving excellent results[25,46].
The test system used to conduct our experiments was equipped with an Intel-i9-9900X CPU, working at 3.5 GHz, 128 GB of RAM, and two 2944-cores NVIDIA RTX 2080. We wrote all code and designed the networks using MathWorks MATLAB 2020a Release together with its Deep Learning Toolbox.
2.4. Performance evaluation
When handling medical data, it is vital to reduce the number of false negatives to the maximum extent possible, particularly when treating an infectious disease such as Covid-19. The consequences of incorrectly diagnosing a patient as Covid-19 negative, which introduces a false negative, include not only inappropriate care and lack of necessary treatment (reflected in cross-contamination among subjects who may have additional pathologies), but also incorrect medications that may harm an infected person. We measured the network classification performances using the validation and test sets. We not only investigated the accuracy but also the precision, recall, and F1-score (Equations Equation 1, Equation 2, Equation 3, Equation 4) and ROC-AUC ([47]). The equations listed below define these metrics, which were computed for each category for both classification scenarios, namely, four and seven classes. TP refers to True Positive classifications, FN denotes False Negative classifications, TN denotes True Negative classification, and FP refers to False Positive classifications.
| Equation 1 |
| Equation 2 |
| Equation 3 |
| Equation 4 |
Considering the importance of reducing the false-negative results, in medical contexts, researchers particularly consider recall, also known as sensitivity. This parameter indicates the performance of evaluating a frame as not containing Covid-19 pneumonia patterns and belonging to either of the classes considered or not being representative of a healthy lung. Nonetheless, precision notifies the reader of the classification performance in terms of detecting, instead of the considered patterns. Consequently, we consider the F1-score as a function of the two former metrics. This parameter yields a better measurement in terms of accuracy considering the trade-off between precision and recall in an unbalanced class distribution.
In conclusion, both recall and F1-score must be considered to minimise the false negatives while maintaining high precision.
3. Results and Discussion
Both the selected architectures steadily approached convergence based on the hyperparameters and training options listed in Table 3. Considering the evaluation metrics discussed in Section 2.4, we present them in terms of the average over the number of groups considered for each classification scenario in Table 5 . The training process involved stochastically splitting the data into training, validation, and test sets; using the training set to optimise the network weights; and at the end of each epoch, exploiting the validation set to assess the models’ accuracies and losses. On completion of the training process, we evaluated the aforementioned metrics for the training, test, and validation sets. We considered the network weights at the end of each training process, regardless of the number of epochs selected for optimisation, as listed in Table 3. We did not doublecheck a particular epoch exhibiting promising performances with the validation set during optimisation because all series of training approaches converged steadily when the number of epochs (listed in Table 3) for each network elapsed. This methodology was repeated seven times for each classification scenario to avoid bias in the final results. Hence, we describe these metrics using their mean values and standard deviations in Table 5. Furthermore, each hyperparameter was extensively tuned to obtain recall and F1-score levels exceeding 90%, indicating a high and reliable balance over both the precision and recall metrics, which is essential when handling unbalanced datasets, such as ours. As presented in Table 5, this resulted in both networks behaving remarkably well in each scenario and with excellent results achieved by ResNet50. In addition, in each experiment, we could obtain recall levels of over 97% on average, thereby verifying the soundness of the classification performances in predicting whether a frame without Covid-19 pneumonia patterns belongs to either of the classes considered, or is not representative of a healthy lung.
Table 5.
- Classification Performance results for test and validation sets: Accuracy, precision, recall, F1-Score and ROC-AUC.
| Metric | Four Classes |
Seven Classes |
||
|---|---|---|---|---|
| ResNet18 | ResNet50 | ResNet18 | ResNet50 | |
| Training Accuracy | ||||
| Training Precision | ||||
| Training Recall | ||||
| Training F1-Score | ||||
| Training ROC-AUC | ||||
| Test Accuracy | ||||
| Test Precision | ||||
| Test Recall | ||||
| Test F1-Score | ||||
| Test ROC-AUC | ||||
| Test Accuracy | ||||
| Validation Accuracy | ||||
| Validation Precision | ||||
| Validation Recal | ||||
| Validation F1-Score | ||||
| Validation ROC-AUC | ||||
Therefore, we must highlight the reliability and validity of our results listed in Table 5 in terms of the collected measurements, averaged over the number of classes for all experiments and repeated seven times each.
