Dear Editor,
In this Journal, the emergence of novel variants with potential to escape vaccine-induced immunity has received commentary.1
The emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants of concern (VOC) and variants of interest (VOI) are challenging the management of the evolving pandemic across countries. The VOI labelled as Eta (WHO Classification),2 combines relevant spike mutations detected in several VOC, such as the same 3 deletions of the Alpha lineage (69del, 70del, 144del), the E484K mutation found in the Gamma and Beta lineages as well as in some Alpha isolates and the ubiquitous D614G. In addition, three mutations (A67V, Q677H and F888L) are unique to Eta variant and it is currently unknown whether they favor escape from natural or vaccine induced immunity to the wild type lineage (B.1), as shown for other variants.3 To test this hypothesis, we measured the serum neutralizing antibody (NtAb) response to Eta variant, as well as to other viral variants, in a cohort of health care workers (HCWs) including both previously infected (n = 15) and uninfected individuals (n = 15) vaccinated with two doses of the BNT162b2 COVID-19 mRNA vaccine. The study was approved by the Ethics Committee of the University of Milan (protocol n. 23/21) and conducted in compliance with Good Clinical Practice guidelines and the Declaration of Helsinki. The previously infected group was tested at baseline (T0inf) and 17±6 days after receiving the second vaccine dose (T2inf); the uninfected HCWs were tested 18±4 days after the second dose vaccination (T2uninf). The infected group had median age [IQR] of 38 (31–52) years, included 8 females and was infected during the first wave of the pandemic. The uninfected group had a median age of 38 (29–59) years with 11 females.
NtAb titers were determined by a microneutralization live virus assay performed in VERO E6 cells using the quantification of cell viability as readout system, as previously described.4 NtAb titers were expressed as median (IQR) and were defined as the reciprocal value of the sample dilution that showed a 50% protection of virus-induced cytopathic effect (ID50). Sera with ID50 titres ≥10 were defined as SARS-CoV-2 neutralizing, while sera with ID50 <10 were defined as negative and scored as 5 for statistical analysis. Fifteen, 14 and 11 individuals at T0inf, T2inf and T2uninf, respectively, had also a quantitative anti-spike protein Ab determination, performed by the SARS-CoV-2 IgG II Quant assay (Abbott). The viral isolates used in the microneutralization live virus assay were sequenced by NGS and the full-lenght SARS-CoV-2 genome was submitted to GISAID (http://gisaid.org/) to assign the right variant (Accession numbers: EPI_ISL_2,472,896, EPI_ISL_1,085,167, EPI_ISL_2,472,918 and EPI_ISL_2,472,916 for the wild type, Alpha, Gamma and Eta variants, respectively). Statistical analyses were performed using IBM SPSS Statistics, version 20. The non-parametric Friedman test and Wilcoxon Signed Rank Sum test was used to analyze changes in paired data. The non-parametric Mann-Whitney test was used to compare unpaired data. Spearman analysis was used to measure the correlation between NtAb titres against the different variants.
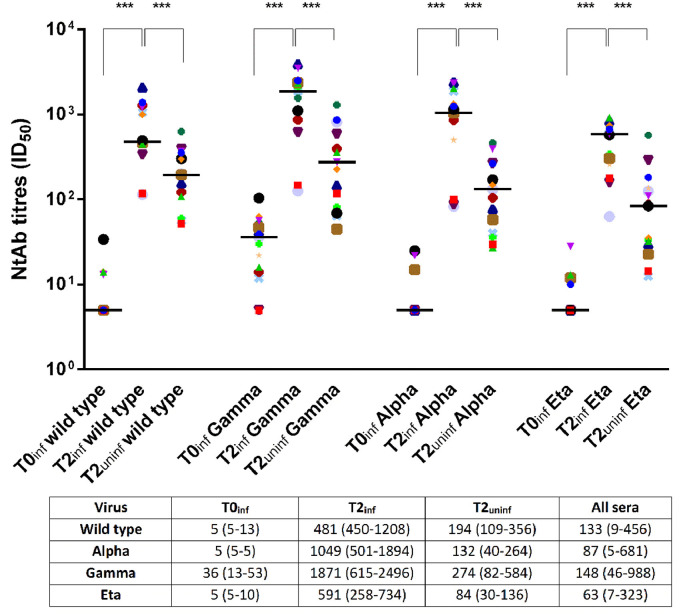
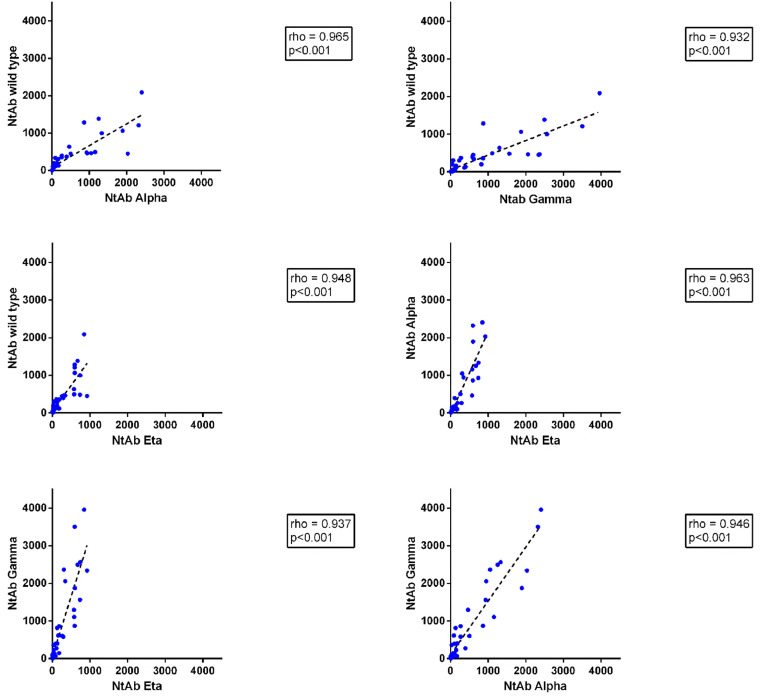
In previously infected HCWs, NtAb titres to all viral variants significantly increased after vaccination (mean T2inf/T0inf ratio 119±66; p<0.001). Notably, 2 to 12 subjects, depending on the reference virus, were negative at T0inf but all of them seroconverted following vaccination. As expected, the NtAb titer after vaccination was higher in the previously infected compared with the uninfected group (mean T2inf/T2uninf ratio 6 ± 2; p <0.001 (Fig. 1 ). Overall, median NtAb titres to the Eta variant (63 [7–323] ID50) correlated well with those to the wild type (133 [9–456]), Gamma (148 [46–988]) and Alpha (87 [5-681]) (p<0.001 for all comparisons) and high correlation was indeed observed between NtAb titres to any pair of virus variants (Fig. 2 ). Of note, NtAb titres to Eta variant were significantly lower with respect to those obtained for each variant (p<0.001). Anti-spike protein antibodies, as measured by enzyme immunoassay, were highly correlated with NtAb titres to B.1 (rho = 0.934), P.1 (rho = 0.914), B.1.1.7 (rho = 0.913) and B.1.525 (rho = 0.918) viruses (p<0.001 for all comparisons). Also, a significant increase was observed when comparing the anti-spike Ab median titres at T2inf and at T0inf (27,763 [18,282–46,108] vs. 1.7 [0.5–4.4]; p = 0.001).