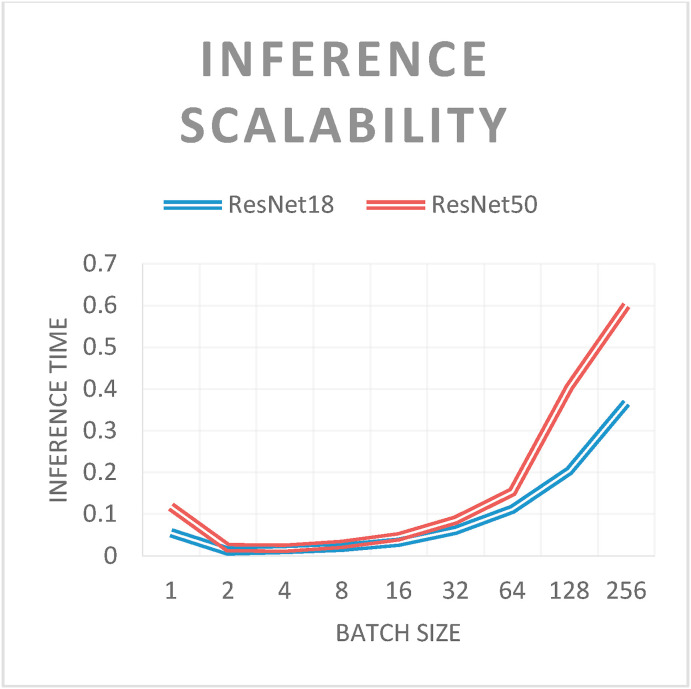
Network scalability was evaluated during inference. Experiments were performed with batch sizes ranging from 1 to 256 (i.e., each network classified between 1 and 256 images for the inference process). As expected, for both networks, the inference times increased with the batch size (see Fig. 5 ). The only exception was the inference of a single image. In this case, the inference time of a batch containing a single image was greater than that of others up to a batch size of 64. This is because it is possible to group multiple images into a single tensor and adopt efficient computational routines to perform the inference. Finally, the inference times of ResNet50 were greater than those of ResNet18, which could be attributed to their network structures. As explained previously, ResNet50 has a deeper and more complex structure than that of ResNet18.
Fig. 5.
- Inference scalability: processing times [s] according to batch size.
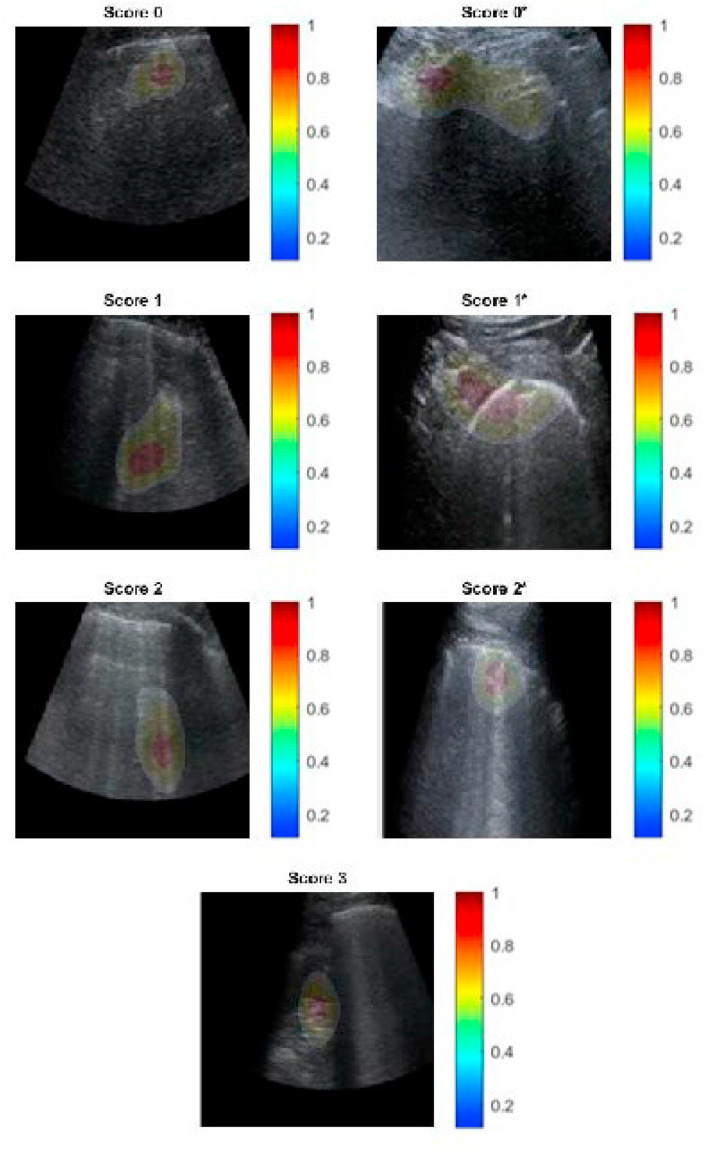
Furthermore, to validate the network results, we used both CAM and Grad-CAM methodologies for each experiment. The physicians evaluated whether the ResNets correctly highlighted B-lines, pleural line irregularities, or other patterns examined in the LUS score subsection when ranking a frame, which is the procedure adopted by physicians to assess patients' health conditions. Fig. 6 depicts the behaviour of ResNet50 in a scenario for which we also evaluated the pleural line. For simplicity and integrity, we present only the CAM results and not the Grad-CAM results, starting from the lowest score, indicating that the considered subject is healthy, and approaching the highest score, indicating that the patient should be urgently treated. The residual architecture correctly and precisely highlights all patterns, namely A- and B-lines, small or broad consolidations, and damage to the pleural line. When considering the scenario containing fewer classes, the pleural line is not taken into account by the statistical models, which, at most, uses it to assess a subjects’ healthiness, specifically when analysing Score 0 frames and the reverb contained in its A-lines.
Fig. 6.
ResNet50 Class Activation Mapping, seven class scenario: both severity scoring, B-lines and pleural line consolidations and irregularities are correctly highlighted along with tissue-like patterns for Score 3.
We have described all the existing state-of-the-art studies in the Introduction section, highlighting both their strengths and weaknesses. However, we compared our study mainly to three recent studies on the application of DL methodologies to LUS data to diagnose Covid-19 pneumonia and evaluate the severity of lung engagement[32,34,35]. The authors of the first study [32] exploited LUS data assigned with a severity score for frame-level classification meeting F1-score levels ranging from 65.1% to 71.4% when considering either the test set results or the average value over three different settings: test set, test set with transition frames dropped, and inter-doctor adjustments. Exploiting a spatial transform network[36] developed in 2015, they proposed a novel scoring methodology[3], concerning already validated and researched scales for evaluating lung health conditions[20], which authors from the third study adopted as well. In all cases, we obtained a performance improvement of 27.15% in terms of the best average performance in comparison with our worst-case outline.