Fig. 1.
Neutralizing antibody (NtAb) titres to four SARS-CoV-2 variants (wild type, Gamma, Alpha and Eta) in 15 previously infected subjects at baseline (T0inf) and after two doses of vaccine (T2inf) and in 15 uninfected subjects after two doses of vaccine (T2uninf). Asterisks indicate significance levels: ***, p<0.001. Median (IQR) titres of neutralizing antibody are reported below.
Fig. 2.
Spearman correlation between NtAb titres to each pair of the SARS-CoV-2 variants used in the study. Data were cumulated for all sera tested at T0inf, T2inf and T2uninf.
Overall, in our small cohort of previously infected or uninfected vaccinated-HCWs it appears that cross-neutralization among different viral variants remains substantial, following natural or artificial immunization with the wild type lineage. However, neutralization of Eta variant is significantly reduced with respect to other variants. Indeed, NtAb titres could be ranked with the definite order Gamma>wild type=Alpha>Eta. In vitro correlates of protection against the Eta variant has been investigated in uninfected vaccinated individuals only in two different works delivering inconsistent results. Indeed, Liu et al.5 observed a modest reduction, while Zani et al.6 reported an increase in Eta variant NtAb titres with respect to the wild type variant. Of note, NtAb studies published so far have used different combination of strategies (e.g., live virus vs. pseudoparticles), viral variants, cell lines and readouts, in the absence of standardized methods and reference viral strains and neutralizing sera.7, 8, 9, 10 For example, the full-length sequencing of the isolates used in the assay should be always reported and submitted to public repositories. Most importantly, while NtAb studies certainly provide a solid basis to infer cross-protection among vaccines and virus variants, the in vivo correlates of in vitro data remain to be established and must be defined through accurate and continuous monitoring of vaccine induced reduction of morbidity and mortality in the context of molecular surveillance of SARS-CoV-2 lineages.
References
- 1.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J Infect. 2021;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira F., Tosta S., Lima M.M., Reboredo de Oliveira da Silva L., Nardy V.B., Gómez M.K.A. Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B1.525 variant of interest in Brazil: case report. J Med Virol. 2021 doi: 10.1002/jmv.27086. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janik E., Niemcewicz M., Podogrocki M., Majsterek I., Bijak M. The Emerging Concern and Interest SARS CoV-2 Variants. Pathogens. 2021;10(6):633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicenti I., Gatti F., Scaggiante R., Boccuto A., Zago D., Basso M. Single-dose BNT162b2 mRNA COVID-19 vaccine significantly boosts neutralizing antibody response in health care workers recovering from asymptomatic or mild natural SARS-CoV-2 infection. Int J Infect Dis. 2021;108:176–178. doi: 10.1016/j.ijid.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;(June) doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- 6.Zani A., Caccuri F., Messali S., Bonfanti C., Caruso A. Serosurvey in BNT162b2 vaccine-elicited neutralizing antibodies against authentic B.1, B.1.1.7, B.1.351, B.1.525 and P.1 SARS-CoV-2 variants. Emerg Microbes Infect. 2021;7:1–6. doi: 10.1080/22221751.2021.1940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betton M., Livrozet M., Planas D., Fayol A., Monel B., Védie B. Sera neutralizing activities against SARS-CoV-2 and multiple variants six month after hospitalization for COVID-19. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab308. ; ciab308. Epub ahead of print. PMID: 33851216; PMCID: PMC8083257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C. Sensitivity of infectious SARS-CoV-2 B1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747–751. doi: 10.1016/j.chom.2021.04.007. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]