In contrast, the authors of the second study [34] proposed a shallow architecture developed from scratch to address both binary Covid-19 detection and severity classification. They exploited the same ranking scale adopted by the authors in the first study. However, we not only exceeded their results concerning the first clustering problem they proposed but also attained a performance improvement of more than 40% in the assessment of lung engagement. Finally, the authors from the last study [35] compared a wide variety of already established deep architectures to prove the accuracy of the transfer learning applied to Covid-19 heterogeneous data, specifically considering CT scans, ultrasound, and CXR. However, they focused on the classification task of determining whether a lung is healthy or has been diagnosed with either pneumonia or Covid-19. Despite the promising results presented, the authors suggested that their images were of inconsistent quality. They based their study on non-validated data from public online repositories; therefore, they did not assess the reliability of the gathered medical examinations. They achieved excellent classification performances, which we have been able to meet, resulting in a more complex problem based on reliable data given by Fondazione IRCCS Policlinico San Matteo. Moreover, they obtained F1-score results ranging from 66% to 99% for the different architectures considered, which is analogous to what we researched, as listed in Table 5. Nonetheless, our metrics are above 97% in all cases.
Regarding the aforementioned works, we optimised the networks for several epochs, specifically between 12 and 15, depending on the specific experiment. Having fine-tuned pre-trained networks with CUDA in a multi-GPU environment, owing to the CNN libraries developed at NVIDIA, the process resulted in training times ranging between 17 and 89 min. Therefore, the number of epochs and the overall training time are considerably lower than those reported by other authors[6,28,32,34]. The computations were spread across two NVIDIA RTX 2080, with 2994-cores each, and we could process a mini-batch, or more, of images every second.
In conclusion, previous studies on the application of DL in Covid-19 detection have presented some drawbacks. Less than half of the studies have not exploited transfer learning; moreover, although authors prefer already proven architectures, they have relied upon unreliable data sources of poor quality, without assessment by a qualified physician. In addition, only a few studies have exploited LUS for diagnosing patients with Covid-19, from among which only the three studies that we compared our results to have assessed illness severity or exploited transfer learning. The first two studies focused on the exploitation of a novel scoring methodology without employing transfer learning; they assessed the lung health conditions and attempted to apply image classification networks with small tweaks to address the classification of small clip portions. Moreover, the authors from the second work did not exploit recently collected LUS data from subjects who contracted Covid-19 but instead collected clips performed at the Yale-New Haven Hospital since 2012.
Therefore, we propose a simple yet effective methodology to address the application of DL to LUS data and Covid-19 detection. We employed already proven and pre-trained residual networks in two configurations. Moreover, we adopted an existing and validated ranking scale, which we extended to hierarchically structure the labels that need to be detected. This helps differentiate the cardiogenic from non-cardiogenic causes of B-lines[33], and the early detection of ARDS pneumonia symptoms enables the timely treatment of patients. To the best of our knowledge, no extended scoring methodology has been proposed to date for assessing the pleural line together with existing patterns. In addition, we acknowledged the challenges faced by our colleagues[32] regarding having several physicians perform the LUS examinations. Hence, we further validated our collection of clips, focusing on data augmentation and hyperparameter tuning to exploit the advantages of transfer learning and obtain the results presented in this paper.
4. Conclusions
We designed and engineered a highly reliable diagnostic instrument to meet the significantly increasing demand for affordable and efficient Covid-19 detection systems by exhausted medical personnel. With close collaboration with Fondazione IRCCS Policlinico San Matteo's ED, we could base our studies on highly reliable and validated LUS data.
We employed modern DL methodologies, including deep residual networks, data augmentation processes, and transfer learning, to rank subjects’ lungs using[20] scoring methodologies, which we extended by adding information regarding pleural line conditions. We not only alleviated the severe drawbacks arising from data heterogeneity (modest sensitivity leading to lack of treatment for infected patients, and cross-contamination problems), but also improved currently accessible state-of-the-art performances[32,34,35] in Covid-19 detection employing LUS data.
This study provides an approach for overcoming the dataset problems discussed by other authors [32] concerning the scoring inconsistencies between ultrasounds due to different physicians scoring different lungs of the same stage. In addition, the Fondazione IRCCS Policlinico San Matteo ED reviewed every exam to homogeneously assign lungs of the same disease stage with the same score.
Because ultrasound imaging technologies require strong expertise to achieve diagnostic reliability – high sensitivity and overall accuracy – in this study, we developed a DL-based system to automatically detect Covid-19 pneumonitis marks in LUS images and rate their severity based on two standardised scoring methods with innovative, reliable, and revolutionary results.
Drclaration of competing interest
We declare that neither the manuscript nor any parts of its content are currently under consideration or published in another journal and that there is no conflict of interest in submitting our paper to your journal.
Acknowledgements
The authors extend their gratitude to Professor Lucia Sacchi, Kevin Cruzata, and GERICO (Gruppo Esteso Ricerca Coronarovirus) Lung US Pavia Study Group: Ilaria Martino, Barbarah Guglielmana, Ilaria Zunino, Federica Quaglia, Pietro Pettenazza, Serena Pioli di Marco, Anna Giulia Falchi, Claudia Alfano, Elisa Mossolani, Lorenzo Demitry, Massimiliano Sciarrini, Caterina Zattera, Igor Maisak, Michele Tassi, Stefano Galati, Capozza, Ilaria Melara, Benedetta Chiodi, Damiano Vignaroli, Lorenzo Scattaglia, Giulia Bissichini, and Marco Bazzini.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104742.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Li Q., et al. Early transmission dynamics in wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi H., et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soldati G., et al. Proposal for international standardization of the use of lung ultrasound for patients with COVID-19: a simple, quantitative, reproducible method. J. Ultrasound Med. 2020 doi: 10.1002/jum.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonsenso D., Pata D., Chiaretti A. COVID-19 outbreak: less stethoscope, more ultrasound. The Lancet Respiratory Medicine. 2020;8 e27 doi: 10.1016/S2213-2600(20)30120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya S., et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: a survey. Sustain. Cities Soc. 2021;65:102589. doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiosa G., Gianfreda R., Pastorino M., Davio P. Role of lung ultrasound in identifying COVID-19 pneumonia in patients with negative swab during the outbreak. Emerg. Care J. 2020;16 [Google Scholar]
- 8.Chen M., et al. Clinical applications of detecting IgG, IgM or IgA antibody for the diagnosis of COVID-19: a meta-analysis and systematic review. Int. J. Infect. Dis. 2021;104:415–422. doi: 10.1016/j.ijid.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J.J.Y., et al. Influenza Other Respi. 2021. Diagnostic performance of COVID‐19 serological assays during early infection: a systematic review and meta‐analysis of 11 516 samples; p. 12841. Viruses 00. irv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J.L., et al. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir. Med. 2020;168:105980. doi: 10.1016/j.rmed.2020.105980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niederman M.S., et al. Guidelines for the management of adults with community-acquired pneumonia. Am. J. Respir. Crit. Care Med. 2001;163:1730–1754. doi: 10.1164/ajrccm.163.7.at1010. [DOI] [PubMed] [Google Scholar]
- 12.Chavez M.A., et al. Lung ultrasound for the diagnosis of pneumonia in adults: a systematic review and meta-analysis. Respir. Res. 2014;15 doi: 10.1186/1465-9921-15-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagano A., et al. Lung ultrasound for diagnosis of pneumonia in emergency department. Intern. Emerg. Med. 2015;10:851–854. doi: 10.1007/s11739-015-1297-2. [DOI] [PubMed] [Google Scholar]
- 14.Bourcier J.E., et al. Performance comparison of lung ultrasound and chest x-ray for the diagnosis of pneumonia in the. Am. J. Emerg. Med. 2014;32:115–118. doi: 10.1016/j.ajem.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 15.McDermott C., et al. Sonographic diagnosis of COVID-19: a review of image processing for lung ultrasound. Front. Big Data. 2021;4:2. doi: 10.3389/fdata.2021.612561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poggiali E., et al. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID-19) pneumonia? Radiology. 2020;295:200847. doi: 10.1148/radiol.2020200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiala M.J. A brief review of lung ultrasonography in COVID-19: is it useful? Ann. Emerg. Med. 2020 doi: 10.1016/j.annemergmed.2020.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai T., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongodi S., et al. Modified lung ultrasound score for assessing and monitoring pulmonary aeration. Ultraschall der Med. 2017;38:530–537. doi: 10.1055/s-0042-120260. [DOI] [PubMed] [Google Scholar]
- 21.Litjens G., et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017:42 60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Shin H.C., et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imag. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei X., et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020:1–5. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: classification and segmentation. Comput. Biol. Med. 2020;126:104037. doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozturk T., et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imag. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 28.Islam M.M., Karray F., Alhajj R., Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Alencar J.C.G., et al. Lung ultrasound score predicts outcomes in COVID-19 patients admitted to the emergency department. Ann. Intensive Care. 2021;11:6. doi: 10.1186/s13613-020-00799-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buda N., Segura-Grau E., Cylwik J., Wełnicki M. Lung ultrasound in the diagnosis of COVID-19 infection - a case series and review of the literature. Adv. Med. Sci. 2020;65:378–385. doi: 10.1016/j.advms.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhammad G., Shamim Hossain M. COVID-19 and non-COVID-19 classification using multi-layers fusion from lung ultrasound images. Inf. Fusion. 2021;72:80–88. doi: 10.1016/j.inffus.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy S., et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imag. 2020 doi: 10.1109/TMI.2020.2994459. 1–1. [DOI] [PubMed] [Google Scholar]
- 33.Arntfield R., et al. Development of a convolutional neural network to differentiate among the etiology of similar appearing pathological b lines on lung ultrasound: a deep learning study. BMJ Open. 2021;11:45120. doi: 10.1136/bmjopen-2020-045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baloescu C., et al. Automated lung ultrasound B-line assessment using a deep learning algorithm. IEEE Trans. Ultrason. Ferroelectrics Freq. Contr. 2020;67:2312–2320. doi: 10.1109/TUFFC.2020.3002249. [DOI] [PubMed] [Google Scholar]
- 35.Horry M.J., et al. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaderberg M., Simonyan K., Zisserman A., Kavukcuoglu K. Spatial transformer networks. Adv. Neural Inf. Process. Syst. 2015;28 [Google Scholar]
- 37.Yosinski J., Clune J., Bengio Y., Lipson H. 2014. How Transferable Are Features in Deep Neural Networks? Undefined. [Google Scholar]
- 38.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Vols 2016-Decem 770–778. IEEE Computer Society; 2016. Deep residual learning for image recognition. [Google Scholar]
- 39.Lichtenstein D.A. Lung Ultrasound in the Critically Ill. Springer International Publishing; 2016. The pleural line; pp. 61–64. [DOI] [Google Scholar]
- 40.Secco G., et al. LUNG ultrasound IN covid-19: a useful diagnostic tool. Emerg. Care J. 2020;16 [Google Scholar]
- 41.Deng J., et al. ImageNet: A Large-Scale Hierarchical Image Database. Institute of Electrical and Electronics Engineers (IEEE); 2010. pp. 248–255. [DOI] [Google Scholar]
- 42.Kingma D.P., Lei Ba, Adam J. undefined; 2015. A METHOD FOR STOCHASTIC OPTIMIZATION. [Google Scholar]
- 43.Monshi M.M.A., Poon J., Chung V., Monshi F. M. CovidXrayNet. Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med. 2021;133:104375. doi: 10.1016/j.compbiomed.2021.104375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J. Big Data. 2019;6:1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selvaraju R.R., et al. Proceedings of the IEEE International Conference on Computer Vision. Institute of Electrical and Electronics Engineers Inc.; 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization. Octob 618–626. [Google Scholar]
- 46.Van Sloun R.J.G., Demi L. Localizing B-lines in lung ultrasonography by weakly supervised deep learning, in-vivo results. IEEE J. Biomed. Heal. Informatics. 2020;24:957–964. doi: 10.1109/JBHI.2019.2936151. [DOI] [PubMed] [Google Scholar]
- 47.Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006;27:861–874. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.